Abstract
Rapid assessment of prostate core biopsy pathology at the point-of-procedure could provide benefit in a variety of clinical situations. Even with advanced trans-rectal ultrasound guidance and saturation biopsy protocols, prostate cancer can be missed in up to half of all initial biopsy procedures. In addition, collection of tumor specimens for downstream histological, molecular, and genetic analysis is hindered by low tumor yield due to inability to identify prostate cancer grossly. However, current point-of-procedure pathology protocols such as frozen section analysis (FSA) are destructive, and too time- and labor-intensive to be practical or economical. Ex vivo microscopy of the excised specimens, stained with fast-acting fluorescent histology dyes, could be an attractive non-destructive alternative to FSA. In this work, we report the first demonstration of video-rate structured illumination microscopy (VR-SIM) for rapid high-resolution diagnostic imaging of prostate biopsies in realistic point-of-procedure timeframes. Large mosaic images of prostate biopsies stained with acridine orange are rendered in seconds, and contain excellent contrast and detail, exhibiting close correlation with corresponding H&E histology. A clinically-relevant review of VR-SIM images of 34 unfixed and uncut prostate core biopsies by two independent pathologists resulted in an area under the ROC curve (AUC) of 0.82–0.88, with a sensitivity ranging from 63–88% and a specificity ranging from 78–89%. When biopsies contained more than 5% tumor content, the sensitivity improved to 75–92%. The image quality, speed, minimal complexity, and ease of use of VR-SIM could prove to be features in favor of adoption as an alternative to destructive pathology at the point-of-procedure.
Keywords: prostate cancer, biopsy, microscopy, fluorescence, point-of-care
Introduction
When a patient presents with elevated prostate specific antigen (PSA) and/or an abnormal digital rectal examination (DRE), the next step in the work up for prostate cancer is core needle biopsy under trans-rectal ultrasound (TRUS) guidance. Prostate cancer is not always reliably visualized with ultrasound, so the TRUS is primarily used to direct the needle into various anatomic regions of the prostate; for example, into each sextant of the gland (1, 2). Sextant biopsy techniques have been shown to miss up to 50% of small tumors (3). This has led to the more widespread adoption of “saturation biopsy” techniques where 8–22 cores are obtained in a single biopsy session focusing on the transition zone or using a perineal template. However, lack of suitable tools to locate cancer within the prostate means that, even with saturation biopsy protocol, initial biopsy may fail to reveal cancers in up to 30% of men (3, 4). Furthermore, up to 14% of men may ultimately need 4 saturation biopsies for a definitive diagnosis (5). Thus, about 70,000 men annually will undergo 2 or more repeat prostate biopsy procedures. In addition, with the transrectal approach, there is always the risk of infection and/or sepsis after the biopsy procedure. With approximately 1,000,000 prostate biopsy procedures performed each year to diagnose approximately 240,000 cases of prostate cancer, there is a clear need to reduce the number of repeat biopsies to prevent unnecessary costs while ensuring that all significant cancers are diagnosed and treated as soon as possible.
Just as prostate cancer is difficult to visualize by TRUS in vivo, it is also difficult to identify grossly in the resected specimen, which hampers collection of cancerous tissue for biospecimen banking. Current protocols for acquisition of prostate tissue for biobanking generally involve random needle biopsy sampling of the resected prostate gland. The sampled tissue is examined histologically at a later time, typically via frozen section, to determine the presence of cancer. This process is inefficient, since many of the cores will not contain cancer, and it will be too late to return to the prostatectomy specimen to obtain additional fresh samples. Additionally, since the tissue that is examined by frozen section is shaved off of the core, it may not be representative of the tissue in the rest of the core – that is, the frozen section slides may have cancer, but the remainder of the core does not, or vice versa. This is less than optimal since the accuracy of subsequent molecular or genetic analysis depends on the amount of cancerous tissue present in the specimen (6). Further, some tissue will be destroyed and lost in the cryostat during preparation of the frozen section slides.
An ideal technique would be to rapidly and non-destructively assess the biopsy tissue at the point of acquisition, so that additional biopsies could be harvested immediately if needed. Currently available techniques for assessing a core needle biopsy on-site – frozen section and touch preparation – have limitations including tissue destruction and low sensitivity, and are also too time-consuming to be practical. Currently, the only routine clinical method used to image biopsies non-destructively at the point of care is X-ray imaging, which is occasionally used on breast core needle biopsy specimens to ensure targeted calcifications are present in the sampled tissue (7). This is a rapid and non-destructive technique, but the contrast mechanism and resolution of X-ray imaging does not allow for microscopic analysis of the tissue, and imaging of microcalcifications is not relevant in prostate. Optical microscopy approaches, on the other hand, are well-suited for on-site microscopic analysis of core needle biopsy specimens, as they allow rapid ex vivo imaging of intact, minimally-processed tissues. Confocal microscopy, for example, is attractive for diagnostic ex vivo imaging because it produces images that closely recapitulate tissue architecture which is visualized on traditional histology slides by using sources of contrast that display histological features similar to traditional histological stains (8–10). Additionally it provides high lateral resolution (necessary to see subcellular diagnostic features) and high axial resolution (necessary to obtain an optical section in lieu of a physical section). A number of groups have successfully demonstrated this concept in biopsies of the skin, breast, and brain (8–16).
Another optical sectioning microscopy method, structured illumination microscopy (SIM), has been gaining increased attention in recent years. SIM is a wide-field optical sectioning technique that uses patterned illumination to preferentially modulate and retain the in-focus object information separately from the out-of-focus background (17). The disadvantage of SIM compared to confocal microscopy is that it has limited ability to obtain high-quality optical sections deep into tissue (18). However, its distinct advantage is that it is a light-efficient wide-field technique, and its speed is decoupled from the field of view (since all pixels are acquired in parallel and the pixel resolution is determined by the camera specifications) (19). With the use of 4.2 megapixel, high speed scientific CMOS cameras and fast ferroelectric spatial light modulators for rapid pattern switching, we have recently demonstrated that we can achieve video-rate SIM (VR-SIM) for optical sectioning (20). We previously demonstrated high-quality images of large tissue surfaces using this method to achieve area-throughput rates for mosaicking microscopy of 4.4 cm2/min with 1.3 μm lateral resolution (using a 7 mm/s automated microscope stage), which corresponds to an effective pixel sampling rate of 17.6 megapixels/sec including stage automation.
In this work we demonstrate the potential of VR-SIM for accurate, high-throughput, non-destructive diagnostic imaging of fluorescently stained prostate biopsies in point-of-care timeframes. We demonstrate high-resolution, high-contrast imaging of full intact biopsy surfaces with average pixel counts of 265 megapixels and average imaging times around 30 seconds or less per biopsy, which brings rapid diagnostic imaging of multiple biopsies fully into the realm of practicality for the clinical workflow.
Materials and Methods
Instrumentation
We previously reported on the development of VR-SIM system to image phantoms and biological tissue (20). Briefly, VR-SIM utilizes incoherent fluorescence structured illumination, and is constructed around a modular automated epi-fluorescence microscope platform (RAMM, Applied Scientific Instrumentation), which incorporates a 7 mm/s motorized XY specimen stage and a motorized Z objective positioner. Light from a 470 nm LED (Thorlabs) is collected by an aspheric condenser lens, transmitted through a polarizing beam splitter (PBS, Moxtek), and is imaged onto a ferroelectric liquid-crystal-on-silicon spatial light modulator (SLM, Model 3DM, Forth Dimension Displays), which is placed in a plane conjugate to the sample plane. Light reflected from the SLM is reflected by the PBS through a clean-up polarizer, the illumination tube lens, and the bandpass excitation filter with 475 nm center wavelength (FITC-Ex01-Clin-25, Semrock). The filtered excitation light is reflected into the imaging objective (Nikon, Plan Apo 10X 0.45 NA) by a 500 nm edge dichroic beamsplitter (FITC-Di01-Clin-25×36, Semrock), and forms an image of the SLM onto the sample. Fluorescence from the sample is collected by the objective, transmitted through the dichroic mirror and the 515 nm longpass emission filter (FITC-LP01-Clin-25, Semrock), and imaged by a Nikon tube lens onto a scientific CMOS camera (Orca Flash 4.0 v2, Hamamatsu). Synchronization and control of the LED, SLM, stage, objective, and camera was achieved via custom LabVIEW software and electronic triggering circuits developed in our laboratory. The self-contained VR-SIM module is mounted to the RAMM base, and the entire system fits comfortably on a custom 4′W × 3′D wheeled lab bench.
Therefore, the VR-SIM system is configured to feature a single-frame field-of-view (FOV) of 1.3 mm × 1.3 mm, at 4.2 megapixel resolution. The 0.45 NA objective lens gives a diffraction-limited optical resolution of 0.75 μm at 550 nm, however, at 10X magnification the sampling resolution of the sCMOS is 0.65 μm/pixel at the sample. Thus, by the Nyquist criterion, the lateral image resolution is 2(0.65) = 1.3 μm and is limited not by diffraction, but by the sCMOS pixel size.
Incoherent SIM for optical sectioning was performed using the square-law detection algorithm described by Neil et al. [4]. The method involves modulating the in-focus plane of the tissue with a sinusoidal grid projected onto the specimen through the illumination system. The grid is highly contrasted only at the focal plane, and is demodulated with defocus at a rate dependent on the spatial frequency of the pattern and the numerical aperture of the illumination objective. The attenuation of the pattern with defocus defines the depth gating or optical sectioning thickness of the image, with higher frequency patterns giving thinner optical sections at the expense of reduced image intensity (20), and vice versa. Commonly, three sequential images are taken with the pattern phase-shifted by one-third of the grid period between each image. The portions of the specimen that are out-of-focus and not highly contrasted by the illumination pattern are common to the three images, and are rejected by the square law detection algorithm in Equation 1, leaving only the highly-modulated information present in the optical section defined by the pattern frequency and objective numerical aperture. Optically-sectioned images are computed immediately after acquisition of the three phase-stepped patterned images in custom LabVIEW control software.
| [Equation 1] |
where ISIM is the recovered optically sectioned image, and x1, x2, and x3 are the three sequentially patterned images, respectively. For SIM imaging in this work, we used a sinusoidal grid illumination pattern with a normalized spatial frequency ν = 0.035 for a good balance between signal to background ratio (SBR) and optical sectioning thickness, as determined previously (20). Specifically, this illumination frequency gave a measured surface optical section thickness (half-width half-max of the axial defocus curve) in tissue-simulating phantoms (20, 21) of 24 μm. With the specific SLM pixel pitch and illumination optics used in this study, the maximum attainable normalized spatial frequency is 0.21, which corresponds to a measured optical section thickness of 6.2 μm; even thinner optical sections are possible as the normalized spatial frequency is increased to 1.
Prostate Biopsy Imaging
The prostate biopsies were previously flash-frozen, de-identified samples obtained from the Louisiana Cancer Research Center (LCRC) Biospecimen Core. LCRC tissue bank staff obtain 5-mm diameter punch core biopsies from intact radical prostatectomy specimens of patients giving informed consent under an IRB-approved protocol, within 20 minutes of surgical resection. The biopsies are halved longitudinally: one half is frozen in optimal cutting temperature (OCT) compound for cryopreservation, and the remaining half is fixed in formalin for formalin fixed paraffin embedded (FFPE) processing, where a single section is taken from the sample and used to provide a diagnosis of the biopsy for curation by the LCRC pathologist (called ‘tissue bank diagnosis’ in this manuscript). In this study we used the cryopreserved portions of the biopsies. The frozen prostate biopsy was thawed, and the surface was sprayed with 2% acridine orange solution (A9231, Sigma-Aldrich) diluted to 0.02% with 95% glacial acetic acid + 5% phosphate buffered saline (PBS) (pH 2.8). The biopsy was immediately rinsed by immersion in 20% glacial acetic acid + 80% 1X PBS for 5 seconds. The biopsy was then blotted dry with lab tissue and 2% acridine orange solution diluted to 0.5% in 1X PBS was sprayed on the surface. Immediately after the staining, the biopsy was rinsed by immersion in PBS for 5 seconds and then blotted dry. The stained biopsy was placed on a glass slide and gentle pressure was applied to the opposite (upper) side of the biopsy, so that the lower surface of the biopsy adhered to the glass slide. The glass slide was mounted on the stage of the VR-SIM system for imaging. The entire staining and positioning process took approximately 2–3 minutes, and can be parallelized for multiple biopsies by staining them en masse in a tissue cassette, and positioning the collection of biopsies on a standard large glass slide.
Test images for every biopsy were taken prior to imaging to adjust the intensity of the LED illumination. The integration time used for all biopsies was kept constant at 100 ms to maximize signal while not unnecessarily exposing the sample to excessive illumination power. The system is capable of acquiring images an order of magnitude faster at 10 ms per frame, or 30 ms per processed SIM frame (20) – however, for this particular study, the samples were small enough that 100 ms integration time was sufficient for fast biopsy imaging. The intensity of the LED illumination was adjusted manually depending on the sample, such that the wide-field fluorescence emission used the entire linear dynamic range of the 16-bit camera without saturation. Mosaics of the tissue were obtained using a serpentine scan approach, with no overlap between frames. Each frame was corrected for non-uniform illumination (i.e. flat-field corrected) using a reference image taken of a fluorescent calibration slide (Chroma). Mosaics were constructed without the use of stitching algorithms, and images were saved as full resolution 16-bit TIFF or BigTIFF format, and were digitally contrast-enhanced using histogram normalization in Fiji (0.04% pixels saturated) to ensure more uniform brightness between images.
Histopathology
Following imaging, the imaged biopsy surface was marked using histological ink, placed in 10% buffered formalin for at least 48 hours, and sent for pathologic processing, where a 10-micron section was cut from the inked biopsy surface, corresponding to the VR-SIM image of the same biopsy. The histotechnician attempted to orient the inked tissue plane parallel to the surface of the paraffin block in order to obtain a section of the approximate tissue plane imaged with VR-SIM. Sections were stained with hematoxylin and eosin, and subsequently digitized on a Nikon Eclipse LV100 with 4x and 10x objectives and a color camera (SPOT Insight).
Blinded Diagnostic Review
Pathologist A reviewed the VR-SIM images and the H&E slides as two separate cohorts. Cohort 1 (‘A’ biopsies) consisted of 20 samples, of which 10 were originally classified as benign and 10 were classified as malignant by the tissue bank as described above. The VR-SIM images and the corresponding H&E slides from this cohort were blinded by independently assigning them random numbers. The study pathologist was unaware of the original tissue bank diagnosis and the number of malignant specimens in the cohort, and did not know which VR-SIM image corresponded with which H&E slide. The pathologist first reviewed the VR-SIM images using Fiji (ImageJ), and could digitally zoom the images to simulate changing microscope objective magnification, although the image resolution was fixed at 1.3 μm. The pathologist then reviewed all of the corresponding H&E slides at a later date, and was free to choose any magnification objective. The pathologist gave a diagnosis ranking for each VR-SIM image ranging from 1 to 5, where 1 indicated most likely to be benign, 2 indicated possibly benign, 3 indicated indeterminate between cancerous or benign, 4 indicated possibly cancerous, and 5 indicated most likely to be malignant. H&E slides were diagnosed as either malignant or non-malignant. In six cases, findings of atypical small acinar proliferation necessitated the use of p63 immunohistochemistry for definitive diagnosis. In all cases, the final diagnosis of the tissue section taken subsequent to VR-SIM imaging was used as the gold standard for comparison since it was the closest approximation to what was actually imaged by the VR-SIM system.
After the review was complete for cohort 1, the VR-SIM images were matched to their respective H&E slides, and the un-blinded set was given to Pathologist A before viewing cohort 2. Cohort 2 (‘B’ biopsies) consisted of an additional 10 benign and 10 malignant samples as determined by the tissue bank. The VR-SIM images and H&E slides from this second cohort were blinded and reviewed in the same fashion as described for cohort 1. Pathologist B had no experience with VR-SIM images prior to this study, and completed a blinded review of both cohorts combined; the blinding and scoring procedures were the same as for Pathologist A.
Statistical analysis
Receiver operating characteristic (ROC) curves were constructed based on the VR-SIM diagnostic rankings to calculate the areas under the ROC curves (AUC) for each reviewer. Sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), and overall accuracy were computed on various values of the cutpoint on the ROC along with associated 95% confidence intervals. Kendall’s tau was computed to evaluate the strength of agreement between reviewers. The Wilcoxon rank-sum test was computed to test for significant differences in diagnostic scores and AUCs between reviewers.
Results
Table 1 contains the study pathologists’ diagnoses and rankings for the H&E sections and the VR-SIM images, respectively; the pixel count of the VR-SIM images; the imaging time of the VR-SIM images; the Gleason score for the malignant biopsies; the tumor content for the malignant biopsies; and, the result of p63 staining conducted on six of the biopsies. The diagnosis of the H&E slides taken after VR-SIM imaging differed from the original tissue bank classification for 12 biopsies. Specifically, in 5 biopsies, cancer was found on the H&E section of the tissues curated as non-malignant by the tissue bank, and in 7 biopsies, no cancer was found on the H&E section of the tissues curated as malignant by the tissue bank. However, since these H&E sections taken subsequent to VR-SIM imaging were taken of the approximate tissue surface imaged by VR-SIM, these diagnoses were used as the gold standard for comparison. Six (6) biopsies were excluded by a consensus panel of non-rater co-authors (DBT, KNE, and JQB) blinded to the diagnosis, because the H&E section was not representative of the VR-SIM image of the biopsy. Reasons for exclusion included: 1) the H&E section was fragmented, and was not representative of the entire length or extent of the biopsy, or 2) the H&E section was taken obliquely, and there was clear discordance in features present in the VR-SIM image and not found in the corresponding locations on the H&E section, or vice versa. The median tumor content for malignant biopsies was 13.5%, with a range of 1–75%. Pathologist A and B disagreed on the diagnosis of 2 biopsies on H&E, therefore, diagnostic accuracy metrics for the VR-SIM biopsy images were computed against the H&E diagnoses from the same pathology rater. Altogether, each pathologist diagnosed 16 malignant biopsies and 18 non-malignant biopsies in the entire cohort retained for analysis. The pixel count of the VR-SIM images ranged from 150 megapixels to 587 megapixels (200 megabytes to 1.1 gigabytes file size), and depended on the size of the imaged biopsies, which ranged from 42.3 mm2 to 236.6 mm2 image area. The total combined pixel count for all 40 images was 10.6 gigapixels, comprised 2,527 individual frames (4,270.6 mm2 total area) and 20.2 gigabytes combined file size, and was collected in 1,265.5 seconds (21.08 minutes).
Table 1.
Sample ID, H&E diagnoses (on section corresponding to VR-SIM imaging) and VR-SIM diagnoses for two pathology raters, # of image pixels, total imaging time in seconds, Gleason score, tumor content (%), and p63 status for all biopsies in cohorts 1 (labeled ‘A’) and 2 (labeled ‘B’). For VR-SIM diagnosis rank: 5 = most likely to be malignant, 4 = possibly malignant, 3 = indeterminate, 2 = possibly benign, 1 = most likely to be benign.
| Sample ID | # pixels (millions) | Image time (s) | Pathologist A H&E | Pathologist A VR-SIM | Pathologist B H&E | Pathologist B VR-SIM | Gleason score | Tumor content (%) | p63 |
|---|---|---|---|---|---|---|---|---|---|
| A1 | 205.8 | 24.5 | Mal | 5 | Mal | 5 | 3+3 | 8 | |
| A2 | 134.2 | 16 | Mal | 4 | Mal | 3 | 3+3 | 5 | |
| A3 | 210 | 25 | Nonmal | 2 | Nonmal | 2 | |||
| A4 | 294 | 35 | Nonmal | 3 | Nonmal | 2 | |||
| A5 | 210 | 25 | Nonmal | 3 | Nonmal | 1 | |||
| A6** | 588 | 72 | Nonmal | 2 | Mal | 3 | 3+3 | 5 | |
| A7 | 340.2 | 40.5 | Nonmal | 2 | Nonmal | 1 | |||
| A8 | 126 | 15 | Excluded due to misregistration between H&E section and VR-SIM image | ||||||
| A9 | 420 | 50 | Excluded due to misregistration between H&E section and VR-SIM image | ||||||
| A10 | 420 | 50 | Mal | 5 | Mal | 2 | 3+4 | 25 | |
| A11** | 205.8 | 24.5 | Mal | 4 | Nonmal | 2 | 3+3 | <5 | |
| A12 | 205.8 | 24.5 | Mal | 5 | Mal | 5 | 3+4 | 15 | |
| A13 | 205.8 | 24.5 | Mal | 5 | Mal | 4 | 3+3 | 15 | |
| A14 | 205.8 | 24.5 | Excluded due to misregistration between H&E section and VR-SIM image | ||||||
| A15 | 205.8 | 24.5 | Excluded due to misregistration between H&E section and VR-SIM image | ||||||
| A16 | 420 | 50 | Mal | 5 | Mal | 4 | 4+4 | 35 | |
| A17 | 340.2 | 40.5 | Mal | 5 | Mal | 4 | 3+3 | 8 | |
| A18 | 268.8 | 32 | Mal | 5 | Mal | 2 | 3+4 | 40 | |
| A19 | 205.8 | 24.5 | Mal | 5 | Mal | 5 | 3+4 | 45 | |
| A20 | 268.8 | 32 | Mal | 5 | Mal | 4 | 3+3 | 12 | |
| B1 | 420 | 50 | Nonmal | 3 | Nonmal | 2 | |||
| B2 | 340.2 | 40.5 | Nonmal | 1 | Nonmal | 2 | |||
| B3 | 340.2 | 40.5 | Nonmal | 2 | Nonmal | 3 | |||
| B4 | 268.8 | 32 | Nonmal | 2 | Nonmal | 3 | + | ||
| B5 | 105 | 12.5 | Nonmal | 1 | Nonmal | 2 | |||
| B6 | 420 | 50 | Mal | 4 | Mal | 4 | 3+3 | 1 | − |
| B7 | 205.8 | 24.5 | Nonmal | 1 | Nonmal | 2 | |||
| B8 | 205.8 | 24.5 | Nonmal | 1 | Nonmal | 2 | |||
| B9 | 205.8 | 24.5 | Mal | 1 | Mal | 2 | 3+3 | 1 | |
| B10 | 268.8 | 32 | Excluded due to misregistration between H&E section and VR-SIM image | ||||||
| B11 | 205.8 | 24.5 | Nonmal | 4 | Nonmal | 2 | |||
| B12 | 420 | 50 | Nonmal | 4 | Nonmal | 2 | |||
| B13 | 151.2 | 18 | Mal | 4 | Mal | 4 | 3+4 | 20 | |
| B14 | 268.8 | 32 | Nonmal | 1 | Nonmal | 2 | + | ||
| B15 | 268.8 | 32 | Nonmal | 4 | Nonmal | 2 | + | ||
| B16 | 205.8 | 24.5 | Mal | 4 | Mal | 2 | 3+3 | 12 | − |
| B17 | 268.8 | 32 | Mal | 2 | Mal | 5 | 4+3 | 75 | |
| B18 | 205.8 | 24.5 | Nonmal | 3 | Nonmal | 4 | + | ||
| B19 | 151.2 | 18 | Nonmal | 4 | Nonmal | 4 | |||
| B20 | 205.8 | 24.5 | Excluded due to misregistration between H&E section and VR-SIM image | ||||||
Indicates biopsies where the diagnosis of malignant or non-malignant differed between the two pathology raters on H&E.
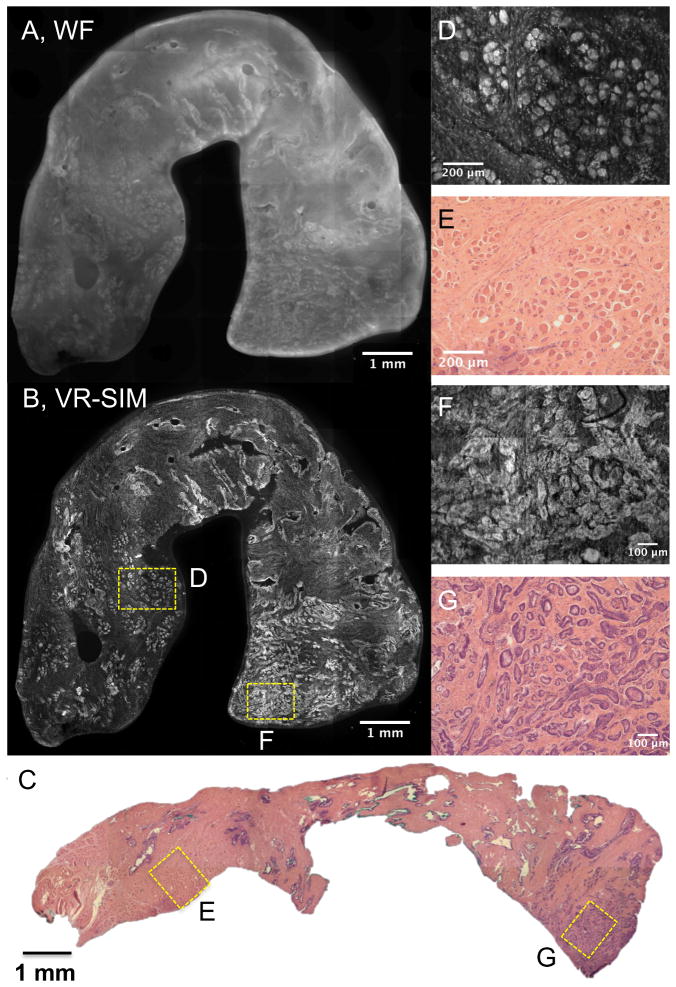
Figure 1 contains VR-SIM images of a biopsy (sample A1) confirmed to contain prostate adenocarcinoma, and demonstrates that VR-SIM increases the useful contrast of thick tissues compared to standard widefield imaging, and can produce images that are comparable to gold standard histopathology. The VR-SIM mosaic image was collected with 0.1 s exposure time (3.3 Hz SIM frame rate), which means the total imaging time for each VR-SIM frame was 0.3 s. Using our current microscope scan stage rated at 7 mm/sec, the time for the stage to move in-between frame captures was 0.2 s. In Figure 1, the VR-SIM image is composed of 49 individual frames (24.5 s total imaging time), at 2048 × 2048 pixels per frame, for a total image resolution of 205.5 megapixels (0.65 μm/pixel). The widefield (WF) image of Figure 1A contains considerable out-of-focus fluorescence from stain which has penetrated into the biopsy, as well as scattered fluorescence by the thick tissue, which ultimately degrades contrast and prevents fine features from being observed in the image. However, in Figure 1B the utility of the use of VR-SIM is apparent: the VR-SIM image in Figure 1B shows superior contrast, resolution, and detail compared to the widefield image due to rejection of out-of-focus background. Figure 1C contains an image of the corresponding H&E section of the whole biopsy. As is observed in both the zoomed-out VR-SIM (1B) and H&E (1C) images of the biopsy, the microarchitecture of the biopsy consists of 3 distinct “zones” from left to right. Zooming into the images allows the morphology of each zone to be identified and compared to standard histology. The left-most zone is characterized by evenly-spaced skeletal muscle fibers in cross-section, interspersed by fibrous stroma. This is observed in the VR-SIM image (Figure 1D) as bright circular structures that correspond to the muscle fibers, interspersed with slightly elongated small bright features corresponding to fibroblast nuclei. Bright spots at the periphery of the muscle fibers correspond to the muscle fiber nuclei. In the corresponding H&E histology, shown in Figure 1E, the same structural arrangements are observed, with the distinction that the cell nuclei and muscle fibers/stroma are highlighted by differences in color rather than intensity (cell nuclei are stained purple, whereas muscle fibers and collagen are stained pink). In this way, areas of a biopsy that appear purple in a low-magnification view such as in Figure 1C correspond to areas with many cell nuclei. This is true of the biopsy in Figure 1C, where a clear focus of purple is observed in the right-most zone of the biopsy. Zooming into this image (Figure 1G) shows a focus of small crowded prostate adenocarcinoma glands. This is observed as a very bright area in the lower right end of the biopsy in the low-digital-zoom VR-SIM image of Figure 1B. Digitally zooming into the VR-SIM image as in Figure 1F shows that the glandular morphology seen in the H&E section is preserved in the VR-SIM images, supporting a positive identification of prostate adenocarcinoma in the biopsy on blinded review.
Figure 1.
VR-SIM images and subsequent H&E slide images of Biopsy A1 confirmed as malignant. A) Wide-field (i.e., without SIM) image of the entire biopsy, B) VR-SIM mosaic image of the entire biopsy, comprising 205.5 megapixels. C) Digital image of the corresponding H&E section. D) VR-SIM and E) H&E zoom images of the regions of interest marked by the correspondingly labeled boxes in B and C, depicting an area of normal skeletal muscle and fibrous stroma. F) VR-SIM and G) H&E zoom images of the regions of interest marked by the correspondingly labeled boxes in B and C, depicting an area of malignant glands.
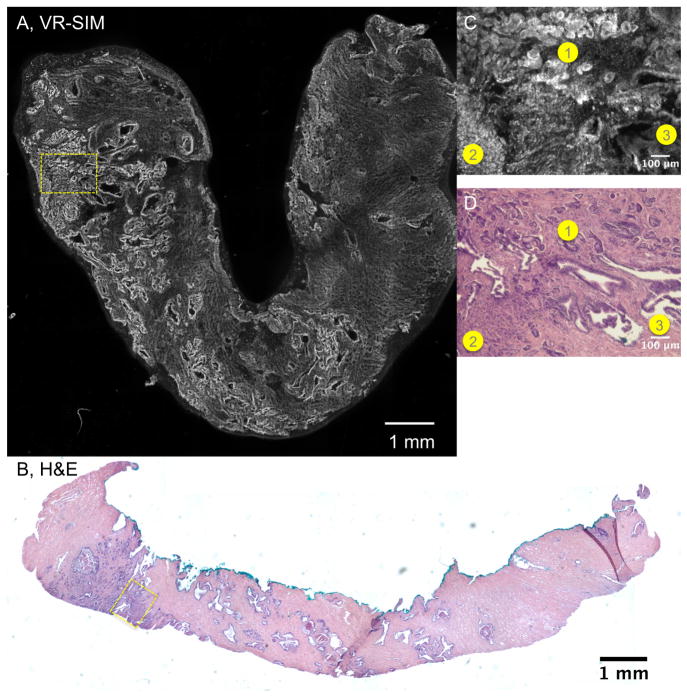
Figure 2 contains another VR-SIM image and corresponding H&E histology image of a biopsy containing prostate adenocarcinoma (sample A19). As in Figure 2, observation of the architecture of the biopsy in the VR-SIM image is achieved by considering the brightness and structure of the features. In this VR-SIM image (Figure 2A), the left end of the biopsy is characterized by a focus of bright, highly textured features, whereas the middle of the biopsy is characterized by larger looping structures, and the right end of the biopsy is characterized by a dim, wavy, finely textured area. These observations are confirmed in the H&E image of Figure 2B, where the left end of the biopsy contains prostate adenocarcinoma, the middle of the biopsy contains mostly normal glands and concretions, and the right end of the biopsy contains fibrous stroma. These observations of the area corresponding to prostate cancer in the VR-SIM and H&E images are seen more closely in the digital zooms of Figure 2C and 2D, respectively. In this case, in the VR-SIM image, 3 distinct zones of tissue architecture are observed (labeled as 1, 2, and 3 in the image). Area 1 consists of small rounded glands, Area 2 consists of many cell nuclei that are not arranged in well-formed glands, and Area 3 consists of larger looping glandular structures. These features are also observed in the H&E image of Figure 2D, where Area 1 is shown to contain an area of prostate cancer corresponding to Gleason pattern 3, Area 2 is shown to contain an area of higher grade prostate cancer corresponding to Gleason pattern 4, and Area 3 is shown to contain an area of normal or benign glands.
Figure 2.
VR-SIM images and subsequent H&E slide images of biopsy A19 confirmed as malignant. A) VR-SIM mosaic image of the entire biopsy, comprising 205.5 megapixels, B) Digital image of the corresponding H&E section. Zooms of the dashed yellow boxes in A and B are shown in C and D, respectively. Corresponding areas of interest between the VR-SIM image and the H&E image are denoted by numbered circles (1 = Gleason grade 3 cancer, 2 = Gleason grade 4 cancer, and 3 = benign glands).
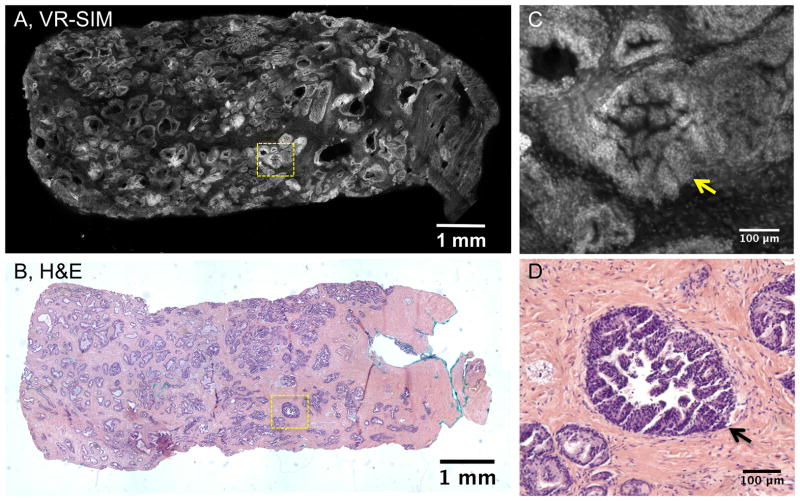
Figure 3 contains images of a biopsy (sample B18) that was noted to contain high grade prostatic intraepithelial neoplasia (PIN), widely thought to be a potential precursor of prostate adenocarcinoma, and similar to carcinomas in situ in other organs. The right side of the biopsy contains an area of dim staining and wavy appearance with no bright structures. The overall appearance of the tissue architecture as seen in the VR-SIM image is also observed in the H&E image of Figure 3B, with many glands of varying size and shape throughout the biopsy. The dimly-stained area on the right of the VR-SIM image corresponds to an area of pink stroma without significant glandular features on the right of Figure 3B. Figure 3C contains a digital zoom of a structure enclosed by the dashed yellow box in Figure 3A. This structure corresponds to high grade PIN with micropapillary architecture. Individual cell nuclei are clearly observed in the image. The corresponding feature in the H&E image is shown in Figure 3D with excellent concordance between the VR-SIM and H&E architecture.
Figure 3.
VR-SIM images and subsequent H&E slide images of biopsy B18 classified as non-malignant, but containing high-grade prostatic intraepithelial neoplasia (HGPIN). A) VR-SIM mosaic image of the entire biopsy, comprising 205.5 megapixels, and B) Digital image of the corresponding H&E section. Zooms of the dashed yellow boxes in A and B are shown in C and D, respectively. C) A close-up of a gland containing PIN (yellow arrow) in the VR-SIM image, with D) the corresponding H&E image.
Figure 4 contains individual ROIs for additional features noted in the VR-SIM biopsy images. Figure 4A contains a digital zoom of corpora amylacea imaged with VR-SIM, along with corresponding H&E histology in Figure 4B. The corpora amylacea are characterized by bright smooth florescence in VR-SIM images, whereas they are characterized in H&E as pink to orange colored structures, often with concentric laminations. Figure 4 also contains VR-SIM images of perineural invasion (Figure 4C and 4D) and nerve ganglions (Figure 4E and 4F). The finding of perineural invasion, marked in the VR-SIM image by a small focus of brightly stained nuclei at the periphery of a nerve bundle, is indicative of cancer cells surrounding and tracking along a nerve fiber, and is associated with a higher risk of extraprostatic extension.
Figure 4.
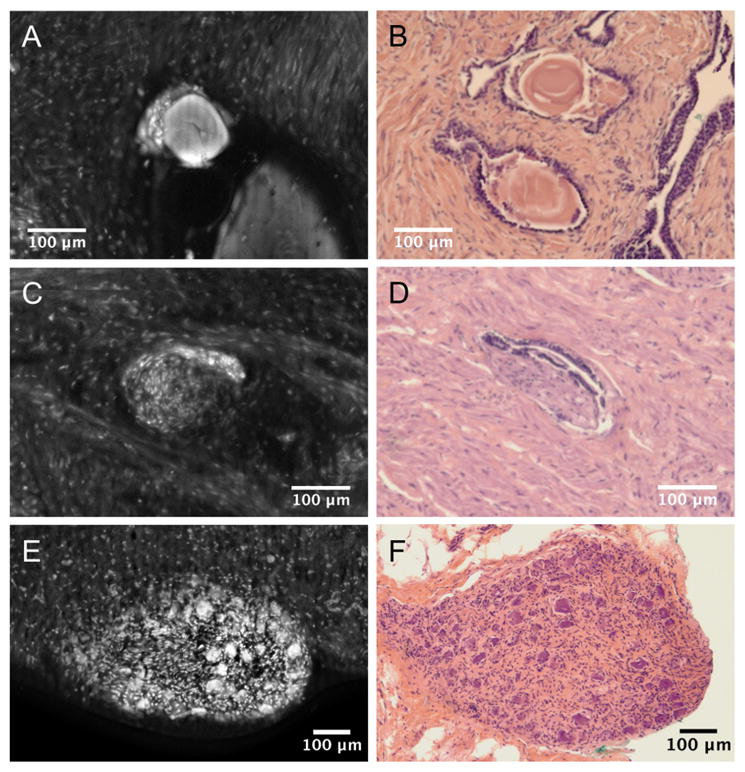
Examples of commonly observed features in prostate biopsies (A, C, E), along with the corresponding H&E section histology (B, D, F). A & C: Corpora amylacea, C & D: Perineural invasion, E & F: Nerve ganglion.
Table 2 contains the diagnostic classification results for whole-biopsy imaging with VR-SIM as compared to gold-standard histopathology. Agreement between the two reviewers was slightly weak (Kendall’s tau = 0.42), but scores were within one point for 25 of the 34 biopsies (73.5%) with no systematic difference in the scores between the two reviewers (p = 0.09, Wilcoxon signed rank test). For Pathologist A, both sensitivity and specificity were maximized for scores of 4 or greater, with 14 of 16 positive samples and 14 of 18 negative samples correctly classified using this threshold. The area under the curve for the ROC was 0.88 (95% CI = [0.75, 1]). For Pathologist B, sensitivity was maximized for a threshold of 3 but specificity was maximized for a threshold of 4, with an AUC of 0.82 (95% CI = [0.69, 0.95]). Using a threshold of 3, 12 of 16 positive samples and 14 of 18 negative samples were correctly identified, while for a threshold of 4 Pathologist B correctly identified only 10 of the positive samples but 16 of the negative samples. There was no significant difference in the AUC between the two reviewers.
Table 2.
Results of the receiver operative curve (ROC) analysis for VR-SIM against standard H&E histopathology.
| False Positives | False Negatives | Sensitivity (95% CI) | Specificity (95% CI) | PPV | NPV | Accuracy | AUC (95% CI) | |
|---|---|---|---|---|---|---|---|---|
| Pathologist A (16 M, 18 NM) | 4 | 2 | 87.5% (60.4%, 97.8%) | 77.8% (51.9%, 92.6%) | 77.8% | 87.5% | 82.4% | 0.88 (0.75, 1) |
| Pathologist B (16 M, 18 NM) | 2 | 6 | 62.5% (35.9%, 83.7%) | 88.9% (63.9%, 98.1%) | 83.3% | 72.7% | 76.5% | 0.82 (0.69, 0.95) |
M = malignant, NM = non-malignant.
Discussion
Here we present a novel use of a microscopy system that can rapidly produce high-resolution, non-destructive diagnostic-quality images of entire core biopsy surfaces within a practical point-of-care time frame. This is the first demonstration to our knowledge of the use of SIM for surface imaging of stained unfixed whole biopsy specimens, and more generally the first application of acridine orange staining and rapid optical sectioning microscopy of prostate cancer specimens. We demonstrate that VR-SIM is able to produce images that have excellent correlation with standard histology images using simple topical fluorescent contrast agents. The presented microscopy method can be used to distinguish between cancerous and benign features of prostate biopsies, even in tissues with limited adenocarcinoma content. In this study, the primary objective was to demonstrate the quality of VR-SIM imaging compared to gold standard histopathology and to obtain an initial estimate of the diagnostic performance, not necessarily to optimize speed. However, even with the relatively large biopsies used in this work taken with a 5-mm diameter biopsy punch, compared to the smaller 18G needles used clinically, we were able to image entire biopsies in 12.5–50 seconds at 0.65 μm/pixel resolution, at a consistent imaging speed of 8.4 megapixels per second. The strength of the system reported here is that compared to other fluorescence ex vivo imaging modalities, such as confocal microscopy, VR-SIM is faster and lower complexity; for example, it uses inexpensive LED illumination and requires no moving parts other than the sample and objective stages. In addition, the light efficiency of VR-SIM allowed the use of a non-immersion, long-working-distance 10X objective to image the biopsies through a standard thickness microscope slide and still achieve high throughput and 1.3 μm lateral resolution. These benefits reduce complexity of sample preparation for imaging, compared to methods that require the use of expensive, short working distance, high-NA water-immersion objectives to maximize light throughput. Perhaps most importantly, all of these images are collected non-destructively, leaving the entire tissue volume available for subsequent processing and analysis.
In this study, we did not conduct an a priori examination of the VR-SIM images and H&E sections in order to pre-select smaller regions of interest (ROIs) for pathologic review, in which tissue features were clearly concordant between the VR-SIM image and the H&E slide. Rather, we chose instead to provide images of the entire biopsy surfaces to the pathologists, as this is ultimately a more clinically-relevant test of the utility of the method. However, this is a more challenging experimental design in terms of pathologic diagnosis, and co-registration between images of the fresh biopsy and subsequent H&E sections taken after standard histopathology processing. We used histological inks to mark the entire surface of the biopsy imaged by VR-SIM; however, since the biopsy is subsequently fixed in formalin and embedded in paraffin for cutting, distortions of the tissue and difficulties in orienting it in the paraffin block means that it is difficult to know the exact angle used to cut the specimen, sometimes resulting in a significantly different area displayed in the resulting H&E slide. This is the reason for the relatively high number of sampling exclusions in this study (6 out of 40 biopsies were rejected due to gross discordance between the VR-SIM image of the surface and the plane of tissue obtained in the H&E section). However, this is a limitation only when attempting to match the image of the biopsy surface with a thin physical section. In eventual clinical practice, with VR-SIM the area of each optical section would be maximized compared to H&E, since the tissue is imaged in its fresh state and there is no concern for inadvertently taking oblique sections as commonly occurs in standard histopathology. We have also found that the minimal tissue handling involved in this method enables full images of fragile samples to be obtained before they can be broken and fragmented during subsequent handling and fixation steps (in fact, no samples were fragmented during the VR-SIM imaging procedure, although some were later fragmented during H&E processing). Finally, we have found that due to the speed of the system, it is practical to re-orient or grossly cut the biopsy and take multiple images of different aspects of the tissue to maximize diagnostic information, for example, collecting images of both the front and back of the core.
The preliminary diagnostic results presented here (AUC 0.82 – 0.88) demonstrate the potential of VR-SIM as an intermediate non-destructive diagnostic assessment of biopsies at the point of care. Pathologist A correctly classified 28/34 biopsies, whereas Pathologist B correctly classified 26/34 biopsies (54/68 combined calls correct), with no significant difference detected between the areas under the ROC curve for the two reviewers. Three biopsies with low tumor content (≤5%) had reviewer rankings within one point of each other and were associated with half of the total number of false negative calls (4/8) for both reviewers combined. For these samples, differences in sampling between the VR-SIM optical section and the physical thin section obtained after imaging cannot be definitively ruled out as contributing to these false negative errors. Excluding these low tumor volume samples improved the sensitivity to 92% for Pathologist A and 75% for Pathologist B. One sample (B19) was overcalled by both reviewers accounting for 2/6 combined false positive calls, and was attributed to a cluster of benign glands that appeared more crowded and suggestive of malignancy than on the corresponding H&E, perhaps related to an area of adenosis that was not readily apparent on the H&E section. Another sample (B18, Figure 3) contained prostatic hyperplasia with HGPIN present in the sample, which is a known mimicker of prostate adenocarcinoma on H&E and accounted for 1 false positive call, with the other reviewer ranking the biopsy as ‘3 - indeterminate’ constituting a true negative at a ranking threshold of 4. The thickness of the VR-SIM optical section compared to the H&E slide in this case may have contributed to the false positive, since it was not as easy to appreciate the double layer of cells indicative of benign glands on the VR-SIM image as in the H&E section.
The preceding errors were generally related to either low tumor volume or the presence of known mimics of adenocarcinoma, and accounted for half of the overall number of incorrect calls. In the remainder of the biopsies accounting for the other half of the total incorrect calls (4 false negatives and 3 false positives), the rankings were quite disparate with one reviewer assigning a score of 4 or 5 and the other assigning a score of 2 (i.e., there were no misclassified samples in common between reviewers for these 7 biopsies). These differences can be attributed to differences in their interpretation of foci of glandular features on the VR-SIM images that were not obviously malignant or non-malignant, which may have also been related to artifacts due to freezing. This interpretation dilemma differentiated the performance of reviewers depending on whether they sacrificed specificity for sensitivity in order to identify even small volume tumor, or sacrificed sensitivity for specificity in order to maintain high positive predictive value. The former approach would be preferable in a diagnostic scenario where it is critical to avoid false negatives, whereas the latter approach would be preferable in assessment of robust tumor tissue adequacy for downstream analysis where false positives should be minimized. In addition, we note that Pathologist A was involved throughout the development of the VR-SIM system and was experienced with viewing VR-SIM images prior to the blinded study, whereas Pathologist B had not reviewed VR-SIM images prior to the blinded review. Pathologist A also benefited from an intermediate review of VR-SIM images versus histopathology after review of Cohort A but before Cohort B, whereas Pathologist B reviewed both Cohorts combined with no intermediate unblinded review. Thus, the differences in performance between reviewers could be expected to be reduced by 1) imaging fresh biopsies free of frozen artifact and 2) providing additional pathologist experience and training by administering a standardized and robust training set, containing biopsies which capture the full spectrum of normal, benign, premalignant, and malignant subtypes in prostate biopsies and over a large range of tumor content. Further improvements in the technology to decrease the optical section thickness while retaining high signal-to-noise ratio (20) would also be expected to more closely approximate thin H&E sections and reduce these interpretation dilemmas. Incidentally in this study magnification and resolution were not cited by the pathology reviewers as factors hindering interpretation, but future work will also evaluate whether using higher magnification and NA objective lenses improves performance.
Notwithstanding, these results are encouraging, as they represent the potential of the method for whole-biopsy review in realistic clinical scenarios. Prostate cancer can be challenging to diagnose even on the gold-standard H&E, particularly when adenocarcinoma content is limited, or mimickers of adenocarcinoma are present, both of which are common. The cohort of biopsies imaged in this study approximated these situations well, since the median cancer content was only 13.5%, and 6/34 biopsies were indeterminate on H&E review due to adenocarcinoma mimics, requiring the use of p63 immunohistochemistry to confirm the diagnoses. Optical methods for point of care pathology have gained increased attention as methods that can greatly improve the efficiency of cancer diagnosis (22). VR-SIM thus represents a practical advancement for ex vivo microscopy that is capable of rapid viewing and diagnosis of biopsy tissues. Ultimately, the high image quality, speed, and ease of use of the system could be features in favor of adoption for rapid on-site biopsy diagnostic screening and digital pathology consultation using web-enabled gigapixel image viewers. The use of this technology is aimed at point-of-care tissue triage, including but not limited to asking the clinician for collection of additional biopsy samples in the setting of low tumor volume, as well as for directed molecular studies of tumor-containing biopsies. Given these scenarios, VR-SIM demonstrates itself as a powerful tool for the pathologist and clinician in its ability to quickly and accurately define the presence of ample tumor. The present study is limited by the small number of samples, and thus the reported diagnostic metrics can be considered an estimate only. Future validation studies will employ fresh tissues to eliminate potential deleterious effects due to previously frozen samples, will employ more samples to more accurately determine diagnostic performance, and will include a standardized training set and more reviewers to more robustly determine the effect of inter-rater variation.
Acknowledgments
Financial support: J.Q. Brown, National Cancer Institute grant R21 CA159936 (Brown PI). Authors M. Wang and T. Schlichenmeyer were also supported in part by this grant. K. Elfer was supported by an NSF Graduate Research Fellowship. D. Tulman was supported by an NSF IGERT grant to Tulane University DGE-1144646.
Financial support for this work was provided in part by the National Cancer Institute (NIH grant R21 CA159936) and an institutional grant from Tulane University. D. Tulman acknowledges fellowship support from a National Science Foundation IGERT DGE-1144646. K. Elfer acknowledges support from a National Science Foundation Graduate Fellowship. We gratefully acknowledge Dr. Walter Lee Murfee and Dr. Richard Sweat for the use of and assistance with the Nikon microscope and color camera for histological slide imaging. Finally, we are thankful for assistance from Jennifer Cvitanovic, Celeste Faia, Alex Alfortish, Dr. Krzystof Moroz, the Tulane Cancer Center, and the Louisiana Cancer Research Consortium Biospecimen Core Laboratory, for providing assistance with procurement of tissue specimens, and for providing access to the specimens at a discounted fee.
Footnotes
Conflicts of interest: The authors have no conflicts to disclose.
References
- 1.Epstein JI, Netto GJ, Epstein JI. Biopsy interpretation of the prostate. 4. Philadelphia: Wolters Kluwer Health/Lippincott Williams & Wilkins; 2008. p. x.p. 358. [Google Scholar]
- 2.Wein AJ, Kavoussi LR, Campbell MF. In: Campbell-Walsh urology. 10. Wein Alan J, Kavoussi Louis R, et al., editors. Philadelphia, PA: Elsevier Saunders; 2012. [Google Scholar]
- 3.Levy DA, Jones JS. Management of rising prostate-specific antigen after a negative biopsy. Current urology reports. 2011;12(3):197–202. doi: 10.1007/s11934-011-0177-1. [DOI] [PubMed] [Google Scholar]
- 4.Ukimura O, Coleman JA, de la Taille A, Emberton M, Epstein JI, Freedland SJ, et al. Contemporary role of systematic prostate biopsies: indications, techniques, and implications for patient care. Eur Urol. 2013;63(2):214–30. doi: 10.1016/j.eururo.2012.09.033. [DOI] [PubMed] [Google Scholar]
- 5.Campos-Fernandes JL, Bastien L, Nicolaiew N, Robert G, Terry S, Vacherot F, et al. Prostate cancer detection rate in patients with repeated extended 21-sample needle biopsy. Eur Urol. 2009;55(3):600–6. doi: 10.1016/j.eururo.2008.06.043. [DOI] [PubMed] [Google Scholar]
- 6.Muley T, Herth F, Schnabel P, Dienemann H, Meister M. From tissue to molecular phenotyping: Pre-analytical requirements Heidelberg experience. Transl Lung Cancer Res. 2012;1(2):111–21. doi: 10.3978/j.issn.2218-6751.2011.12.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krupinski EA, Borders M, Fitzpatrick K. Processing stereotactic breast biopsy specimens: impact of specimen radiography system on workflow. The breast journal. 2013;19(4):455–6. doi: 10.1111/tbj.12132. [DOI] [PubMed] [Google Scholar]
- 8.Dobbs JL, Ding H, Benveniste AP, Kuerer HM, Krishnamurthy S, Yang W, et al. Feasibility of confocal fluorescence microscopy for real-time evaluation of neoplasia in fresh human breast tissue. J Biomed Opt. 2013;18(10):106016. doi: 10.1117/1.JBO.18.10.106016. [DOI] [PubMed] [Google Scholar]
- 9.Gareau D, Bar A, Snaveley N, Lee K, Chen N, Swanson N, et al. Tri-modal confocal mosaics detect residual invasive squamous cell carcinoma in Mohs surgical excisions. J Biomed Opt. 2012;17(6):066018. doi: 10.1117/1.JBO.17.6.066018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gareau DS, Karen JK, Dusza SW, Tudisco M, Nehal KS, Rajadhyaksha M. Sensitivity and specificity for detecting basal cell carcinomas in Mohs excisions with confocal fluorescence mosaicing microscopy. Journal of Biomedical Optics. 2009;14(3) doi: 10.1117/1.3130331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abeytunge S, Li Y, Larson B, Peterson G, Seltzer E, Toledo-Crow R, et al. Confocal microscopy with strip mosaicing for rapid imaging over large areas of excised tissue. J Biomed Opt. 2013;18(6):61227. doi: 10.1117/1.JBO.18.6.061227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bini J, Spain J, Nehal K, Hazelwood V, DiMarzio C, Rajadhyaksha M. Confocal mosaicing microscopy of human skin ex vivo: spectral analysis for digital staining to simulate histology-like appearance. Journal of Biomedical Optics. 2011;16(7) doi: 10.1117/1.3596742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gareau DS. Feasibility of digitally stained multimodal confocal mosaics to simulate histopathology. J Biomed Opt. 2009;14(3):034050. doi: 10.1117/1.3149853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gareau DS, Li YB, Huang B, Eastman Z, Nehal KS, Rajadhyaksha M. Confocal mosaicing microscopy in Mohs skin excisions: feasibility of rapid surgical pathology. Journal of Biomedical Optics. 2008;13(5) doi: 10.1117/1.2981828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Patel R, Khan A, Wirth D, Kamionek M, Kandil D, Quinlan R, et al. Multimodal optical imaging for detecting breast cancer. Journal of Biomedical Optics. 2012;17(6) doi: 10.1117/1.JBO.17.6.066008. [DOI] [PubMed] [Google Scholar]
- 16.Snuderl M, Wirth D, Sheth SA, Bourne SK, Kwon CS, Ancukiewicz M, et al. Dye-Enhanced Multimodal Confocal Imaging as a Novel Approach to Intraoperative Diagnosis of Brain Tumors. Brain Pathol. 2013;23(1):73–81. doi: 10.1111/j.1750-3639.2012.00626.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neil MAA, Juskaitis R, Wilson T. Method of obtaining optical sectioning by using structured light in a conventional microscope. Optics Letters. 1997;22(24):1905–7. doi: 10.1364/ol.22.001905. [DOI] [PubMed] [Google Scholar]
- 18.Mertz J. Optical sectioning microscopy with planar or structured illumination. Nat Methods. 2011;8(10):811–9. doi: 10.1038/nmeth.1709. [DOI] [PubMed] [Google Scholar]
- 19.Fiolka R, Shao L, Rego EH, Davidson MW, Gustafsson MGL. Time-lapse two-color 3D imaging of live cells with doubled resolution using structured illumination. P Natl Acad Sci USA. 2012;109(14):5311–5. doi: 10.1073/pnas.1119262109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schlichenmeyer TC, Wang M, Elfer KN, Brown JQ. Video-rate structured illumination microscopy for high-throughput imaging of large tissue areas. Biomed Opt Express. 2014;5(2):366–77. doi: 10.1364/BOE.5.000366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fu HL, Mueller JL, Javid MP, Mito JK, Kirsch DG, Ramanujam N, et al. Optimization of a widefield structured illumination microscope for non-destructive assessment and quantification of nuclear features in tumor margins of a primary mouse model of sarcoma. PLoS ONE. 2013;8(7):e68868. doi: 10.1371/journal.pone.0068868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boppart SA, Richards-Kortum R. Point-of-care and point-of-procedure optical imaging technologies for primary care and global health. Science translational medicine. 2014;6(253):253rv2. doi: 10.1126/scitranslmed.3009725. [DOI] [PMC free article] [PubMed] [Google Scholar]