SUMMARY
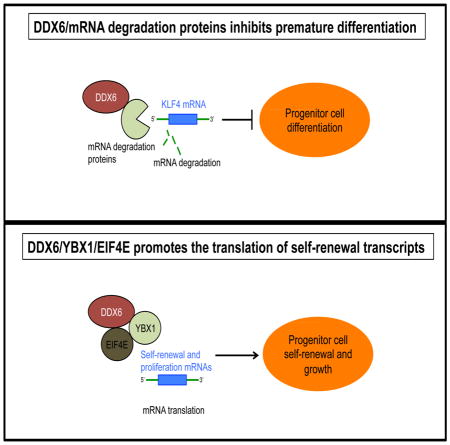
In adult tissues, stem and progenitor cells must balance proliferation and differentiation to maintain homeostasis. How this is done is unclear. Here, we show that the DEAD box RNA helicase, DDX6 is necessary for maintaining adult progenitor cell function. DDX6 loss results in premature differentiation and decreased proliferation of epidermal progenitor cells. To maintain self-renewal, DDX6 associates with YBX1 to bind the stem loops found in the 3′ untranslated regions (UTRs) of regulators of proliferation/self-renewal (CDK1, EZH2) and recruit them to EIF4E to facilitate their translation. To prevent premature differentiation of progenitor cells, DDX6 regulates the 5′UTR of differentiation inducing transcription factor, KLF4 and degrades its transcripts through association with mRNA degradation proteins. Our results demonstrate that progenitor function is maintained by DDX6 complexes through two distinct pathways which include the degradation of differentiation inducing transcripts as well as promoting the translation of self-renewal and proliferation mRNAs.
Keywords: Stem Cell, Differentiation, Epidermis, Skin Differentiation, DDX6, RCK, KLF4, YBX1, YB1, EIF4E, Translation, Initiation, Progenitor Cells, mRNA Degradation, mRNA Decay, CDK1, EZH2, HMGB2, ACTL6A, Myoblasts, Muscle, Post-transcriptional Regulation, Keratinocytes, Self-renewal, Proliferation
Graphical Abstract
INTRODUCTION
Adult stem and progenitor cells act throughout life to restore lost cells due to turnover or injury. The epidermis is an example of a dynamic tissue where differentiated cells are constantly lost from the tissue surface to be replaced from the progeny of stem and progenitor cells that reside in the basal layer of the epithelium and serves as a good model to understand the mechanisms governing stem and progenitor cell self-renewal. Once epidermal progenitor cells commit to differentiation, they stop proliferation, express early differentiation proteins such as keratins 1 and 10, detach from the basement membrane and migrate upwards to form the spinous layer (Sen, 2011). Upon migration to the outer epidermal layers such as the granular layer and then the stratum corneum, these cells express terminal differentiation proteins such as filaggrin, loricrin, and the late cornified envelope proteins, which are necessary for skin barrier function (Segre, 2006). Due to the high turnover rate of the epidermis, the stem and progenitor cells must tightly balance proliferation and differentiation to maintain homeostasis. Diseases that involve alterations in epidermal growth and differentiation impact every one in five people (Lopez-Pajares et al., 2013). Thus, understanding the mechanisms regulating stem and progenitor cell self-renewal and differentiation is of utmost importance. We and others have shown that maintenance of epidermal stem and progenitors cells in a self-renewing and undifferentiated state requires transcriptional regulators such as p63, DNMT1, UHRF1, EZH2, ACTL6a, HDAC1, HDAC2, and ING5 (Bao et al., 2013; Ezhkova et al., 2009; LeBoeuf et al., 2010; Mulder et al., 2012; Sen et al., 2010; Yang et al., 1999). In order to switch from a progenitor to a differentiated cell, epigenetic factors such as the histone demethylase, JMJD3 and transcription factors such as KLF4, GRHL3, and ZNF750 are necessary to induce and maintain the differentiated phenotype (Segre et al., 1999; Sen et al., 2012; Sen et al., 2008; Ting et al., 2005).
Transcriptional regulators such as epigenetic and transcription factors that either maintain progenitor function or promote differentiation have been well described as discussed above. However, it is not known whether there are post-transcriptional mechanisms in place to selectively promote the translation of these self-renewal transcripts while suppressing the expression of differentiation inducing ones to maintain progenitor cell function.
In an attempt to identify regulators of this process, we focused on a small RNA interference (RNAi) screen targeting RNA binding and processing factors and found RCK/p54 (DDX6) to have a prominent role in progenitor cell maintenance. DDX6 is an evolutionarily conserved member of the DEAD-box RNA helicase family that contributes to RNA metabolism (Weston and Sommerville, 2006). With its ATP-dependent RNA unwinding activity, DDX6 is thought to remodel messenger ribonucleoproteins (mRNPs) and to regulate post-transcriptional gene expression.
It is necessary for proper meiotic development in Xenopus, Drosophila, and Caenorhabditis by regulating the translation of mRNPs (Weston and Sommerville, 2006). More recently, DDX6 has been reported to associate with AGO1, AGO2, and the CCR4-NOT complex to promote microRNA (miRNA) mediated gene repression (Chen et al., 2014a; Chu and Rana, 2006; Mathys et al., 2014). DDX6 has also been shown to localize and associate with mRNA degradation proteins (DCP2, DCP1a, EDC3) in processing (P) bodies which may contribute to a role for DDX6 in regulating mRNA storage, translation, and degradation (Arribas-Layton et al., 2013; Franks and Lykke-Andersen, 2008). However, not all of DDX6’s function has been attributed to mRNA repression as DDX6 was recently shown to promote the translation of hepatitis C virus (Scheller et al., 2009). It is currently not known whether DDX6 has any role in progenitor cell maintenance.
Here, we show that DDX6 complexes maintain progenitor cell fate through the mRNA degradation and translation pathways by degrading differentiation inducing transcripts or promoting the translation of self-renewal/proliferation mRNAs.
RESULTS
DDX6 Sustains the Proliferative Capacity of Epidermal Progenitor Cells
To identify genes that regulate progenitor cell function, a small RNAi screen targeting 19 RNA binding or processing factors was performed (Figure S1A). Of the genes targeted, only knockdown of DDX6 had impacts on both proliferation and differentiation (Figure S1B–D). Knockdown of DDX6 using two distinct shRNAs [DDX6i(A) and DDX6i(B)] inhibited proliferation by ~ 75% in primary human epidermal progenitor cells and resulted in increased expression of differentiation gene IVL (Figure S1A–D).
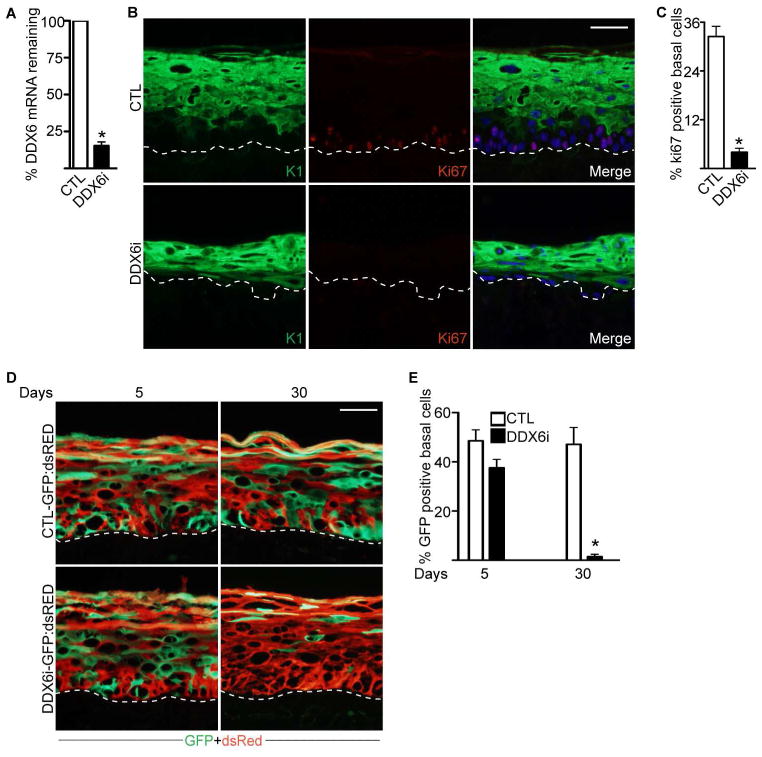
To test the role of DDX6 in a setting which recapitulates the 3D structure and differentiation gene expression program of human epidermis (Khavari, 2006; Sen et al., 2008), DDX6 expression was depleted in regenerated human epidermal tissue (Figure 1A). DDX6 knockdown tissue was extremely hypoplastic with a dramatic decrease in basal layer cells and increased expression of differentiation protein K1 in the normally undifferentiated basal layer (Figure 1B). The hypoplastic tissue suggests that DDX6 is necessary to sustain progenitor function by preventing premature differentiation and maintaining the proliferative capacity of the basal layer. In line with this, loss of DDX6 diminished the proliferative capacity of the basal layer cells to less than 5% (Figure 1A–C). DDX6i cells accumulated in G0/G1 and decreased their S and M phase of the cell cycle (Figure S2A). DDX6 knockdown cells also increased their rate of apoptosis suggesting that the hypoplastic tissue may be the result of increased apoptosis, premature differentiation, and loss of proliferative capacity of epidermal progenitor cells (Figure 1A–C and S2A–B).
Figure 1. DDX6 loss results in premature differentiation of human epidermal tissue.
(A) Epidermal progenitor cells transduced with either control (CTL) or DDX6 shRNAs (DDX6i) were used to regenerate human epidermis by placing the genetically modified cells on devitalized human dermis. RNA was isolated from CTL or DDX6i tissue and RT-QPCR was used to determine the extent of DDX6 knockdown. QPCR results were normalized to GAPDH levels. Error bars=SD, n=3. (B) Staining for differentiation protein keratin 1 (K1) is shown in green and proliferation marker Ki67 in red. Hoechst staining in blue marks the nuclei. The dashed lines denote basement membrane zone (Scale bar=40μm; n=3 regenerated human epidermis per shRNA construct). (C) Quantification of Ki67 positive cells in the basal layer of the epidermis. 500 basal cell nuclei were counted for each regenerated epidermis. Error bars=SD; n=3. (D) In-vivo human epidermal progenitor competition assay. GFP expressing cells were knocked down for DDX6 (DDX6i) or control (CTL) and mixed at a 1:1 ratio with dsRed expressing keratinocytes. The mixed cells were used to regenerate human epidermis and grafted on immune deficient mice and harvested at days 5 and 30 post-grafting. GFP expressing cells are shown in green while dsRed expressing cells are shown in red. Scale bar=40μm; n=4 grafted mice per shRNA construct per timepoint. (E) Quantification of GFP positive cells in the basal layer. Error bars=SD, n=4. *= p<0.05 (T-test) for Figure 1A, C and E.
DDX6 Controls Epidermal Self-renewal through Cell Autonomous Mechanisms
To determine whether DDX6 is necessary for progenitor cell function in-vivo as well as whether DDX6 is acting through cell or non-cell autonomous mechanisms we used the progenitor cell competition assay we previously developed (Mistry et al., 2012; Sen et al., 2010). Primary human epidermal cells were first transduced with retroviral vectors encoding green fluorescent protein (GFP) and then knocked down for either control (CTL: control shRNA) or DDX6. The cells were mixed at a 1:1 ratio with control cells expressing red fluorescent protein (dsRed) and used to regenerate human epidermis on immune compromised mice. Initially, CTL-GFP and DDX6i-GFP cells were present in the basal layer and contributed to all layers of the epidermis (Figure 1D–E). However, by 30 days post-grafting of the tissue on mice, less than 3% of DDX6i-GFP cells were found in the basal layer whereas the vast majority of the DDX6i-GFP cells were found in the upper differentiated layers of the epidermis (Figure 1D–E). The CTL-dsRed cells were also unable to rescue the DDX6i-GFP progenitor cell self-renewal/proliferation defects. In contrast, CTL-GFP cells were found at a similar percentage to CTL-dsRed cells in the basal layer as well as the rest of the epidermis. To rule out off-target RNAi effects, competition assays were performed using DDX6i-GFP cells that have been transduced with retroviruses encoding DDX6 or control beta-galactosidase (LacZ). The exogenous DDX6 (open reading frame) can’t be targeted by the DDX6 shRNA since the shRNA targets the 3′UTR of DDX6. Expression of DDX6 but not LacZ rescued the loss of DDX6i-GFP cells from both the basal layer and the rest of the epidermis (Figure S2C–D). These data suggest that DDX6 is necessary to maintain self-renewal of the epidermis through cell autonomous mechanisms.
DDX6 Controls a Gene Expression Program that Inhibits Differentiation while Maintaining Proliferation
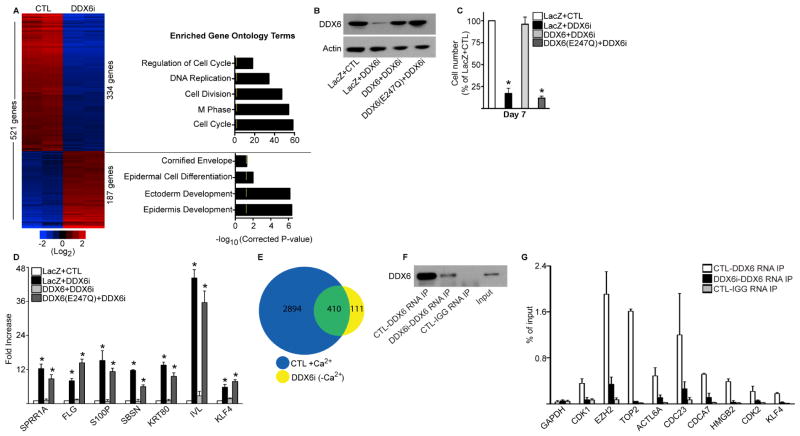
Global gene expression profiling was performed on CTL and DDX6i cells with 521 genes changing significantly upon DDX6 loss (Figure 2A: Left Panel and Table S1). 334 genes with decreased expression were enriched for Gene Ontology (GO) terms such as cell cycle and DNA replication (Figure 2A: Right Panel). This loss of proliferative capacity could be rescued by the expression of wildtype DDX6 in DDX6i cells (Figure 2B–C). In contrast, a single amino acid substitution (DDX6 E247Q) in the DEAD box RNA helicase domain (DEAD to DQAD) predicted to inactivate the RNA unwinding activity of DDX6 failed to rescue the proliferation defects in DDX6i cells (Figure 2B–C)(Qi et al., 2012). 187 upregulated genes were enriched for GO terms such as cornified envelope and epidermal cell differentiation suggesting the DDX6i cells had prematurely differentiated (Figure 2A: Right Panel). This included upregulation of differentiation genes encoding structural and transcription factors such as IVL, SPRR1A, SBSN, FLG, and KLF4 which could be reduced to control levels upon exogenous expression of wildtype but not mutant (E247Q) DDX6 (Figure 2B and 2D). To determine the extent to which DDX6 regulated genes are involved in epidermal differentiation, the genes were compared to our previously generated set of 3,304 genes that were significantly changed during calcium-induced differentiation (Sen et al., 2010). 78.7% (410/521) of the DDX6 regulated genes are differentially expressed differentiation genes (Figure 2E). This suggests that DDX6 sustains progenitor function by suppression of the differentiation program.
Figure 2. DDX6 suppresses the differentiation program by binding to self-renewal, proliferation, and differentiation mRNAs.
(A) Heat map (left panel) of the 521 genes that change significantly upon DDX6 knockdown. Gene ontology analysis (right panel) of the genes with increased or decreased values upon DDX6 knockdown. Yellow mark in bar graphs demark p value=0.5. (B) Western blot for expression of wildtype and mutant DDX6 in DDX6 knockdown cells. The mutant DDX6 (E247Q) contains a single amino acid change in the DEAD box domain (DEAD to DQAD) predicted to abolish the RNA helicase function of the protein. (C) Cells generated in (B) were seeded at 100,000 cells and counted 7 days later. Cell number is represented as a percent of control (LacZ+CTL). Error bars=SD, n=3. *= p<0.05 (T-test), compared to LacZ+CTL samples. (D) Expression of wildtype DDX6 but not DDX6 (E247Q) prevents the premature induction of differentiation genes in DDX6i cells. RT-QPCR on differentiation gene expression. Error bars=SD, n=3. *= p<0.05 (T-test), compared to LacZ+CTL samples. (E) Overlap of differentiation regulated genes with DDX6i genes. 3,304 genes (blue; CTL+Ca2+) change significantly during calcium (+Ca2+) induced epidermal differentiation. These 3,304 genes were overlapped with the 521 genes that change when DDX6 is knocked down in growth conditions (DDX6i (−Ca2+)). Shown in the overlap (410 genes:green) are DDX6 regulated genes that are also differentiation regulated. (F) Western blot for the RNA immunoprecipitations (RNA IP) that were performed using an antibody against DDX6 or IgG on CTL or DDX6i cells. Input=5%. (G) RNA IP was performed as indicated in (F) and RT-QPCR was used to determine the levels of binding of DDX6 to tested transcripts. Binding was calculated as a percent of input. N=3, Error bars=SD.
DDX6 Binds to mRNAs Critical for Proliferation, Self-renewal, and Differentiation
The ability of DDX6 to maintain progenitor function suggests that it may bind to and regulate transcripts that are involved in self-renewal, proliferation, and differentiation. To test this, RNA immunoprecipitations (RNA IP) were performed under native conditions using a DDX6 antibody on cell lysates from control and DDX6i cells (Figure 2F–G). Notably, strong binding of DDX6 to transcripts that are necessary for self-renewal (EZH2, ACTL6A)(Bao et al., 2013; Ezhkova et al., 2009), proliferation (CDK1, CDK2, HMGB2, TOP2, CDC23, CDCA7)(Bermejo et al., 2009; Campbell and Rudnicki, 2013; Gill et al., 2013; Van Hoof et al., 2009) and differentiation (KLF4)(Segre et al., 1999) could be detected in control but not DDX6i cells (Figure 2G). These transcripts were specifically bound to DDX6 as no binding could be detected for GAPDH (Figure 2G).
DDX6 Promotes the Translation of Self-renewal and Proliferation Transcripts
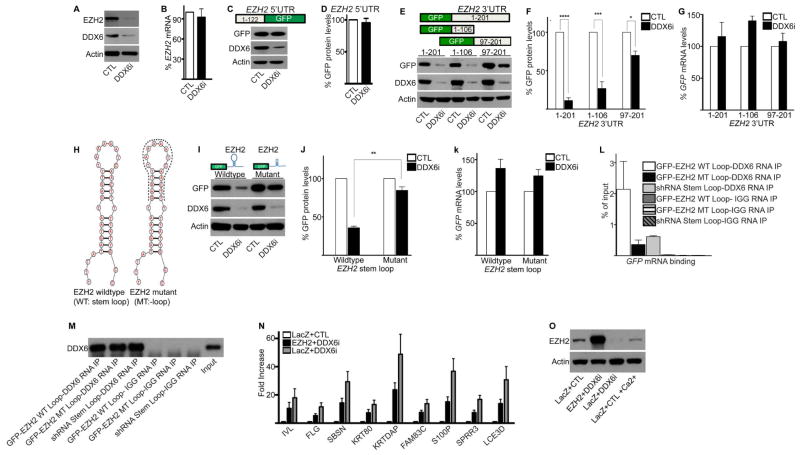
Because EZH2 is necessary for self-renewal of adult and embryonic stem cells, we decided to determine the effects of DDX6 binding to EZH2 transcripts (Ezhkova et al., 2009; Juan et al., 2011; Shen et al., 2008). Since DDX6 has been reported to have roles in translation and mRNA degradation (Scheller et al., 2009; Weston and Sommerville, 2006), EZH2 mRNA and protein levels were measured in control and DDX6i cells. Loss of DDX6 resulted in decreased protein levels of EZH2 whereas there were minimal effects on EZH2 transcripts suggesting that DDX6 may promote the translation of EZH2 (Figure 3A–B). To determine where the DDX6 responsive elements are in the EZH2 transcript, the 5′ UTR (1-122) and 3′ UTR (1-201) of EZH2 was fused immediately upstream or downstream respectively of a green fluorescent protein (GFP) reporter (Figure 3C and 3E). Loss of DDX6 did not impact GFP protein levels in the GFP reporter construct that had been fused to the 5′UTR (1-122) of EZH2 (Figure 3C–D). In contrast, knockdown of DDX6 resulted in reduced protein levels of GFP in constructs that had been linked to the 3′UTR (1-201) of EZH2 (Figure 3E–F). Consistent with regulation on the translational level, GFP transcript levels were unchanged or increased upon DDX6 knockdown (Figure 3G). Removal of 95bp (construct 1-106) from the 3′ end of the EZH2 3′ UTR did not alter DDX6 regulated protein expression of GFP thus resulting in a loss of GFP expression upon DDX6 knockdown (Figure 3E–F). In contrast, removal of 96bp (construct 97-201) from the 5′ end of the EZH2 3′ UTR significantly reduced DDX6 regulation of GFP protein levels (Figure 3E–F). This suggests that the DDX6 regulatory elements are found in the 5′ end of the EZH2 3′UTR. Secondary RNA structure analysis predicted a 38bp stem loop in the first 106bp of the EZH2 3′UTR (Figure 3H). Upon fusion of this stem loop sequence to the GFP reporter construct, the reporter became responsive to DDX6 levels and resulted in decreased GFP protein levels upon DDX6 depletion (Figure 3H–J). Removal of the 18bp that is central to loop formation diminished DDX6 mediated regulation of GFP (Figure 3H–J). Again, the GFP mRNA levels do not dramatically change in the wildtype (WT) or mutant (MT) stem loop constructs due to loss of DDX6 (Figure 3K). To determine if DDX6 binds directly to the stem loop structure in the 3′UTR of EZH2, RNA IP (using DDX6 antibodies) was performed with cells expressing WT or MT stem loop reporter constructs (Figure 3L–M). Binding of DDX6 to GFP mRNA was dependent on an intact EZH2 stem loop structure suggesting that DDX6 recognizes the stem loop structure to promote translation of the transcript (Figure 3L). To test whether DDX6 can recognize any stem loop structure non-specifically, the stem loop structure of the pSuper shRNA was cloned downstream of GFP in the reporter construct (Brummelkamp et al., 2002). Notably, there is minimal binding of DDX6 to the GFP transcripts containing the shRNA stem loop suggesting that DDX6 binding to the EZH2 stem loop is specific (Figure 3L). The importance of EZH2 in promoting self-renewal predicts that part of DDX6’s effects on progenitor function may be mediated through the regulation of EZH2. To test this, EZH2 (containing only the open reading frame) or control LACZ was expressed through retroviruses in control or DDX6i cells. Expression of EZH2 partially rescued the premature differentiation gene induction in DDX6i cells (Figure 3N–O).
Figure 3. DDX6 maintains progenitor function by facilitating the translation of EZH2 through a stem loop in its 3′ UTR.
(A) Western blot for the protein levels of EZH2 and DDX6 in CTL and DDX6i cells. (B) RT-QPCR for the mRNA levels of EZH2 from samples in (A). Error bars=SD, n=3. (C) The 5′UTR (122bp) of EZH2 was fused upstream of the GFP open reading frame in the pLEGFP retroviral vector. Western blotting was performed for GFP on CTL or DDX6i cells expressing this construct. (D) Quantification of samples shown in (C). Error bars=SD, n=3. (E) The 201bp, 3′UTR of EZH2 was fused downstream of the GFP open reading frame in the pLEGFP retroviral vector. Mutant constructs in which the 5′ and 3′ regions of the 3′UTR were deleted are shown in the diagram. These constructs were transduced into keratinocytes and then knocked down for CTL or DDX6. Western blotting was performed to determine GFP and DDX6 protein levels. Representative results are shown, n=3. (F) Quantification of the samples shown in (E). Error bars=SD, n=3. *= p<0.05, ***= p<0.0005, ****=p<0.00005 (T-test). (G) RT-QPCR of GFP mRNA levels for samples shown in (E). (H) Predicted stem loop structure in the first 106bp of the 3′UTR of EZH2 (left panel). Dotted lines in the right panel indicate sequences that were removed to generate the EZH2 mutant construct. (I) Wildtype or mutant stem loop expressing cells were knocked down for CTL or DDX6 and Western blotted for GFP or DDX6 expression. (J) Quantification of GFP expression for samples shown in (I). ** p<0.005. (K) RT-QPCR for GFP mRNA levels for samples shown in (I). (L) RNA IP was performed on extracts isolated from cells expressing the wildtype (WT) or mutant (MT) EZH2 stem loop-GFP using antibodies targeting DDX6 or IGG. DDX6 RNA IP was also performed on keratinocytes expressing the stem loop structure from the pSuper p53 shRNA. RT-QPCR was used to determine the levels of DDX6 binding to GFP mRNA. Error bars=SD, n=3. (M) Efficiency of the DDX6 IPs for samples shown in (L). (N) Keratinocytes expressing LacZ or EZH2 were knocked down for CTL or DDX6. RT-QPCR was used to assay the levels of differentiation genes. Error bars=SD, n=3. (O) Western blot for expression of EZH2 for samples shown in (N).
Our RNA IP results also suggest that DDX6 binding to other proliferation and self-renewal genes may be regulated in the same manner as EZH2. Similar to the 5′UTR of EZH2, there were no impacts on GFP protein levels upon DDX6 knockdown when the 5′UTR of CDK1 was cloned upstream of the GFP reporter (Figure S3A–B). This suggests that the DDX6 regulatory elements are in the 3′UTRs of self-renewal and proliferation mRNAs. To test this, the 3′UTRs of CDK1, HMGB2, and ACTL6a were analyzed to determine if similar stem loop structures could be found. All three genes contained stem loop structures similar to EZH2 (Figure S3C). Each of these stem loops as well as mutants (with loops removed) were fused downstream of the GFP reporter. DDX6 loss led to decreased GFP protein expression with minimal changes in transcript levels of cells expressing wildtype stem loops (Figure S3D–F). In contrast, deletion of the loop abrogated DDX6’s ability to regulate its protein expression suggesting that DDX6 facilitates the translation of self-renewal and proliferation transcripts through binding secondary/tertiary stem loop structures rather than through a specific sequence motif (Figure S3D–F).
DDX6 Prevents Premature Differentiation of Progenitor Cells by Targeting and Destabilizing KLF4 mRNA
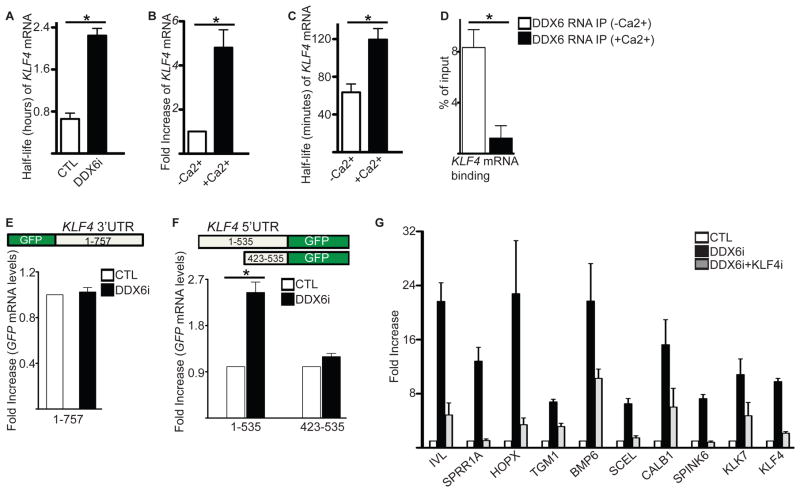
The RNA IP results also showed that DDX6 bound to KLF4 mRNA which encodes a transcription factor (Segre et al., 1999) necessary for terminal epidermal differentiation which may be mediating part of DDX6’s loss of function differentiation inducing phenotype (Figure 2G). Loss of DDX6 increased the levels of KLF4 transcripts by ~6 fold and increased its stability by more than 3 fold (Figure 2D and Figure 4A). KLF4 mRNA levels also increased and became more stable during differentiation due to loss of DDX6 binding during differentiation (Figure 4B–D). This differential binding and destabilization by DDX6 allows KLF4 levels to be kept low in progenitor cells and rapidly increased upon differentiation. To identify the DDX6 regulatory regions of KLF4, the 5′ or 3′UTR was fused to the GFP reporter construct. Loss of DDX6 increased GFP mRNA levels in the reporters fused with the 535 bp KLF4 5′UTR (1-535) whereas no changes was found with the 757 bp KLF4 3′UTR(Figure 4E–F). Analysis of the 5′UTR of KLF4, showed that it contained CpG rich regions. CpG rich 5′UTR containing mRNAs were recently identified by microarray based mRNA stability assays to destabilize transcripts and thus have short half-lives (Sharova et al., 2009). Removal of the CpG rich region in the reporter assays (construct 423-535) prevented the increase in GFP mRNA levels upon DDX6 knockdown (Figure 4F). To determine if the increased differentiation gene expression seen in DDX6i cells is mediated through increased KLF4 levels, KLF4 and DDX6 were simultaneously knocked down. Knockdown of DDX6 resulted in increased levels of KLF4 transcripts, which could be restored similar to control levels upon double knockdown with KLF4 (Figure 4G). The mRNA levels of differentiation induced genes were increased in DDX6i cells but were partially restored to control levels in KLF4 and DDX6 double knockdown cells (Figure 4G). These results suggest that DDX6 prevents the premature expression of differentiation genes in progenitor cells by destabilizing KLF4 transcripts.
Figure 4. DDX6 prevents premature differentiation in progenitor cells by destabilizing KLF4 transcripts through GC rich regions in its 5′ UTR.
(A) CTL and DDX6i cells grown in proliferating conditions were treated with actinomycin D to determine the half-life of KLF4 mRNA. RT-QPCR was used to determine the levels of KLF4. Error bars=SD, n=3. *= p<0.05 (T-test). (B) RT-QPCR measurements on levels of KLF4 in proliferating (−Ca2+) and differentiated (+Ca2+, day 3) keratinocytes. Error bars=SD, n=3. *= p<0.05 (T-test). (C) Cells grown in +/− Ca2+ conditions were treated with actinomycin D to determine the half-life of KLF4 mRNA. Error bars=SD, n=3. *= p<0.05 (T-test). (D) RNA IP was performed on extracts isolated from +/− Ca2+ treated cells using a DDX6 antibody. RT-QPCR was used to determine DDX6 binding to KLF4 transcripts. Error bars=SD, n=3. *= p<0.05 (T-test). (E) The 3′UTR of KLF4 was fused downstream of GFP in the pLEGFP retroviral vector. Keratinoctyes expressing this construct were knocked down for CTL or DDX6 and levels of GFP mRNA expression quantitated by RT-QPCR. (F) The 5′ UTR of KLF4 was fused upstream of GFP in the pLEGFP retroviral vector (1-535). A mutant construct where 422bp of high GC rich content was deleted from the 5′ end of the 5′UTR was also generated (423-535). Wildtype and mutant construct expressing keratinocytes were knocked down for CTL or DDX6 and GFP mRNA expression quantitated by RT-QPCR. Error bars=SD, n=3. *= p<0.05 (T-test). (G) Double knockdown of KLF4 and DDX6 can partially prevent the increase in differentiation gene expression seen in DDX6i cells. RT-QPCR was used to assay the levels of differentiation genes. Error bars=SD, n=3.
DDX6 Associates with RNA Binding Proteins and Mediators of the mRNA Degradation and Translation Pathways
To understand how DDX6 maintains progenitor function through both the translation and mRNA degradation pathways, mass spectrometry was used to identify the proteins associated with it (Figure S4A–C and Table S2). Nonspecific interactions were subtracted out by performing DDX6 IP from DDX6 knockdown cells as well as control IgG pulldowns. 48 proteins were found to be associated with DDX6 which were enriched for GO terms such as RNA binding, regulation of mRNA stability, and translation (Figure S4B–C and Table S2). These include RNA binding proteins (YBX1, IGF2BP2, FXR1), proteins involved in mRNA degradation (EDC3, LSM family, DCP1A, PATL1), and mediators of translation (EIF4E, RPL23A, RPS14).
EIF4E is Necessary for the Translation of Self-renewal and Proliferation mRNAs
Since EIF4E is one of the rate limiting steps of translation initiation (Raught and Gingras, 1999), association of DDX6 with EIF4E may allow for selective translation of bound proliferation and self-renewal transcripts to maintain progenitor function. Supporting this, knockdown of EIF4E inhibited the protein expression of EZH2 and CDK1 with minimal impacts on DDX6 or Actin levels (Figure 5A–B). Similar to DDX6 loss, knockdown of EIF4E also inhibited cell proliferation (Figure 5C). While EIF4E can bind to all mRNAs containing a 5′ cap, the association of EIF4E with self-renewal/proliferation transcripts was dependent on the presence of DDX6 suggesting additional modes of regulation (Figure 5D–E). The stem loop reporter constructs for CDK1, ACTL6a, HMGB2, and EZH2 were also regulated by EIF4E as EIF4E knockdown resulted in decreased GFP protein expression with minimal changes in GFP transcript levels (Figure 5F–H). Removal of the loop abrogated EIF4E regulation of translation suggesting that translation of self-renewal and proliferation transcripts are dependent on the loop structure (Figure 5F–H).
Figure 5. EIF4E is necessary for the translation of self-renewal and proliferation transcripts.
(A) Protein levels of EIF4E, DDX6, EZH2, CDK1, and Actin upon knockdown of EIF4E. Cells were knocked down for EIF4E using two distinct shRNAs: EIF4E(A)i and EIF4E(B)i. Representative blots are shown, n=3. (B) Quantification of the proteins levels of EZH2, DDX6, and CDK1 in CTL or EIF4Ei cells. Samples were normalized to actin expression. ***=p<0.0005, ****=p<0.00005 (T-test). (C) Keratinocytes transduced with shRNAs targeting EIF4E or control (CTL) were counted over a period of 11 days to determine impacts on cell proliferation. Error bars=SD, n=2. (D) RNA IP was performed using an antibody against EIF4E or IgG on CTL or EIF4Ei cells. RT-QPCR was used to determine the levels of EIF4E binding to target transcripts. Error bars=SD, n=3. *= p<0.05 (T-test), CTL-EIF4E RNA IP vs CTL-IGG RNA IP. (E) RNA IP was performed using an EIF4E antibody or IgG on CTL or DDX6i cells. *= p<0.05 (T-test), CTL-EIF4E RNA IP vs CTL-IGG RNA IP. (F) Cells expressing wildtype or mutant stem loop sequences of CDK1, EZH2, ACTL6A, and HMGB2 fused downstream of GFP were knocked down for CTL or EIF4E. GFP and EIF4E protein expression is shown. n=3. (G) Quantification of GFP expression for samples shown in (F). *= p<0.05, ***=p<0.0005. (H) RT-QPCR for GFP mRNA levels for samples shown in (F). Error bars=SD, n=3.
EDC3 Regulates the mRNA Levels of KLF4
Our mass spectrometry data also showed that DDX6 associates with mRNA degradation components such as the enhancer of mRNA decapping (EDC3) which has been shown in yeast to promote mRNA decapping and degradation (Badis et al., 2004). Thus the association of DDX6 with EDC3 may mediate the degradation of KLF4 mRNA. Supporting this, knockdown of EDC3 also resulted in an increase in expression of KLF4 transcripts and differentiation genes (Figure S5A). Notably, no changes to EZH2 or CDK1 protein levels were detected in EDC3 knockdown cells suggesting that the mRNA degradation and translation pathways are separate (Figure S5B). Knockdown of EDC3 also resulted in an increase in GFP mRNA levels in constructs that were fused to the full length KFL4 5′UTR GFP constructs (Figure S5C).
YBX1 Recruits DDX6 and EIF4E to target mRNAs
The association of DDX6 with different RNA binding proteins suggests that they may be recruiting DDX6 to specific transcripts (Figure S4C). To test this, the RNA binding proteins that we identified by mass spectrometry to associate with DDX6 were knocked down ≥ 75% by RT-QPCR (Figure 6A). Of the factors tested, only knockdown of YBX1 reduced the protein levels of EZH2 and CDK1 and none had any impacts on KLF4 mRNA levels (Figure 6B–C). In progenitor cells, YBX1 bound to self-renewal and proliferation transcripts which was abrogated in YBX1i cells (Figure 6D). Interestingly, YBX1 was able to bind self-renewal and proliferation transcripts even in the absence of DDX6 whereas loss of YBX1 abrogated DDX6’s ability to bind to the transcripts (Figure 6E–F). These effects were not due to loss of DDX6 protein levels as YBX1 depletion did not impact DDX6 expression (Figure 6B). Similarly, loss of YBX1 prevents EIF4E binding to the same mRNAs while having no impact on EIF4E protein expression (Figure 6G and 6B). These results suggest that YBX1 is recruiting the mRNAs to DDX6 and EIF4E (Figure 6E–G). Knockdown of YBX1 also resulted in diminished GFP protein expression in EZH2 and CDK1 stem-loop constructs whereas removal of the loop abolished this regulation (Figure 6H–I). Minimal changes to GFP transcripts levels were detected suggesting regulation on the translational level (Figure 6J).
Figure 6. YBX1 recruits DDX6 and EIF4E to facilitate the translation of self-renewal and proliferation transcripts.
(A) Knockdown of each gene shown on X axis. QPCR results were normalized to GAPDH levels. Error bars=SD, n=3. (B) RNA binding proteins were knocked down and impacts on EZH2, CDK1, YBX1, DDX6, and EIF4E expression were assayed by Western blotting. (C) Impacts on KLF4 mRNA levels were determined upon knockdown of the RNA binding proteins. Error bars=SD, n=3. (D) RNA IP was performed using an antibody against YBX1 or IgG on CTL or YBX1i cells. RT-QPCR was used to determine the levels of YBX1 binding to transcripts. Error bars=SD, n=3. *= p<0.05 (T-test), CTL-YBX1 RNA IP vs YBX1i-YBX1i RNA IP samples. (E) RNA IP was performed using an antibody against YBX1 or IgG on CTL or DDX6i cells. n.s.= not significant (p>0.05 T-test), CTL-YBX1 RNA IP vs DDX6i-YBX1 RNA IP. (F) RNA IP was performed using an antibody against DDX6 or IgG on CTL or YBX1i cells. *= p<0.05 (T-test), CTL-DDX6 RNA IP vs YBX1i-DDX6 RNA IP. (G) RNA IP was performed using an antibody against EIF4E or IgG on CTL or YBX1i cells. *= p<0.05 (T-test). (H) EZH2 and CDK1 wildtype and mutant stem loop-GFP expressing keratinocytes were knocked down for YBX1 or CTL. Protein levels of GFP and YBX1 were determined by Western blotting. (I) Quantification of GFP expression from samples in (H). Error bars=SD, n=3: *= p<0.05, ***=p<0.0005. (J) GFP mRNA levels were determined by RT-QPCR from cells isolated in (H). Error bars=SD, n=3.
DDX6 Associates with Mediators of mRNA Degradation and Translation in Separate Complexes
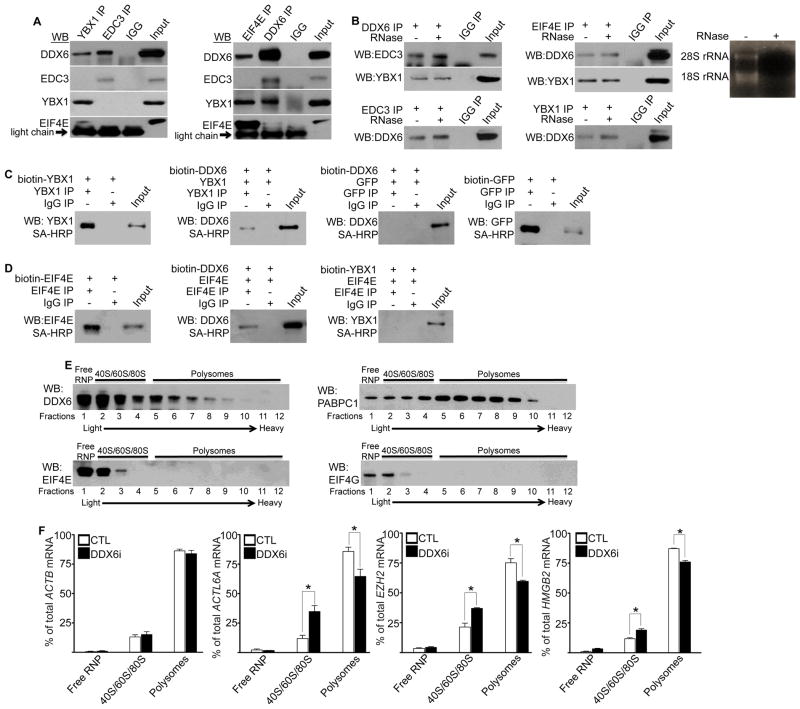
Our data suggests that DDX6 acts to facilitate translation of self-renewal and proliferation transcripts while also being able to mediate degradation of pro-differentiation mRNAs through association with mediators of each pathway. To determine if DDX6 is part of two distinct pathways or whether the translation and degradation pathways intersect, co-IPs were performed. DDX6 was able to associate with components of both pathways including YBX1, EIF4E, and EDC3 (Figure 7A). YBX1 was found to co-precipitate with EIF4E and DDX6 but not EDC3 (Figure 7A). EIF4E could pull down DDX6 and YBX1 but not EDC3 (Figure 7A). Similarly, EDC3 could only associate with DDX6 and not with EIF4E or YBX1 (Figure 7A). These associations are not dependent on RNA as RNase A treatment failed to disrupt the interaction between DDX6 and YBX1, EIF4E or EDC3 (Figure 7B). To determine whether the association between DDX6, YBX1, and EIF4E is direct, these proteins were in-vitro transcribed and translated. Recombinant DDX6, YBX1, and EIF4E were biotinylated so that the proteins could be detected by streptavidin linked to horseradish peroxidase (SA-HRP). Biotin labeled DDX6 was mixed with either unlabeled YBX1 or GFP. An antibody against YBX1 or GFP was used to IP each protein and then SA-HRP was used to determine if DDX6 was pulled down with the protein. YBX1 but not GFP could co-IP biotin labeled DDX6 suggesting that the interaction is direct (Figure 7C). Biotin labeled YBX1 and GFP were used as positive controls for the efficiency of the IPs using their respective antibodies (Figure 7C). IgG IPs were also used as controls for specificity (Figure 7C). To test if EIF4E directly interacted with both DDX6 and YBX1, both proteins were biotin labeled and an EIF4E antibody used to pull down unlabeled EIF4E with either labeled YBX1 or DDX6 (Figure 7D). Interestingly, EIF4E directly interacted with DDX6 but not YBX1 suggesting that the interaction between YBX1 and EIF4E detected in-vivo may be bridged via other proteins such as DDX6 (Figure 7D). These results suggest that DDX6 directly associates with EIF4E and YBX1.
Figure 7. DDX6 associates with mRNA translation and degradation proteins in distinct complexes and is necessary for translation of self-renewal and proliferation transcripts.
(A) IPs were performed using YBX1, EDC3, EIF4E, or DDX6 antibodies and blotted (WB) for the indicated proteins. Representative results are shown, n=3. (B) IPs were treated +/− RNase A. Pulldowns were performed using YBX1, EDC3, EIF4E, or DDX6 antibodies and blotted for the indicated proteins. Ribosomal RNA (28S and 18S rRNA) digestion with RNase A was used as a positive control. 5% of the cell lysate was used as input. (C) In-vitro transcribed/translated YBX1, DDX6, or GFP were biotin labeled. Biotin labeled DDX6 was mixed at a 1:1 ratio with unlabeled YBX1 or GFP. An YBX1 or GFP antibody was used to IP YBX1 or GFP. Streptavidin conjugated horseradish peroxidase (SA-HRP) was used to determine if biotin labeled DDX6 was brought down with YBX1 or GFP. IgG IP was used as a negative control. Biotin labeled YBX1 and GFP were IP’d using their respective antibodies and detected by SA-HRP to determine the efficiency of the pulldown. Input=5%. (D) Biotin labeled DDX6 or YBX1 were mixed at equal ratios with EIF4E. An antibody against EIF4E was used to IP EIF4E. SA-HRP was used to detect whether biotin labeled DDX6 or YBX1 was brought down with EIF4E. Input=5%. (E) Cell lysates were fractionated through sucrose density gradients. Ribosomal fractions (40S, 60S, 80S and polysomes) were determined by OD 254nm measurements and ribosomal RNA staining. The distribution of each protein in the different fractions was visualized by Western blotting (WB). (F) Lysates from CTL or DDX6 knockdown cells were fractionated through sucrose density gradients. RT-QPCR was used to determine the distribution of ACTL6A, EZH2, HMGB2, and ACTB mRNA in either the polysome, 40S/60S/80S, or free RNP. Results were calculated as a percentage of the total RNA. Error bars=SD, n=3: *= p<0.05 (T-test).
DDX6 Associates with Fractions Containing Polysomes and is Necessary for the Translation of Target mRNAs
To determine if DDX6 has a direct role in promoting translation by associating with fractions containing polysomes, cell extracts were fractionated on a sucrose density gradient. DDX6 could be found in the lighter fractions containing translation initiation factors EIF4E and EIF4G (Figure 7E). This suggests the interaction between DDX6 and EIF4E occurs in the lighter fractions. DDX6 could also be found in heavier fractions associated with polysomes suggesting a direct role in actively translating mRNA. Notably, known polysome associated protein, PABPC1 was also found in similar fractions as DDX6 and was found in our mass spectrometry data to associate with DDX6 (Figure 7E and Table S2) (Lemieux and Bachand, 2009). To test if knockdown of DDX6 had any impacts on self-renewal and proliferation transcripts associating with polysomes, RT-QPCR was performed on ACTL6A, EZH2, and HMGB2 on control and DDX6i fractionated cell extracts. Loss of DDX6 shifted the transcripts from the polysomal fractions to lighter fractions (containing 40S/60S/80S) suggesting that DDX6 is necessary for the loading of these mRNAs onto polysomes (Figure 7F). The loss of association of self-renewal and proliferation transcripts with polysomes upon DDX6 knockdown is specific as no changes in actin (ACTB) mRNA association with polysomal fractions were detected in DDX6i cells (Figure 7F).
DDX6 Maintains Muscle Progenitor Cell Function through the Translation of Self-Renewal and Proliferation mRNAs
To determine whether DDX6 is a global regulator of progenitor status in adult progenitor cells, DDX6 was knocked down in human muscle progenitors. Loss of DDX6 resulted in increased expression of muscle differentiation genes such as transcription factors (MYOG), structural genes (MYL1, MYL2, MYL3, TNNT3) and enzymes (TTN, PTGIS) that are essential to muscle differentiation (Figure S6A). DDX6 loss also inhibited cell proliferation due to cells withdrawing from the S phase and accumulating in G0/G1 phase of the cell cycle (Figure S6B–C). Strikingly, DDX6 bound to the same self-renewal and proliferation mRNAs in both cell types (Figure S6D). DDX6 was also required to facilitate the translation of GFP reporters that have been linked to the 3′ UTRs of CDK1, ACTL6A, HMGB2, and EZH2 (Figure S6E–F). Again, no significant changes were detected in GFP mRNA levels upon DDX6 depletion (Figure S6G). These results suggest that DDX6 maintains adult progenitor function through a conserved mechanism.
DISCUSSION
Here, we examined the role of DDX6 in the epidermis and found it to maintain epidermal progenitor function by two distinct mechanisms.
First, DDX6 associates with the mRNAs of genes critical for proliferation and self-renewal to promote their translation. DDX6 associates with stem loop structures in the 3′UTRs of CDK1, HMGB2, ACTL6a, and EZH2 to promote translation. The loop is critical to DDX6 regulation as removal of it in reporter constructs diminished DDX6’s ability to bind and regulate its protein expression. It is also interesting to note that the stem loop structure itself has a translation inhibitory effect in the absence of DDX6. Only upon availability and binding of DDX6 are the transcripts bearing these stem loops translated. This may potentially be a mechanism to keep stem cells quiescent in-vivo where the DDX6 complex may not be able to target these mRNAs for translation until signals are sent for the cells to divide/self-renewal.
In this study, we also determined whether part of the impacts of DDX6 on self-renewal and differentiation is mediated through EZH2. EZH2 is a global regulator of self-renewal in both embryonic and adult stem cells (Ezhkova et al., 2009; Juan et al., 2011; Shen et al., 2008). Importantly, expression of exogenous EZH2 could partially inhibit the upregulation of differentiation induced genes seen in DDX6i cells. It is unlikely that the DDX6 loss of function phenotype could be fully rescued since the phenotype is likely due to a combinatorial loss of translation of both proliferation and self-renewal genes as well as the points mentioned immediately below.
Second, DDX6 also binds to the mRNAs of differentiation inducing factors such as KLF4 to promote their degradation. KLF4 is a transcription factor necessary for activating the epidermal differentiation gene expression program as well as being necessary for the conversion of fibroblasts to keratinocyte-like cells (Chen et al., 2014b; Mistry et al., 2014; Segre et al., 1999). Thus, regulation of this key factor is essential to epidermal cell fate. Strikingly, knockdown of DDX6 in epidermal progenitor cells resulted in stabilization of the half-life of KLF4 transcripts which led to an increase in its mRNA levels. DDX6 regulates the GC rich regions in the 5′ UTR of KLF4 to promote its degradation. Removal of the GC rich 5′ UTR sequence of KLF4 in GFP reporter constructs prevented DDX6 regulation of the transcript. However it is still unclear which specific sequences within the 5′UTR of KLF4 triggers DDX6 mediated degradation since the GC-rich region comprises ~400 nucleotides of sequence. Importantly, part of the effect DDX6 has on differentiation is mediated through KLF4 since double knockdown of DDX6 and KLF4 could partially prevent the induction of differentiation genes seen in DDX6i cells. This suggests that DDX6 prevents premature differentiation in progenitor cells by targeting and degrading KLF4 transcripts. Upon induction of differentiation, DDX6 no longer binds to KLF4 transcripts which leads to an increase in half-life as well as an increase in its mRNA levels. The increase in KLF4 levels then allows it to transcriptionally activate the differentiation program.
To our knowledge, a mechanism that promotes the translation of genes involved in the regulation of proliferation and self-renewal while degrading differentiation inducing transcripts in stem or progenitor cells has not been described previously. Prior reports have identified RNA regulators such as STAU1 which associates with a long non-coding RNA, TINCR, to stabilize differentiation related mRNAs to promote epidermal differentiation (Kretz et al., 2013). In embryonic stem cells, Thoc2 and Thoc5 maintain self-renewal by promoting the nuclear export of pluripotency transcripts such as Nanog and Sox2(Wang et al., 2013). Since DDX6 acts at the nexus between the mRNA translation and degradation pathway as well as behaves mechanistically distinct from previously described mechanisms of promoting self-renewal or differentiation, we performed mass spectrometry to determine its associated proteins. DDX6 has been previously described to be part of a ribonucleoprotein complex containing mRNA decay proteins such as EDC3, DCP1a, and the LSM family of proteins (Arribas-Layton et al., 2013; Franks and Lykke-Andersen, 2008). These proteins were also identified in our data. This explains DDX6’s impacts on the stability of KLF4 mRNA. Supporting this, knockdown of EDC3 also resulted in increased KLF4 levels and differentiation gene expression. These results suggest DDX6 and EDC3 mediate the degradation of KLF4 in order to prevent premature differentiation of progenitor cells.
Our mass spectrometry data also identified several RNA binding proteins associated with DDX6. Of the 6 tested only YBX1 knockdown mimicked loss of DDX6 function with decreased protein expression of both CDK1 and EZH2. YBX1 has been shown to promote the translation of mRNAs involved in epithelial to mesenchymal transition (EMT) in cancer cells through the recognition of stem loop structures found in their 5′ UTRs (Evdokimova et al., 2009). Here we demonstrate that the function of YBX1 in epidermal progenitor cells is to recruit DDX6 and EIF4E to self-renewal and proliferation transcripts through the recognition of stem loop structures in the 3′UTR to promote their translation. In contrast, YBX1 has been shown in cancer cell lines to block the translation of proliferation transcripts (Evdokimova et al., 2006; Evdokimova et al., 2009). This difference in targeting mechanism (5′UTR versus 3′UTR) as well as impacts on proliferation transcripts may potentially be due to the differences between transformed and primary cells.
Our model for YBX1 recruiting DDX6 and EIF4E to self-renewal and proliferation transcripts is supported by our RNA IP results. These results demonstrate that in the absence of YBX1, neither DDX6 nor EIF4E could efficiently associate with their target mRNAs whereas loss of DDX6 has no impact on YBX1 transcript binding. In addition there is a direct interaction between YBX1 and DDX6 as well as between DDX6 and EIF4E. Once YBX1 recruits the transcripts to EIF4E and DDX6, EIF4E promotes the translation initiation of the mRNAs. DDX6 is then necessary for the transition of the targeted mRNAs to polysomes. This is supported by the cell lysate fractionation experiments where knockdown of DDX6 shifted the mRNAs of self-renewal and proliferation transcripts from polysomal to lighter fractions suggesting that DDX6 is essential for the loading of targeted transcripts onto polysomes. DDX6 may play a role in translation by unwinding the secondary structures of its target transcripts through its DEAD box helicase domain. Supporting the importance of the helicase activity of DDX6, point mutations that abolished the helicase activity of DDX6 (DEAD to DQAD) failed to rescue the proliferation and differentiation defects seen in DDX6 knockdown cells.
Our results also suggest that the mRNA degradation and translation pathways are distinct with the exception of DDX6 being critical to both pathways. In support of this, loss of EDC3 had no impacts on the protein levels of CDK1 or EZH2 while YBX1 knockdown had no effect on KLF4 mRNA levels. Furthermore, YBX1 and EIF4E did not associate with EDC3. These results suggest that YBX1 directs DDX6 and EIF4E towards translation of stem loop containing proliferation and self-renewal transcripts while DDX6 associates with mediators of mRNA decay such as EDC3 to destabilize differentiation inducing mRNAs such as KLF4 in separate complexes.
In conclusion, we have addressed the fundamental question of how the expression of self-renewal and proliferation proteins is maintained and how expression of differentiation promoting mRNAs is kept low in adult progenitor cells. DDX6 complexes achieve this by targeting specific transcripts to the mRNA degradation and translation pathways.
EXPERIMENTAL PROCEDURES
The Supplemental Experimental Procedures section which includes a detailed description of the protocols as well as a list of retroviral constructs, siRNAs, antibodies, and primer sequences is available online.
Retroviral transduction
The retroviral constructs (3ug) were transfected using Fugene 6 (Roche) into amphotrophic phoenix cells to knockdown or overexpress genes. Viral supernatants were collected 48 hours post-transfection and used to infect primary human keratinocytes or myoblasts. Cells were incubated in the viral supernatants and centrifuged at 1000 rpm for 1 hour with the addition of polybrene (5ug/ml). Cells were transduced on two consecutive days with the shRNA retroviral constructs and then selected using puromycin (2ug/ml). Cells were transduced once for the LZRS retroviral overexpression constructs and pLEGFP reporter constructs. For all reporter assays (pLEGFP): cells were first transduced with the pLEGFP reporter and subsequently split into two dishes to be transduced with control or shRNA knockdown (DDX6i, YBX1i, or EIF4Ei). This was done to ensure that control and shRNA knockdown groups contained equal levels of transduced reporter construct.
Cell culture
Primary human epidermal keratinocytes were derived from neonatal foreskin and cultured in growth medium (Life Technologies, KSFM) as previously described (Sen et al., 2010; Sen et al., 2008). Cells were induced to differentiate by the addition of 1.2mM calcium for 3 days in full confluence. Primary human skeletal myoblasts were purchased from Cell Applications and cultured according to manufacturer’s protocols. Phoenix cells were cultured in DMEM with 10% fetal calf serum.
Retroviral constructs
Retroviral constructs to knockdown genes were generated by cloning oligos into the pSuper retroviral vector as previously described (Mistry et al., 2012). The full-length open reading frames of DDX6 and EZH2 were cloned into the LZRS retroviral vector. The full length 3′UTRs (as well as any truncations/stem loops) of CDK1, ACTL6a, HMGB2, EZH2, and KLF4 were cloned into the pLEGFP retroviral construct immediately downstream of GFP. The 5′UTR (as well as any truncations) of KLF4 were cloned into the pLEGFP construct immediately upstream of GFP.
Western blotting and immunofluorescence
20ug of proteins were used for western blotting. For immunofluorescence, 7μm thick sections taken from regenerated human epidermis were used.
Microarray analysis
Cells knocked down for DDX6 or control were harvested one week after puromycin selection. Microarray analysis using Affymetrix HG-U133 2.0 plus arrays was performed on duplicate samples. Significantly changed genes were identified as previously described (Mistry et al., 2012; Mistry et al., 2015; Sen et al., 2010). Data was deposited in GEO with accession number: GSE62417.
Regenerated human epidermis
To generate human epidermis as organotypic cultures, 1million control or DDX6 knockdown cells were seeded on devitalized human dermis (Sen et al., 2008). To regenerate human epidermis on immune compromised mice, cells were first seeded on devitalized human dermis and then grafted onto SCID mice as previously described (Mistry et al., 2012).
Supplementary Material
Highlights.
DDX6 is necessary for epidermal progenitor cell function.
YBX1 recruits DDX6 and EIF4E to self-renewal mRNAs to promote their translation.
DDX6 binds stem loop structures in the 3′UTRs of mRNAs to promote polysome loading.
DDX6 associates with EDC3 to destabilize KLF4 mRNA to prevent differentiation.
Acknowledgments
We thank N. Ling and J. Li for pre-submission review and helpful discussions. This work was supported by grants from the California Institute of Regenerative Medicine (CIRM:RB4-05779) and the National Institutes of Health (NIH R01AR066530-01A1).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arribas-Layton M, Wu D, Lykke-Andersen J, Song H. Structural and functional control of the eukaryotic mRNA decapping machinery. Biochim Biophys Acta. 2013;1829:580–589. doi: 10.1016/j.bbagrm.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badis G, Saveanu C, Fromont-Racine M, Jacquier A. Targeted mRNA degradation by deadenylation-independent decapping. Mol Cell. 2004;15:5–15. doi: 10.1016/j.molcel.2004.06.028. [DOI] [PubMed] [Google Scholar]
- Bao X, Tang J, Lopez-Pajares V, Tao S, Qu K, Crabtree GR, Khavari PA. ACTL6a enforces the epidermal progenitor state by suppressing SWI/SNF-dependent induction of KLF4. Cell Stem Cell. 2013;12:193–203. doi: 10.1016/j.stem.2012.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermejo R, Capra T, Gonzalez-Huici V, Fachinetti D, Cocito A, Natoli G, Katou Y, Mori H, Kurokawa K, Shirahige K, et al. Genome-organizing factors Top2 and Hmo1 prevent chromosome fragility at sites of S phase transcription. Cell. 2009;138:870–884. doi: 10.1016/j.cell.2009.06.022. [DOI] [PubMed] [Google Scholar]
- Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- Campbell PA, Rudnicki MA. Oct4 interaction with hmgb2 regulates akt signaling and pluripotency. Stem Cells. 2013;31:1107–1120. doi: 10.1002/stem.1365. [DOI] [PubMed] [Google Scholar]
- Chen Y, Boland A, Kuzuoglu-Ozturk D, Bawankar P, Loh B, Chang CT, Weichenrieder O, Izaurralde E. A DDX6-CNOT1 complex and W-binding pockets in CNOT9 reveal direct links between miRNA target recognition and silencing. Mol Cell. 2014a;54:737–750. doi: 10.1016/j.molcel.2014.03.034. [DOI] [PubMed] [Google Scholar]
- Chen Y, Mistry DS, Sen GL. Highly rapid and efficient conversion of human fibroblasts to keratinocyte-like cells. J Invest Dermatol. 2014b;134:335–344. doi: 10.1038/jid.2013.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu CY, Rana TM. Translation repression in human cells by microRNA-induced gene silencing requires RCK/p54. PLoS Biol. 2006;4:e210. doi: 10.1371/journal.pbio.0040210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evdokimova V, Ruzanov P, Anglesio MS, Sorokin AV, Ovchinnikov LP, Buckley J, Triche TJ, Sonenberg N, Sorensen PH. Akt-mediated YB-1 phosphorylation activates translation of silent mRNA species. Mol Cell Biol. 2006;26:277–292. doi: 10.1128/MCB.26.1.277-292.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evdokimova V, Tognon C, Ng T, Ruzanov P, Melnyk N, Fink D, Sorokin A, Ovchinnikov LP, Davicioni E, Triche TJ, et al. Translational activation of snail1 and other developmentally regulated transcription factors by YB-1 promotes an epithelial-mesenchymal transition. Cancer Cell. 2009;15:402–415. doi: 10.1016/j.ccr.2009.03.017. [DOI] [PubMed] [Google Scholar]
- Ezhkova E, Pasolli HA, Parker JS, Stokes N, Su IH, Hannon G, Tarakhovsky A, Fuchs E. Ezh2 orchestrates gene expression for the stepwise differentiation of tissue-specific stem cells. Cell. 2009;136:1122–1135. doi: 10.1016/j.cell.2008.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks TM, Lykke-Andersen J. The control of mRNA decapping and P-body formation. Mol Cell. 2008;32:605–615. doi: 10.1016/j.molcel.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill RM, Gabor TV, Couzens AL, Scheid MP. The MYC-associated protein CDCA7 is phosphorylated by AKT to regulate MYC-dependent apoptosis and transformation. Mol Cell Biol. 2013;33:498–513. doi: 10.1128/MCB.00276-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juan AH, Derfoul A, Feng X, Ryall JG, Dell’Orso S, Pasut A, Zare H, Simone JM, Rudnicki MA, Sartorelli V. Polycomb EZH2 controls self-renewal and safeguards the transcriptional identity of skeletal muscle stem cells. Genes Dev. 2011;25:789–794. doi: 10.1101/gad.2027911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khavari PA. Modelling cancer in human skin tissue. Nat Rev Cancer. 2006;6:270–280. doi: 10.1038/nrc1838. [DOI] [PubMed] [Google Scholar]
- Kretz M, Siprashvili Z, Chu C, Webster DE, Zehnder A, Qu K, Lee CS, Flockhart RJ, Groff AF, Chow J, et al. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature. 2013;493:231–235. doi: 10.1038/nature11661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBoeuf M, Terrell A, Trivedi S, Sinha S, Epstein JA, Olson EN, Morrisey EE, Millar SE. Hdac1 and Hdac2 act redundantly to control p63 and p53 functions in epidermal progenitor cells. Dev Cell. 2010;19:807–818. doi: 10.1016/j.devcel.2010.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemieux C, Bachand F. Cotranscriptional recruitment of the nuclear poly(A)-binding protein Pab2 to nascent transcripts and association with translating mRNPs. Nucleic Acids Res. 2009;37:3418–3430. doi: 10.1093/nar/gkp207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Pajares V, Yan K, Zarnegar BJ, Jameson KL, Khavari PA. Genetic pathways in disorders of epidermal differentiation. Trends Genet. 2013;29:31–40. doi: 10.1016/j.tig.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathys H, Basquin J, Ozgur S, Czarnocki-Cieciura M, Bonneau F, Aartse A, Dziembowski A, Nowotny M, Conti E, Filipowicz W. Structural and biochemical insights to the role of the CCR4-NOT complex and DDX6 ATPase in microRNA repression. Mol Cell. 2014;54:751–765. doi: 10.1016/j.molcel.2014.03.036. [DOI] [PubMed] [Google Scholar]
- Mistry DS, Chen Y, Sen GL. Progenitor Function in Self-Renewing Human Epidermis is Maintained by the Exosome. Cell Stem Cell. 2012;11:127–135. doi: 10.1016/j.stem.2012.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mistry DS, Chen Y, Wang Y, Sen GL. Transcriptional profiling of SNAI2 regulated genes in primary human keratinocytes. Genomics data. 2015;4:43–46. doi: 10.1016/j.gdata.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mistry DS, Chen Y, Wang Y, Zhang K, Sen GL. SNAI2 controls the undifferentiated state of human epidermal progenitor cells. Stem Cells. 2014;32:3209–3218. doi: 10.1002/stem.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder KW, Wang X, Escriu C, Ito Y, Schwarz RF, Gillis J, Sirokmany G, Donati G, Uribe-Lewis S, Pavlidis P, et al. Diverse epigenetic strategies interact to control epidermal differentiation. Nat Cell Biol. 2012;14:753–763. doi: 10.1038/ncb2520. [DOI] [PubMed] [Google Scholar]
- Qi MY, Wang ZZ, Zhang Z, Shao Q, Zeng A, Li XQ, Li WQ, Wang C, Tian FJ, Li Q, et al. AU-rich-element-dependent translation repression requires the cooperation of tristetraprolin and RCK/P54. Mol Cell Biol. 2012;32:913–928. doi: 10.1128/MCB.05340-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raught B, Gingras AC. eIF4E activity is regulated at multiple levels. Int J Biochem Cell Biol. 1999;31:43–57. doi: 10.1016/s1357-2725(98)00131-9. [DOI] [PubMed] [Google Scholar]
- Scheller N, Mina LB, Galao RP, Chari A, Gimenez-Barcons M, Noueiry A, Fischer U, Meyerhans A, Diez J. Translation and replication of hepatitis C virus genomic RNA depends on ancient cellular proteins that control mRNA fates. Proc Natl Acad Sci U S A. 2009;106:13517–13522. doi: 10.1073/pnas.0906413106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segre JA. Epidermal barrier formation and recovery in skin disorders. J Clin Invest. 2006;116:1150–1158. doi: 10.1172/JCI28521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segre JA, Bauer C, Fuchs E. Klf4 is a transcription factor required for establishing the barrier function of the skin. Nat Genet. 1999;22:356–360. doi: 10.1038/11926. [DOI] [PubMed] [Google Scholar]
- Sen GL. Remembering one’s identity: the epigenetic basis of stem cell fate decisions. FASEB J. 2011 doi: 10.1096/fj.11-182774. [DOI] [PubMed] [Google Scholar]
- Sen GL, Boxer LD, Webster DE, Bussat RT, Qu K, Zarnegar BJ, Johnston D, Siprashvili Z, Khavari PA. ZNF750 Is a p63 Target Gene that Induces KLF4 to Drive Terminal Epidermal Differentiation. Dev Cell. 2012;22:669–677. doi: 10.1016/j.devcel.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen GL, Reuter JA, Webster DE, Zhu L, Khavari PA. DNMT1 maintains progenitor function in self-renewing somatic tissue. Nature. 2010;463:563–567. doi: 10.1038/nature08683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen GL, Webster DE, Barragan DI, Chang HY, Khavari PA. Control of differentiation in a self-renewing mammalian tissue by the histone demethylase JMJD3. Genes Dev. 2008;22:1865–1870. doi: 10.1101/gad.1673508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharova LV, Sharov AA, Nedorezov T, Piao Y, Shaik N, Ko MS. Database for mRNA half-life of 19 977 genes obtained by DNA microarray analysis of pluripotent and differentiating mouse embryonic stem cells. DNA research: an international journal for rapid publication of reports on genes and genomes. 2009;16:45–58. doi: 10.1093/dnares/dsn030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Liu Y, Hsu YJ, Fujiwara Y, Kim J, Mao X, Yuan GC, Orkin SH. EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol Cell. 2008;32:491–502. doi: 10.1016/j.molcel.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting SB, Caddy J, Hislop N, Wilanowski T, Auden A, Zhao LL, Ellis S, Kaur P, Uchida Y, Holleran WM, et al. A homolog of Drosophila grainy head is essential for epidermal integrity in mice. Science. 2005;308:411–413. doi: 10.1126/science.1107511. [DOI] [PubMed] [Google Scholar]
- Van Hoof D, Munoz J, Braam SR, Pinkse MW, Linding R, Heck AJ, Mummery CL, Krijgsveld J. Phosphorylation dynamics during early differentiation of human embryonic stem cells. Cell Stem Cell. 2009;5:214–226. doi: 10.1016/j.stem.2009.05.021. [DOI] [PubMed] [Google Scholar]
- Wang L, Miao YL, Zheng X, Lackford B, Zhou B, Han L, Yao C, Ward JM, Burkholder A, Lipchina I, et al. The THO complex regulates pluripotency gene mRNA export and controls embryonic stem cell self-renewal and somatic cell reprogramming. Cell Stem Cell. 2013;13:676–690. doi: 10.1016/j.stem.2013.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston A, Sommerville J. Xp54 and related (DDX6-like) RNA helicases: roles in messenger RNP assembly, translation regulation and RNA degradation. Nucleic Acids Res. 2006;34:3082–3094. doi: 10.1093/nar/gkl409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang A, Schweitzer R, Sun D, Kaghad M, Walker N, Bronson RT, Tabin C, Sharpe A, Caput D, Crum C, et al. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature. 1999;398:714–718. doi: 10.1038/19539. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.