Abstract
MicroRNAs (miRNAs) constitute a large family of small non-coding RNAs encoded by the genomes of most organisms. They regulate gene expression through post-transcriptional mechanisms to attenuate protein output in various genetic networks. The discovery of miRNAs has transformed our understanding of gene regulation and sparked intense efforts intended to harness their potential as diagnostic markers and therapeutic tools. Over the last decade a flurry of studies have shed light on placental miRNAs but have also raised many questions regarding the scope of their biological action. Moreover, the recognition that miRNAs of placental origin are continually released in the maternal circulation throughout pregnancy suggested that circulating miRNAs might serve as biomarkers for placental function during pregnancy. While this generated much enthusiasm, recently recognized challenges have delayed the application of miRNA-based biomarkers and therapeutics in clinical practice. In this review, we summarize key findings in the field and discuss current knowledge related to miRNAs in the context of placental biology.
Keywords: Placenta, trophoblast, miRNAs, exosomes
1. Introduction
The recent finding of pervasive transcription across the genomes of all kingdoms of life challenges some long-held ideas regarding the genome and its regulation. A consequence of this widespread transcription is the production of numerous RNA transcripts with relatively unknown functions. A large fraction of these transcribed RNAs are not translated into proteins but exhibit regulatory functions that are increasingly recognized as critical factors in development and homeostasis. A major class of these non-coding RNAs, and one of the best studied, is the family of small regulatory RNAs called microRNAs (miRNAs). miRNAs were originally described in the nematode Caenorhabditis elegans and were later found in the genomes of protists, plants, animals, and viruses, with the notable exception of bacteria. MiRNAs are single-strand RNA molecules of 20 to 24 nucleotides (nt) that usually repress gene expression by guiding an RNA-induced silencing complex (RISC) containing Ago proteins to a target RNA, which they bind through imperfect base-pairing. Gene expression is then attenuated to a variable degree by inhibition of the mRNA translation and transcript destabilization, resulting in reduced protein synthesis. Interestingly, while most miRNAs exert a modest effect on individual targets,1,2 perturbations in miRNA expression levels can have marked biological consequences. Indeed, a growing list of miRNAs have been implicated in the pathogenesis of human diseases, including but not limited to cancer, cardiovascular pathology, liver and kidney diseases, and psychiatric disorders.3–7 Tissue expression of these miRNAs is commonly quantified using PCR, northern blot, microarrays and RNA sequencing.
To date, the biological database miRBase, which was developed by the Griffiths-Jones lab at the Faculty of Life Sciences, University of Manchester,8 contains more than 2500 entries for human miRNAs, although that number might be an overestimation as some of the species represent computer-based predictions without experimental validation.9,10 Different cell types express common and unique miRNA species, and miRNA expression patterns are influenced by developmental and pathological states. The human placenta expresses a distinct miRNA repertoire, characterized by the fact that a large proportion of miRNAs are derived from the two largest clusters of miRNAs in humans, the chromosome 14 miRNA cluster (C14MC) and the chromosome 19 miRNA cluster (C19MC).11 Although the functions of placental miRNAs are largely unknown, recent research has begun to shed light on their role in placental biology, as detailed below. Likewise, the finding that placental miRNAs are released into the maternal circulation has raised the exciting prospect of using miRNA expression profiles as non-invasive markers of placental dysfunction. In this review, we briefly describe how miRNAs are produced and summarize recent developments in our understanding of the biological action of miRNAs in the human placenta.
2. The discovery of miRNAs
MicroRNAs were discovered in the nematode Caenorhabditis elegans by the groups of Victor Ambros and Gary Ruvkun while studying a pair of developmental genes. One of these genes, lin-14, controls stage-specific cell lineages during larval development, and was known to be itself regulated by lin-4 gene. Ln-4 does not encode a protein product but instead gives rise to a small RNA transcript of 22 nt with complementarity to 3’-untranslated regions of the lin-14 mRNA.12 These 3’-untranslated regions contain short conserved elements complementary to parts of the lin-4 transcript.13 These pioneering studies suggested that small antisense RNAs could bind and inhibit specific mRNAs that contain complementary sequences to the small RNA. A second small regulatory RNA called let-7 has been known for years, yet was identified as an antisense RNA only in 2000.14 The discovery of many regulatory, 22-nt RNAs led to the term microRNA (miRNA).15 With the expanded list of miRNA species came the need for better nomenclature.16 Currently, each miRNA is assigned the prefix “miR”, followed by a number that represents the order of naming. MiRNAs are commonly preceded by three letters that denote the organism (e.g. “hsa-miR-121 is the human miR-121 and mmu-miR-121 is the mouse version). Additional information on the miRNA naming system as well as miRNA sequences can be found in miRNA database such as miRBase.8
3. MiRNA biogenesis
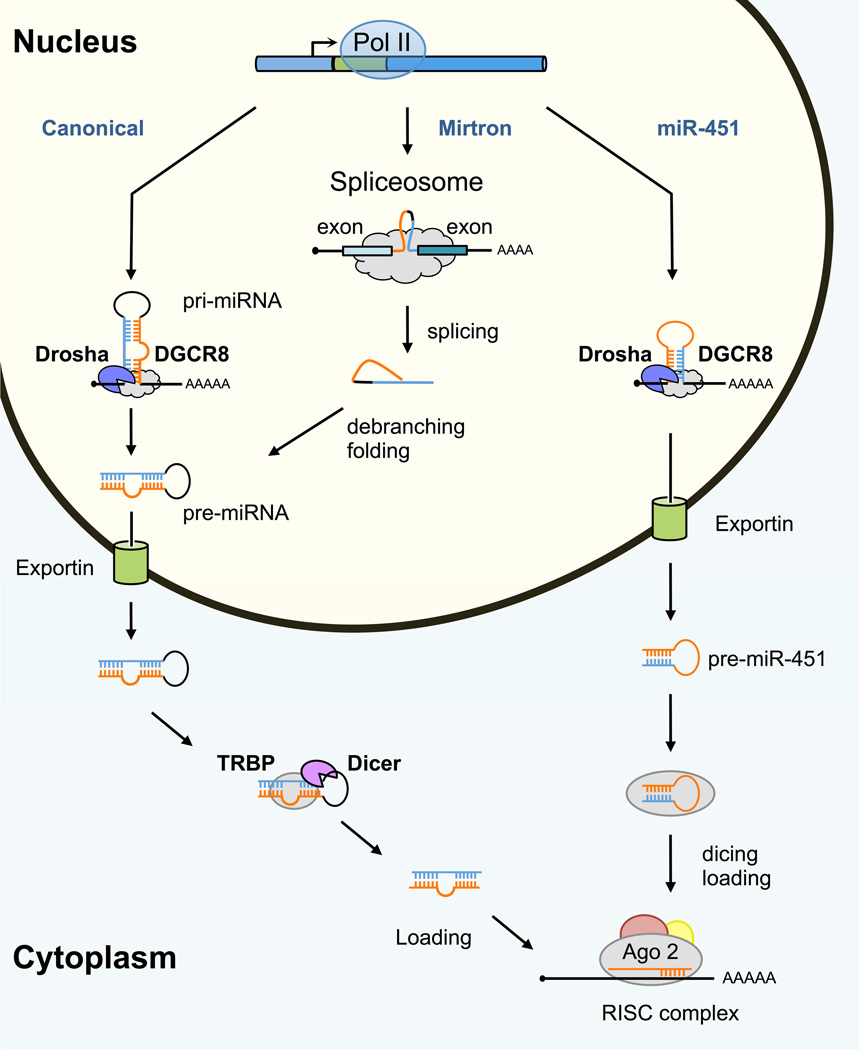
MiRNAs are encoded in the genome and are typically transcribed as long primary miRNA (pri-miRNA) by RNA polymerase II17 before undergoing a multi-step process leading to mature miRNAs (Fig. 1). The pri-miRNA contains partially self-complementary regions that fold back and form hairpin structure, which harbors the mature miRNA sequence. In the nucleus, the hairpin structure (65–80 nt long) is released from the pri-miRNA by endonucleolytic cleavage executed by the Microprocessor, a protein complex consisting of an RNase III endonuclease (Drosha), guided by the RNA-binding protein DGCR8.18–20 The stem-loop precursor (pre-miRNA) is then actively transported across the nuclear membrane by the RanGTPase-dependent Exportin-5, where it is further cleaved by the cytoplasmic form of the RNase III endonuclease Dicer in partnership with the RNA-binding protein TPBP.21–24 Cleavage of the pre-miRNA releases a small RNA duplex of 20–24 nt that is loaded on an effector protein complex containing Argonaute proteins and referred to as the RISC.25–27
Figure 1. The biogenesis of miRNAs.
In the canonical pathway, miRNA precursors transcribed from exonic, intronic, or intergenic sequences are recognized by the RNA-binding protein DGCR8 and cleaved by the RNase III Drosha. This cleavage generates a 55- to 70-nt pre-miRNA hairpin that is exported to the cytoplasm, where it undergoes a second cleavage by the RNase III enzyme Dicer that removes the terminal loop. The miRNA duplex is then loaded into the RISC complex containing Ago2 in mammals. Some miRNAs derive from mirtron loci, which produce short intronic hairpins that are excised by the splicing machinery. Mirtron intermediates are then linearized by the lariat-debranching enzyme and exported to the cytoplasm where they are further processed by Dicer. Some mirtrons are produced with extended 5’ or 3’ end regions (“tailed mirtrons”) and trimmed before their export and cleavage by Dicer. In the case of miR-451, processing of the primary transcript by Drosha releases an unusually short hairpin that cannot be cleaved. Instead, it is directly loaded onto the RISC complex and cleaved by Ago2 to release a 30-nt-long precursor that is likely processed and trimmed by Poly(A)-specific ribonuclease (PARN) to produce mature miR-451.
At this stage, the double-stranded RNA is unwound, and the strand with the lowest internal stability at the 5’ end (the guide strand) is retained to form the mature RISC complex, while the other strand (the passenger strand) is discarded.28,29 While this canonical pathway produces most mammalian miRNAs, alternative routes were recently described. For example, mirtrons are derived from introns of mRNA coding genes after processing by the spliceosome complex and, therefore, bypass the need of Drosha.30–35 Other small RNA species, including tRNAs and snoRNAs, can also give rise to miRNAs33,36,37 although their function as genuine miRNAs remains unclear.9,10 Most non-canonical miRNAs are relatively infrequent in mammals, with the exception of miR-451, an abundant miRNA that is produced without involving Dicer and only depends on the slicer activity of Ago2 for its processing.38–40 Lastly, we emphasize that multiple auxiliary proteins may contribute to miRNA processing at different stages, as described in recent reviews.35,41–43
4. The biological functions of miRNAs
MicroRNAs are involved in virtually every cellular process, modulating regulatory pathways that control development, differentiation, and organ function in health and disease. For example, the liver-specific miR-122 participates in the regulation of many genes associated with cholesterol and lipid metabolism, and is also a target of the hepatitis C virus (HCV), which uses it for its own replication (review in5). Several miRNAs species, such as miR-1 and miR-133 family, are expressed in the heart where they regulate heart development and cardiovascular diseases.44,45
While research in this area continues to progress, the physiological targets of most miRNAs and the mechanisms underlying their action remain largely unknown. Because target recognition involves base-pairing between the miRNA and the targeted mRNA, target prediction algorithms have been designed to identify interacting pairs. However, while in plants there is often a near perfect complementarity between the miRNA and its targets, the complementarity in animals is incomplete, usually reduced to a stretch of six nucleotides at the 5’-end of the miRNA (called the seed).46 Reflecting this limited level of miRNA-mRNA complementarity, target-prediction algorithms now incorporate additional parameters, such as thermodynamic stability, evolutionary conservation, and binding site structural accessibility (review in47). Despite increasing sophistication, these search algorithms remain plagued by a high rate of false positive predictions (~60%).1,2,48–50 Therefore, algorithmic predictions are commonly used in combination with direct biochemical methods, based on the crosslinked immunoprecipitation of Ago2 complexes, to isolate miRNAs bound to target mRNAs.51–54
Other experimental methods rely on genomic screens, in which a specific miRNA is either deleted or overexpressed under the assumption that the corresponding target genes will be identified as they are de-repressed or silenced. With the recent development of genome editing, it has become possible to introduce specific mutations in miRNA binding sites and validate the functional interaction of miRNA-mRNA in a physiologic context and without the need to create gene knockouts or the overexpression of factors that are prone to non-specific effects.55
The complexity of miRNA-based gene regulation is furthered by the promiscuous interaction of most miRNAs with mRNA targets, where each miRNA has the potential to interact with and regulate a multitude of target mRNAs. It is estimated that a large fraction of the genome is regulated by miRNAs.50,56–59 Notably, miRNA-mediated repression is rather modest, commonly with a less-than-twofold change in target protein levels.1,2 Notwithstanding, global depletion of miRNAs, achieved by disrupting a factor such as Dicer,60 or Ago2,61 now known to be involved in the processing of other types of RNAs, results in lethality in mammalian models. To date, many gain- and loss-of-function studies designed to identify the biological functions of miRNA species have been conducted in animal models such as Caenorhabditis elegans or in mice. Interestingly, many of these experiments show that, despite their high regulatory potential, deletion of a miRNA gene often fails to produce any obvious defect.62–64 This is likely explained, at least in part, by the high level of redundancy among miRNAs, reflecting gene duplication that resulted in homologous families of miRNAs (review in65).
5. MiRNAs and the placenta
The human placenta is a rapidly evolving organ that harbors a rich and diverse transcriptome. It is estimated that 66% of all human proteins are expressed in the placenta.66 Not surprisingly, the human placenta also expresses numerous types of miRNA species, with a fraction of these species being specific to trophoblasts.67,68 Early genetic inactivation experiments in the mouse, which centered on genetic inactivation of genes coding critical miRNA biosynthetic enzymes, such as Dicer60 or Ago2,69,70 failed to shed light on miRNA function in the placenta due to early post-implantation embryonic lethality. Others have used more targeted murine gene knockout experiments to elucidate miRNA function in the placenta. In a study that centered on the role of the abundant embryonic H19 transcript, it was reported that miR-675 is expressed from the H19 transcript exclusively in the placenta.71 Interestingly, the levels of miR-675 increased toward the second half of gestation, which corresponds to the reduced pace of placental growth. Consistent with this association, miR-675 exhibits anti-proliferative activities, likely by silencing of the insulin-like growth factor 1 receptor (Igf1r), and a targeted deletion of miR-675 causes placental overgrowth.71
Abundant expression of a specific miRNA does not imply functional significance. For example, the large miR-379/miR-410 cluster (also called C14MC in humans) can be deleted without obvious placental consequences, even though these mutant mice display a partially penetrant neonatal lethality.72 The miR-379/miR-410–C14MC cluster is highly expressed in the placenta73 and produces up to 77 mature miRNAs in the mouse (63 in humans) and, thus, carries a large regulatory potential. To date, the function of the human C19MC miRNAs in the placenta remains unknown.
MiRNAs from the miR-17~92 cluster are among of the best-studied miRNAs and have been shown to play a role in development and during tumorigenesis.74 Members of the miR-17~92 cluster regulate the differentiation of primary human trophoblasts by targeting pivotal placental proteins such as the CYP19A1 (aromatase) and the transcription factor GCM1.75 Curiously, targeted deletion of this family of miRNAs in the mouse does not seem to cause placental abnormalities.76 Other trophoblast-regulatory miRNA candidates were identified using computational methods and were subsequently validated in cell lines. For example, miR-378a-5p and miR-376c enhance trophoblast proliferation and invasion through targeting the Nodal signaling pathway.77,78 In contrast, miR-155 inhibits trophoblast invasion by targeting CYR61 and cyclin D1, thus potentially contributing to the development of preeclampsia.79,80 Other miRNA species, such as miR-210 and miR-125b-1–3p, have been implicated in the inhibition of trophoblast proliferation and invasion and thereby contribute to placental disorders.81–84 Other miRNA species have been associated with placental physiology and pathology, and were reviewed elsewhere (see85). However, their role in placental function has not been firmly established.
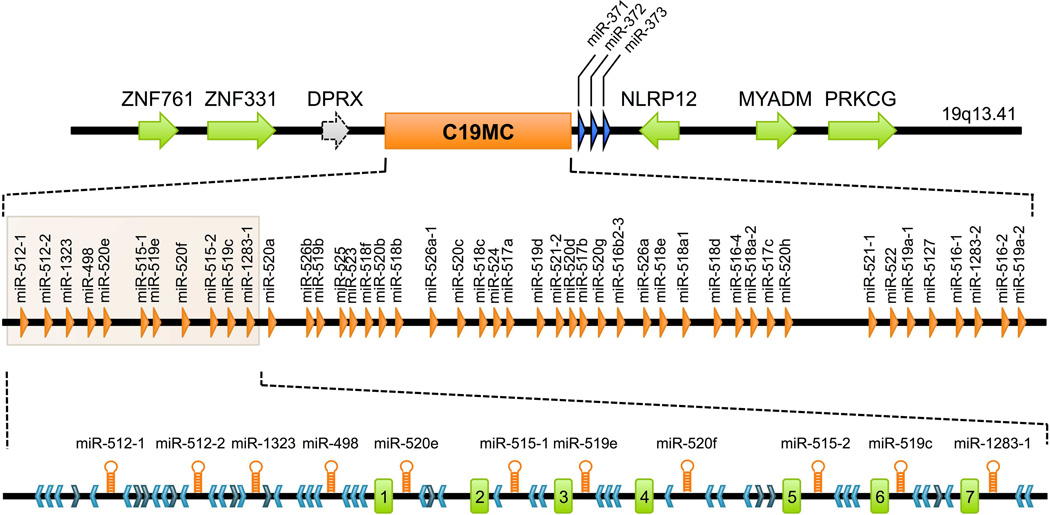
One of the most intriguing families of trophoblast-specific miRNAs is the C19MC. This cluster contains 46 intronic miRNA genes that are scattered over 100 kb of genomic DNA and produce 58 mature miRNA species that are expressed almost exclusively in the placenta. Unlike other miRNAs cited earlier, the C19MC miRNAs are primate-specific with no orthologous regions in the mouse genome. The genomic structure of C19MC is also unique, as intronic C19MC miRNA sequences are flanked by numerous Alu sequences and short exons with highly repetitive DNA elements, representing non-coding RNAs of unknown function (Fig. 2).86 A number of recent observations implicate C19MC miRNAs in the regulation of cell proliferation, invasion, and differentiation. For example, C19MC miRNAs are found in human embryonic stem cells, where they contribute to the control of stemness, and their expression declines rapidly upon differentiation.87–89
Figure 2. The genomic organization of the C19MC miRNA cluster.
The figure depicts the genomic region of the C19MC cluster, with the middle section showing the position of the miRNA genes. At the bottom, an enlargement of the 5′ end of the region details the structural organization of the cluster elements. The light blue chevrons represent Alu elements in an antisense orientation relative to the miRNA genes, while the dark blue chevrons correspond to sense Alu sequences. The seven first-expressed exons are represented by green rectangles numbered 1–7. The figure was modified from Bortoloin-Cavaille.86
Although fine spatial mapping of C19MC miRNA expression in different trophoblast subpopulations is not yet available, laser-capture microdissection experiments suggest that C19MC expression is reduced in extravillous trophoblasts compared to villous trophoblasts.90 Furthermore, forced expression of the C19MC miRNAs attenuates the migration of human extravillous trophoblasts in vitro.90 Surprisingly, expression of the C19MC miRNAs “off context” produces the opposite effect and suggests an oncogenic potential. Indeed, reactivation of the C19MC locus has been observed in several types of malignancies.91–97
Perhaps the most unexpected finding is related to the role of C19MC miRNAs in viral resistance. Primary human trophoblasts, which express high levels of C19MC miRNAs,98 are resistant to infection by a diverse and unrelated panel of DNA and RNA viruses. This viral resistance can be transferred to non-placental cells upon transfection with a BAC plasmid expressing the C19MC miRNAs99. Mechanistically, the process involves autophagy, as C19MC miRNAs stimulate autophagy in non-placental cells, and genomic or pharmacologic inhibition of autophagy attenuates the antiviral response.99,100 Together, these findings attest to the complex, context-dependent action of C19MC miRNAs in the placenta, and highlight the need for additional mechanistic studies designed to elucidate the function and regulation of C19MC miRNAs in the human placenta.
6. Extracellular miRNAs
Although transcribed miRNAs accumulate in the nuclear and cytoplasmic compartments within cells, recent data clearly highlight their release to the extracellular space, including plasma and other extracellular fluids.101 In the context of pregnancy, Chim et al. detected placental miRNAs in the plasma of pregnant women, with a rapid decline in the plasma levels of these species following delivery.102 Differences in plasma expression profiles have been attributed to physiological or pathological conditions, including cancer.103–108
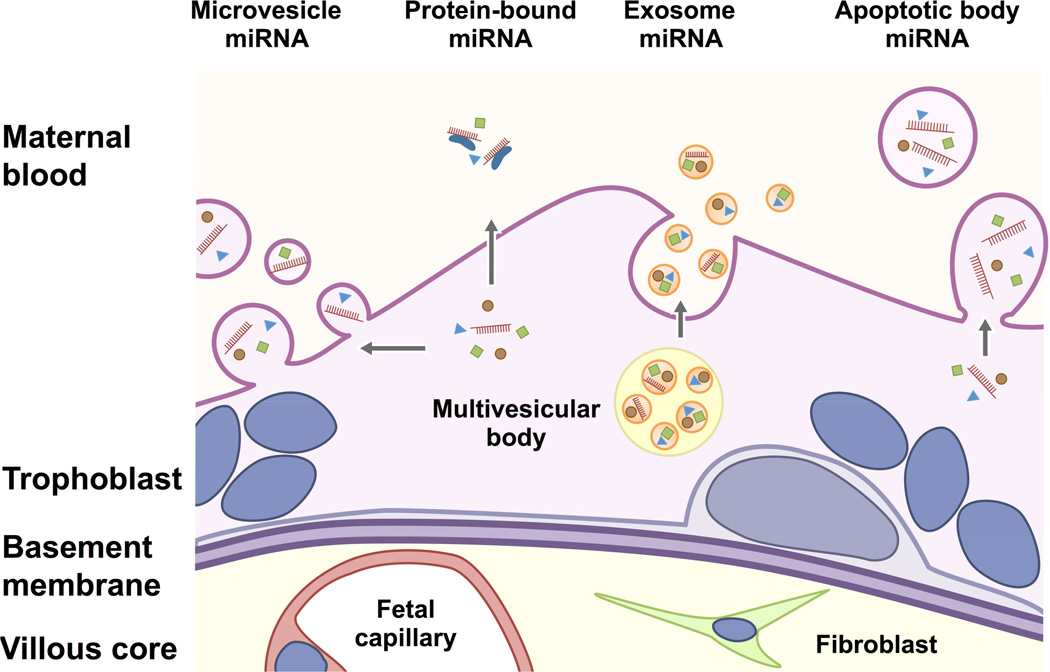
Circulating miRNAs of placental origin are thought to derive primarily from the trophoblast layer, which lines the human placental villi. Extracellular trophoblastic miRNAs are present in at least two forms: vesicular miRNAs or non-vesicular, protein-bound miRNAs. These miRNA-binding proteins include Ago2, high-density lipoproteins, and nucleophosmin 1.109–112 Vesicular miRNAs are shielded from degradation by circulating RNase by the lipid bilayer, which enhances their stability in the plasma. Three major forms of extracellular vesicles commonly found in the blood are classified by their size and include exosomes (40–150 nm), microvesicles (0.1–1 µm) and apoptotic blebs (1–5 µm) (Fig. 3).
Figure 3. A schematic of extracellular miRNAs derived from human trophoblasts.
MiRNAs can be released from the trophoblast layer in different forms: microvesicle-enveloped form; apoptotic body–enveloped form; nano-sized, exosome-encapsulated form; and RNA-binding, protein-bound form. Exosomes are formed by budding in intraluminal vesicles to form multivesicular bodies (MVB). Some MVBs will fuse with the plasma membrane and release their intraluminal vesicles or exosomes into the extracellular space. In contrast, microvesicles are produced directly by budding and the detachment of membrane vesicles from the plasma membrane. Apoptotic bodies (blebs) derive from cells undergoing apoptotic fragmentation and the formation of membrane-enclosed vesicles also called apoptosomes. The figure was modified from Ouyang.156
The mechanisms of miRNA loading, sorting, and vesicle-based trafficking are poorly understood. Key players in this process include the endosomal sorting complex required for transport (ESCRT)113 and several other associated proteins, such as programmed cell death 6 interacting protein (PDCD6IP; also known as ALIX)114 and tumor susceptibility gene 101 protein (TSG101).115 Moreover, while vesicular miRNA can affect target cells,116–119 the number of miRNA molecules packaged within exosomes and other vesicles, the tissue targets of circulating miRNA-containing vesicles, and their uptake mechanisms by recipient cells are largely unknown.120
Lastly, the recent discovery of an interaction between exosomal miRNAs and pattern recognition receptors from the family of toll-like receptors within target cell endosomes suggests a novel pathway for recognition and amplification of miRNA signals.121,122
7. The clinical implications of placental miRNA
The discovery of an altered miRNA landscape in human diseases, along with technological advances in miRNA measurement and delivery, fueled the search for novel miRNA-based diagnostic and therapeutic tools. To date, a number of miRNAs have been identified as important mediators of cardiac fibrosis, and technologies are currently being evaluated to either boost or inhibit miRNA function as a way to attenuate cardiac disease (review in123). Another example is miR-122, a liver-enriched miRNA that shows great promise as a biomarker of liver injury and also as a potential therapeutic agent in cases of hepatitis C viral infection.124 In the field of pregnancy, our knowledge of placental miRNAs and their biological function is currently rudimentary, and clinical applications await additional research. The discovery of the rich representation of placental miRNAs in the circulation of pregnant women102 stimulated attempts to identify, non-invasively, miRNA biomarkers for placental dysfunction and related clinical syndromes. Women with preeclampsia, for example, exhibit altered expression levels of several miRNAs in the placenta and plasma, with miR-210 exhibiting a relatively consistent up-regulation.81,83,125–132 MiR-210 is typically induced by HIF-1α, an important regulator of cellular hypoxic response in different contexts.133–135 Elevated levels of placental miR-210 may contribute to the pathogenesis of preeclampsia by inhibiting trophoblast invasion,82 stimulating mitochondrial respiration targeting,127,136,137 or over-stimulating the immune system by inhibition of the STAT6/Interleukin-4 Pathway.138 Several potential molecular targets of miR-210 in the context of preeclampsia include ephrin-A3 and homeobox-A981 and potassium channel modulatory factor 1 (KCMF1).83 In addition to miR-210, differential expression of C19MC miRNA members has been reported,131,139,140 including increased expression of C19MC miRNA members in extravillous trophoblasts from women with preeclampsia. Notably, preeclampsia is associated with reduced invasion of extravillous trophoblasts in vitro,141 and this observation is consistent with the recent finding of reduced migration of human extravillous trophoblast cell line by over-expression of C19MC miRNAs.90 Aberrant C19MC miRNA expression patterns have also been observed in placentas from pregnancies complicated by fetal growth restriction (FGR).142–144 Other studies characterized changes in the miRNA landscape in association with placental disorders and related diseases, such as fetal growth restriction, preterm birth, and the like. Unfortunately, many of the findings are inconsistent and lack validation. Additional information can be found in recent reviews.129,145,146
Despite intense effort by many research groups, the promise of using circulating miRNAs as biomarkers has not yet been realized; studies were not designed to determine miRNA use as diagnostic tests and, in most cases, miRNA changes are not validated even when similar research designs are deployed. This conundrum is also observed in other fields, with serious inconsistencies between profiles of circulating miRNAs in breast cancer.147 One of the reasons for the inconsistency is that the vast majority of circulating miRNA is derived from blood components and endothelial cells, while tissue-specific miRNAs contribute a small fraction of all detected miRNA molecules.148 Other factors include a lack of methodological standardization, including sample processing, profiling technologies, and bioinformatics analysis.149–153 Extensive integration of multiple datasets comprising miRNA profiles combined with profiles of other relevant molecules, such as lncRNAs, might increase assay specificity and lead to the development of clinically useful tests. Another promising direction might be the isolation and characterization of sub-populations of miRNA-containing extracellular vesicles.153 The repertoire and the exact classification of these vesicles are still imperfect, yet their use as in vivo biopsies of their tissue of origin may offer a unique view into the transcriptional landscape during disease. The knowledge of tissue-specific protein surface markers may be used to capture sub-populations of circulating extracellular vesicles and profile their RNA cargo.
The presence of placental miRNA in the maternal circulation suggests that these miRNAs may participate in feto-placental-maternal communication, influencing local and distant target tissues. Initial recent reports suggested that miRNAs were packaged within plasma vesicles, which communicate among cells.116 Similarly, tumor miRNAs, packaged within diverse vesicle types, are believed to influence metastasis, angiogenesis and the tumor environment (review in 154). The notion that trophoblast-derived exosomes that contain C19MC miRNAs may confer viral resistance to recipient cells is consistent with these observations.99 A different group recently showed the transfer of miR-517a from trophoblasts to maternal NK cells, and subsequent inhibition of the target mRNA PRKG1.155 However, the mechanisms underlying these observations remain unclear, and the small number of miRNA molecules that are packaged within vesicles120 may suggest a more complex mechanism of miRNA-mediated intercellular communication.
8. Conclusions and perspectives
The discovery of miRNAs and their widespread influence on gene expression has advanced our understanding of gene regulatory networks and brought new insight into the molecular mechanisms that control development and homeostasis. In the field of placental biology and perinatal medicine, work remains to be done in defining the placental miRNA landscape and in characterizing miRNA function and regulation. Currently, at least 500 different miRNAs species are known to be expressed in placental trophoblasts,85 yet the biological significance of most miRNAs is currently unknown. While limited by relevant in vivo experimental models of humans, a better understanding of the biology of placental miRNAs is likely to elucidate some of the key molecular circuits that control placental development and function in health and disease. The evaluation of miRNA species in maternal plasma extracellular vesicles as biomarkers of placental health is particularly germane to the field of perinatal biology and obstetrics and is likely to provide greatly needed means to dynamically assess placental health in real time. At the present time, and despite intense efforts, miRNA biomarkers for placental conditions remain unknown. Several technical and conceptual challenges must be addressed before the promise of miRNAs in diagnostics and therapeutics during pregnancy can be realized.
Acknowledgements
The authors thank Lori Rideout for assistance during preparation of the manuscript and Bruce Campbell for editing. The project was supported by Pennsylvania Department of Health Research Formula Funds (to JFM), NIH R01HD065893 and NIH R21HD071707 (to YS), NIH R01AI081759 (to CBC), NIH R01HD075665 (to CBC and YS).
Glossary
- Argonaute
A family of proteins characterized by a specific structural organization and a critical role in the silencing process by miRNAs. Argonaute 2 (Ago2), for example, is a part of the RNA-induced silencing complex (RISC) and is responsible for the cleavage of the target mRNA.
- Canonical pathway
The prototypical pathway of a biological process. Non-canonical refers to pathways that deviate from the canonical pathway or represent a less frequent or less known alternative.
- Dicer
An RNA-specific enzyme that cleaves a pre-miRNA (and other types of double-stranded RNAs) into 21–24-nucleotide long double stranded RNAs with a 2-base overhang at the 3’ end.
- Drosha
A nuclear RNA-specific enzyme that processes newly transcribed primary miRNA to produce a ~70 base pairs transcript with a hairpin shape, called pre-miRNA.
- Endonucleolytic cleavage
The enzymatic cleavage of nucleic acid molecules through the hydrolysis of internal covalent bonds between nucleotides.
- Exosomes
Small vesicles (50 −150 nm) that are released into the extracellular environment when endosomal multivesicular bodies fuse with the plasma membrane.
- Exportin 5
A nuclear envelope protein that mediates the nuclear export of pre-miRNAs to the cytoplasm. This process is assisted by the protein cofactor Ran-GTP (see below).
- Microprocessor
A protein complex consisting of a catalytic core made of the Drosha nuclease and the RNA-binding protein DGCR8 (DiGeorge syndrome critical region 8).
- Mirtrons
A subpopulation of miRNAs that are located in the introns of genes and produced by an alternative synthesis pathway, independent of the Drosha enzymatic complex.
- Ran GTPase
A member of the family of GTPase enzymes that is involved in many nucleo-cytoplasmic transport pathways by regulating the interactions of protein carriers with their cargo.
- RNA polymerase
II (RNA pol II or RNAP II): An enzyme that orchestrates the transcription of DNA into RNA or miRNA molecules.
- RNase III
An RNA-specific endonuclease that cleaves double-stranded RNA molecules. Drosha and Dicer are members of this family.
- Stem-loop
A secondary structure in DNA or RNA molecules that occurs when a strand folds and form intramolecular base pairing with another section of the same strand, creating a U-shape structure.
- Spliceosome
A large ribonucleoprotein complex involved in the removal of introns from unprocessed mRNAs in eukaryotic cells.
- TRBP
A double strand RNA binding protein that is an essential interacting partner of Dicer in the biogenesis of miRNAs.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure: CBC and YS are named inventors on a pending patent application describing the use of C19MC microRNAs as therapeutics.
Previous presentation: Not applicable.
References
- 1.Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 3.Lujambio A, Lowe SW. The microcosmos of cancer. Nature. 2012;482:347–355. doi: 10.1038/nature10888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Latronico MV, Condorelli G. MicroRNAs and cardiac pathology. Nat Rev Cardiol. 2009;6:419–429. doi: 10.1038/nrcardio.2009.56. [DOI] [PubMed] [Google Scholar]
- 5.Szabo G, Bala S. MicroRNAs in liver disease. Nat Rev Gastroenterol Hepatol. 2013;10:542–552. doi: 10.1038/nrgastro.2013.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Trionfini P, Benigni A, Remuzzi G. MicroRNAs in kidney physiology and disease. Nat Rev Nephrol. 2015;11:23–33. doi: 10.1038/nrneph.2014.202. [DOI] [PubMed] [Google Scholar]
- 7.Issler O, Chen A. Determining the role of microRNAs in psychiatric disorders. Nat Rev Neurosci. 2015;16:201–212. doi: 10.1038/nrn3879. [DOI] [PubMed] [Google Scholar]
- 8.Griffiths-Jones S. The microRNA Registry. Nucleic Acids Res. 2004;32:D109–D111. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chiang HR, Schoenfeld LW, Ruby JG, et al. Mammalian microRNAs: experimental evaluation of novel and previously annotated genes. Genes Dev. 2010;24:992–1009. doi: 10.1101/gad.1884710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42:D68–D73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morales-Prieto DM, Chaiwangyen W, Ospina-Prieto S, et al. MicroRNA expression profiles of trophoblastic cells. Placenta. 2012;33:725–734. doi: 10.1016/j.placenta.2012.05.009. [DOI] [PubMed] [Google Scholar]
- 12.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 13.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 14.Reinhart BJ, Slack FJ, Basson M, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 15.Ruvkun G. Molecular biology. Glimpses of a tiny RNA world. Science. 2001;294:797–799. doi: 10.1126/science.1066315. [DOI] [PubMed] [Google Scholar]
- 16.Ambros V, Bartel B, Bartel DP, et al. A uniform system for microRNA annotation. RNA. 2003;9:277–279. doi: 10.1261/rna.2183803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee Y, Kim M, Han J, et al. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- 19.Lee Y, Jeon K, Lee JT, Kim S, Kim VN. MicroRNA maturation: stepwise processing and subcellular localization. EMBO J. 2002;21:4663–4670. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zeng Y, Yi R, Cullen BR. Recognition and cleavage of primary microRNA precursors by the nuclear processing enzyme Drosha. EMBO J. 2005;24:138–148. doi: 10.1038/sj.emboj.7600491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 22.Grishok A, Pasquinelli AE, Conte D, et al. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106:23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- 23.Hutvagner G, Mclachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 24.Ketting RF, Fischer SE, Bernstein E, Sijen T, Hannon GJ, Plasterk RH. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 2001;15:2654–2659. doi: 10.1101/gad.927801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Filipowicz W. RNAi: the nuts and bolts of the RISC machine. Cell. 2005;122:17–20. doi: 10.1016/j.cell.2005.06.023. [DOI] [PubMed] [Google Scholar]
- 26.Sontheimer EJ. Assembly and function of RNA silencing complexes. Nat Rev Mol Cell Biol. 2005;6:127–138. doi: 10.1038/nrm1568. [DOI] [PubMed] [Google Scholar]
- 27.Tomari Y, Zamore PD. Perspective: machines for RNAi. Genes Dev. 2005;19:517–529. doi: 10.1101/gad.1284105. [DOI] [PubMed] [Google Scholar]
- 28.Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 29.Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 30.Berezikov E, Chung WJ, Willis J, Cuppen E, Lai EC. Mammalian mirtron genes. Mol Cell. 2007;28:328–336. doi: 10.1016/j.molcel.2007.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell. 2007;130:89–100. doi: 10.1016/j.cell.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007;448:83–86. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev. 2008;22:2773–2785. doi: 10.1101/gad.1705308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Flynt AS, Greimann JC, Chung WJ, Lima CD, Lai EC. MicroRNA biogenesis via splicing and exosome-mediated trimming in Drosophila. Mol Cell. 2010;38:900–907. doi: 10.1016/j.molcel.2010.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang JS, Lai EC. Alternative miRNA biogenesis pathways and the interpretation of core miRNA pathway mutants. Mol Cell. 2011;43:892–903. doi: 10.1016/j.molcel.2011.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ender C, Krek A, Friedlander MR, et al. A human snoRNA with microRNA-like functions. Mol Cell. 2008;32:519–528. doi: 10.1016/j.molcel.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 37.Cazalla D, Xie M, Steitz JA. A primate herpesvirus uses the integrator complex to generate viral microRNAs. Mol Cell. 2011;43:982–992. doi: 10.1016/j.molcel.2011.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cheloufi S, Dos Santos CO, Chong MM, Hannon GJ. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature. 2010;465:584–589. doi: 10.1038/nature09092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cifuentes D, Xue H, Taylor DW, et al. A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science. 2010;328:1694–1698. doi: 10.1126/science.1190809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang JS, Maurin T, Robine N, et al. Conserved vertebrate mir-451 provides a platform for Dicer-independent, Ago2-mediated microRNA biogenesis. Proc Natl Acad Sci U S A. 2010;107:15163–15168. doi: 10.1073/pnas.1006432107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 42.Tran N, Hutvagner G. Biogenesis and the regulation of the maturation of miRNAs. Essays Biochem. 2013;54:17–28. doi: 10.1042/bse0540017. [DOI] [PubMed] [Google Scholar]
- 43.Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 44.Liu N, Olson EN. MicroRNA regulatory networks in cardiovascular development. Dev Cell. 2010;18:510–525. doi: 10.1016/j.devcel.2010.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Romaine SP, Tomaszewski M, Condorelli G, Samani NJ. MicroRNAs in cardiovascular disease: an introduction for clinicians. Heart. 2015 doi: 10.1136/heartjnl-2013-305402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oulas A, Karathanasis N, Louloupi A, et al. Prediction of miRNA targets. Methods Mol Biol. 2015;1269:207–229. doi: 10.1007/978-1-4939-2291-8_13. [DOI] [PubMed] [Google Scholar]
- 48.Rajewsky N. microRNA target predictions in animals. Nat Genet. 2006;(38 Suppl):S8–S13. doi: 10.1038/ng1798. [DOI] [PubMed] [Google Scholar]
- 49.Cloonan N, Brown MK, Steptoe AL, et al. The miR-17–5p microRNA is a key regulator of the G1/S phase cell cycle transition. Genome Biol. 2008;9:R127. doi: 10.1186/gb-2008-9-8-r127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Krek A, Grun D, Poy MN, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 51.Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460:479–486. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hafner M, Landthaler M, Burger L, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Konig J, Zarnack K, Rot G, et al. iCLIP--transcriptome-wide mapping of protein-RNA interactions with individual nucleotide resolution. J Vis Exp. 2011 doi: 10.3791/2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Helwak A, Kudla G, Dudnakova T, Tollervey D. Mapping the human miRNA interactome by CLASH reveals frequent noncanonical binding. Cell. 2013;153:654–665. doi: 10.1016/j.cell.2013.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bassett AR, Azzam G, Wheatley L, et al. Understanding functional miRNA-target interactions in vivo by site-specific genome engineering. Nat Commun. 2014;5:4640. doi: 10.1038/ncomms5640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 57.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 58.Xie X, Lu J, Kulbokas EJ, et al. Systematic discovery of regulatory motifs in human promoters and 3’ UTRs by comparison of several mammals. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stark A, Brennecke J, Bushati N, Russell RB, Cohen SM. Animal MicroRNAs confer robustness to gene expression and have a significant impact on 3’UTR evolution. Cell. 2005;123:1133–1146. doi: 10.1016/j.cell.2005.11.023. [DOI] [PubMed] [Google Scholar]
- 60.Bernstein E, Kim SY, Carmell MA, et al. Dicer is essential for mouse development. Nat Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- 61.Liu J, Carmell MA, Rivas FV, et al. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 62.Miska EA, Alvarez-Saavedra E, Abbott AL, et al. Most Caenorhabditis elegans microRNAs are individually not essential for development or viability. PLoS Genet. 2007;3:e215. doi: 10.1371/journal.pgen.0030215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Alvarez-Saavedra E, Horvitz HR. Many families of C. elegans microRNAs are not essential for development or viability. Curr Biol. 2010;20:367–373. doi: 10.1016/j.cub.2009.12.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Park CY, Jeker LT, Carver-Moore K, et al. A resource for the conditional ablation of microRNAs in the mouse. Cell Rep. 2012;1:385–391. doi: 10.1016/j.celrep.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Berezikov E. Evolution of microRNA diversity and regulation in animals. Nat Rev Genet. 2011;12:846–860. doi: 10.1038/nrg3079. [DOI] [PubMed] [Google Scholar]
- 66.Uhlen M, Fagerberg L, Hallstrom BM, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 67.Landgraf P, Rusu M, Sheridan R, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Luo SS, Ishibashi O, Ishikawa G, et al. Human villous trophoblasts express and secrete placenta-specific microRNAs into maternal circulation via exosomes. Biol Reprod. 2009;81:717–729. doi: 10.1095/biolreprod.108.075481. [DOI] [PubMed] [Google Scholar]
- 69.Liu H, Chen Y, Lv J, et al. Quantitative epigenetic co-variation in CpG islands and co-regulation of developmental genes. Sci Rep. 2013;3:2576. doi: 10.1038/srep02576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Morita S, Horii T, Kimura M, Goto Y, Ochiya T, Hatada I. One Argonaute family member, Eif2c2 (Ago2), is essential for development and appears not to be involved in DNA methylation. Genomics. 2007;89:687–696. doi: 10.1016/j.ygeno.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 71.Keniry A, Oxley D, Monnier P, et al. The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and Igf1r. Nat Cell Biol. 2012;14:659–665. doi: 10.1038/ncb2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Labialle S, Marty V, Bortolin-Cavaille ML, et al. The miR-379/miR-410 cluster at the imprinted Dlk1-Dio3 domain controls neonatal metabolic adaptation. EMBO J. 2014;33:2216–2230. doi: 10.15252/embj.201387038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Seitz H, Royo H, Bortolin ML, Lin SP, Ferguson-Smith AC, Cavaille J. A large imprinted microRNA gene cluster at the mouse Dlk1-Gtl2 domain. Genome Res. 2004;14:1741–1748. doi: 10.1101/gr.2743304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mogilyansky E, Rigoutsos I. The miR-17/92 cluster: a comprehensive update on its genomics, genetics, functions and increasingly important and numerous roles in health and disease. Cell Death Differ. 2013;20:1603–1614. doi: 10.1038/cdd.2013.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kumar P, Luo Y, Tudela C, Alexander JM, Mendelson CR. The c-Myc-regulated microRNA-17~92 (miR-17~92) and miR-106a~363 clusters target hCYP19A1 and hGCM1 to inhibit human trophoblast differentiation. Mol Cell Biol. 2013;33:1782–1796. doi: 10.1128/MCB.01228-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ventura A, Young AG, Winslow MM, et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132:875–886. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Luo L, Ye G, Nadeem L, et al. MicroRNA-378a-5p promotes trophoblast cell survival, migration and invasion by targeting Nodal. J Cell Sci. 2012;125:3124–3132. doi: 10.1242/jcs.096412. [DOI] [PubMed] [Google Scholar]
- 78.Fu G, Ye G, Nadeem L, et al. MicroRNA-376c impairs transforming growth factor-beta and nodal signaling to promote trophoblast cell proliferation and invasion. Hypertension. 2013;61:864–872. doi: 10.1161/HYPERTENSIONAHA.111.203489. [DOI] [PubMed] [Google Scholar]
- 79.Zhang Y, Diao Z, Su L, et al. MicroRNA-155 contributes to preeclampsia by down-regulating CYR61. Am J Obstet Gynecol. 2010;202:466 e1–466 e7. doi: 10.1016/j.ajog.2010.01.057. [DOI] [PubMed] [Google Scholar]
- 80.Dai Y, Qiu Z, Diao Z, et al. MicroRNA-155 inhibits proliferation and migration of human extravillous trophoblast derived HTR-8/SVneo cells via down-regulating cyclin D1. Placenta. 2012;33:824–829. doi: 10.1016/j.placenta.2012.07.012. [DOI] [PubMed] [Google Scholar]
- 81.Zhang Y, Fei M, Xue G, et al. Elevated levels of hypoxia-inducible microRNA-210 in pre-eclampsia: new insights into molecular mechanisms for the disease. J Cell Mol Med. 2012;16:249–259. doi: 10.1111/j.1582-4934.2011.01291.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Anton L, Olarerin-George AO, Schwartz N, et al. miR-210 inhibits trophoblast invasion and is a serum biomarker for preeclampsia. Am J Pathol. 2013;183:1437–1445. doi: 10.1016/j.ajpath.2013.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Luo R, Shao X, Xu P, et al. MicroRNA-210 contributes to preeclampsia by downregulating potassium channel modulatory factor 1. Hypertension. 2014;64:839–845. doi: 10.1161/HYPERTENSIONAHA.114.03530. [DOI] [PubMed] [Google Scholar]
- 84.Li Q, Pan Z, Wang X, Gao Z, Ren C, Yang W. miR-125b-1–3p inhibits trophoblast cell invasion by targeting sphingosine-1-phosphate receptor 1 in preeclampsia. Biochem Biophys Res Commun. 2014;453:57–63. doi: 10.1016/j.bbrc.2014.09.059. [DOI] [PubMed] [Google Scholar]
- 85.Morales-Prieto DM, Ospina-Prieto S, Schmidt A, Chaiwangyen W, Markert UR. Elsevier Trophoblast Research Award Lecture: origin, evolution and future of placenta miRNAs. Placenta. 2014;(35 Suppl):S39–S45. doi: 10.1016/j.placenta.2013.11.017. [DOI] [PubMed] [Google Scholar]
- 86.Bortolin-Cavaille ML, Dance M, Weber M, Cavaille J. C19MC microRNAs are processed from introns of large Pol-II, non-protein-coding transcripts. Nucleic Acids Res. 2009;37:3464–3473. doi: 10.1093/nar/gkp205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bar M, Wyman SK, Fritz BR, et al. MicroRNA discovery and profiling in human embryonic stem cells by deep sequencing of small RNA libraries. Stem Cells. 2008;26:2496–2505. doi: 10.1634/stemcells.2008-0356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Laurent LC. MicroRNAs in embryonic stem cells and early embryonic development. J Cell Mol Med. 2008;12:2181–2188. doi: 10.1111/j.1582-4934.2008.00513.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ren J, Jin P, Wang E, Marincola FM, Stroncek DF. MicroRNA and gene expression patterns in the differentiation of human embryonic stem cells. J Transl Med. 2009;7:20. doi: 10.1186/1479-5876-7-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Xie L, Mouillet JF, Chu T, et al. C19MC microRNAs regulate the migration of human trophoblasts. Endocrinology. 2014;155:4975–4985. doi: 10.1210/en.2014-1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Fornari F, Milazzo M, Chieco P, et al. In hepatocellular carcinoma miR-519d is up-regulated by p53 and DNA hypomethylation and targets CDKN1A/p21, PTEN, AKT3 and TIMP2. J Pathol. 2012;227:275–285. doi: 10.1002/path.3995. [DOI] [PubMed] [Google Scholar]
- 92.Huang Q, Gumireddy K, Schrier M, et al. The microRNAs miR-373 and miR-520c promote tumour invasion and metastasis. Nat Cell Biol. 2008;10:202–210. doi: 10.1038/ncb1681. [DOI] [PubMed] [Google Scholar]
- 93.Li M, Lee KF, Lu Y, et al. Frequent amplification of a chr19q13.41 microRNA polycistron in aggressive primitive neuroectodermal brain tumors. Cancer Cell. 2009;16:533–546. doi: 10.1016/j.ccr.2009.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rippe V, Dittberner L, Lorenz VN, et al. The two stem cell microRNA gene clusters C19MC and miR-371-3 are activated by specific chromosomal rearrangements in a subgroup of thyroid adenomas. PLoS One. 2010;5:e9485. doi: 10.1371/journal.pone.0009485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kleinman CL, Gerges N, Papillon-Cavanagh S, et al. Fusion of TTYH1 with the C19MC microRNA cluster drives expression of a brain-specific DNMT3B isoform in the embryonal brain tumor ETMR. Nat Genet. 2014;46:39–44. doi: 10.1038/ng.2849. [DOI] [PubMed] [Google Scholar]
- 96.Zhao JJ, Yang J, Lin J, et al. Identification of miRNAs associated with tumorigenesis of retinoblastoma by miRNA microarray analysis. Childs Nerv Syst. 2009;25:13–20. doi: 10.1007/s00381-008-0701-x. [DOI] [PubMed] [Google Scholar]
- 97.Tan SM, Kirchner R, Jin J, et al. Sequencing of captive target transcripts identifies the network of regulated genes and functions of primate-specific miR-522. Cell Rep. 2014;8:1225–1239. doi: 10.1016/j.celrep.2014.07.023. [DOI] [PubMed] [Google Scholar]
- 98.Donker RB, Mouillet JF, Chu T, et al. The expression profile of C19MC microRNAs in primary human trophoblast cells and exosomes. Mol Hum Reprod. 2012;18:417–424. doi: 10.1093/molehr/gas013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Delorme-Axford E, Donker RB, Mouillet JF, et al. Human placental trophoblasts confer viral resistance to recipient cells. Proc Natl Acad Sci U S A. 2013;110:12048–12053. doi: 10.1073/pnas.1304718110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bayer A, Delorme-Axford E, Sleigher C, et al. Human trophoblasts confer resistance to viruses implicated in perinatal infection. Am J Obstet Gynecol. 2015;212:71 e1–71 e8. doi: 10.1016/j.ajog.2014.07.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Weber JA, Baxter DH, Zhang S, et al. The microRNA spectrum in 12 body fluids. Clin Chem. 2010;56:1733–1741. doi: 10.1373/clinchem.2010.147405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chim SS, Shing TK, Hung EC, et al. Detection and characterization of placental microRNAs in maternal plasma. Clin Chem. 2008;54:482–490. doi: 10.1373/clinchem.2007.097972. [DOI] [PubMed] [Google Scholar]
- 103.Lawrie CH, Gal S, Dunlop HM, et al. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br J Haematol. 2008;141:672–675. doi: 10.1111/j.1365-2141.2008.07077.x. [DOI] [PubMed] [Google Scholar]
- 104.Chen X, Ba Y, Ma L, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 105.Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ji X, Takahashi R, Hiura Y, Hirokawa G, Fukushima Y, Iwai N. Plasma miR-208 as a biomarker of myocardial injury. Clin Chem. 2009;55:1944–1949. doi: 10.1373/clinchem.2009.125310. [DOI] [PubMed] [Google Scholar]
- 107.Wang GK, Zhu JQ, Zhang JT, et al. Circulating microRNA: a novel potential biomarker for early diagnosis of acute myocardial infarction in humans. Eur Heart J. 2010;31:659–666. doi: 10.1093/eurheartj/ehq013. [DOI] [PubMed] [Google Scholar]
- 108.Wang K, Zhang S, Marzolf B, et al. Circulating microRNAs, potential biomarkers for drug-induced liver injury. Proc Natl Acad Sci U S A. 2009;106:4402–4407. doi: 10.1073/pnas.0813371106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Arroyo JD, Chevillet JR, Kroh EM, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A. 2011;108:5003–5008. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Turchinovich A, Weiz L, Langheinz A, Burwinkel B. Characterization of extracellular circulating microRNA. Nucleic Acids Res. 2011;39:7223–7233. doi: 10.1093/nar/gkr254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol. 2011;13:423–433. doi: 10.1038/ncb2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wang K, Zhang S, Weber J, Baxter D, Galas DJ. Export of microRNAs and microRNA-protective protein by mammalian cells. Nucleic Acids Res. 2010;38:7248–7259. doi: 10.1093/nar/gkq601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Raiborg C, Stenmark H. The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature. 2009;458:445–452. doi: 10.1038/nature07961. [DOI] [PubMed] [Google Scholar]
- 114.Baietti MF, Zhang Z, Mortier E, et al. Syndecan-syntenin-ALIX regulates the biogenesis of exosomes. Nat Cell Biol. 2012;14:677–685. doi: 10.1038/ncb2502. [DOI] [PubMed] [Google Scholar]
- 115.Nabhan JF, Hu R, Oh RS, Cohen SN, Lu Q. Formation and release of arrestin domain-containing protein 1-mediated microvesicles (ARMMs) at plasma membrane by recruitment of TSG101 protein. Proc Natl Acad Sci U S A. 2012;109:4146–4151. doi: 10.1073/pnas.1200448109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 117.Chiba M, Kimura M, Asari S. Exosomes secreted from human colorectal cancer cell lines contain mRNAs, microRNAs and natural antisense RNAs, that can transfer into the human hepatoma HepG2 and lung cancer A549 cell lines. Oncol Rep. 2012;28:1551–1558. doi: 10.3892/or.2012.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Morel L, Regan M, Higashimori H, et al. Neuronal exosomal miRNA-dependent translational regulation of astroglial glutamate transporter GLT1. J Biol Chem. 2013;288:7105–7116. doi: 10.1074/jbc.M112.410944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Umezu T, Ohyashiki K, Kuroda M, Ohyashiki JH. Leukemia cell to endothelial cell communication via exosomal miRNAs. Oncogene. 2013;32:2747–2755. doi: 10.1038/onc.2012.295. [DOI] [PubMed] [Google Scholar]
- 120.Chevillet JR, Kang Q, Ruf IK, et al. Quantitative and stoichiometric analysis of the microRNA content of exosomes. Proc Natl Acad Sci U S A. 2014;111:14888–14893. doi: 10.1073/pnas.1408301111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Fabbri M, Paone A, Calore F, et al. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc Natl Acad Sci U S A. 2012;109:E2110–E2116. doi: 10.1073/pnas.1209414109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Lehmann SM, Kruger C, Park B, et al. An unconventional role for miRNA: let-7 activates Toll-like receptor 7 and causes neurodegeneration. Nat Neurosci. 2012;15:827–835. doi: 10.1038/nn.3113. [DOI] [PubMed] [Google Scholar]
- 123.Thum T. Noncoding RNAs and myocardial fibrosis. Nat Rev Cardiol. 2014;11:655–663. doi: 10.1038/nrcardio.2014.125. [DOI] [PubMed] [Google Scholar]
- 124.Bandiera S, Pfeffer S, Baumert TF, Zeisel MB. miR-122--a key factor and therapeutic target in liver disease. J Hepatol. 2015;62:448–457. doi: 10.1016/j.jhep.2014.10.004. [DOI] [PubMed] [Google Scholar]
- 125.Pineles BL, Romero R, Montenegro D, et al. Distinct subsets of microRNAs are expressed differentially in the human placentas of patients with preeclampsia. Am J Obstet Gynecol. 2007;196:261 e1–261 e6. doi: 10.1016/j.ajog.2007.01.008. [DOI] [PubMed] [Google Scholar]
- 126.Lee DC, Romero R, Kim JS, et al. miR-210 targets iron-sulfur cluster scaffold homologue in human trophoblast cell lines: siderosis of interstitial trophoblasts as a novel pathology of preterm preeclampsia and small-for-gestational-age pregnancies. Am J Pathol. 2011;179:590–602. doi: 10.1016/j.ajpath.2011.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Muralimanoharan S, Maloyan A, Mele J, Guo C, Myatt LG, Myatt L. MIR-210 modulates mitochondrial respiration in placenta with preeclampsia. Placenta. 2012;33:816–823. doi: 10.1016/j.placenta.2012.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ishibashi O, Ohkuchi A, Ali MM, et al. Hydroxysteroid (17-beta) dehydrogenase 1 is dysregulated by miR-210 and miR-518c that are aberrantly expressed in preeclamptic placentas: a novel marker for predicting preeclampsia. Hypertension. 2012;59:265–273. doi: 10.1161/HYPERTENSIONAHA.111.180232. [DOI] [PubMed] [Google Scholar]
- 129.Kleinrouweler CE, Van Uitert M, Moerland PD, Ris-Stalpers C, Van Der Post JA, Afink GB. Differentially expressed genes in the pre-eclamptic placenta: a systematic review and meta-analysis. PLoS One. 2013;8:e68991. doi: 10.1371/journal.pone.0068991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Enquobahrie DA, Abetew DF, Sorensen TK, Willoughby D, Chidambaram K, Williams MA. Placental microRNA expression in pregnancies complicated by preeclampsia. Am J Obstet Gynecol. 2011;204:178 e12–178 e21. doi: 10.1016/j.ajog.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Xu P, Zhao Y, Liu M, et al. Variations of microRNAs in human placentas and plasma from preeclamptic pregnancy. Hypertension. 2014;63:1276–1284. doi: 10.1161/HYPERTENSIONAHA.113.02647. [DOI] [PubMed] [Google Scholar]
- 132.Zhu XM, Han T, Sargent IL, Yin GW, Yao YQ. Differential expression profile of microRNAs in human placentas from preeclamptic pregnancies vs normal pregnancies. Am J Obstet Gynecol. 2009;200:661 e1–661 e7. doi: 10.1016/j.ajog.2008.12.045. [DOI] [PubMed] [Google Scholar]
- 133.Chan SY, Loscalzo J. MicroRNA-210: a unique and pleiotropic hypoxamir. Cell Cycle. 2010;9:1072–1083. doi: 10.4161/cc.9.6.11006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Devlin C, Greco S, Martelli F, Ivan M. miR-210: More than a silent player in hypoxia. IUBMB Life. 2011;63:94–100. doi: 10.1002/iub.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Ivan M, Huang X. miR-210: fine-tuning the hypoxic response. Adv Exp Med Biol. 2014;772:205–227. doi: 10.1007/978-1-4614-5915-6_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Colleoni F, Padmanabhan N, Yung HW, et al. Suppression of mitochondrial electron transport chain function in the hypoxic human placenta: a role for miRNA-210 and protein synthesis inhibition. PLoS One. 2013;8:e55194. doi: 10.1371/journal.pone.0055194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Myatt L, Muralimanoharan S, Maloyan A. Effect of preeclampsia on placental function: influence of sexual dimorphism, microRNA’s and mitochondria. Adv Exp Med Biol. 2014;814:133–146. doi: 10.1007/978-1-4939-1031-1_12. [DOI] [PubMed] [Google Scholar]
- 138.Kopriva SE, Chiasson VL, Mitchell BM, Chatterjee P. TLR3-induced placental miR-210 down-regulates the STAT6/interleukin-4 pathway. PLoS One. 2013;8:e67760. doi: 10.1371/journal.pone.0067760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Hromadnikova I, Kotlabova K, Ondrackova M, et al. Circulating C19MC microRNAs in preeclampsia, gestational hypertension, and fetal growth restriction. Mediators Inflamm. 2013;2013:186041. doi: 10.1155/2013/186041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Yang S, Li H, Ge Q, Guo L, Chen F. Deregulated microRNA species in the plasma and placenta of patients with preeclampsia. Mol Med Rep. 2015 doi: 10.3892/mmr.2015.3414. [DOI] [PubMed] [Google Scholar]
- 141.Anton L, Olarerin-George AO, Hogenesch JB, Elovitz MA. Placental Expression of miR-517a/b and miR-517c Contributes to Trophoblast Dysfunction and Preeclampsia. PLoS One. 2015;10:e0122707. doi: 10.1371/journal.pone.0122707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Higashijima A, Miura K, Mishima H, et al. Characterization of placenta-specific microRNAs in fetal growth restriction pregnancy. Prenat Diagn. 2013;33:214–222. doi: 10.1002/pd.4045. [DOI] [PubMed] [Google Scholar]
- 143.Wang D, Chen X, Song Y, Lv Q, Lai L, Li Z. Disruption of imprinted gene expression and DNA methylation status in porcine parthenogenetic fetuses and placentas. Gene. 2014;547:351–358. doi: 10.1016/j.gene.2014.06.059. [DOI] [PubMed] [Google Scholar]
- 144.Guo L, Tsai SQ, Hardison NE, et al. Differentially expressed microRNAs and affected biological pathways revealed by modulated modularity clustering (MMC) analysis of human preeclamptic and IUGR placentas. Placenta. 2013;34:599–605. doi: 10.1016/j.placenta.2013.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Sadovsky Y, Mouillet JF, Ouyang Y, Bayer A, Mishima T, Coyne CB. The function of trophomiRs and other microRNAs in the human placenta. In: Bianchi DW, Norwitz ER, editors. Reproductive Medicine. Woodbury, NY: Cold Spring Harbor Laboratory Press; 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Zhao Z, Moley KH, Gronowski AM. Diagnostic potential for miRNAs as biomarkers for pregnancy-specific diseases. Clin Biochem. 2013;46:953–960. doi: 10.1016/j.clinbiochem.2013.01.026. [DOI] [PubMed] [Google Scholar]
- 147.Leidner RS, Li L, Thompson CL. Dampening enthusiasm for circulating microRNA in breast cancer. PLoS One. 2013;8:e57841. doi: 10.1371/journal.pone.0057841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Williams Z, Ben-Dov IZ, Elias R, et al. Comprehensive profiling of circulating microRNA via small RNA sequencing of cDNA libraries reveals biomarker potential and limitations. Proc Natl Acad Sci U S A. 2013;110:4255–4260. doi: 10.1073/pnas.1214046110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Pritchard CC, Cheng HH, Tewari M. MicroRNA profiling: approaches and considerations. Nat Rev Genet. 2012;13:358–369. doi: 10.1038/nrg3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Watson AK, Witwer KW. Do platform-specific factors explain microRNA profiling disparities? Clin Chem. 2012;58:472–474. doi: 10.1373/clinchem.2011.175281. author reply 74–5. [DOI] [PubMed] [Google Scholar]
- 151.Kirschner MB, Van Zandwijk N, Reid G. Cell-free microRNAs: potential biomarkers in need of standardized reporting. Front Genet. 2013;4:56. doi: 10.3389/fgene.2013.00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Moldovan L, Batte KE, Trgovcich J, Wisler J, Marsh CB, Piper M. Methodological challenges in utilizing miRNAs as circulating biomarkers. J Cell Mol Med. 2014;18:371–390. doi: 10.1111/jcmm.12236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Witwer KW. Circulating microRNA biomarker studies: pitfalls and potential solutions. Clin Chem. 2015;61:56–63. doi: 10.1373/clinchem.2014.221341. [DOI] [PubMed] [Google Scholar]
- 154.Miller IV, Grunewald TG. Tumour-derived exosomes: Tiny envelopes for big stories. Biol Cell. 2015 doi: 10.1111/boc.201400095. [DOI] [PubMed] [Google Scholar]
- 155.Kambe S, Yoshitake H, Yuge K, et al. Human exosomal placenta-associated miR-517a-3p modulates the expression of PRKG1 mRNA in Jurkat cells. Biol Reprod. 2014;91:129. doi: 10.1095/biolreprod.114.121616. [DOI] [PubMed] [Google Scholar]
- 156.Ouyang Y, Mouillet JF, Coyne CB, Sadovsky Y. Review: placenta-specific microRNAs in exosomes - good things come in nano-packages. Placenta. 2014;(35 Suppl):S69–S73. doi: 10.1016/j.placenta.2013.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]