Abstract
Purpose
A new approach has been developed to quantify cell sizes and intracellular volume fractions using temporal diffusion spectroscopy with diffusion-weighted acquisitions.
Theory and Methods
Temporal diffusion spectra may be used to characterize tissue microstructure by measuring the effects of restrictions over a range of diffusion times. Oscillating gradients have been used previously to probe variations on cellular and subcellular scales, but their ability to accurately measure cell sizes larger than 10 μm is limited. By combining measurements made using oscillating gradient spin echo (OGSE) and a conventional pulsed gradient spin echo (PGSE) acquisition with a single, relatively long diffusion time, we can accurately quantify cell sizes and intracellular volume fractions.
Results
Based on a two compartment model (incorporating intra- and extracellular spaces), accurate estimates of cell sizes and intracellular volume fractions were obtained in vitro for (i) different cell types with sizes ranging from 10 to 20 μm, (ii) different cell densities, and (iii) before and after anti-cancer treatment.
Conclusion
Hybrid OGSE-PGSE acquisitions sample a larger region of temporal diffusion spectra and can accurately quantify cell sizes over a wide range. Moreover, the maximum gradient strength used was lower than 15 G/cm, suggesting that this approach is translatable to practical MR imaging.
Keywords: cell size, volume fraction, diffusion, MRI, oscillating gradient, diffusion time
Introduction
Cell size plays an important role in affecting the functional properties of cells from the molecular to the organismal level, such as cellular metabolism (1), proliferation (2) and tissue growth (3). For cancer diagnosis and prognosis, measurements of cell size have been widely used to differentiate cancer types (4) and monitor treatment-induced apoptosis (5). In addition to cell size, other microstructural information on a subcellular length scale, such as nuclear sizes, also provides insights into the functional properties of cancer cells. For example, nuclear size has been used to diagnose tumors (6) and distinguish low- from high-grade tumors (7). Conventionally, both cellular (e.g. cell size and cellularity) and subcellular (e.g. nuclear size and nuclear-to-cytoplasm ratio) information is obtained via invasive biopsy. However, biopsy is limited by various clinical complications, including pain, hemorrhage, infection, and even death (8). Therefore, a non-invasive technique with the capability of detecting tissue microstructural information on both cellular and subcellular length scales would be of great interest for clinical and research applications.
Diffusion-weighted MRI (DWI) provides a non-invasive way to map the diffusion properties of tissue water molecules that are affected by restrictions and hindrances to free movement, and is thereby able to provide information on tissue microstructure. Conventionally, an effective mean diffusion rate, the apparent diffusion coefficient (ADC), is obtained by using motion-sensitizing pulsed gradient spin echo (PGSE) pulse sequences to probe water displacements. ADC values are potentially affected by various tissue properties, including cellularity (9,10), cell size (11), nuclear size (12), and membrane permeability (13). ADC values therefore reflect overall diffusion effects and are not specific for any single parameter such as cell size. More elaborate methods have also been developed to map mean compartment size in porous media or tissues. Diffusion-diffraction effects (14) have been reported to successfully characterize the mono-dispersed cell sizes of erythrocytes (15), but fail in most real biological tissues due to the heterogeneous distribution of cell sizes. Q-space imaging generates a displacement probability distribution function that can provide apparent compartment sizes in biological tissues. Unfortunately, estimates of cell size are biased in practice because of methodological limitations (for example, the short-gradient pulse approximation (16) is usually not met, and only relatively low gradient strengths are available on regular MRI systems (17)) and the inherent complexity of real biological tissues (e.g. the existence of multiple compartments (17) and coexisting inflammation (18)). To overcome these problems, multi-compartment models that mimic real tissues have been developed to more accurately characterize cell size. For example, models that include more than one compartment and/or the effects of membrane permeability have been used to assess axon sizes in bovine optic nerve (19), to characterize axon size distributions in vitro (20) and in vivo (21), and to quantify mean axon size in the corpus callosum of human (22) and monkey brain (23). In addition, as an extension of the PGSE method, the double-PGSE sequence has been used to measure cell size and anisotropy in fixed yeast cells (24). However, despite these successes, subcellular microstructural information has not been obtainable by practical PGSE-based methods. One reason is that PGSE sequences typically employ a diffusion time of 20–80 ms in vivo due to hardware limitations. Thus, the characteristic diffusion length probed by water molecules is on the order of 5–20 μm, which is similar to the distance between hindering cell membranes of typical eukaryotic cells. As a result, PGSE measurements are sensitive to cellularity in biological tissues, but cannot isolate the effects of subcellular structures (< 5 μm). Consequently, conventional PGSE methods are suited to characterize cell sizes and cell densities in biological tissues non-invasively, but do not in practice provide information on subcellular microstructure.
Oscillating gradient spin echo (OGSE) sequences have emerged as a novel means to obtain much shorter effective diffusion times by replacing bipolar diffusion-sensitizing gradients in PGSE methods with oscillating gradients (25–27). Previous reports have shown that OGSE measurements, even at moderately low frequencies, are capable of detecting restrictions to water diffusion over a short spatial scale (< 5 μm) which is usually not accessible by conventional PGSE methods (25). Moreover, by varying the oscillation frequency (f), temporal diffusion spectra can be obtained to provide more comprehensive information about biological tissues at both cellular and subcellular length scales (28,29). OGSE sequences have been successfully implemented and used to measure abnormal brain metabolites (30) and treatment-induced variations in organelles (31), nuclear size changes after anti-cancer therapy (32) and hypoxia-ischemia (33,34). Recently, there has been increasing interest in using OGSE methods to measure axon sizes (35–37). However, despite these successes in probing microstructure, the ability of OGSE sequences that use harmonic waveforms to probe relatively large cell sizes is, ironically, limited by the maximum achievable diffusion time within the constraints of practical echo times. In typical cosine-modulated OGSE methods, the effective diffusion time Δdiff = 1/(4f) = δ/(4N), where f is the frequency, δ is the duration of diffusion gradient, and N is the number of oscillations in each gradient (25). Hence, even for a very long gradient duration of 100 ms, the maximum achievable Δdiff = 25 ms, corresponding to a diffusion distance ≈ 10 μm if the diffusion coefficient is 2 μm2/ms. Such a relatively short displacement significantly decreases the ability to quantify larger cell sizes (e.g. 10 – 20 μm), which is typical in tumors. Moreover, long gradient durations and lower frequencies in practice may not be used because of signal losses caused by T2 relaxation. As an example, as shown in our recent histology-validated measurements using OGSE to measure mean axon diameters, dimensions in the range 1 to 5 μm were measured accurately, but the axon diameters larger than 6 μm were significantly underestimated (37).
For a general time-dependent gradient, the diffusion-weighted MR signal can be expressed as (25)
| (1) |
where F(ω) is the Fourier transform of the time integral of the diffusion gradient, and D(ω) is the frequency-dependent diffusion tensor that contains microstructural information. With appropriately designed gradient waveform (e.g. cosine-modulated), F(ω) can serve as a well-defined sampling function and hence D(ω), the temporal diffusion spectrum, can be obtained. However, due to hardware limitations, it is impossible to achieve all frequencies in practice, and hence only a segment of a diffusion spectrum can be acquired. For a conventional PGSE sequence, in which δ is the gradient pulse width and Δ is the diffusion time, , which therefore samples low frequency regions of the spectrum that are difficult to sample with oscillating waveforms. In Figure 1, the spectrum is divided into two regions by the grey band, representing the readily accessible spectral ranges for PGSE (left) and OGSE (right) methods, separately. For the diffusion spectra of restricted water diffusion inside impermeable spheres (Figure 1), the rise in apparent diffusion coefficient in response to an increase in the frequency is the key to extract cell size (37). In order to measure the size of spheres smaller than 10 μm, an appropriate observation window can be chosen in the OGSE-accessible region to capture the rising portion of the curve. However, as the cell size becomes larger, the spectra in the right region are almost flat and the observation window needs to extend to the PGSE-accessible region. In the current study, 40 and 80 Hz OGSE measurements and PGSE with a diffusion time of 52 ms (labeled by vertical dash line in Figure 1) were combined, corresponding to an observation window with Δdiff from 3.13 to 52 ms. We hypothesized that such a combination would cover a broader segment of the temporal diffusion spectrum and enable the quantification of larger cell sizes (up to 20 μm). In the rest of the paper, this hypothesis is elucidated and validated using both computer simulations and well-characterized cell culture experiments.
Figure 1.
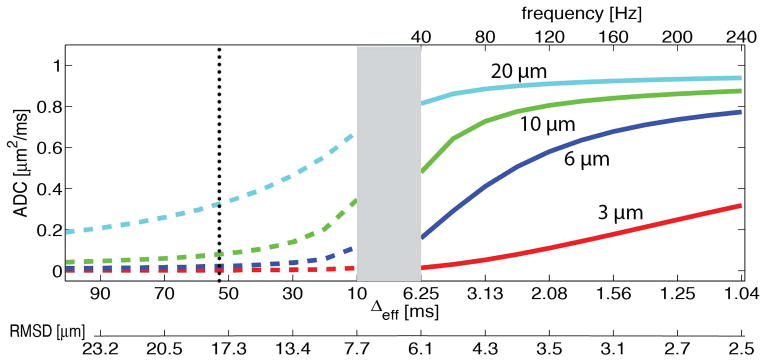
Diffusion spectra of restricted water diffusion inside impermeable spheres with diameters of 3, 6, 10, and 20 μm. The grey band divides the spectrum into two regions, representing the readily accessible spectral ranges for PGSE (left) and OGSE (right) methods, separately.
Methods
Diffusion signal modeling
We assume that the diffusion-weighted signals of cell samples can be expressed as the sum of signals arising from intra- and extracellular spaces, namely,
| (2) |
where fin is the water volume fraction of intracellular space, and Sin and Sex are the signal magnitudes per volume from the intra- and extracellular spaces, respectively. The water exchange between intra- and extracellular spaces is omitted, as suggested in the CHARMED model (38).
Cells are modeled as spheres despite the significant variations of cell shape in realistic tissues. The analytical expressions of OGSE signals in some typical geometrical structures, e.g. cylinders and spheres, have been derived previously (39). For OGSE measurements of diffusion within impermeable spheres using cosine-modulated gradient waveforms, the intracellular diffusion signal can be expressed as
| (3) |
where Din is the intracellular diffusion coefficient, f is the oscillation frequency, δ is the gradient duration, Δ is the separation of two diffusion gradients, λn, Bn are structure dependent parameters containing sphere diameter. The accuracy of Eq.(3) has been validated by computer simulations (39) and phantom experiments (40). Note that, if f→0, a cosine-modulated OGSE pulse degenerates into a conventional PGSE pulse and hence Eq.(3) becomes
| (4) |
Therefore, Eqs.(3) and (4) describe intracellular diffusion signals obtained using OGSE and PGSE methods, respectively, and the microstructural parameters (i.e. λn and Bn, as well as sphere diameter) can be fit simultaneously using these equations.
Currently, only a narrow range of frequencies (less than 160 Hz) may be sampled with clinically achievable gradient strengths (< 15 G/cm). Under such circumstances, the ADC values for the extracellular space have been reported to show a linear dependence on the oscillating-gradient frequency (37). As a result, the extracellular diffusion signal can be modeled as a linear function of gradient frequency, namely,
| (5) |
where Dex0 is a constant and βex is the slope of extracellular diffusion coefficient with respect to frequency f, which contains information on structural dimensions but may not be simply assigned to any specific morphological feature in the tissue. If f→0, a cosine-modulated OGSE pulse degenerates into a conventional PGSE pulse and Eq.(5) becomes:
| (6) |
Simulation
Biological tissue was modeled as a two-compartment system, consisting of intracellular and extracellular spaces, without any water exchange between these two compartments. Cells were modeled as a collection of densely packed spheres on a face-centered cube (FCC) (12). The following diffusion and morphological parameters were used in the simulation: Din= 1 μm2/ms, Dex= 2 μm2/ms, cell diameter = 2.5, 5, 10, 15, 20, and 25 μm, intra-cellular volume fraction = 43%, 51%, and 62%. A finite difference method was employed, and further details of the computational aspects of the simulations have been reported elsewhere (41). Diffusion-weighted signals were simulated for both OGSE and PGSE sequences with the same parameters set in the cell culture experiments.
Cell Preparation
Two sets of cell experiments, designated as “A” and “B”, were performed. Experiment “A” was designed to measure cell sizes for different types of cells at different densities, while experiment “B” was designed to characterize the changes in the tumor cell size before and after anti-cancer treatment. In experiment “A”, murine erythroleukemia (MEL) and human promyelocytic leukemia K562 cells, purchased from American Type Culture Collection (ATCC, Manassas, VA), were cultured in DMEM medium supplemented with 10% FBS, 50 U/ml penicillin, and 50 μg/ml streptomycin (Invitrogen, CA) under standard culture conditions in a humidified incubator maintained at 5% CO2 and 37 °C. Cells were spread every three days by 1:10 dilution and cell density was limited to be no more than 0.8 ×106 cells/ml. All cell samples were collected, washed with PBS, and then fixed with 4% paraformaldehyde in PBS for over 2 hours. After fixation, cells were washed, and transferred to a 0.65ml Eppendorf tube. In order to obtain different cell densities, tubes were divided into three groups and centrifuged (Bio-rad microcentrifuge) at three different centrifugal forces (200, 1000, and 6000 g) for 2 minutes to result in three different cell densities. The liquid on the top was carefully removed, and the residual cell pellets, whose net weights were around 150 to 200 mg, were used for NMR measurements.
In experiment “B”, nab-paclitaxel (Abraxane) treated MDA-MBA-231 cells were used to mimic the effects of anti-cancer therapy on cell sizes. Abraxane is an FDA-approved mitotic inhibitor drug used in the treatment of breast, lung and pancreatic cancers (43, 44). It interrupts cell division during the mitotic (M) phase of the cell cycle when two sets of fully formed chromosomes are supposed to separate into daughter cells. Cells are thereby trapped in the M phase and then undergo apoptosis. Note that cells significantly increase their sizes during the M phase, and hence the cell size can be used a surrogate biomarker to monitor the efficacy of Abraxane treatment. Human breast cancer cells MDA-MBA-231 are highly metastatic and show a positive response to Abraxane treatment. MDA-MB-231 cells were cultured in the same way as MEL and K562 cells. After that, a half of the MDA-MB-231 cells were treated with 200nM of Abraxane for 24 hours. All cell samples were collected, washed with PBS, and then fixed with 4% paraformaldehyde in PBS for over 2 hours. After fixation, cells were washed, and transferred to a 0.65ml Eppendorf tube. Tubes were centrifuged (Bio-rad microcentrifuge) at 1000g for 2 minutes.
NMR measurements
All measurements were performed on a 7.0-T, 16-cm bore Varian DirectDrive™ spectrometer (Varian Inc. Palo alto, CA). As described in ‘cell preparation’ section, MEL and K562 cells were prepared at three different cell densities (low, medium and high) with 4 samples at each density. Nine b-values evenly distributed between 0 and 2000 s/mm2 were used in both PGSE and OGSE measurements. The OGSE pulse sequence substitutes the two bipolar diffusion-sensitizing gradients in PGSE with two apodized cosine-modulated gradients. Details of the OGSE pulse sequence can be found in (42). For PGSE experiments, diffusion gradient durations were δ=4 ms, and separation Δ=52 ms. The OGSE method measured frequencies at 40, 80, and 120 Hz with δ/Δ=25/30 ms. Note that the echo times (TE=60 ms) for both PGSE and OGSE measurements were the same to minimize relaxation effects.
Data analysis
The PGSE and OGSE diffusion signals were fit to Eq. (1)-(5) with five unknown variables: cell diameter d, intracellular diffusion coefficient Din, intracellular volume fraction fin, and extracellular parameters Dex0 and βex (see Eq.(5)), using the lsqcurvefit function in Matlab (Mathworks, Natick, MA). The constraints for fitting parameters were: 0 ≤ d ≤ 40 μm, 0≤ fin ≤ 1, 0 ≤ Din ≤3.0 μm2/ms, 0 ≤ Dex0 ≤ 3.0 μm2/ms, and 0 ≤ βex ≤ 2 μm2. Randomly-generated initial parameters values were used. To ensure the global minimum was reached, the fitting was repeated 100 times for each sample, and the analyses corresponding to the smallest fitting residual were chosen as the final results.
Statistical Analysis
The correlations between fitted and preset intracellular volume fractions was assessed using Kendall’s tau correlation coefficient (43). The differences in MR-derived cell sizes and intracellular volume fractions among the cell samples having different densities (Figure 5) were summarized using means and standard deviations, and compared by one-way ANOVA. The differences in the other three fitted parameters (Din, Dex0, and βex) among the cell samples having different densities (Figure 6) were summarized using means and standard deviations, and compared by one-way ANOVA. All the tests were two-sided and a p-value of 0.05 or less was taken to indicate statistical significance. Statistical analyses were performed using OriginPro 9.0 (OriginLab. Northampton, MA).
Figure 5.
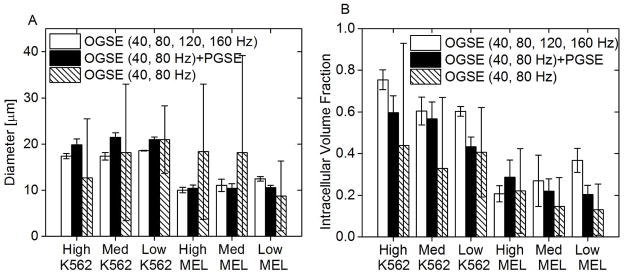
Diameter (left) and intracellular volume fractions (right) fitted from three combinations of OGSE and PGSE signals for K562 and MEL cells at three different densities: high, medium, and low. It is noted that varying cell density does not affect the fitted cell size for the same type of cells regardless of the data combination (p>0.05).
Figure 6.
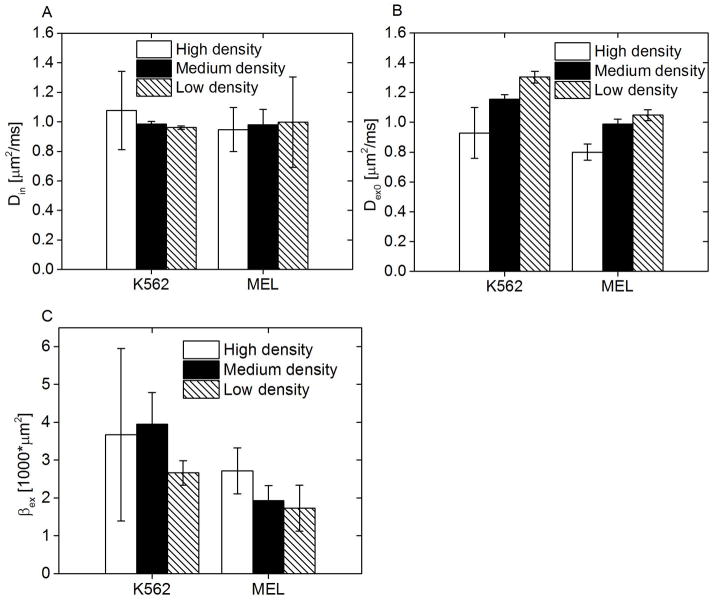
Fitted Din, Dex0, and βex for K562 and MEL cells at different cell densities: low, medium, and high.
Results
Simulation
For model systems mimicking tightly packed cells of diameter ranging from 2.5 to 25 μm and various intracellular volumes (43%, 51%, and 62%), both the OGSE signals with frequencies ranging from 40 to 160 Hz and the PGSE signals with a single long diffusion time of 48 ms were obtained from simulations. The size and intracellular volume fraction were fitted from three different combinations of the simulated data, including OGSE at 40 and 80 Hz, OGSE at 40 and 80 Hz + PGSE, and OGSE at 40, 80, 120, and 160 Hz. The cutoff frequencies, 80 and 160 Hz, are determined by two different maximum gradient strengths, 15 and 40 G/cm, corresponding to readily achievable gradient strengths on regular animal scanners and those with advanced human gradient coils (44), respectively. Figure 2A shows the fitted mean sizes of cells at three different densities. The OGSE method employing a maximum gradient strength of 15 G/cm is sufficient to extract the sizes of the cells smaller than 8 μm, while it underestimates the sizes of the cells larger than 8 μm. For cells larger than 8 μm, the accuracy of size measurement can be significantly improved by incorporating either PGSE or more OGSE data with more frequencies. Correlations between fitted and preset intracellular volume fractions are shown in Figure 2B and were measured via Kendall’s concordance coefficient. The calculated value of Kendall’s coefficient was 0.6 (p=0.0007), 0.75 (p<0.0001), and 0.72 (p<0.0001) for OGSE at 40 and 80 Hz, OGSE at 40 and 80 Hz + PGSE, and OGSE at 40, 80, 120, and 160 Hz, respectively. The fact that our proposed combination has a strong correlation demonstrates that this approach is a useful tool for the quantification of the intracellular volume fraction.
Figure 2.
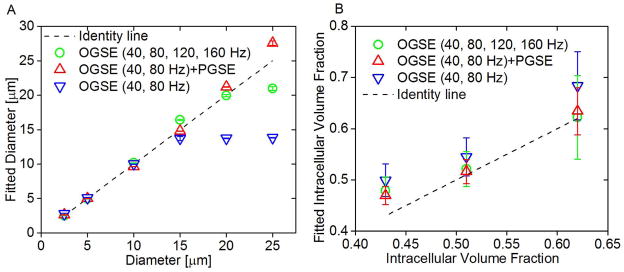
(A) Correlation between fitted diameters and preset diameters in simulations. Fitted diameters generated from three different combinations of OGSE and PGSE signals, mean ± std (n=3, with three different intracellular volume fractions: 43%, 51%, and 62%) vs. preset diameters. The dot line represents the identity line. (B) Fitted intracellular volume fraction generated from three different combinations of OGSE and PGSE signals, mean ± std (n=6, with cell size ranging from 2.5 to 25 μm) vs. preset intracellular volume fractions. The dot line represents the identity line.
Cell size measurements with different cell types and densities
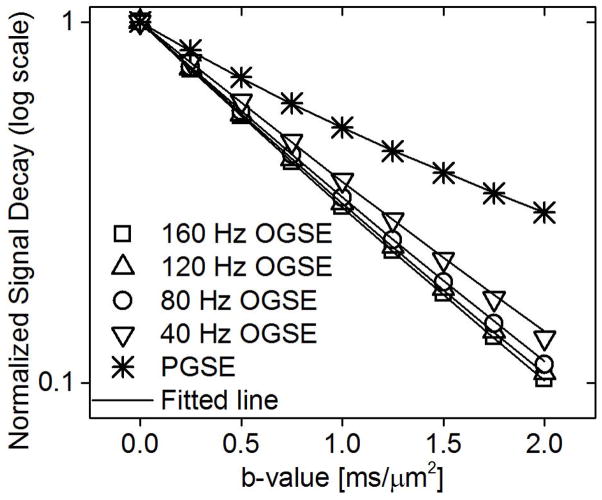
Representative OGSE and PGSE signals for a sample K562 cell pellet with medium density are shown in Figure 3. As expected, the diffusion-weighted signals decay faster as the effective diffusion time decreases. The PGSE signals were significantly higher than OGSE signals, indicating that lower effective diffusion rates were obtained at longer diffusion times. The solid lines represent the fits from Eq (1).
Figure 3.
Typical OGSE and PGSE signals for the K562 cell pellet which was centrifuged at a centrifugal force of 1000g. The solid line represents the fit using Eq(1)–(5).
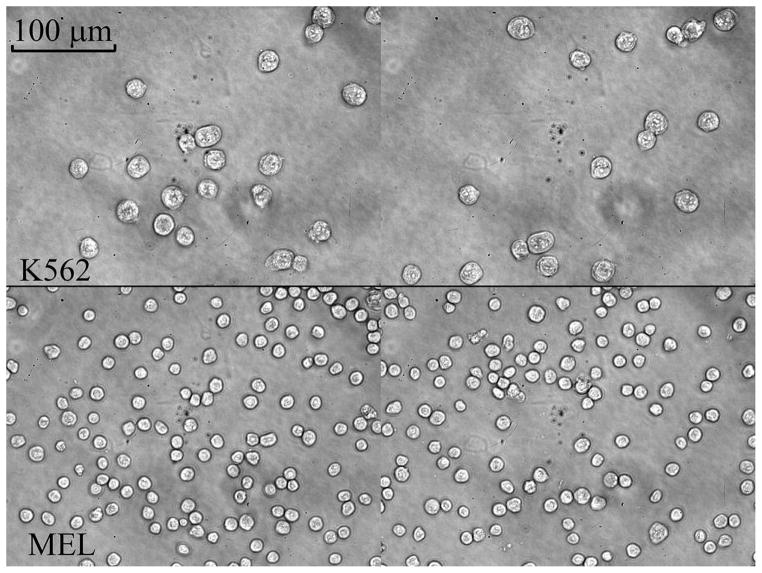
Figure 4 shows typical 40x microscope images for K562 (top) and MEL (bottom) cells. It is evident that K562 cells are much larger than MEL cells. The area of each cell was calculated from these light microscope images using an auto-segmentation program written in Matlab, and then converted to a diameter assuming each cell is a sphere. The area-weighted diameters (29) for K562 and MEL, which were determined from approximate 1000 cells, were 20.94±1.08 and 11.74±1.30 μm, respectively.
Figure 4.
Typical 40x light microscopy pictures for K562 (top) and MEL (bottom) cells.
Figure 5 compares the diameters (A) and intracellular volume fractions (B) fitted from three combinations of diffusion-weighted signals for K562 and MEL cells at three different densities. The two parameters extracted from 40 and 80 Hz OGSE signals have dramatic variations among the samples having the same cell types and the same densities, indicating that the combination of 40 and 80 Hz OGSE signals does not generate reliable results under these experimental conditions.
Using the high-frequency OGSE data, the mean diameters for K562 and MEL cells at three densities are close to the microscope-derived diameters. However, one-way ANOVA revealed that the diameters for the samples at low density are significantly larger than those from the high (p= 0.04 and 0.005 for K562 and MEL, respectively) and medium densities (p= 0.03 and 0.04 for K562 and MEL, respectively). Similarly, the mean intracellular volume fractions for MEL cells at high density are significantly lower than those for the low density (p=0.004), suggesting that this combination of OGSE signals does not estimate the intracellular volume fraction reliably.
By contrast, by combining the PGSE and low-frequency OGSE signals, the fitted cell sizes are not only consistent with microscope-derived sizes (Table 1) but also insensitive to the variations in cell density (p>0.05), indicating this approach is accurate for size quantification and robust to varying cell densities. Moreover, the intracellular volume fractions for cell samples of high density are significantly higher than that for low-density samples (p=0.02 and 0.03 for K562 and MEL, respectively). The medium-density samples were not differentiated from the other two densities (p>0.05).
Table 1.
Fitted and microscope-derived diameters for K562 and MEL cells at different cell densities: low, medium, and high.
| Density | Light microscope | |||
|---|---|---|---|---|
| high | medium | low | ||
| K562 | 19.82±1.30 | 21.5±0.98 | 20.96±0.60 | 20.94±1.08 |
| MEL | 10.42±0.72 | 10.46±0.98 | 10.6±0.48 | 11.74±1.30 |
In Figure 6, the other three parameters (Din, Dex0, and βex) obtained from the combination of PGSE and low-frequency OGSE signals are displayed. Din remains almost constant for different cell densities and cell types. Dex0 increases with the decreasing cell density (p<0.05), possibly because of the increased extracellular tortuosity. βex does not show any apparent correlation with the cell density (p>0.05).
Cell size measurements with anti-cancer treatment
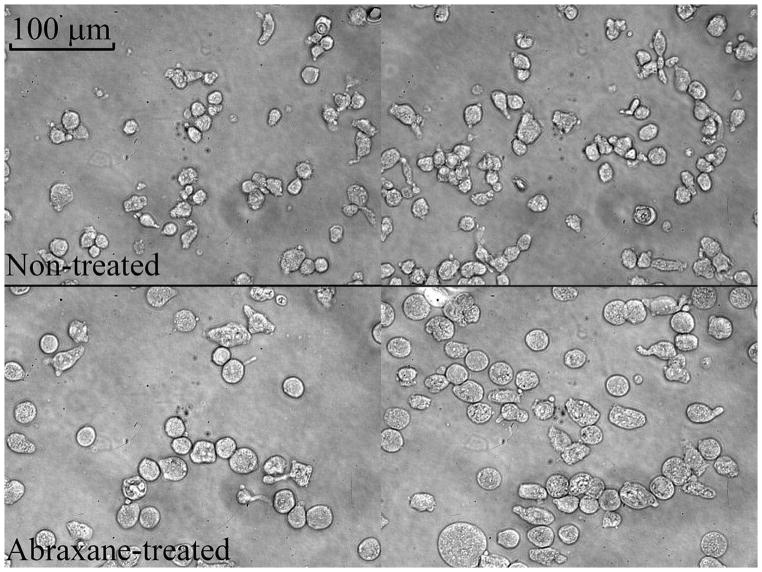
Abraxane is used to trap cells in the M phase and then undergo apoptosis. It is known that cells significantly increase their sizes during the M phase. Our microscope observations (Figure 7) confirm that Abraxane-treated cells are larger on average than the untreated MDA-MBA-231 cells. MRI-derived cell sizes are close to the area-weighted diameters calculated from light microscopy (table 2), indicating that the proposed approach is capable of quantifying increases in cell sizes in response to anti-cancer treatment. Table 3 shows the other four parameters (Din, fin, Dex0, and βex) for Abraxane-treated and non-treated cells. The intracellular volume fraction became larger after treatment most likely due to the increase of individual cell size, although the packing density for large cells was lower than small cells. The extracellular diffusion coefficient (Dex0) increased in the cell samples with larger cells, presumably due to the increased mean restricting distances in the extracellular spaces. Note that the changes of Din may not reflect the true values because the applied gradient frequencies were not high enough to sensitize the intracellular free diffusion (40).
Figure 7.
Typical 40x light microscopy pictures for non-treated (top) and Abraxane-treated (bottom) MDA-MBA-231 cells. It is noted that most cells are not spherical.
Table 2.
Fitted and microscope-derived diameters for non-treated and Abraxane-treated MDA-MBA-231 cells.
| Fitted | Light microscope | |
|---|---|---|
| Non-treated | 14.9±1.58 | 15.72±1.84 |
| Abraxane-treated | 22.24±0.36 | 22.48±0.9 |
Table 3.
Fitted Din, Intracellular volume fraction, Dex0, and βex for non-treated and Abraxane-treated MDA-MBA-231 cells.
| Din (μm2/ms) | Intracellular volume fraction (%) | Dex0 (μm2/ms) | βex (1000*μm2) | |
|---|---|---|---|---|
| Non-treated | 0.62±0.018 | 42±6.8 | 0.84±0.047 | 4.2±0.39 |
| Abraxane-treated | 0.76±0.0012 | 59±2.3 | 1.00±0.029 | 4.17±0.87 |
Discussion
Various microstructural features of tumors, such as the cellularity, cell size, and nuclear size, play an important role in the diagnosis and prognosis of cancer. Conventionally, these parameters are obtainable only from invasive biopsy. Diffusion-weighted MRI provides a means to probe cellular characteristics of biological tissues non-invasively, and hence is potentially a viable approach for non-invasively obtaining critical information about cancer. Although PGSE and OGSE sequences are often conceived of as different, the theory of temporal diffusion spectroscopy shows how the information obtainable from different sequences is related. While OGSE implementation allow probing of the high frequency regions of a diffusion spectrum, PGSE has some advantages in practice for assessing low frequency values of the spectrum, a region that appears critical for accurately estimating the sizes of larger scale structures. PGSE methods are sensitive to cell size and cellularity, but relatively insensitive to subcellular microstructures. By contrast, OGSE methods are more sensitive to features that restrict diffusion at a subcellular scale, such as nuclear size, but in practice are not well suited to measuring larger structures. Our PGSE measurements with a single long diffusion time (52 ms) sample the diffusion spectra close to f → 0, extending the range of OGSE spectral data. By sampling a sufficient range of frequencies, a larger range of cell sizes (6 – 20 μm) can be accurately measured. This combination extends the ability of diffusion measurements to probe microstructural parameters over a broader range of length scales, from subcellular scales to large cell sizes. This more comprehensive information may have significant potential in cancer imaging.
Although it is possible to implement both conventional PGSE or OGSE methods to characterize cell dimensions and subcellular scales, respectively, this strategy is time-consuming and hence impractical in clinics. Previous studies have reported one hour scanning for PGSE-based methods to obtain mean axon diameter of human corpus callosum (22) and 2.5 hours to obtain the mean cell size of mouse tumors (45). Multi-frequency and multi-b-value OGSE measurements are also time-consuming especially with a limited signal-to-noise ratio (SNR) (37). In the current study, we report a novel approach to combine OGSE and PGSE measurements with a single, long diffusion time. Such an approach significantly improves the ability of OGSE methods to accurately measure large cell size (up to 20 μm) with slightly increased scanning time. Therefore, both cellular and subcellular information can be obtained by such an approach in limited scanning time, which may be of crucial interest in practice.
Both the simulations and cell culture experiments demonstrate that the temporal diffusion data obtained with combined PGSE and OGSE measurements enable an accurate in vitro estimation of cell size ranging from 10–20 μm using a maximum gradient strength of 15 G/cm. This gradient strength is available with the recently developed Connectome gradient coil, which provides a gradient strength up to 30 G/cm for human heads. Even for normal gradient coils with relatively low gradient strength, e.g. < 8 G/cm per channel, on regular MRI machines, it is still possible to employ our method. For example, a gradient duration δ = 25 ms was used in this work. Since the b value is proportional to δ3 and G2, a gradient strength of 7.5 G/cm can result in the same b values used in the current study with δ = 40 ms, a typical gradient duration for human OGSE studies (46,47). This means that our proposed approach is directly translatable to current human imaging, although it will benefit significantly from more advanced gradient coils.
It is also shown that incorporating more high-frequency OGSE data improves the accuracy of cell size estimates for large cells. The high-frequency OGSE data are considered to have higher sensitivity to detect small dimensions, so it is interesting that they helped make the measurements of larger cell sizes more accurate. As discussed previously, the rise in apparent diffusion coefficient in response to an increase in the frequency is the key to extract cell size (37). As shown in the curves with 10 – 20 μm in Figure 1, ADCs kept increasing at 120 and 160 Hz, indicating that these OGSE frequencies are still sensitize to large cell size ranging from 10 20 μm. When more and more frequencies that sensitize the cell size were included in the cell size fitting, more accurate results were obtained even if those relatively higher frequencies had lower sensitivities to large cell size. However, this broader frequency range method shows larger errors compared with our approach in measuring large cell size. For example, as shown in figure 5A, our approach led to 5.34%, 2.67%, and 0.08% deviations from the microscopy-derived diameters for K562 cells at high, medium, and low densities, respectively, while the broader frequency method led to deviations of 16.97%, 17.16% and 11.36%. More importantly, the acquisitions of high-frequency OGSE data covering the same range of b-value need very high gradient strength (40 G/cm), which limits the clinical use of this approach.
The water exchange between intra and extracellular spaces was assumed negligible in the current study. This assumption has been used in previous OGSE studies (28,32,48), because the effective diffusion time of the OGSE measurement is usually much shorter (< 5 ms) compared with the intracellular lifetime of water molecules (49,50). However, the precise effect of water exchange on diffusion measurements in vivo remains unclear. The incorporation of PGSE measurements with a long diffusion time makes this method more likely to be affected by water exchange than typical OGSE methods. In addition, membranes of tumor cells may have altered permeability resulting from either treatment or tumor development (51). Therefore, the influence of water exchange effects on the proposed approach needs to be further investigated, especially for in vivo studies. A more complex model which can account for water exchange between intra and extracellular spaces, such as the Karger model (52), will be investigated in a future study.
The current study used a simplified model which describes cells as homogenous spheres without nuclei. Interestingly, cell sizes fitted by this sphere model are reasonably accurate for non-spherical MDA-MBA-231 cells (Figure 7), suggesting that this method may not be very sensitive to variations in cell shape. This is encouraging since cells in real biological tissues are usually not spherical. On the other hand, it has been found in computer simulations and in vivo studies that a change of nuclear size can affect high-frequency OGSE signals (12,28). A change of nuclear size is widely observed in many biological processes, such as necrosis and apoptosis induced by cancer treatment. Therefore, it will be valuable to develop a more comprehensive model and broader range of measurements to be able to quantitatively characterize cell size, cell density, and nuclear size. If successful, this could provide more quantitative tissue characterization of tumors.
Fitted intracellular volume fractions also show a good correlation with cell densities. However, it is technically demanding to perform the histology validation for in vitro cell pellets. As an alternative, it has been reported that the intracellular volume fraction can be extracted from changes of diffusion-weighted signal in the extracellular space, resulting from the administration of Gadolinium-based contrast agent which changes the T1 of the extracellular space only (51). Efforts to compare intracellular volume fractions generated from these two methods are in progress.
Conclusion
Temporal diffusion spectroscopy combining a single long diffusion time PGSE and low-frequency OGSE measurements, was developed for accurately measuring relatively large cell sizes (10–20 μm). Using this method, accurate cellular sizes were obtained with a weak gradient strength of 15 G/cm, in different cancer cells and at three different cell densities. Size changes in breast cancer cells MDA-MBA-231 in response to chemotherapy were also detected. These findings were confirmed by light microscopy and confirm the potential of this method for providing microstructural information non-invasively to assist better diagnosis and prognosis of cancer.
Acknowledgments
This work was funded by NIH Grants K25CA168936, R01CA109106, R01CA173593, and P50CA128323.
References
- 1.Kozlowski J, Konarzewski M, Gawelczyk AT. Cell size as a link between noncoding DNA and metabolic rate scaling. P Natl Acad Sci USA. 2003;100(24):14080–14085. doi: 10.1073/pnas.2334605100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baserga R. Is cell size important? Cell cycle. 2007;6(7):814–816. doi: 10.4161/cc.6.7.4049. [DOI] [PubMed] [Google Scholar]
- 3.Savage VM, Allen AP, Brown JH, Gillooly JF, Herman AB, Woodruff WH, West GB. Scaling of number, size, and metabolic rate of cells with body size in mammals. P Natl Acad Sci USA. 2007;104(11):4718–4723. doi: 10.1073/pnas.0611235104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sun L, Sakurai S, Sano T, Hironaka M, Kawashima O, Nakajima T. High-grade neuroendocrine carcinoma of the lung: Comparative clinicopathological study of large cell neuroendocrine carcinoma and small cell lung carcinoma. Pathol Int. 2009;59(8):522–529. doi: 10.1111/j.1440-1827.2009.02402.x. [DOI] [PubMed] [Google Scholar]
- 5.Brauer M. In vivo monitoring of apoptosis. Prog Neuro-Psychoph. 2003;27(2):323–331. doi: 10.1016/S0278-5846(03)00026-5. [DOI] [PubMed] [Google Scholar]
- 6.Zink D, Fischer AH, Nickerson JA. Nuclear structure in cancer cells. Nat Rev Cancer. 2004;4(9):677–687. doi: 10.1038/nrc1430. [DOI] [PubMed] [Google Scholar]
- 7.Hsu CY, Kurman RJ, Vang R, Wang TL, Baak J, Shih IM. Nuclear size distinguishes low- from high-grade ovarian serous carcinoma and, predicts outcome. Hum Pathol. 2005;36(10):1049–1054. doi: 10.1016/j.humpath.2005.07.014. [DOI] [PubMed] [Google Scholar]
- 8.Tobkes AI, Nord HJ. Liver-Biopsy - Review of Methodology and Complications. Digest Dis. 1995;13(5):267–274. doi: 10.1159/000171507. [DOI] [PubMed] [Google Scholar]
- 9.Gauvain KM, McKinstry RC, Mukherjee P, Perry A, Neil JJ, Kaufman BA, Hayashi RJ. Evaluating pediatric brain tumor cellularity with diffusion-tensor imaging. Am J Roentgenol. 2001;177(2):449–454. doi: 10.2214/ajr.177.2.1770449. [DOI] [PubMed] [Google Scholar]
- 10.Sugahara T, Korogi Y, Kochi M, Ikushima I, Shigematu Y, Hirai T, Okuda T, Liang LX, Ge YL, Komohara Y, Ushio Y, Takahashi M. Usefulness of diffusion-weighted MRI with echo-planar technique in the evaluation of cellularity in gliomas. Jmri-J Magn Reson Im. 1999;9(1):53–60. doi: 10.1002/(sici)1522-2586(199901)9:1<53::aid-jmri7>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 11.Szafer A, Zhong JH, Gore JC. Theoretical-Model for Water Diffusion in Tissues. Magnet Reson Med. 1995;33(5):697–712. doi: 10.1002/mrm.1910330516. [DOI] [PubMed] [Google Scholar]
- 12.Xu J, Does MD, Gore JC. Sensitivity of MR diffusion measurements to variations in intracellular structure: effects of nuclear size. Magnetic resonance in medicine: official journal of the Society of Magnetic Resonance in Medicine/Society of Magnetic Resonance in Medicine. 2009;61(4):828–833. doi: 10.1002/mrm.21793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu J, Does MD, Gore JC. Dependence of temporal diffusion spectra on microstructural properties of biological tissues. Magnetic resonance imaging. 2011;29(3):380–390. doi: 10.1016/j.mri.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Callaghan PT, Coy A, Macgowan D, Packer KJ, Zelaya FO. Diffraction-Like Effects in Nmr Diffusion Studies of Fluids in Porous Solids. Nature. 1991;351(6326):467–469. [Google Scholar]
- 15.Torres AM, Michniewicz RJ, Chapman BE, Young GA, Kuchel PW. Characterisation of erythrocyte shapes and sizes by NMR diffusion-diffraction of water: correlations with electron micrographs. Magnetic resonance imaging. 1998;16(4):423–434. doi: 10.1016/s0730-725x(98)00005-8. [DOI] [PubMed] [Google Scholar]
- 16.Mitra PP, Halperin BI. Effects of Finite Gradient-Pulse Widths in Pulsed-Field-Gradient Diffusion Measurements. J Magn Reson Ser A. 1995;113(1):94–101. [Google Scholar]
- 17.Ong HH, Wright AC, Wehrli SL, Souza A, Schwartz ED, Hwang SN, Wehrli FW. Indirect measurement of regional axon diameter in excised mouse spinal cord with q-space imaging: simulation and experimental studies. NeuroImage. 2008;40(4):1619–1632. doi: 10.1016/j.neuroimage.2008.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Y, Wang Q, Haldar JP, Yeh FC, Xie MQ, Sun P, Tu TW, Trinkaus K, Klein RS, Cross AH, Song SK. Quantification of increased cellularity during inflammatory demyelination. Brain: a journal of neurology. 2011;134:3587–3598. doi: 10.1093/brain/awr307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Assaf Y, Cohen Y. Assignment of the water slow-diffusing component in the central nervous system using q-space diffusion MRS: implications for fiber tract imaging. Magnetic resonance in medicine: official journal of the Society of Magnetic Resonance in Medicine/Society of Magnetic Resonance in Medicine. 2000;43(2):191–199. doi: 10.1002/(sici)1522-2594(200002)43:2<191::aid-mrm5>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 20.Assaf Y, Blumenfeld-Katzir T, Yovel Y, Basser PJ. AxCaliber: a method for measuring axon diameter distribution from diffusion MRI. Magnetic resonance in medicine: official journal of the Society of Magnetic Resonance in Medicine/Society of Magnetic Resonance in Medicine. 2008;59(6):1347–1354. doi: 10.1002/mrm.21577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barazany D, Basser PJ, Assaf Y. In vivo measurement of axon diameter distribution in the corpus callosum of rat brain. Brain: a journal of neurology. 2009;132(Pt 5):1210–1220. doi: 10.1093/brain/awp042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alexander DC, Hubbard PL, Hall MG, Moore EA, Ptito M, Parker GJ, Dyrby TB. Orientationally invariant indices of axon diameter and density from diffusion MRI. NeuroImage. 2010;52(4):1374–1389. doi: 10.1016/j.neuroimage.2010.05.043. [DOI] [PubMed] [Google Scholar]
- 23.Dyrby TB, Sogaard LV, Hall MG, Ptito M, Alexander DC. Contrast and stability of the axon diameter index from microstructure imaging with diffusion MRI. Magnet Reson Med. 2013;70(3):711–721. doi: 10.1002/mrm.24501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shemesh N, Ozarslan E, Basser PJ, Cohen Y. Accurate noninvasive measurement of cell size and compartment shape anisotropy in yeast cells using double-pulsed field gradient MR. Nmr Biomed. 2012;25(2):236–246. doi: 10.1002/nbm.1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gore JC, Xu JZ, Colvin DC, Yankeelov TE, Parsons EC, Does MD. Characterization of tissue structure at varying length scales using temporal diffusion spectroscopy. Nmr Biomed. 2010;23(7):745–756. doi: 10.1002/nbm.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gross B, Kosfeld R. Anwendung der spin-echo-methode der messung der selbstdiffusion. Messtechnik. 1969;77:171–177. [Google Scholar]
- 27.Schachter M, Does MD, Anderson AW, Gore JC. Measurements of restricted diffusion using an oscillating gradient spin-echo sequence. Journal of magnetic resonance. 2000;147(2):232–237. doi: 10.1006/jmre.2000.2203. [DOI] [PubMed] [Google Scholar]
- 28.Xu J, Li K, Smith RA, Waterton JC, Zhao P, Chen H, Does MD, Manning HC, Gore JC. Characterizing tumor response to chemotherapy at various length scales using temporal diffusion spectroscopy. PloS one. 2012;7(7):e41714. doi: 10.1371/journal.pone.0041714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu J, Xie J, Jourquin J, Colvin DC, Does MD, Quaranta V, Gore JC. Influence of cell cycle phase on apparent diffusion coefficient in synchronized cells detected using temporal diffusion spectroscopy. Magnetic resonance in medicine: official journal of the Society of Magnetic Resonance in Medicine/Society of Magnetic Resonance in Medicine. 2011;65(4):920–926. doi: 10.1002/mrm.22704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marchadour C, Brouillet E, Hantraye P, Lebon V, Valette J. Anomalous diffusion of brain metabolites evidenced by diffusion-weighted magnetic resonance spectroscopy in vivo. Journal of cerebral blood flow and metabolism: official journal of the International Society of Cerebral Blood Flow and Metabolism. 2012;32(12):2153–2160. doi: 10.1038/jcbfm.2012.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Colvin DC, Jourquin J, Xu JZ, Does MD, Estrada L, Gore JC. Effects of Intracellular Organelles on the Apparent Diffusion Coefficient of Water Molecules in Cultured Human Embryonic Kidney Cells. Magnet Reson Med. 2011;65(3):796–801. doi: 10.1002/mrm.22666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Colvin DC, Loveless ME, Does MD, Yue Z, Yankeelov TE, Gore JC. Earlier detection of tumor treatment response using magnetic resonance diffusion imaging with oscillating gradients. Magnetic resonance imaging. 2011;29(3):315–323. doi: 10.1016/j.mri.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aggarwal M, Burnsed J, Martin LJ, Northington FJ, Zhang J. Imaging neurodegeneration in the mouse hippocampus after neonatal hypoxia-ischemia using oscillating gradient diffusion MRI. Magnetic resonance in medicine: official journal of the Society of Magnetic Resonance in Medicine/Society of Magnetic Resonance in Medicine. 2014;72(3):829–840. doi: 10.1002/mrm.24956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu D, Martin LJ, Northington FJ, Zhang J. Oscillating gradient diffusion MRI reveals unique microstructural information in normal and hypoxia-ischemia injured mouse brains. Magnetic resonance in medicine: official journal of the Society of Magnetic Resonance in Medicine/Society of Magnetic Resonance in Medicine. 2014;72(5):1366–1374. doi: 10.1002/mrm.25441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shemesh N, Alvarez GA, Frydman L. Measuring small compartment dimensions by probing diffusion dynamics via Non-uniform Oscillating-Gradient Spin-Echo (NOGSE) NMR. Journal of magnetic resonance. 2013;237:49–62. doi: 10.1016/j.jmr.2013.09.009. [DOI] [PubMed] [Google Scholar]
- 36.Siow B, Drobnjak I, Ianus A, Christie IN, Lythgoe MF, Alexander DC. Axon radius estimation with Oscillating Gradient Spin Echo (OGSE) Diffusion MRI diffusion fundamentals. 2013;18:1–6. [Google Scholar]
- 37.Xu J, Li H, Harkins KD, Jiang X, Xie J, Kang H, Does MD, Gore JC. Mapping mean axon diameter and axonal volume fraction by MRI using temporal diffusion spectroscopy. NeuroImage. 2014;103C:10–19. doi: 10.1016/j.neuroimage.2014.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Assaf Y, Freidlin RZ, Rohde GK, Basser PJ. New modeling and experimental framework to characterize hindered and restricted water diffusion in brain white matter. Magnetic resonance in medicine: official journal of the Society of Magnetic Resonance in Medicine/Society of Magnetic Resonance in Medicine. 2004;52(5):965–978. doi: 10.1002/mrm.20274. [DOI] [PubMed] [Google Scholar]
- 39.Xu J, Does MD, Gore JC. Quantitative characterization of tissue microstructure with temporal diffusion spectroscopy. Journal of magnetic resonance. 2009;200(2):189–197. doi: 10.1016/j.jmr.2009.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li H, Gore JC, Xu J. Fast and robust measurement of microstructural dimensions using temporal diffusion spectroscopy. Journal of magnetic resonance. 2014;242:4–9. doi: 10.1016/j.jmr.2014.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu J, Does MD, Gore JC. Numerical study of water diffusion in biological tissues using an improved finite difference method. Physics in medicine and biology. 2007;52(7):N111–126. doi: 10.1088/0031-9155/52/7/N01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Does MD, Parsons EC, Gore JC. Oscillating gradient measurements of water diffusion in normal and globally ischemic rat brain. Magnet Reson Med. 2003;49(2):206–215. doi: 10.1002/mrm.10385. [DOI] [PubMed] [Google Scholar]
- 43.Conover WJ. Practical nonparametric statistics. viii. New York: Wiley; 1999. p. 584. [Google Scholar]
- 44.Setsompop K, Kimmlingen R, Eberlein E, Witzel T, Cohen-Adad J, McNab JA, Keil B, Tisdall MD, Hoecht P, Dietz P, Cauley SF, Tountcheva V, Matschl V, Lenz VH, Heberlein K, Potthast A, Thein H, Van Horn J, Toga A, Schmitt F, Lehne D, Rosen BR, Wedeen V, Wald LL. Pushing the limits of in vivo diffusion MRI for the Human Connectome Project. NeuroImage. 2013;80:220–233. doi: 10.1016/j.neuroimage.2013.05.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Panagiotaki E, Walker-Samuel S, Siow B, Johnson SP, Rajkumar V, Pedley RB, Lythgoe MF, Alexander DC. Noninvasive quantification of solid tumor microstructure using VERDICT MRI. Cancer Res. 2014;74(7):1902–1912. doi: 10.1158/0008-5472.CAN-13-2511. [DOI] [PubMed] [Google Scholar]
- 46.Baron CA, Beaulieu C. Oscillating gradient spin-echo (OGSE) diffusion tensor imaging of the human brain. Magnetic resonance in medicine: official journal of the Society of Magnetic Resonance in Medicine/Society of Magnetic Resonance in Medicine. 2014;72(3):726–736. doi: 10.1002/mrm.24987. [DOI] [PubMed] [Google Scholar]
- 47.Van AT, Holdsworth SJ, Bammer R. In Vivo Investigation of Restricted Diffusion in the Human Brain with Optimized Oscillating Diffusion Gradient Encoding. Magnet Reson Med. 2014;71(1):83–94. doi: 10.1002/mrm.24632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu JZ, Xie JP, Jourquin J, Colvin DC, Does MD, Quaranta V, Gore JC. Influence of Cell Cycle Phase on Apparent Diffusion Coefficient in Synchronized Cells Detected Using Temporal Diffusion Spectroscopy. Magnet Reson Med. 2011;65(4):920–926. doi: 10.1002/mrm.22704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Quirk JD, Bretthorst GL, Duong TQ, Snyder AZ, Springer CS, Ackerman JJH, Neil JJ. Equilibrium water exchange between the intra- and extracellular spaces of mammalian brain. Magnet Reson Med. 2003;50(3):493–499. doi: 10.1002/mrm.10565. [DOI] [PubMed] [Google Scholar]
- 50.Zhao L, Kroenke CD, Song J, Piwnica-Worms D, Ackerman JJ, Neil JJ. Intracellular water-specific MR of microbead-adherent cells: the HeLa cell intracellular water exchange lifetime. Nmr Biomed. 2008;21(2):159–164. doi: 10.1002/nbm.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bailey C, Giles A, Czarnota GJ, Stanisz GJ. Detection of apoptotic cell death in vitro in the presence of Gd-DTPA-BMA. Magnetic resonance in medicine: official journal of the Society of Magnetic Resonance in Medicine/Society of Magnetic Resonance in Medicine. 2009;62(1):46–55. doi: 10.1002/mrm.21972. [DOI] [PubMed] [Google Scholar]
- 52.Karger J, Pfeifer H, Heink W. Principles and application of self-diffusion measurements by nuclear magnetic resonance. Advanced Magnetic Resonance. 1988;12:1–89. [Google Scholar]