Abstract
We have recently shown that two-dimensional (2D) and force-regulated kinetics of TCR–pMHC-I interactions predict responses of CD8+ T cells. To test whether these findings are applicable to CD4+ T cells, we analyzed the in situ 3.L2 TCR–pMHC-II interactions for a well-characterized panel of altered peptide ligands on the T-cell surface using the adhesion frequency assay with a micropipette and the thermal fluctuation and force-clamp assays with a biomembrane force probe. We found that the 2D effective TCR–pMHC-II affinity and off-rate correlate with, but better predict the T-cell response than, the corresponding measurements with the surface plasmon resonance in three dimensions (3D). The 2D affinity of the CD4 for MHCII was very low, approaching the detection limit, making it 1-2 orders of magnitude lower than the affinity of CD8 for MHC-I. In addition, the signal-dependent cooperation between TCR and co-receptor for pMHC binding previously observed for CD8 was not observed for CD4. Interestingly, force elicited TCR–pMHC-II catch-slip bonds for agonists but slip-only bonds for antagonists, thereby amplifying the power of discrimination between APLs. These results show that the force-regulated 2D binding kinetics of the 3.L2 TCR for pMHC-II determine functions of CD4+ T cells.
INTRODUCTION
TCR–pMHC interaction has intrigued many because of its low 3D affinity (Kd ~1-100μM) but high specificity and sensitivity (1-3). One of the most widely used methods to characterize the TCR–pMHC interaction is surface plasmon resonance (SPR) analysis with soluble molecules under flow-governing conditions (2-6). These analyses show that the affinity and/or dissociation off-rate predict the T-cell response, with the agonist pMHC having a higher affinity and/or slower off-rate than the antagonist (4). However, the cellular environment where the TCR resides differs from that where SPR experiments are performed. The native TCR–pMHC binding occurs across the junctional gap between two surfaces, i.e., at a two-dimensional (2D) interface, whereas in SPR soluble molecules capable of three-dimensional (3D) movement bind surface-immobilized counter-molecules. While attempts have been made to account for this difference by physical considerations (7) and mathematical models (8, 9), a more critical problem may be the use of engineered TCR constructs in SPR experiments, which ignores potential effects of other subunits of the TCR complex and their interactions with other cellular components (e.g., co-receptors or other membrane and cytoplasmic proteins) or movements of cellular structures (e.g., cell migration, actin retrograde flow, and membrane undulation) that could actively restrict or promote TCR binding to pMHC in a force dependent manner (10-12). Thus, analyzing 2D TCR–pMHC binding kinetics may yield important insights to T-cell biology as direct measurement is made in situ in the physiological membrane environment (13-16).
Two types of techniques have recently been used to measure the 2D kinetics of TCR–pMHC interactions: one based on fluorescent imaging, e.g., Förster resonance energy transfer (FRET) (14) or tracking molecular diffusion (17, 18), and the other based on mechanical assays, e.g., adhesion frequency (12, 13, 15, 16, 19-23), thermal fluctuation (12, 13, 16, 24), force-clamp (12, 24), and flow chamber (25) assays. Huppa et al. (14) studied FRET between lipid bilayer-anchored pMHC class II (pMHC-II) and CD4+ T blasts. The FRET signal change between the fluorophore-labeled MCC:I-Ek and 5c.c7 TCR allowed the calculation of binding parameters. Compared to 3D, their results showed much faster 2D binding and unbinding kinetics with no change when the CD4 was blocked. The same system was studied by Axmann et al. (17) using diffusion analysis and O'Donoghue et al. (18) using fluorescence imaging combined with diffusion analysis, but reported drastically different results. The former paper reported fast off-rates in agreement with the results of Huppa et al. (14), whereas the latter paper reported much slower off-rates.
Using the adhesion frequency and thermal fluctuation assays, our group found that 2D kinetics of TCR binding to a panel of peptides bound to MHC class I (MHC-I) were faster, had a much larger dynamic range, and better correlated with T-cell activation than their 3D counterparts in several CD8+ systems (13, 15, 16). Furthermore, the characteristics of mouse CD8 binding to H-2Kb and H-2Db and human CD8 binding to HLA-A2 were measured to define the basal CD8 affinity for MHC class I (16, 19). Moreover, an intriguing cooperation between TCR and CD8 was observed for >1s contacts with pMHC-I, suggesting the formation of trimeric TCR–pMHC-I–CD8 bonds to abruptly increase the bond number (16, 20, 23). In addition, we recently showed that mechanical force impacts TCR–pMHC-I dissociation kinetics in a peptide-specific manner to further amplify T-cell discrimination (24) and to trigger calcium signaling in CD8+ T-cells (22, 24). However, it is not known how much of these observations are applicable to pMHC-II-restricted TCRs on CD4+ T cells.
In this study, we used mechanical based assays to analyze in situ kinetics of 3.L2 TCR binding to a well-characterized panel of peptides presented by MHC-II at zero force and in a range of constant forces. Consistent with the MHC-I-restricted TCR systems, the zero-force 2D kinetics of this MHC-II-restricted TCR correlated better to the T-cell functions than the 3D kinetics. With applied force on the TCR–pMHC-II bonds, agonist and weak agonist behaved as catch-slip bonds where bond lifetime first increased, reached a maximum, then decreased with increasing force (24, 26). In contrast, antagonists behaved as slip-only bonds where bond lifetime decreased monotonically with increasing force (24, 26). This peptide-specific force-dependency of TCR–pMHC-II dissociation increased the dynamic range of the bond lifetime differences. CD4 binding was barely detectible and did not display cooperation with TCR in pMHC-II binding. Our results show that in situ kinetics and force-regulated bond dissociation discriminate ligands and determine functions in CD4+ T cells.
MATERIALS AND METHODS
Mice and cell preparation
Transgenic 3.L2 mice were housed at the Emory University Department of Animal Resources facility and experiments followed a protocol approved by the Institutional Animal Care and Use Committee of Emory University. The mouse expressed I-Ek restricted 3.L2 TCR specific to murine hemoglobulin epitope 64-76 (Hb). Naive T cells were purified via magnetic negative selection from 6–8 week old mouse spleens using either CD4+ or CD8+ T-cell isolation kit (Miltenyi Biotec) according to the manufacturer's instructions. Cells were washed and stored at room temperature for up to 24 hrs in R10 media, which consists of RPMI 1640 (Cellgro) supplemented with 10% FBS (Cellgro), 2mM L-glutamine (Cellgro), 0.01M HEPES buffer (Cellgro), 100μg/ml gentamicin (Cellgro), and 2×10−5M 2-β-mercaptoethanol (2-BM) (Sigma-Aldrich).
Protein preparation
Recombinant pMHC monomers were from the National Institutes of Health Tetramer Core Facility at Emory University. These consisted of the wild-type peptide Hb, or an altered peptide ligand, covalently tethered to a C-terminally biotinylated I-Ek (27). The peptides used in this study, with sequence and relative activity given in parentheses, included: Hb (GKKVITAFNEGLK, agonist), T72 (GKKVITAFTEGLK, weak agonist), I72 (GKKVITAFIEGLK, antagonist), and A72 (GKKVITAFAEGLK, weak antagonist). Briefly, agonist provides the maximal level of functional response, weak agonist induces maximal levels with a more than 10-fold higher peptide concentration, and antagonists block agonist activity with low (antagonist) or high (weak antagonist) concentrations without inducting response by themselves. Ovalbumin (OVA) peptide 257-264 (SIINFEKL) bound to H-2Kb and moth cytochrome c (MCC) peptide 88-103 (ANERADLIAYLKQATK) bound to I-Ek were prepared in the same way for irrelevant control. For 3D SPR measurement, single-chain TCR (only the variable region connected by a flexible linker) was expressed in E. coli and refolded from inclusion bodies as described previously (28).
Preparation of pMHC coated surfaces
Detailed procedures have been described previously (13, 19, 20). In brief, human RBCs were isolated from the whole blood of healthy volunteers according to a protocol approved by the Institutional Review Board of the Georgia Institute of Technology. RBCs were biotinylated, conjugated with streptavidin, and incubated with biotinylated pMHC for adhesion frequency assay with a micropipette. In the case of BFP experiments, silanized beads were covalently linked to streptavidin-maleimide (Sigma-Aldrich) then conjugated with subsaturating biotinylated pMHC.
Site density measurement
Site densities of the TCR, CD4, and pMHC were measured by flow cytometry (13, 19, 20) using three fluorescent antibodies: a PE-conjugated anti-mouse Vβ8.3 TCR (1B3.3, BD Pharmingen), a PE-conjugated anti-mouse CD4 (GK1.5, eBioscience), and a PE-conjugated anti-mouse I-Ek (17-3-3, Santa Cruz ; 14-4-4s, BD Pharmingen). Antibodies were used at 10μg/ml concentration in 100μl of FACS buffer (PBS without calcium and magnesium, 5mM EDTA, 1% BSA, 25mM HEPES, 0.02% sodium azide) at 4°C for 30 min; fluorescent intensities were measured by the BD LSR II flow cytometer (BD Biosciences); and site densities were calculated by comparing to the BD QuantiBRITE PE standard beads (BD Biosciences).
Adhesion frequency assay
Detailed procedures have been described previously (13, 19, 20). In brief, two micropipettes holding a T-cell on one side and a pMHC-coated RBC on the other (Fig. 1A) were controlled to make repeated contacts with a constant area (Ac). From the presence or absence of RBC membrane deflection upon retraction after a contact time (tc), the adhesion events in 50 contact cycles were enumerated to calculate adhesion frequency (Pa). The Pa vs. tc data (Fig. 1B) were fitted by the least mean square method to the following probabilistic kinetic equation (29)
| (1) |
to determine the effective 2D affinity (AcKa) and off-rate (koff) using the independently measured receptor (mr) and ligand (ml) densities.
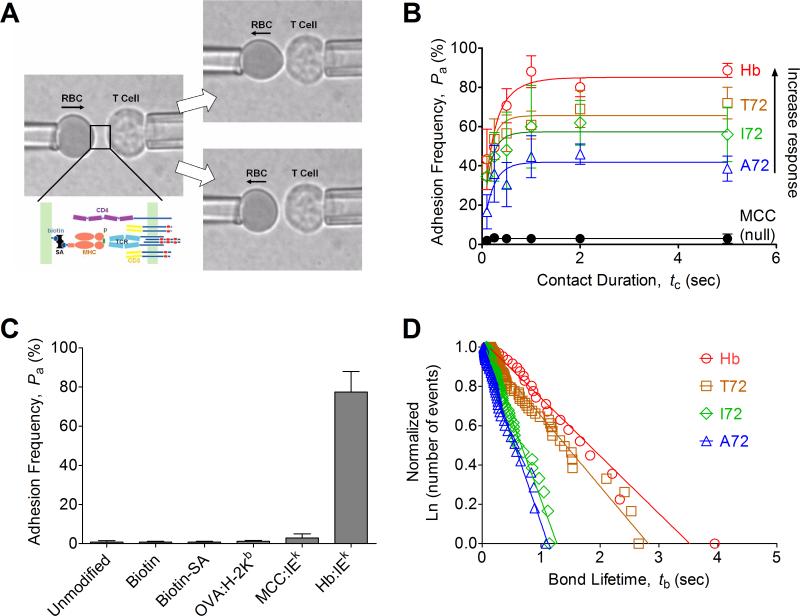
FIGURE 1. Micropipette adhesion frequency assay and thermal fluctuation assay.
(A) Schematic of micropipette apparatus. A T-cell and a pMHC-coated RBC are held by two apposing micropipettes before the contact. Below the left photomicrograph, the TCR complex and CD4 are drawn on the T-cell surface, whereas the pMHC coupled via biotin–streptavidin interaction are drawn on the RBC surface. The RBC was brought to contact the T-cell for certain contact duration and retract to observe the presence, signified by the elongation (upper right), or absence, signified by the lack of elongation (lower right), of the soft RBC membrane at the end of the contact. (B) The adhesion frequency assay was conducted for a panel of cognate peptides (Hb, T72, I72, A72) and an irrelevant peptide (MCC) with representative data (from three independent experiments) shown along with the following molecule densities (TCR:pMHC in molecules/μm2): Hb (185:33), T72 (179:67), I72 (171:114), A72 (158:186), and MCC (136: 4483, 315 for CD4). Each point represents mean ± SEM (n=3-5 cell pairs each contacted 50 times to estimate an adhesion frequency) (C) Controls for nonspecific adhesion were performed at 5s contact duration between 3.L2 T cell and unmodified RBCs, biotinylated RBCs without further coupling to streptavidin, biotinylated RBCs linked with streptavidin without further coating of pMHC, biotinylated RBCs linked with streptavidin coated with pMHC-I (OVA:H-2Kb) or noncognate pMHC-II (MCC:I-Ek), which were compared to biotinylated RBCs linked to streptavidin coated with cognate pMHC-II (Hb:I-Ek). Each bar is presented as mean ± SEM (n=5 cell pairs each contacted 50 times to estimate an adhesion frequency). (D) The thermal fluctuation assay was performed as described in Methods. Ranked bond lifetime distributions for each ligand were linearized by ln(# of events with a lifetime > tb)/ln(total # of events) versus tb plots to allow us to estimate koff from the negative slope of the line. The numbers of bond lifetimes are 22 (Hb), 66 (T72), 65 (I72), and 46 (A72). The high levels of goodness-of-fit as assessed by R2 (> 0.97 for all ligands) support the use of the first order kinetics model.
Non-specific adhesion to 3.L2 T cells was controlled using 5-sec contacts to (1) unmodified RBCs (Pa < 1%), (2) biotinylated RBCs (Pa < 1% for 800μM biotinylated concentration), (3) biotinylated RBCs linked to streptavidin (Pa < 1% for 4335 molecules/μm2), (4) biotinylated RBCs linked to streptavidin and coated with irrelevant pMHC-I (Pa ~ 1% for 1548 molecules/μm2 of OVA257-264:H-2Kb), (5) biotinylated RBCs linked to streptavidin and coated with irrelevant pMHC-II (Pa < 5% for 4140 molecules/μm2 of MCC88-103:I-Ek), and (6) biotinylated RBCs linked to streptavidin and coated with antigenic pMHC-II (Pa ~ 75% for 144 molecules/μm2 of TCR and 36 molecules/μm2 of Hb64-76:I-Ek) (Fig. 1C).
Thermal fluctuation assay
Detailed procedures have been described previously (30, 31). In brief, the bond formation and dissociation was detected by the reduction and resumption of the thermal fluctuations of the BFP bead. A bond lifetime (tb) spans from the instant of bond association to the instant of bond dissociation. Modeling the kinetic process as a single-step first order dissociation of a single monomeric TCR–pMHC-II bond, the probability Pb of a bond formed at time 0 to remain intact at time tb is
| (2) |
Taking a natural log linearizes the exponential function on right hand-side of this equation. Plotting the data as ln(# of events with a lifetime > tb) versus tb allows us to estimate koff from the negative slope of the line (Fig. 1D).
Force-lifetime measurement
Detailed procedures have been described previously (24). In brief, bond lifetimes were measured by the force-clamp assay, which loads a bond to a preset force and holds the force constant to measure how long the bond last under that force. More than 300 lifetime events were measured for each ligand in a force range of 5-40pN segregated into 5-7 bins. To affirm >95% single-bond events, pMHC-II densities on the beads were adjusted to compensate their affinities so as to keep the adhesion frequency <20% (29, 32).
Surface plasmon resonance measurement
Detailed procedures have been described previously (28). In brief, concentration series of scTCR covering at least two orders of magnitude were injected in a Biacore 2000 instrument using flow rate of 30μL/min over CM5 sensor chips (GE Healthcare) that were coupled with pMHC-II monomer. All sensorgrams were corrected for nonspecific binding by subtracting the response from a surface coupled with CLIP/I-Ek, and corrected for bulk flow effects by subtracting the response obtained from plain buffer injection.
Sensorgrams were fitted to a 1:1 Langmuir binding model using BiaEvaluation version 4.1 to derive the 3D koff, kon, and Ka (= 1/Kd = kon/koff). Kd and maximum response (RMax) values from equilibrium binding analysis were obtained by plotting the equilibrium response (Req) at each concentration and fitting these data to a one-step binding model using GraphPad Prism version 6.0a for Macintosh (GraphPad Software, San Diego, CA). Scatchard plots were generated to confirm 1:1 binding stoichiometry by graphing Req/[TCR] versus Req.
Blocking experiment
T cells were incubated with 20μg/ml of a CD4 blocking antibody (GK1.5, eBioscience) (14, 33) or 50μg/ml of an anti-3.L2 clonotypic antibody (CAb) (34) in 100μl for 30min in 4°C and then tested in the experiment chamber with the presence of the same concentrations of the respective antibodies.
RESULTS
CD4 has low affinity to I-Ek
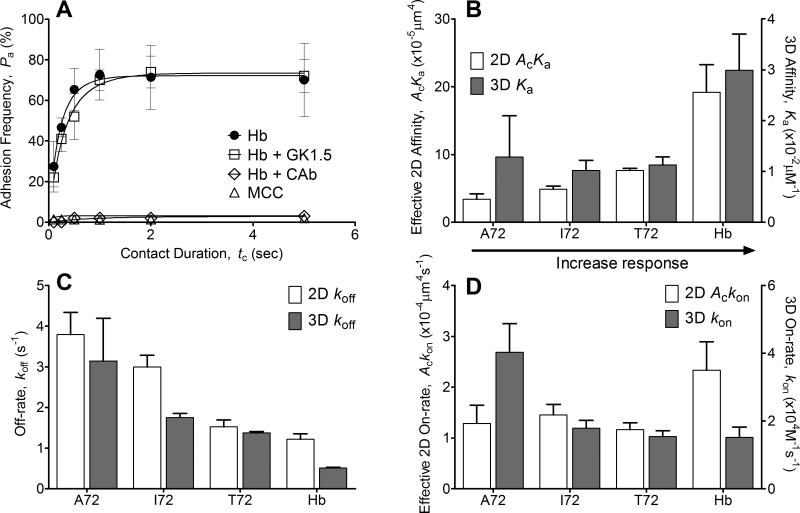
It has previously been shown by tetramer staining (35) and FRET experiments (14) that blocking CD4 does not affect CD4+ T-cell binding to pMHC-II. To confirm this result we used the adhesion frequency assay for it has proven to possess higher sensitivity (21). Treatment with the anti-3.L2 clonotypic antibody (CAb) completely abolished the ~70% steady-state adhesion frequency measured without treatment (Fig. 2A). In sharp contrast, treatment with the anti-CD4 antibody (GK1.5) resulted in a binding curve indistinguishable to the curve without treatment. Both curves show single-stage binding, with a transient increase followed by a plateau, which can be well fitted by Eq. 1 (Fig. 2A) and return comparable effective 2D affinity and off-rate values (Table 1). This is in sharp contrast to binding of pMHC-I to CD8+ T cells, which exhibits a two-stage curve consisting of an initial plateau followed by a second, higher plateau after a ~1-s delay that is not predicted by Eq. 1 but characteristic of the cooperative TCR–pMHC-I–CD8 trimolecular binding, with the second stage blockable by anti-CD8 antibody (16, 20, 23). These results indicate that CD4 binding is much weaker than TCR, and the two molecules do not cooperate to bind pMHC-II to increase the bond number under our experimental conditions.
FIGURE 2. Comparison of 2D binding parameters with 3D counterparts.
(A) Lack of CD4 binding to I-Ek. Treatment with the TCR blocking antibody CAb abolished the adhesion frequency observed without blocking, but treatment with the CD4 blocking antibody (GK1.5) did not affect the binding curve. The noncognate ligand MCC:I-Ek had some binding at a longer contact duration (5s) with a very high site density of >4,000 molecules/μm2, but overall binding was very low (<5%). The molecule densities used to generate these representative data are (TCR:pMHC molecules/μm2): Hb (116:34), Hb + GK1.5 (116:34), Hb + CAb (163:28), and MCC (136:4483, 315 for CD4). Each point represents mean ± SEM (n≥3 cell pairs each contacted 50 times to estimate an adhesion frequency). (B-D) Comparison between 2D vs. 3D affinity (B), off-rate (C), and on-rate (D). 2D measurements were from adhesion frequency assay and thermal fluctuation assay whereas 3D measurements were from SPR. Each 2D kinetic parameter is presented as mean ± SEM (n≥3 sets of adhesion frequency vs. contact time curve).
Table I.
2D kinetics and binding affinities of the 3.L2 TCR–pMHC-II interactions.
| Peptide | Classification of Ligand | Effective 2D affinity, AcKa (μm4) | Effective 2D on-rate, Ackon (μm4s−1) | 2D off-rate, koff (s−1)* | 2D off-rate, koff (s−1)** | Effective 2D affinity, AcKa (μm4)*** |
|---|---|---|---|---|---|---|
| Hb | Agonist | 19.2 ± 4.1 × 10−5 | 2.33 ± 0.56 × 10−4 | 1.22 ± 0.14 | 2.51 ± 0.43 | 15.6 ± 0.5 × 10−5 |
| T72 | Weak agonist | 7.6 ± 0.3 × 10−5 | 1.16 ± 0.14 × 10−4 | 1.52 ± 0.17 | 4.58 ± 1.51 | 5.2 ± 0.7 × 10−5 |
| I72 | Antagonist | 4.8 ± 0.5 × 10−5 | 1.45 ± 0.20 × 10−4 | 2.99 ± 0.29 | 3.31 ± 0.73 | 2.7 ± 0.3 × 10−5 |
| A72 | Weak antagonist | 3.4 ± 0.8 × 10−5 | 1.29 ± 0.36 × 10−4 | 3.79 ± 0.55 | 5.28 ± 0.67 | 1.54 ± 0.3 × 10−5 |
| Hb (GK1.5) | Agonist | 22.5 ± 1.2 × 10−5 | 3.16 ± 0.42 × 10−4 | 1.41 ± 0.17 | 1.89 ± 0.31 |
2D off-rate were measured by thermal fluctuation assay (*) and adhesion frequency assay (**).
Effective 2D affinity for CD8+ 3.L2 TCR were measured by adhesion frequency assay (***).
To directly measure CD4 binding, we coated a noncognate MCC88-103:I-Ek to RBCs at a high density of over 4,000 molecules/μm2 and tested binding of these RBCs to 3.L2 T cells at a long 5-s contact time. The resulting low adhesion frequency translated to an effective 2D affinity of AcKa < 7.0 × 10−8 μm4, 1-2 orders of magnitude lower than that of CD8–H-2Kb (2.8-5.7 × 10−6 μm4) and CD8–H-2Db (0.1-0.5 × 10−6 μm4) (19). This indicates that under zero-force, I-Ek binding to CD4+ T-cells is largely bimolecular with the TCR but not CD4.
Zero-force kinetics predict ligand potency
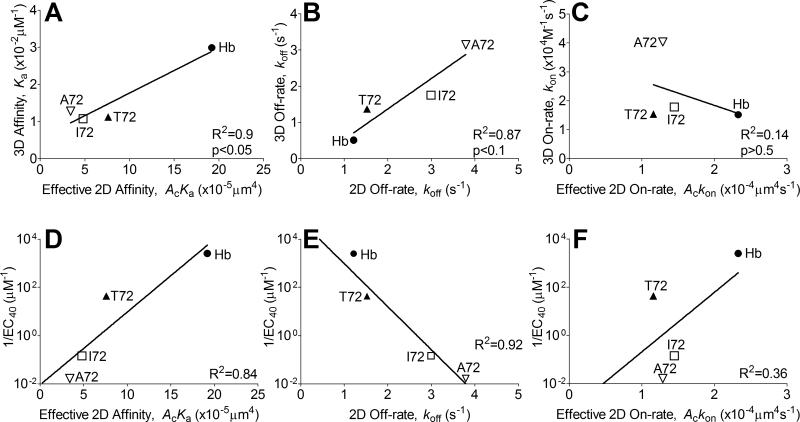
The 3.L2 TCR was tested using a series of well-characterized ligands, including the wild-type peptide Hb and three APLs with a single amino acid alteration at residue 72 that possess a range of potency to stimulate T-call activation (6, 36). To obtain mid-ranged adhesion frequencies amendable to fitting by Eq. 1, different densities for Hb (agonist), T72 (weak agonist), I72 (antagonist), and A72 (weak antagonist) were coated on the RBCs to compensate for their distinct affinities. Figure 1B shows binding curves resulted from using these RBCs to test naïve 3.L2 T cells by the adhesion frequency assay. Equation 1 was fit to these data to evaluate the effective 2D binding affinities for all ligands (Table I), including the weak antagonist A72 with the lowest potency that was not previously detectable using SPR (6). The measured values span over a 6-fold range (AcKa ~ 3.4-19.2 × 10−5 μm4) with the agonist Hb being the highest and the weakest antagonist A72 the lowest (Fig. 2B). This is a broader dynamic range than the 3D affinities for the same TCR–pMHC-II interactions measured by SPR (Fig. 2B), but the gain is much smaller than the previously studied MHC-I systems (13, 15, 16). The 2D and 3D affinities show positive correlation (p<0.05, Fig. 3A), but the high R2 value (0.9) may be due to their much higher values for the agonist Hb:I-Ek than the other three APLs, which have similar values in 3D but are better resolved by the 2D analysis.
FIGURE 3. Correlating 2D kinetics with 3D counterparts and with T-cell response.
(A-C) Correlation between 2D vs. 3D kinetics. The effective 2D affinity correlates highly (R2=0.9, p<0.05) to the 3D affinity due solely to the much higher values of the agonist pMHC-II than the other three APLs (A). The off-rates correlate well (R2=0.87, p<0.1) between 2D and 3D (B). The effective 2D on-rates poorly correlate (R2=0.14, p>0.5) between 2D and 3D (C). The mean values from both 2D (Table I) and 3D (Table II) measurements were compared. (D-F) Correlation of 2D kinetics with T-cell response. The effective 2D affinity correlates well (R2=0.84) with the reciprocal of the ligand concentration required to produce 40% B cell apoptosis (1/EC40) (D). The 2D off-rate highly (R2=0.92), but negatively correlates with 1/EC40 (E). The effective 2D on-rate shows a poor correlation (R2=0.36) (F). The parameters used for the 2D kinetics are the mean values from the adhesion frequency and thermal fluctuation assays. The EC40 values are from Ref. (36).
The 2D off-rates can also be derived from fitting the adhesion frequency data with Eq. 1 (Table 1). In addition, the thermal fluctuation assay was also used (Fig. 1D), as its increased temporal resolution allows us to obtain more accurate off-rate values from analysis of single-bond lifetime measurements (30, 31) (Table 1). Our previous study on the CD8+ OT1 T cells finds drastically different 2D and 3D off-rate values for the same ligands that show opposite ranked orders between 2D and 3D for the same panel of ligands, with the ligand potency correlating positively with 2D off-rate but negatively with 3D off-rate. By comparison, the off-rates measured in the present study on the CD4+ 3.L2 T cells have similar values in both 2D and 3D, show the same ranked orders, and correspond to ligand potency ranging from the Hb to A72 (Fig. 2C). As such, the 2D and 3D off-rates are well correlated (R2=0.87, p<0.1) (Fig. 3B). The 2D effective on-rates were calculated from multiplying the effective 2D affinity by off-rate, Ackon = AcKa × koff (Fig. 2D, Table 1). The 3D on-rates measured by SPR are also plotted on Fig. 2D for comparison, which show poor correlation with the 2D on-rates (R2=0.14, p>0.5) (Fig. 3C). The previously measured peptide potency, i.e., reciprocal of the ligand concentration required to generate 40% of radiolabeled B cell apoptosis (1/EC40) measured by liquid scintillation counting (36), was plotted vs. the 2D binding parameters. These plots show positive correlation with the effective 2D affinity (Fig. 3D), negative correlation with 2D off-rate (Fig. 3E), and lack of correlation with the effective 2D on-rate (Fig. 3F).
3.L2 TCR forms catch bond with agonist and slip bond with antagonist
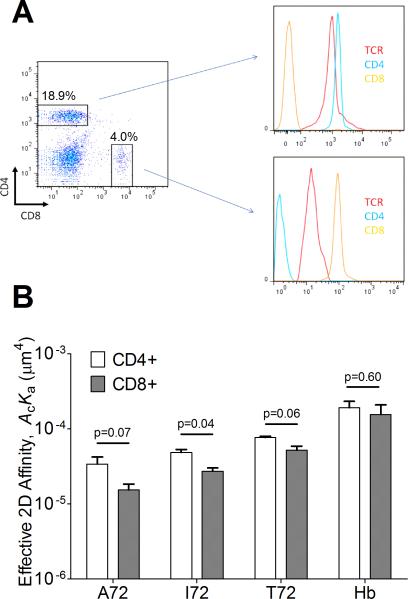
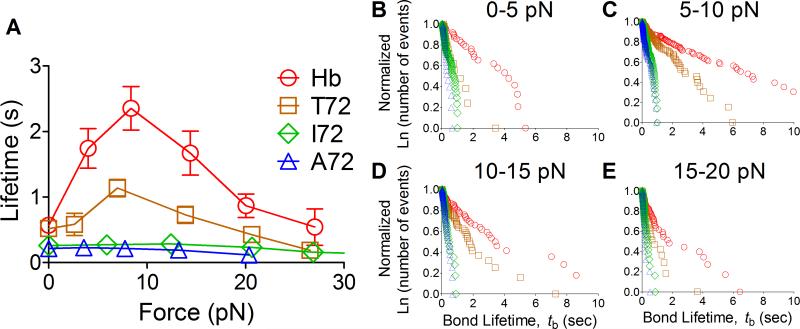
We used a BFP force-clamp assay (24, 37) to measure TCR–pMHC-II bond lifetimes under mechanical force. To focus our studies on the TCR interaction with pMHC-II, we took advantage of a known property of the 3.L2 TCR transgenic mice: a propensity to generate a reasonable number of CD8+, CD4− mature T cells (38) (Fig. 4A). Confirming the minimal contribution of CD4, the CD8+ 3.L2 T cells showed similar effective 2D affinity to CD4+ T cells for the same panel of pMHC-II (Fig. 4B, Table 1). Under force, I72 and A72 (antagonists) bond lifetime decreases monotonically with increasing force (Fig. 5A). In sharp contrast, the more potent ligands displayed bond lifetimes that first increased, reached a maximum around 10pN, and decreased as force continued to increase (Fig. 5A). These characteristics define TCR dissociation from Hb and T72 (agonists) as catch-slip bonds and I72 and A72 (antagonist) as slip-only bonds (24). The bond lifetime distributions were plotted for the four ligands (Fig. 5B-E). It follows from Eq. 2 that the off-rates equal to the negative reciprocal slopes of the data lines. As such, the bond lifetime distributions decay less rapidly for agonists that exhibited catch-slip bonds than the antagonists that showed slip-only bonds. The greatest resolution in bond lifetimes occurred when 5-15pN of force was applied where agonist ligand bond lifetimes were prolonged mostly by the optimal force (Fig. 5C and D).
FIGURE 4. Comparison between CD4+ and CD8+ 3.L2 T cells for TCR–pMHC-II binding.
(A) Flow cytometry scatter plot of 3.L2 T-cells. CD4+ and CD8+ cells were purified with respective CD4 and CD8 negative purification protocols then analyzed for TCR, CD4, and CD8 expressions by flow cytometry. (B) Comparison of effective 2D affinity between CD4+ and CD8+ 3.L2 T cells. No significant difference were observed between most of the two subpopulation. Student's t-test was used for statistical analysis. Each bar represents mean ± SEM (n≥3).
FIGURE 5. Ligand dependent bond lifetime under force.
(A) Mean ± SEM bond lifetimes measured by force-clamp assay are plotted vs. force. The agonist ligands, Hb and T72, show catch-slip bonds, whereas the antagonist ligands, I72 and A72, show slip-only bonds. The number of lifetime measurements for each curve is: Hb (298), T72 (300), I72 (448), and A72 (343). (B-E) Normalized lifetime distributions of bimolecular TCR–pMHC-II bonds with the indicated peptides measured by the force-clamp assay in the indicated force regimes. The shallower the curve, the slower the dissociation, and the greater the number of bonds surviving a given time. As force increases, the separation between agonist and antagonist ligands increases, reaches a maximum at 5-15pN (C and D), and then decreases.
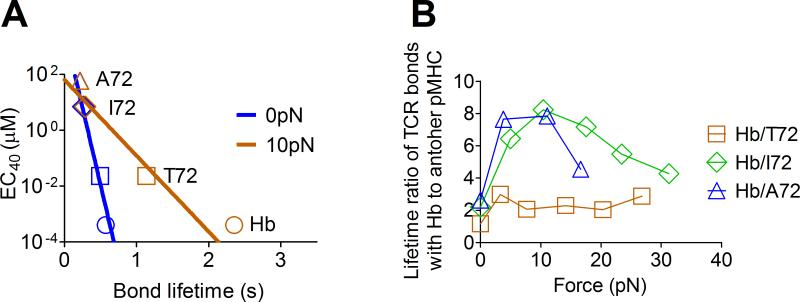
Force enhances ligand discrimination
To examine how force impacts the correlation between bond lifetime and ligand potency, the EC40 values from Fig. 3 was replotted against bond lifetimes at 0 and 10pN (Fig. 6A). Force tilted the EC40 vs. bond lifetime line at zero-force, making it less steep (blue line, Fig. 6A). At 10pN (the force at which the longest bond lifetimes were observed for agonists), the slope of the EC40 vs. bond lifetime line become the shallowest. This expanded the dynamic range of the bond lifetime differences, allowing greater resolution of ligand potency than found at zero-force (Fig. 6A). To further reveal how force amplifies the power of ligand discrimination, we plotted the ratio of bond lifetime of Hb to that of different peptide ligands vs. force (Fig. 6B). The bond lifetime ratios of Hb to I72 (green) and A72 (blue) increased with force to reach maximum at 10-15pN before decreasing. This is different from what was found for T72 (brown), thus increasing the power of discrimination between agonists and antagonists under force.α
FIGURE 6. Force amplifies ligand discrimination.
(A) The ligand concentration required to generate 40% B cell apoptosis (EC40) from Ref. (36) was plotted vs. mean bond lifetime for Hb (circle), T72 (square), I72 (diamond), and A72 (triangle) and fitted by a straight line, one for 0pN (blue) and another for 10pN (brown). Force tilts the EC40 vs. bond lifetime curve to increase the dynamic range in the x-axis by more than one fold. (B) The ratio of bond lifetime of Hb to another peptide was plotted vs. force to show increased power of ligand discrimination around optimal force where bond lifetime for Hb reaches maximum. The increase in the ratios for Hb/I72 (green) and Hb/A72 (blue) but not for Hb/T72 (brown) further indicates separation between agonist and antagonist ligands. The data from force vs. bond lifetime were used to calculate the ratio.
DISCUSSION
In situ TCR–pMHC kinetic analyses using mechanical based assays have mostly been on CD8+ T-cell systems (12, 13, 15, 16, 19, 20, 22-25, 39). The micropipette adhesion frequency assay has been used to measure the 2D affinities of polyclonal CD4+ T cells reactive to self MOG and pathogen GP61 peptides (21), of 2D2 TCR for MOG or NFM peptides, and of SMARTA TCR for GP61 peptide (40, 41), all presented by MHC-II. While these studies showed that the micropipette assay has much higher sensitivity than 3D pMHC-II tetramer staining and the 2D affinity correlates with the T-cell biology, the present paper represents a more systematic study using the mechanical assays to correlate the force-dependent in situ TCR–pMHC-II binding kinetics to corresponding SPR measurements and with CD4+ T-cell function, allowing us to compare similarities and contrast differences between the CD4+ and CD8+ T-cell systems. Although caution should be excused in the data interpretation because only a modest number of altered peptide ligands were tested, our conclusions are strengthened by the combined use of 2D measurements both at zero force and in a range of tensile forces, which is more comprehensive because these measurements account for the impact of cellular environment and the effects of mechanical forces on in situ TCR–pMHC interactions.
Comparisons can be made on the relative ranges between the binding parameters and the T-cell responses, the parameter values, and their ranked orders. TCR–pMHC interactions are highly elegant in that they do not merely result in an all-or-none response; rather, the T-cell “reads” the information presented by the pMHC, transduces it into intracellular signals, and responds to the complex input with a broad set of phenotypic outcomes. For a response range of 6-7 logs measured by functional avidity or peptide concentration required to achieve a certain level of function, the range of binding parameters measured in 3D typically spans one log or less, thereby requires substantial signal amplification that is attributed to the work of intracellular signaling cascades. The in situ 3.L2 TCR affinity range is similar to that of the 42F3 TCR (15), which spans 1 log, but much smaller than that of OT1 TCR (13), which spans 3 logs. Compared to the OT1 system (13), the 3.L2 system show effective 2D affinity and on-rate values within the respective ranges of those of A2 (agonist) and G4 (weak agonist), whereas the 2D off-rate for agonist Hb is below the level of E1 (weak agonist/antagonist). Similar to the CD8+ systems (13, 15, 16), the in situ TCR affinities of the present CD4+ system correlate with, and correspond to the T-cell response better than, the 3D TCR affinities. In contrast to the OT1 TCR whose off-rates are much faster in 2D than 3D and have opposite ranked orders in 2D and 3D (12, 13), the 3.L2 TCR has similar 2D and 3D off-rates that are of the same ranked orders, which correlate well with the T-cell response. Unlike the 2D OT1 TCR on-rates that span a broad 4-log range and correlate well with T-cell responses (13), the 2D 3.L2 TCR on-rates span only one log and correlate poorly with the biology. These comparisons shed lights on the TCR–pMHC interaction parameter ranges and values, and impose important constraints to models that relate TCR recognition to T-cell function.
Unlike the CD8–MHC-I interaction that is readily measurable by the adhesion frequency and thermal fluctuation assays with kinetic parameters comparable to those of TCR interaction with antagonists (16, 19), CD4–MHC-II interaction was barely detectable by the same mechanical based assays, only allowing us to set an upper bound for its effective 2D affinity with unmeasurable off-rate. Despite the much lower affinity of CD8 than that of TCR for agonist pMHC-I, CD8 and TCR cooperate to form a trimeric bond with pMHC-I to synergistically increase the bond number in a signaling-dependent manner (16, 20, 23). This phenomenon was not observed for the 3.L2 TCR in the present CD4+ T-cell system. Future studies will determine what different properties of the CD4 and CD8 coreceptors may cause this distinction, and whether other MHC-II-restricted TCRs or other conditions exist that may enable cooperation between the CD4 and TCR for synergistic MHC-II binding.
In three recent studies, TCR–pMHC-I bond lifetimes have been measured over a range of forces in several systems (24, 25, 39). The role of physical force in regulating TCR–pMHC-I dissociation kinetics and in triggering T-cell activation has been highlighted in the findings that force elicits agonist-specific catch bonds (24, 39) that correlate to the triggering of calcium signals in T-cells (24). Similar to the OT1 system, the present study has demonstrated that the 3.L2 TCR also forms catch-slip bonds with agonist, and slip-only bonds with antagonist, peptides presented by MHC-II. Interestingly, the force ranges where catch bonds are observed are comparable for the OT1 (24), N15 (39), and 3.L2 TCRs, with the bond lifetimes reaching maxima around 10-15pN. Similar to the OT1 system (24), force greatly amplifies the power of antigen discrimination by the 3.L2 TCR in a CD4+ T-cell system. This is despite the fact that 10pN force reverses the ranked orders of the off-rates of the OT1 TCR dissociating from a panel of pMHC-I measured at zero-force, whereas the ranked orders of the off-rates of the 3.L2 TCR dissociating from a panel of pMHC-II remain the same at 0 and 10pN. These findings provide a new parameter dimension relevant to T-cell mechanoimmunology, which is nonexistent for 3D kinetic analysis as 3D binding takes place under stress-free conditions. Future studies will likely determine how TCR engagement may induce intracellular forces that may exert on the TCR–pMHC bonds to regulate their dissociation and further trigger T-cell activation.
Table II. 3D SPR kinetic parameters of the 3.L2 TCR–pMHC-II interactions.
In order to directly compare our 2D parameters with 3D, new 3D kinetic parameters were measured with SPR using single-chain TCR and peptides covalently linked I-Ek as in the 2D measurements. Whereas Hb64-76 and T72 3D SPR measurements were readily calculated by simultaneous modeling of kon and koff, injection spikes coupled with extremely fast kinetics precluded successful curve fitting for I72 and A72. The SPR measurements for these ligands were instead derived by separately modeling the kon and koff using data from the sensorgram corresponding to the highest scTCR concentration injection.
| Analyte / Ligand | kon (M−1s−1) | koff (s−1) | Kd (μM) koff/kon | Half-life (s) | Kd (μM) Equilibrium | N |
|---|---|---|---|---|---|---|
| scTCR / Hb64-76:I-Ek | 15,183 ± 3,017 | 0.508 ± 0.019 | 35.3 ± 0.82 | 1.36 | 28.73 ± 5.88 | 6 |
| scTCR / T72:I-Ek | 15,433 ± 1,711 | 1.37 ± 0.038 | 90.7 ± 1.28 | 0.51 | 73.71 ± 4.23 | 3 |
| scTCR / I72: I-Ek | (28,100 – 14,000)* | (1.63 - 1.95)* | (0.355 - 0.425) | 366 ± 18.7 | 3 | |
| scTCR / A72: I-Ek | (71,000 – 24,100)* | (1.12 - 4.61)* | (0.150 - 0.619) | 1,309 ± 108 | 3 |
3D kon and koff for I72 and A72 were derived by separate modeling using data from the sensorgram corresponding to the highest scTCR concentration injection.
ACKNOWLEDGEMENTS
We thank Laurel Lawrence for maintaining the mouse colony; Larissa Doudy for T-cell purification; Drs. Lindsay Edwards, Joe Sabatino, Baoyu Liu, Wei Chen, and Zhenhai Li for helping with experiment and helpful discussions; and the NIH Tetramer Core Facility at Emory University for providing the MHC monomers.
This work was supported by NIH grants AI38282 and GM096187 (to C.Z.); AI096879 and NS071518 (to B.E.); and AI24157 (to P.A.).
Abbreviations used in this article
- 2D
two-dimensional
- 3D
three-dimensional
- SPR
surface plasmon resonance
- BFP
biomembrane force probe
- pMHC
peptide bound MHC
- Hb
hemoglobin
Footnotes
DISCLOSURES
The authors have no financial conflict of interest.
REFERENCES
- 1.Morris GP, Allen PM. How the TCR balances sensitivity and specificity for the recognition of self and pathogens. Nat Immunol. 2012;13:121–128. doi: 10.1038/ni.2190. [DOI] [PubMed] [Google Scholar]
- 2.Stone JD, Chervin AS, Kranz DM. T-cell receptor binding affinities and kinetics: impact on T-cell activity and specificity. Immunology. 2009;126:165–176. doi: 10.1111/j.1365-2567.2008.03015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davis MM, Boniface JJ, Reich Z, Lyons D, Hampl J, Arden B, Chien Y. Ligand recognition by alpha beta T cell receptors. Annu Rev Immunol. 1998;16:523–544. doi: 10.1146/annurev.immunol.16.1.523. [DOI] [PubMed] [Google Scholar]
- 4.Gascoigne NR, Zal T, Alam SM. T-cell receptor binding kinetics in T-cell development and activation. Expert Rev Mol Med. 2001;2001:1–17. doi: 10.1017/S1462399401002502. [DOI] [PubMed] [Google Scholar]
- 5.Degano M, Garcia KC, Apostolopoulos V, Rudolph MG, Teyton L, Wilson IA. A functional hot spot for antigen recognition in a superagonist TCR/MHC complex. Immunity. 2000;12:251–261. doi: 10.1016/s1074-7613(00)80178-8. [DOI] [PubMed] [Google Scholar]
- 6.Kersh GJ, Kersh EN, Fremont DH, Allen PM. High- and low-potency ligands with similar affinities for the TCR: the importance of kinetics in TCR signaling. Immunity. 1998;9:817–826. doi: 10.1016/s1074-7613(00)80647-0. [DOI] [PubMed] [Google Scholar]
- 7.Dustin ML, Bromley SK, Davis MM, Zhu C. Identification of self through two-dimensional chemistry and synapses. Annu Rev Cell Dev Biol. 2001;17:133–157. doi: 10.1146/annurev.cellbio.17.1.133. [DOI] [PubMed] [Google Scholar]
- 8.Bell GI. Models for the specific adhesion of cells to cells. Science. 1978;200:618–627. doi: 10.1126/science.347575. [DOI] [PubMed] [Google Scholar]
- 9.Wu Y, Vendome J, Shapiro L, Ben-Shaul A, Honig B. Transforming binding affinities from three dimensions to two with application to cadherin clustering. Nature. 2011;475:510–513. doi: 10.1038/nature10183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu C, Jiang N, Huang J, Zarnitsyna VI, Evavold BD. Insights from in situ analysis of TCR-pMHC recognition: response of an interaction network. Immunol Rev. 2013;251:49–64. doi: 10.1111/imr.12016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen W, Zhu C. Mechanical regulation of T-cell functions. Immunological reviews. 2013;256:160–176. doi: 10.1111/imr.12122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu B, Chen W, Natarajan K, Li Z, Margulies DH, Zhu C. The cellular environment regulates in situ kinetics of T-cell receptor interaction with peptide major histocompatibility complex. European journal of immunology. 2015;45:2099–2110. doi: 10.1002/eji.201445358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang J, Zarnitsyna VI, Liu B, Edwards LJ, Jiang N, Evavold BD, Zhu C. The kinetics of two-dimensional TCR and pMHC interactions determine T-cell responsiveness. Nature. 2010;464:932–936. doi: 10.1038/nature08944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huppa JB, Axmann M, Mortelmaier MA, Lillemeier BF, Newell EW, Brameshuber M, Klein LO, Schutz GJ, Davis MM. TCR-peptide-MHC interactions in situ show accelerated kinetics and increased affinity. Nature. 2010;463:963–967. doi: 10.1038/nature08746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Adams JJ, Narayanan S, Liu B, Birnbaum ME, Kruse AC, Bowerman NA, Chen W, Levin AM, Connolly JM, Zhu C, Kranz DM, Garcia KC. T cell receptor signaling is limited by docking geometry to peptide-major histocompatibility complex. Immunity. 2011;35:681–693. doi: 10.1016/j.immuni.2011.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu B, Zhong S, Malecek K, Johnson LA, Rosenberg SA, Zhu C, Krogsgaard M. 2D TCR-pMHC-CD8 kinetics determines T-cell responses in a self-antigen-specific TCR system. Eur J Immunol. 2014;44:239–250. doi: 10.1002/eji.201343774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Axmann M, Huppa JB, Davis MM, Schutz GJ. Determination of interaction kinetics between the T cell receptor and peptide-loaded MHC class II via single-molecule diffusion measurements. Biophys J. 2012;103:L17–19. doi: 10.1016/j.bpj.2012.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O'Donoghue GP, Pielak RM, Smoligovets AA, Lin JJ, Groves JT. Direct single molecule measurement of TCR triggering by agonist pMHC in living primary T cells. eLife. 2013;2:e00778. doi: 10.7554/eLife.00778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang J, Edwards LJ, Evavold BD, Zhu C. Kinetics of MHC-CD8 interaction at the T cell membrane. J Immunol. 2007;179:7653–7662. doi: 10.4049/jimmunol.179.11.7653. [DOI] [PubMed] [Google Scholar]
- 20.Jiang N, Huang J, Edwards LJ, Liu B, Zhang Y, Beal CD, Evavold BD, Zhu C. Two-Stage Cooperative T Cell Receptor-Peptide Major Histocompatibility Complex-CD8 Trimolecular Interactions Amplify Antigen Discrimination. Immunity. 2011;34:13–23. doi: 10.1016/j.immuni.2010.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sabatino JJ, Jr., Huang J, Zhu C, Evavold BD. High prevalence of low affinity peptide-MHC II tetramer-negative effectors during polyclonal CD4+ T cell responses. J Exp Med. 2011;208:81–90. doi: 10.1084/jem.20101574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pryshchep S, Zarnitsyna VI, Hong J, Evavold BD, Zhu C. Accumulation of Serial Forces on TCR and CD8 Frequently Applied by Agonist Antigenic Peptides Embedded in MHC Molecules Triggers Calcium in T Cells. J Immunol. 2014;193:68–76. doi: 10.4049/jimmunol.1303436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Casas J, Brzostek J, Zarnitsyna VI, Hong JS, Wei Q, Hoerter JA, Fu G, Ampudia J, Zamoyska R, Zhu C, Gascoigne NR. Ligand-engaged TCR is triggered by Lck not associated with CD8 coreceptor. Nature communications. 2014;5:5624. doi: 10.1038/ncomms6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu B, Chen W, Evavold BD, Zhu C. Accumulation of Dynamic Catch Bonds between TCR and Agonist Peptide-MHC Triggers T Cell Signaling. Cell. 2014;157:357–368. doi: 10.1016/j.cell.2014.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Robert P, Aleksic M, Dushek O, Cerundolo V, Bongrand P, van der Merwe PA. Kinetics and mechanics of two-dimensional interactions between T cell receptors and different activating ligands. Biophys J. 2012;102:248–257. doi: 10.1016/j.bpj.2011.11.4018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marshall BT, Long M, Piper JW, Yago T, McEver RP, Zhu C. Direct observation of catch bonds involving cell-adhesion molecules. Nature. 2003;423:190–193. doi: 10.1038/nature01605. [DOI] [PubMed] [Google Scholar]
- 27.Kozono H, White J, Clements J, Marrack P, Kappler J. Production of soluble MHC class II proteins with covalently bound single peptides. Nature. 1994;369:151–154. doi: 10.1038/369151a0. [DOI] [PubMed] [Google Scholar]
- 28.Persaud SP, Donermeyer DL, Weber KS, Kranz DM, Allen PM. High-affinity T cell receptor differentiates cognate peptide-MHC and altered peptide ligands with distinct kinetics and thermodynamics. Mol Immunol. 2010;47:1793–1801. doi: 10.1016/j.molimm.2010.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chesla SE, Selvaraj P, Zhu C. Measuring two-dimensional receptor-ligand binding kinetics by micropipette. Biophys J. 1998;75:1553–1572. doi: 10.1016/S0006-3495(98)74074-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen W, Evans EA, McEver RP, Zhu C. Monitoring receptor-ligand interactions between surfaces by thermal fluctuations. Biophys J. 2008;94:694–701. doi: 10.1529/biophysj.107.117895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen W, Zarnitsyna VI, Sarangapani KK, Huang J, Zhu C. Measuring Receptor-Ligand Binding Kinetics on Cell Surfaces: From Adhesion Frequency to Thermal Fluctuation Methods. Cellular and molecular bioengineering. 2008;1:276–288. doi: 10.1007/s12195-008-0024-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Long M, Zhao H, Huang KS, Zhu C. Kinetic measurements of cell surface E-selectin/carbohydrate ligand interactions. Ann Biomed Eng. 2001;29:935–946. doi: 10.1114/1.1415529. [DOI] [PubMed] [Google Scholar]
- 33.Krogsgaard M, Li QJ, Sumen C, Huppa JB, Huse M, Davis MM. Agonist/endogenous peptide-MHC heterodimers drive T cell activation and sensitivity. Nature. 2005;434:238–243. doi: 10.1038/nature03391. [DOI] [PubMed] [Google Scholar]
- 34.Kersh GJ, Donermeyer DL, Frederick KE, White JM, Hsu BL, Allen PM. TCR transgenic mice in which usage of transgenic alpha- and beta-chains is highly dependent on the level of selecting ligand. J Immunol. 1998;161:585–593. [PubMed] [Google Scholar]
- 35.Crawford F, Kozono H, White J, Marrack P, Kappler J. Detection of antigen-specific T cells with multivalent soluble class II MHC covalent peptide complexes. Immunity. 1998;8:675–682. doi: 10.1016/s1074-7613(00)80572-5. [DOI] [PubMed] [Google Scholar]
- 36.Kersh GJ, Allen PM. Structural basis for T cell recognition of altered peptide ligands: a single T cell receptor can productively recognize a large continuum of related ligands. J Exp Med. 1996;184:1259–1268. doi: 10.1084/jem.184.4.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen W, Lou J, Zhu C. Forcing switch from short- to intermediate- and long-lived states of the alphaA domain generates LFA-1/ICAM-1 catch bonds. J Biol Chem. 2010;285:35967–35978. doi: 10.1074/jbc.M110.155770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kao H, Allen PM. An antagonist peptide mediates positive selection and CD4 lineage commitment of MHC class II-restricted T cells in the absence of CD4. J Exp Med. 2005;201:149–158. doi: 10.1084/jem.20041574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Das DK, Feng Y, Mallis RJ, Li X, Keskin DB, Hussey RE, Brady SK, Wang JH, Wagner G, Reinherz EL, Lang MJ. Force-dependent transition in the T-cell receptor beta-subunit allosterically regulates peptide discrimination and pMHC bond lifetime. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:1517–1522. doi: 10.1073/pnas.1424829112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kersh AE, Edwards LJ, Evavold BD. Progression of relapsing-remitting demyelinating disease does not require increased TCR affinity or epitope spread. J Immunol. 2014;193:4429–4438. doi: 10.4049/jimmunol.1401456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rosenthal KM, Edwards LJ, Sabatino JJ, Jr., Hood JD, Wasserman HA, Zhu C, Evavold BD. Low 2-dimensional CD4 T cell receptor affinity for myelin sets in motion delayed response kinetics. PloS one. 2012;7:e32562. doi: 10.1371/journal.pone.0032562. [DOI] [PMC free article] [PubMed] [Google Scholar]