Abstract
BCR-ABL+ acute lymphoblastic leukemia patients have transient responses to current therapies. However, the fusion of BCR to ABL generates a potential leukemia-specific antigen that could be a target for immunotherapy. We demonstrate that the immune system can limit BCR-ABL+ leukemia progression although ultimately this immune response fails. To address how BCR-ABL+ leukemia escapes immune surveillance, we developed a peptide: MHC-II tetramer that labels endogenous BCR-ABL-specific CD4+ T cells. Naïve mice harbored a small population of BCR-ABL-specific T cells that proliferated modestly upon immunization. The small number of naïve BCR-ABL specific T cells was due to negative selection in the thymus, which depleted BCR-ABL specific T cells. Consistent with this observation, we saw that BCR-ABL specific T cells were cross-reactive with an endogenous peptide derived from ABL. Despite this cross-reactivity, the remaining population of BCR-ABL reactive T cells proliferated upon immunization with the BCR-ABL fusion peptide and adjuvant. In response to BCR-ABL+ leukemia, BCR-ABL specific T cells proliferated and converted into regulatory T cells (Treg cells), a process that was dependent on cross-reactivity with self-antigen, TGFβ1, and MHC-II antigen presentation by leukemic cells. Treg cells were critical for leukemia progression in C57Bl/6 mice, as transient Treg cell ablation led to extended survival of leukemic mice. Thus, BCR-ABL+ leukemia actively suppresses anti-leukemia immune responses by converting cross-reactive leukemia-specific T cells into Treg cells.
Introduction
Cancer immunotherapy is an effective clinical approach in malignancies with high rates of non-synonymous mutations (1–4). Most cancer immunotherapy approaches currently focus on neo-antigen specific T cells, which ideally respond to mutations in proteins that drive tumorigenesis (5–8). However, identifying non-synonymous immunogenic mutations in driver genes is not always possible, thereby necessitating the use of either multiple antigens, or cross-reactive self-antigens, to prevent immune escape. This problem is illustrated in B cell acute lymphoblastic leukemia (B-ALL), which has few non-synonymous mutations (9). However, B-ALL is characterized by chromosomal translocations that give rise to fusion proteins encoding neo-antigens that drive transformation (10). We focused on BCR-ABL+ B-ALL, which creates a neo-antigen at the fusion of BCR to ABL. Immunotherapy is an attractive goal in BCR-ABL+ B-ALL because current therapies elicit only transient responses and long-term survival is poor. CD4+ T cells from patients with BCR-ABL+ B-ALL can secrete IFNγ upon ex vivo restimulation with peptides from the BCR-ABL fusion, but these responses are inadequate to eradicate leukemia in vivo (11, 12). To understand why BCR-ABL specific immunity fails to eliminate BCR-ABL+ B-ALL in mice, we identified BCR-ABL specific CD4+ T cells and tracked their responses to leukemia in vivo.
To examine anti-leukemia T cell responses we made use of a BCR-ABL+ B-ALL mouse model that recapitulates the human disease (13). To track anti-leukemia T cell responses, we generated a BCR-ABL peptide (BAp):MHC Class II tetramer reagent. We demonstrate that an adaptive immune response is elicited against BCR-ABL+ B-ALL and this response limits leukemia progression. BAp:I-Ab-specific T cells exist in mice and proliferate in response to immunization with BAp peptide plus an adjuvant. Inoculation with live BCR-ABL+ leukemic cells also resulted in proliferation of BAp:I-Ab-specific T cells. However, these cells were converted into Treg cells and thus unable to eliminate leukemia. Importantly, transient Treg ablation with Foxp3DTR/DTR mice resulted in extended lifespan of leukemic mice, which correlated with increased number of CD44hi, Ly6C+ BAp:I-Ab-specific T cells, suggesting that induction of Treg cells by the leukemia led to decreased priming and Th1-like CD4+ T cell differentiation.
Materials and Methods
Mice
C57BL/6 mice and Cdkn2a−/− (strain 01XF6, B6, 129-Cdkn2atm1Cjs/Nci, (14)) mice came from the National Cancer Institute. Foxp3-GFP (stock# 006772) mice came from Jackson Laboratories (Bar Harbor, ME). Bim−/−, OT-IxRag2−/−, Foxp3DTR/DTR, OT-IIxRag2−/− and SM1xRag2−/− mice were generated locally as previously described (15–19). Mice were housed at the University of Minnesota in specific pathogen free conditions and all experiments were approved by IACUC.
Immunizations
Mice were immunized with Complete Freund’s Adjuvant (CFA)+BAp subcutaneously in the hind flank.
Anti-TGFβ in vivo treatment
Mice were treated with anti-TGFβ (clone 1D11, Bio X Cell) or isotype (clone MOPC21, Bio X Cell) with 1mg i.p. on the same day that the mice were inoculated with leukemia, followed by 200μg i.p. every-other-day for fourteen days.
Diphtheria Toxin Treatment
Mice were treated with 0.2μg/mouse diphtheria toxin (List Biologicals) by i.p. injection every-other-day. Treg depletion was analyzed by monitoring GFP+, CD4+ cells.
Leukemia model
The BCR-ABL+ B Acute Lymphoblastic Leukemia model has been previously described (20). Briefly, Cdkn2a−/− mouse bone marrow cells were transduced with viral supernatant containing a BCR-ABL (P190)-IRES-GFP retrovirus (21). Cells were cultured in RPMI1640+10%FBS+1%Penicillin-Streptomycin+1%L-Glutamine+0.001%beta-mercaptoethanol in a 37_C incubator. 2,500 live cells were adoptively transferred i.v., into host mice without prior irradiation. The SP1 cells were derived from leukemia in a STAT5bCAxPax5+/− mouse as previously described (22). MHC-II−/− leukemia was generated by crossing Arf−/− mice with I-Ab−/− mice to generate Arf−/−, I-Ab−/− mice. Bone marrow from this genotype was transduced with the BCR-ABL IRES GFP retrovirus to generate MHC-II−/−, BCR-ABL+ GFP+ leukemic cells.
Tetramer production
The BAp:I-Ab tetramer and others used herein were produced as described (23). Purified monomer was tetramerized with SA-PE or SA-APC.
Antigen-Specific CD4+ T cell Enrichment Strategy
I-Ab tetramer-binding cell enrichment was performed as described (23). Tetramers loaded with PE and APC were used together, and tetramer double-positive events were analyzed to increase sensitivity and specificity (24). Cells were enriched with either Miltenyi or StemCell anti-APC and anti-PE reagents following manufacturer’s protocols. The enriched fractions were mixed with Accucheck counting beads (LifeTechnologies, Grand Island NY) for cell enumeration.
Histology
Histology was performed as previously described and antibodies are listed below (25).
Antibodies
Antibodies for flow cytometry and histology include CD3 PE, CD4 (RM4-5) PerCPCy5.5, CD8 (53-6.7) BV650, CD11c (N418) PE, FOXP3 (FJK16S) PE, were purchased from BD Biosciences (San Jose, CA); NK1.1 (PK136), CD11b (M1/70), CD11c (N418), B220 (RA3-6B2), and F4/80 in APC-eFluor780; PD1 (J43) FITC, CD73 eFluor450, FR4 PE-Cy7, PDL1 PerCP-eFluor710, MHC-II I-Ab eFluor450, IL7R PE, and all ELISpot antibodies were purchased from eBiosciences (San Diego, CA), and IgM (Fab′) APC was purchased from Jackson Immunoresearch (West Grove, PA). Rat IgG1 (HRPN) PerCP-Cy5.5 Isotype and Rat IgG2a (2A3) violetFluor450 Isotype were purchased from Tonbo Biosciences (San Diego, CA). Cells from enriched fractions were analyzed on an LSR-II Fortessa cytometer (BD Biosciences, San Jose CA) and data was analyzed in FlowJo (Treestar, Ashland OR). Intracellular cytokine staining was done using BD Cytofix/Cytoperm reagents and protocols (BD Biosciences, San Jose CA). Anti-IL10 (JES5-16E3, PE, eBiosciences), TGFB1(LAP) (TW7-16B4, ef710, eBiosciences), IFNγ (XMG1.2, BV421, BD Horizon), and TNFα (MP6-XT22, AF488, eBiosciences) were stained by this procedure.
ELISPOT
C57BL/6 mice were immunized with e1a2 peptide (GEGAFHGDAEALQRPVASDF, 2 mg/ml) or 2W1S peptide (EAWGALANWAVDSA, 0.2 mg/ml) emulsified in Complete Freund’s Adjuvant (CFA) (Sigma-Aldrich, St. Louis, MO) and harvested two weeks later. CD4+ T cells were enriched using a mouse CD4 T cell Isolation Kit (Miltenyi, Bergisch Gladbach Germany). Naïve mouse splenocytes were harvested, irradiated and mixed at a 1:1 ratio with the immunized CD4+ T cells in wells coated with IFN-gamma capture antibody (AN-18) at a concentration of 15μg/mL. Synthetic overlapping peptides that spanned the BCR-ABL p190 fusion (Genscript, Piscataway, NJ) were added to individual wells. 2W1S peptide and Concanavalin A were used as positive controls. Samples were developed using a biotinylated IFN-gamma detection antibody (R4-6A2) and streptavidin-Alkaline Phosphatase with 5-Bromo-4-chloro-3′-indolyphosphate (BCIP) and Nitro-blue tetrazolium (NBT) developing agents and photographed and analyzed with a ImmunoSpot S6 Microanalyzer using the ImmunoCapture 6.3 and ImmunoSpot 5.0 Pro DC software (Cellular Technology Ltd. (Shaker Heights, OH).
Statistics
Standard normality tests suggested departures from normality, so non-parametric tests (Mann-U Whitney test for two groups, Kruskal-Wallace & Dunns’ Test for more than two groups) were used unless otherwise stated. Normality assessments and non-parametric tests were done in GraphPad Prism (LaJolla, CA). Linear regressions and correlation coefficients were estimated in GraphPad Prism.
Results
The adaptive immune system mounts a response to BCR-ABL+ leukemia
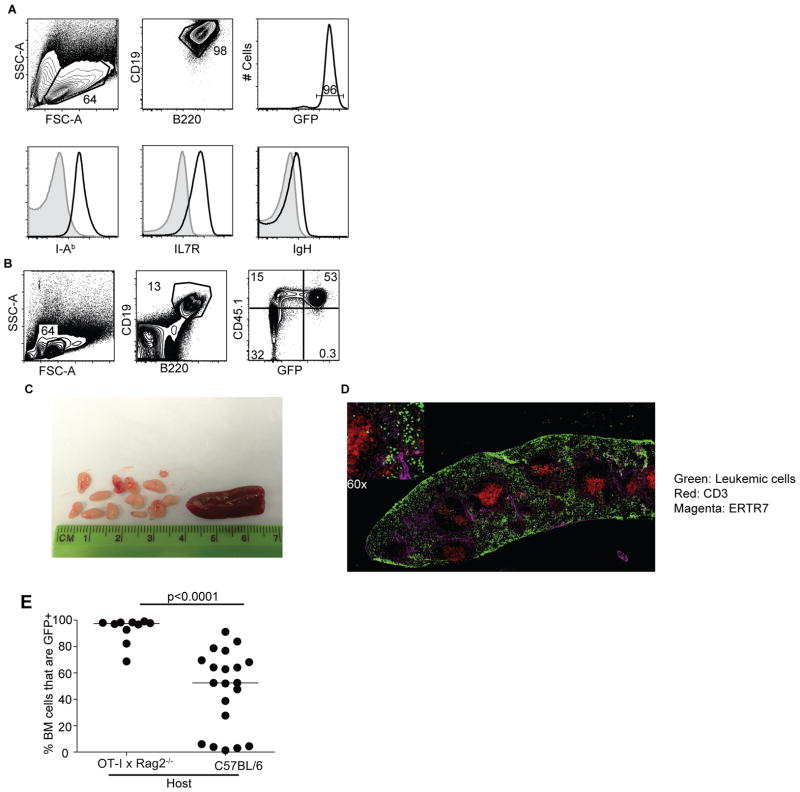
To examine whether BCR-ABL+ leukemia elicited an adaptive immune response in mice, we adoptively transferred Cdkn2a−/− BCR-ABL+ pre-B cells into healthy, immune-competent recipient mice. This model incorporates two genetic alterations common in high-risk human B-ALL: BCR-ABL and loss of the tumor suppressor ARF (26). Further, these cells can be transferred into unconditioned host mice where they faithfully form BALL (20). The leukemic cells express CD19, B220, IL7R, MHC-II, and low levels of immunoglobulin heavy chain (IgH) and therefore closely resemble human progenitor BALL (Fig 1A). In syngeneic (C57BL/6) hosts, leukemia arose in the bone marrow and the splenic red pulp with small numbers of leukemic cells in the lymph nodes (Fig 1B–D).
Figure 1.
BCR-ABL+ B-ALL elicits an adaptive immune response. A. Phenotype of BCR-ABL+ cells grown in vitro. Cells were analyzed after 3 weeks in culture, and express CD19, B220, GFP, I-Ab, IL7R, and Ig Heavy Chain. Isotype controls are in grey and experimental stains are in black; shown are representative stains. B. BCR-ABL+ leukemic cell phenotype in vivo. Live lymphocytes are gated for CD19+ B220+, and CD45.1+GFP+ events are leukemic cells. C. Representative lymph nodes and spleen from a mouse with leukemia. D. Representative histology of a leukemic mouse spleen at 14 days post-transfer. Leukemic cells are GFP+(Green) and are localized in the red pulp (ERTR7+, magenta). T cells are stained with CD3 in Red. E. GFP+ cells in BM as a percentage of the CD19+B220+ cells in C57BL/6 and OT-IxRag2−/− hosts. Twenty days after leukemic cell transfer BM cells were harvested from WT or OT-IxRag2−/− hosts and CD19+B220+GFP+ cells were gated as in A. GFP+ cells in BM as a percentage of the CD19+B220+ cells are graphed. P-value from Mann-Whitney U Test. Data shown from three separate experiments.
To determine if the adaptive immune system had any control over BCR-ABL+ B-ALL progression we compared leukemic cell counts (referred to hereafter as leukemic burden) in C57BL/6 and OT-IxRag2−/− mice. OT-IxRag2−/− mice were used because they have T cells but no leukemia-specific adaptive immunity. The percentage of leukemic (CD45.1+, GFP+) B cells in the bone marrow (BM) was significantly higher in OT-IxRag2−/− mice than in C57BL/6 mice (Fig 1E).
BAp:I-Ab-specific T cells in naïve mice
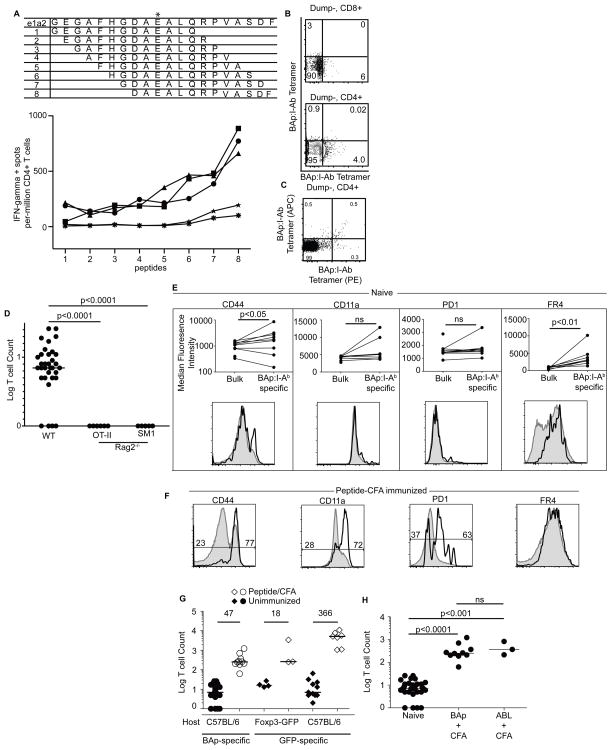
We studied the interaction between BCR-ABL+ B-ALL and CD4+ T cells by tracking leukemia-specific T cells. Since BCR-ABL is a somatic mutation, the peptide spanning the fusion of BCR to ABL could potentially elicit a T cell response. Using an ELISpot assay, we found a MHC-II binding peptide in the BCR-ABL fusion that elicited a CD4+ T cell response (Fig 2A), and constructed a peptide:I-Ab monomer wherein this peptide (BAp) was covalently linked to biotinylated I-Ab (referred to as BAp:I-Ab). We labeled BAp:I-Ab-specific T cells by tetramerizing BAp:I-Ab monomer with Streptavidin-PE or Streptavidin-APC. Using this reagent, we detected ~8 BAp:I-Ab-specific CD4+ T cells per mouse (Fig 2B,D). No BAp:I-Ab-specific T cells were detected in OT-IIxRag2−/− and SM1xRag2−/− mice, where all T cells are specific for an ovalbumin or a flagellin peptide bound to I-Ab, respectively, demonstrating specificity of the BAp:I-Ab tetramer for the BAp:I-Ab-specific T cell TCR (Fig 2D). The BAp:I-Ab-specific T cells typically expressed low levels of canonical activation markers including CD11a and PD1 in naïve mice, however expression of CD44 and FR4 were significantly, albeit modestly, increased on BAp:I-Ab-specific T cells in naïve mice (Fig 2E). Nonetheless, all of these markers were uniformly expressed at high levels following BAp peptide + CFA immunization demonstrating that the BAp:I-Ab-specific T cells responded to cognate peptide immunization (Fig 2C,F). Taken together, these findings show that BAp:I-Ab-specific T cells are capable of responding to BAp antigen.
Figure 2.
BAp:I-Ab-specific T cells in mice. A. Immunized mice expressed IFNγ when exposed to BCR-ABL. Peptides 1–8 on the X-axis are 13aa peptides that progressively span the BCR-ABL e1a2 fusion (top panel, asterisk represents the fusion junction). Mice were immunized with the full e1a2 20aa peptide. Two weeks later, cells were harvested from SLO and separate wells were pulsed with individual peptides 1 through 8 and then stained with anti-IFNgamma. Five mouse repeats are shown. B. Naïve BAp:I-Ab-specific T cells are Dump−, CD8−, CD4+. Dump−CD8+ (top panel) or Dump−CD4+ (bottom panel) T cells were gated on and stained with BAp:I-Ab-APC (Y-axis) and BAp:I-Ab-PE (X-axis). C. Representative flow plot of T cells from a C57BL/6 mouse BAp:I-Ab tetramers labeled with APC and PE, 14 days after immunization with BAp peptide plus adjuvant. D. Enumeration of BAp:I-Ab-specific T cells from C57BL/6, OT-IIxRag2−/−, and SM1xRag2−/−. Shown are 44 mice from 10 experiments, y-axis is the Log(Y+1) of the BAp:I-Ab-specific T cell count. E. Phenotype of BAp:I-Ab-specific T cells from naïve mice (black lines) compared to the bulk CD4+ population from the same mouse (grey lines). Shown are results from three independent experiments whose events were then concatenated for the histograms. All comparisons were made with Mann-Whitney U test (2 groups). F. BAp:I-Ab-specific T cells from mice immunized with peptide plus adjuvant 14 days later (black lines). Grey histograms are bulk CD4+ T cells. Shown are 10 mice from 3 independent experiments, whose events were then concatenated for the histograms. The numbers are percentage of cells falling into each given gate. G. C57BL/6 or Foxp3-GFP mice were immunized with BAp or GFP peptide + CFA. Secondary lymphoid organs were harvested 14 days later and BAp:I-Ab-specific or GFP:I-Ab-specific T cells were enumerated. Y axis is the Log (Y+1) of the BAp:I-Ab-specific T cell count or the GFP:I-Ab-specific T cell count. The number above the line is the fold change from median (indicated by black bars). All fold expansions shown resulted in significantly higher T cell counts than in unimmunized mice; >2 independent experiments were conducted for each immunization shown. H. C57BL/6 mice were immunized with BAp peptide or ABL peptide emulsified in CFA. 14 days later, secondary lymphoid organs were harvested as previously described. Y-axis is the Log (Y+1) of BAp:I-Ab tetramer-binding events; data is from three independent experiments. All comparisons were made with Kruskal-Wallace Test (>2 groups).
We next examined whether BAp:I-Ab specific T cells had comparable proliferative capacity to other antigen-specific naïve T cells. We found that in response to peptide plus adjuvant immunization, BAp:I-Ab-specific T cells proliferated modestly (47-fold, Fig 2G). BAp:I-Ab-specific T cell proliferation resembled that observed with GFP:I-Ab-specific T cells in Foxp3-GFP mice (18-fold), where GFP is a self-antigen. In contrast, immunization with GFP in C57BL/6 mice (in which GFP is a foreign antigen) resulted in a 366-fold expansion of GFP:I-Ab specific T cells (Fig 2G). Thus BAp:I-Ab-specific T cell expansion resembled that seen for T cells that recognized low abundance self-antigens (GFP in Foxp3-GFP mice).
T cell-based cancer immunotherapy is thought to be more effective when T cells recognize cancer cell neo-antigens with minimal cross-reactivity for self-antigens. Thus, we addressed if BAp:I-Ab-specific T cells recognized neo-antigens or cross-reactive self antigens. Recent studies have shown that small peptide:MHC-II specific T cell repertoires are predictive of cross-reactivity with self-antigen(27). The BAp peptide (DAEALQRPVASDF) closely resembles the endogenous ABL peptide (LEEALQRPVASDF) (85% identical, see underlined portion), so we tested if BAp:I-Ab tetramer-binding T cells could recognize the overlapping ABL peptide. When mice were immunized with adjuvant plus ABL peptide, BAp:I-Ab tetramer-binding cells proliferated 46-fold, which is similar to the proliferation seen after immunization with BAp peptide plus adjuvant (Fig 2G,H). Taken together, this evidence suggests that BAp:I-Ab-binding T cells are cross-reactive with the self-peptide ABL. These results may explain the occasional expression of CD44 that we saw on BAp:I-Ab-specific T cells in naïve mice.
BAp:I-Ab specific T cell numbers in naïve mice are controlled by central tolerance
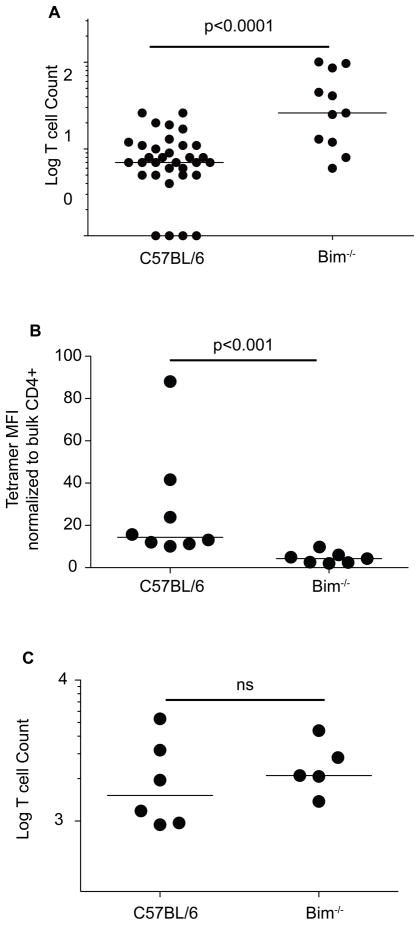
The precursor frequency of BAp:I-Ab-specific T cells was quite low (Fig 2D), thus we examined whether immune central tolerance may be limiting the number of BAp:I-Ab-specific T cells in naïve mice. Bim−/− mice, which have defective thymic negative selection, had significantly (~5 fold) more BAp:I-Ab-specific T cells (Fig 3A) (18, 28, 29). Thus, ~80% of the BAp:I-Ab-specific T cell progenitors were deleted during thymic negative selection.
Figure 3.
BAp:I-Ab-specific T cells are cross-reactive with self-antigen. A. BAp:I-Ab-specific T cells were enumerated from Bim−/− mice and compared to C57BL/6. Y axis is the Log (Y+1) of the BAp:I-Ab-specific T cell count and three independent experiments are shown. B. BAp:I-Ab-specific T cells were harvested from C57BL/6 or Bim−/− mice and BAp:I-Ab tetramer mean fluorescence intensity (PE channel) was measured. Shown are 3 independent experiments, p-value is from Mann-Whitney U Test. C. C57BL/6 or Bim−/− mice were immunized with BAp peptide emulsified in CFA, and BAp:I-Ab-specific T cells were harvested from the spleen and lymph nodes 14 days later and these cells were enumerated. Y axis is the Log (Y+1) of the BAp:I-Ab-specific T cell count. Shown is data from 2 independent experiments, significance was calculated with the Mann-Whitney U test.
We next reasoned that the BAp:I-Ab-specific T cell clones, which were rescued in Bim−/− mice, might have high affinity for cognate BAp:I-Ab. To examine this issue, we analyzed the tetramer mean fluorescence intensity of BAp:I-Ab tetramer binding cells and normalized this value to that found in the matching bulk CD4+ T cell compartment. Interestingly, we found that the tetramer mean fluorescence intensity was significantly higher in the C57BL/6 mice than in the Bim−/− mice (Fig 3B). In addition, when we immunized Bim−/− mice with BAp peptide emulsified in CFA, we saw no increase in the number of BAp:I-Ab-specific T cells recovered from Bim−/− mice (Fig 3C). Thus, Bim−/− mice have an increased number of BAp:I-Ab-specific T cells, but these cells do not appear to bind cognate BAp:I-Ab with higher affinity, nor do these cells accumulate to higher numbers upon immunization.
BAp:I-Ab-specific T cells respond to BCR-ABL+ B-ALL
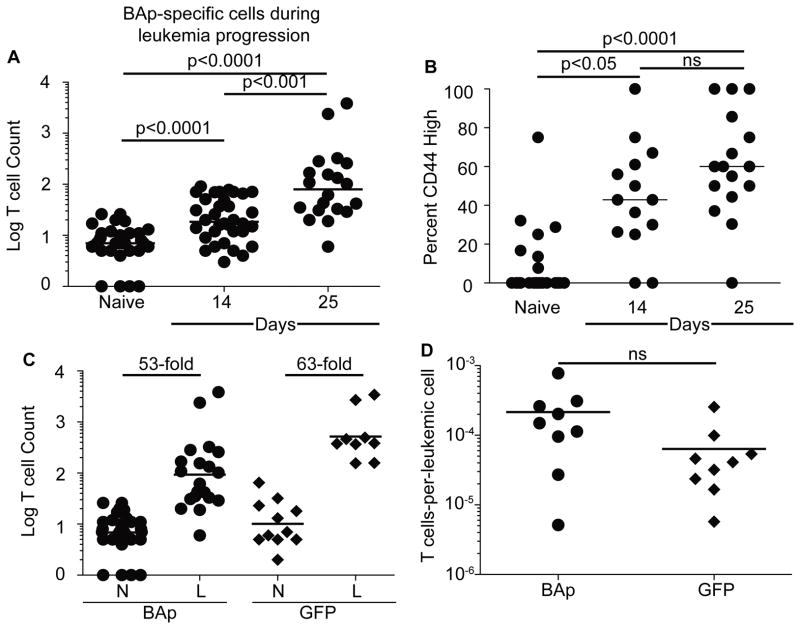
Despite their limited numbers, we hypothesized that BAp:I-Ab-specific T cells might still respond to BCR-ABL+ leukemia. To test this hypothesis, we tracked BAp:I-Ab-specific T cells following adoptive transfer of leukemic cells into C57BL/6 mice and observed a significant accumulation of BAp:I-Ab specific T cells at 14 and 25 days post-transfer of BCR-ABL+ leukemic cells (Fig 4A). CD44 expression on BAp:I-Ab-specific T cells also increased in a stepwise fashion at 14 and 25 days post-transfer, suggesting that these cells were recognizing antigen in leukemic mice (Fig 4B). This response was antigen-specific as BAp:I-Ab-specific T cells did not proliferate or upregulate CD44 in response to a BCR-ABL negative leukemia (data not shown). Thus, BAp:I-Ab-specific cells proliferate but do not eliminate leukemia.
Figure 4.
BAp:I-Ab-specific T cells are cross-reactive with self-antigen in comparison to GFP:I-Ab-specific T cells. A. 2,500 live BCR-ABL+ leukemic cells were transferred into mice. BAp:I-Ab-specific T cells were counted at fourteen days or twenty-five days post-inoculation. Y axis is the Log (Y+1) of the BAp:I-Ab-specific T cell count. Cumulative results from more than three experiments are shown; data is from greater than three independent experiments. B. Percent CD44-high expression on BAp:I-Ab-specific T cells. The data was collected as in A. C. Fold increase in antigen-specific T cell count between naïve (N) mice and leukemic (L) mice. D. Target-to-effector ratio of leukemic mice, either tracking BAp-specific T cells or GFP-specific T cells; three independent experiments shown. All comparisons were made with Mann-Whitney U test (2 groups) or Kruskal-Wallace test (>2 groups).
BCR-ABL+ leukemic cells also expressed a GFP reporter, making GFP a leukemia-specific neo-antigen in leukemic C57BL/6 mice. Because our findings suggest that BAp:I-Ab-specific T cells are cross-reactive, we used a GFP:I-Ab-specific tetramer (30) to examine if neo-antigen specific T cells behave similarly to BAp:I-Ab-specific T cells in response to leukemia. While BAp:I-Ab-specific T cells proliferated as much in response to leukemia (53-fold) as they did in response to BAp peptide plus adjuvant (47-fold), GFP:I-Ab-specific T cells proliferated only 17% as much in response to leukemia (66-fold) as GFP peptide plus adjuvant (366-fold, Fig 4C, 2G). Importantly, the ratio of GFP:I-Ab specific T cells to GFP+ leukemic cells was not different than the BAp:I-Ab-specific T cell-to-leukemic cell ratio (Fig 4D). Thus, the proliferation of neo-antigen specific T cells responding to BCR-ABL+ B-ALL is inhibited.
T cells specific for cross-reactive leukemia antigens are converted into pTreg cells by leukemia
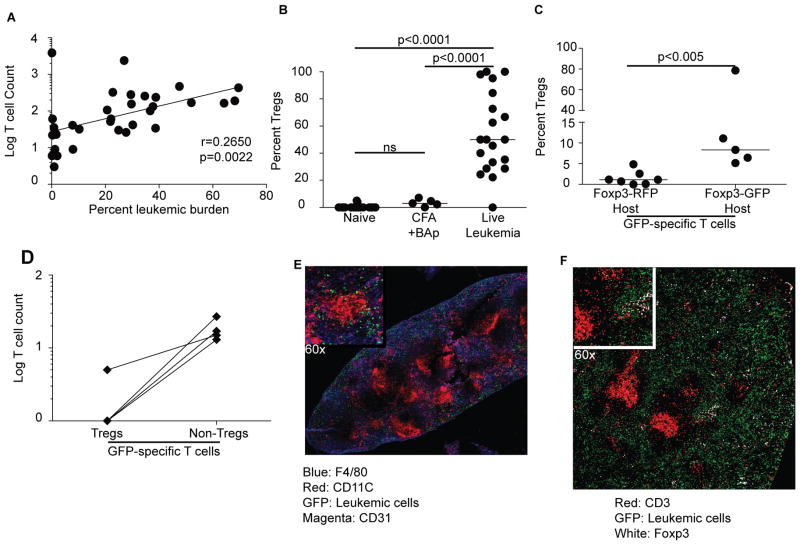
Our studies provide evidence that BAp:I-Ab-specific T cells respond to BCR-ABL+ BALL despite cross-reactivity with self-antigen. However, leukemia still progresses in C57BL/6 mice, which suggests that mechanisms of tolerance inhibit the immune responses by cross-reactive T cells. Indeed, BAp:I-Ab-specific T cells positively correlated with leukemic burden in naïve mice inoculated with leukemia (Fig 5A), suggesting that the BAp:I-Ab-specific T cells might prevent immune responses to B-ALL. A possible explanation was that BAp:I-Ab-specific T cells were regulatory T cells (Treg cells). Both naïve Foxp3-GFP mice and Foxp3-GFP mice immunized with BAp plus adjuvant typically had no or few BAp:I-Ab specific Treg cells whereas the majority of BAp:I-Ab specific T cells in leukemic mice were FOXP3-GFP+ (Fig 5B). This contrast supports the conclusion that B-ALL converts BAp:I-Ab-specific T cells into pTreg cells, and provides a mechanism for host tolerance of leukemia.
Figure 5.
BCR-ABL+ B-ALL induces pTregs from cross-reactive CD4+ T cells. A. Log (Y+1) of the BAp:I-Ab-specific T cell counts (Y-axis) correlates with leukemic burden (depicted as the percent of bone marrow B cells, which are GFP+) on the X-Axis. More than three experiments are shown, Spearman correlation is shown. B. Mice were unimmunized, immunized with CFA+BAp, or injected with 2500 BCR-ABL+ cells; percentage of BAp:I-Ab-specific Tregs were analyzed 14 days post-inoculation. >2 independent experiments for each group are shown, Kruskal-Wallace test performed (>2 groups). C. Percent of BAp:I-Ab-specific T cells that are FOXP3+ Treg cells in Foxp3-dsRed or Foxp3-GFP mice. 2,500 BCR-ABL+ B-ALL cells were transferred into Foxp3-RFP or Foxp3-GFP mice and BAp:I-Ab-specific T cells were enumerated 14 days later. Y-axis is percentage of BAp:I-Ab-specific T cells that are FOXP3+ Treg cells in their respective hosts. >2 independent experiments are shown for each group, Mann-Whitney U test performed (2 groups). D. SLO were harvested from naïve Foxp3-GFP mice, and GFP:I-Ab-specific T cells were enumerated; FOXP3-GFP expression was evaluated on these cells. The Y axis shows the log(Y+1) of GFP:I-Ab-specific T cell count from these mice. E. Representative histology of spleen from a C57BL/6 mouse 14 days after leukemic cell transfer. Blue is F4/80, Red is CD11C, Magenta is CD31, and Green is GFP+(leukemic cells). F. Representative histology from spleen as in E, showing FOXP3+ Treg cells in the splenic red pulp. Red is CD3, White is FOXP3, Green is GFP+(leukemic cells).
We next determined if leukemia-specific Treg polarization was related to the cross-reactivity of BAp:I-Ab-specific T cells with self-antigen. To address this question, we examined both neo-antigen specific T cells and cross-reactive T cells responding to leukemia. The leukemic cells used in these experiments express both GFP and BCR-ABL, thus GFP was a leukemia-specific neo-antigen in C57BL/6 mice and BCR-ABL was a cross-reactive antigen in C57BL/6 mice. We enumerated GFP:I-Ab-specific Treg cells responding to leukemia in Foxp3-RFP mice (where GFP is a non-self antigen) and Foxp3-GFP mice (where GFP is a self-antigen). Most GFP:I-Ab-specific T cells did not polarize into FOX3-RFP+ Treg cells in response to leukemia (1.5% +/− 1.7%). In contrast, significantly more GFP:I-Ab-specific T cells were Treg cells in leukemic FOXP3-GFP mice (8.3% +/− 31.2%, p<0.005; Fig 5C). Importantly, 75% of naïve Foxp3-GFP mice contained no GFP:I-Ab-specific Treg cells (Fig 5D), suggesting that GFP:I-Ab-specific Treg cells seen in leukemic Foxp3-GFP mice were induced Treg cells. Thus, peripheral CD4+ T cells that cross-react with self have increased capacity to become pTreg cells in response to leukemia. Histologically, Treg cells were localized in the splenic red pulp, adjacent to red pulp macrophages and leukemic cells (Fig 5E,F). Red pulp macrophages have been shown to induce Treg cells in vitro (31). Thus, Tregs responding to leukemia antigens are preferentially cross-reactive, and localized in an immune-suppressive microenvironment near the leukemic cells.
Tregs allow leukemia progression in C57BL/6 mice
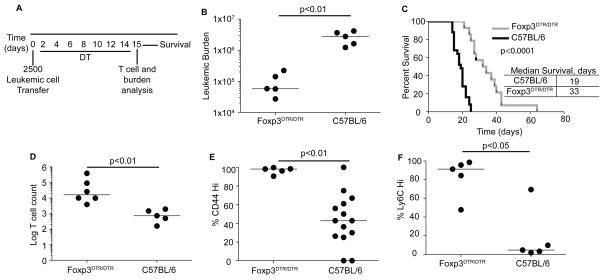
To determine if Treg cells inhibited the immune response to BCR-ABL+ leukemia, we selectively ablated Treg cells using Foxp3DTR/DTR mice (17). Transient depletion of Treg cells led to a 98% decrease in leukemic burden in the BM at fourteen days post-inoculation with leukemia (Fig 6A,B). Depletion of Tregs during the first 14 days following leukemia inoculation also led to an increased survival of mice as compared to untreated C57BL/6 mice (Fig 6C). In parallel BAp:I-Ab-specific T cells proliferated ~30-fold more in DT-treated Foxp3DTR/DTR mice than in DT-treated C57BL/6 mice (Fig 6D).
Figure 6.
Immunity to BCR-ABL+ B-ALL is hindered by Tregs. A. Foxp3DTR/DTR or C57BL/6 mice were inoculated with 2500 leukemic cells and then treated with Diphtheria Toxin (DT) at 0.25μg/kg DT daily, and compared to C57BL/6 mice treated with DT. B. Mice were harvested at 14 days post-inoculation and leukemic cells were enumerated from the spleen and lymph nodes. C. Foxp3DTR/DTR mice were treated as in C and survival was analyzed. P-values from Log-rank test; three independent experiments shown. D. Log(Y+1) of the BAp:I-Ab-specific T cell count in Foxp3DTR/DTR mice and C57BL/6 mice inoculated with leukemia and treated with DT as in A. Three independent experiments are shown. E. Experiments were conducted as in A; shown is the percent CD44hi BAp:I-Ab-specific T cells. F. Experiments were conducted as in A; shown is the percent Ly6Chi BAp:I-Ab-specific T cells. All comparisons were made with Mann-Whitney U test (2 groups).
We next sought mechanistic insight into how the presence of Tregs inhibited the leukemia-specific immune response. We found that the percent of BAp:I-Ab-specific T cells that were CD44hi was significantly higher in DT-treated Foxp3DTR/DTR mice, suggesting that Tregs inhibit priming of BAp:I-Ab-specific T cells (Fig 6E). We reasoned that Ly6C+ Th1-like cells might also be important in controlling leukemia progression (32–34). Indeed, the fraction of CD44+ and Ly6C+ BAp:I-Ab-specific T cells was increased in DT-treated Foxp3DTR/DTR mice, suggesting that Tregs inhibit BAp:I-Ab-specific T cell priming and effector differentiation during the immune response to leukemia (Fig 6F).
Treg conversion in response to leukemia is dependent on TGFβ and antigen presentation by leukemic cells
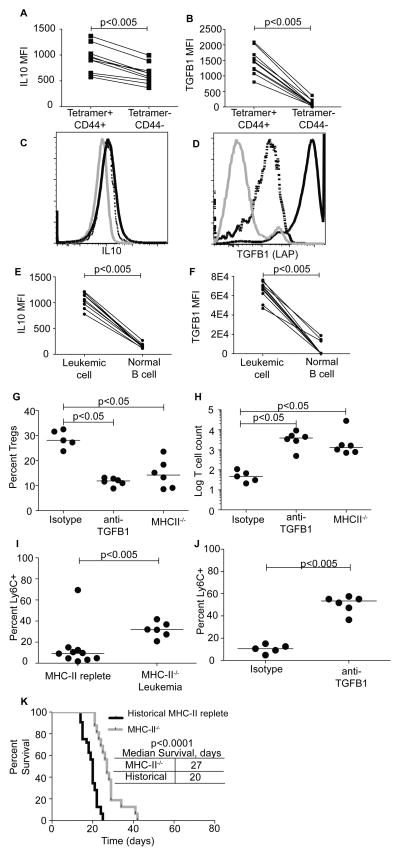
Tregs were clearly important for leukemia progression (Fig 6F). However the mechanism that drives BAp:I-Ab-specific T cells into the Treg lineage in mice with leukemia remains unclear. One possible explanation is that the leukemic cells might produces cytokines that drive Treg conversion. To address this point, we analyzed cytokines production by leukemic cells and by BAp:I-Ab-specific T cells responding to leukemia. When responding to leukemia, both leukemic cells and BAp:I-Ab-specific T cells produced the immune-suppressive cytokines IL10 and TGFβ1 (Fig 7A–F). Since TGFβ1 is important for conversion of naïve CD4+ T cells into peripheral Treg cells (35, 36), we reasoned that TGFβ1 might drive BAp:I-Ab-specific Treg conversion in response to leukemia. When leukemic mice were treated with anti-TGFβ antibody or isotype control, anti-TGFβ treatment led to significantly less BAp:I-Ab-specific Tregs, and significantly more proliferation of BAp:I-Ab-specific T cells than isotype treatment or historical analyses of leukemic mice at the same timepoint (Fig 4A, 7G,H). These results suggested that TGFβ1 was critical for efficient induction of leukemia-specific Tregs in this model.
Figure 7.
BAp:I-Ab-specific Treg conversion depends on TGFβ1 and leukemic cell antigen presentation. A, B. Leukemic cells were gated as CD45.1+, GFP+ and were stained for cytokines. Shown is the median fluorescence intensity of IL10 (A) and TGFβ1 (B). Three independent experiments are shown, comparisons were made with Wilcoxan Matched-Pairs test (2 paired groups). C, D. Shown is the overlay of IL10 (C) and TGFβ1 (D) expression on BAp:I-Ab Tetramer−, CD44− CD4+ T cells (Grey), BAp:I-Ab Tetramer+, CD44+ CD4+ T cells (Dashed), and Leukemic cells (Black). Shown are 10 mice from three independent experiments, whose results were concatenated. E, F. BAp:I-Ab-specific T cells were gated on BAp:I-Ab Tetramer (APC)+, CD44+ and were stained for cytokines. Shown is the median fluorescence intensity of IL10 (E) and TGFβ1 (F). Three independent experiments are shown, comparisons were made with Wilcoxan Matched-Pairs test (2 paired groups). G. Mice were inoculated with MHC-II replete leukemia or MHC-II−/− leukemia as labeled, or treated with Isotype or anti-TGFβ antibody. BAp:I-Ab-specific Tregs were gated as previously described and the percent Tregs was analyzed. Results are from three independent experiments, Kruskal-Wallace and Dunns’ test used to establish significance. H. Mice were treated as in G, and BAp:I-Ab-specific T cell count was analyzed as previously described. Results are from three independent experiments, Kruskal-Wallace and Dunns’ test used to establish significance. I. BAp:I-Ab-specific T cells from mice inoculated with MHC-II−/− leukemia or MHC-II replete leukemia and harvested 14 days later were analyzed for Ly6C expression. Shown are three independent experiments, significance was established with Mann-Whitney U test. J. BAp:I-Ab-specific T cells from mice inoculated with leukemia and treated with anti-TGFβ or isotype control were analyzed for Ly6C expression 14 days later. Shown are three independent experiments, significance was established with Mann-Whitney U test. K. Mice inoculated with MHC-II−/− leukemia or MHC-II replete leukemia were aged until leukemia-induced morbidity and survival to morbidity was analyzed. P-values from Log-rank test; three independent experiments shown.
Leukemic cells and Tregs were also in relatively close contact in the splenic red pulp (Fig 5F). Thus, we reasoned that the leukemic cells might present antigen in the context of MHC-II, and initiate Treg induction by simultaneously providing TGFβ1. To test this hypothesis, we generated MHC-II−/− leukemic cells and inoculated Foxp3-GFP mice with them. When we analyzed BAp:I-Ab-specific Treg induction 14 days later, we found that significantly fewer BAp:I-Ab-specific T cells were Tregs in response to MHC-II−/− leukemia than in response MHC-II replete leukemia (either from historical experiments or during experiments done at the same time with isotype treatment Fig 5B, 7G). In parallel, MHC-II−/− leukemia or anti-TGFβ treatment led to significant increases in Ly6C expression on BAp:I-Ab-specific T cells, suggesting that these cells might be converted into a Th1-like lineage instead of the Treg lineage (Fig 7I,J). Additionally, MHC-II−/− leukemia correlated with significantly more proliferation of BAp:I-Ab-specific T cells in leukemic mice (Fig 7H), again suggesting that the decreased BAp:I-Ab-specific Treg conversion might allow for increased proliferation of the BAp:I-Ab-specific T cells. Consistent with these findings, we observed a significant increase in survival of mice inoculated with MHCII−/− leukemia compared to MHCII-replete leukemia (Fig. 7K). Thus, antigen presentation by the leukemic cells enhances induction of BAp:I-Ab-specific T cells into the Treg lineage, inhibits proliferation of Ly6C+ BAp:I-Ab-specific T cells and reduces survival of leukemic mice.
Discussion
Recent studies demonstrate that augmenting immune responses can be an effective treatment for cancer (6, 37–40). While the immune system recognizes both tumor-associated self-antigens and tumor-specific neo-antigens, cell-based immunotherapy approaches have focused on utilizing neo-antigen specific T cells, which should not be subject to immune tolerance. Many of these approaches utilize mutations arising in the malignancy that may not drive cancer progression, thus potentially allowing immune escape. Further, most of these treatments have been employed in cancers with many non-synonymous mutations. In this study, we examine the immune response to a cancer with few non-synonymous mutations by investigating the adaptive immune response to the oncoprotein BCR-ABL. Our approach was informed by using BAp:I-Ab tetramers, to track cross-reactive leukemia-specific T cells, and GFP:I-Ab tetramers to track neo-antigen specific T cells. A major finding from our studies is that BCR-ABL+ leukemia induces tolerance to both cross-reactive tumor associated self-antigens and tumor-specific neo-antigens. However, the mechanism of tolerance differed depending on whether the tumor-derived antigen was a cross-reactive tumor antigen or a neo-antigen. While leukemia induced conversion of BAp:I-Ab-specific T cells into Treg cells, it inhibited proliferation of GFP:I-Ab-specific T cells without driving Treg induction. We speculate that cross-reactive antigen-specific Treg cells are one mechanism by which leukemias induce tolerance to tumor-specific neo-antigens.
The murine host may also allow for immune tolerance to leukemia. In our model, the majority of BAp:I-Ab specific T cells are clonally deleted in the thymus, which may result in too few BAp:I-Ab specific T cells to effectively clear leukemia. For example, previous studies have shown that increasing the number of antigen-specific CD4+ T cells enhanced anti-tumor immunity (5). Though we could recover more BAp:I-Ab-specific T cells in Bim−/− mice, our data do not support that the increased number of these cells correlates with increased functionality. In support of this, immunization of Bim−/− mice led to the same number of BAp:I-Ab-specific T cells as seen in C57BL/6 mice that were immunized in the same way. Thus, central tolerance may remove BAp:I-Ab-specific T cells but other mechanisms also exist that limit functional immune responses to cancer.
A key question in cancer immunotherapy is whether T cell tolerance can be prevented or reversed. Our findings demonstrate that BCR-ABL specific tolerance can be limited by Treg depletion, which correlates with increased expression of Ly6C on the BAp:I-Ab-specific T cells. Ly6C is expressed on Th1-like effector cells that produce high levels of IFNγ, IL2, and Granzyme B (33). Importantly, Th1 cells are classically thought to be critical for tumor control (41). Thus our data suggest that LY6C+FOXP3− BAp:I-Ab-specific T cells are useful biomarkers of a robust immune response to leukemia.
Our studies clearly demonstrate that Treg induction is an important mechanism for suppressing anti-leukemia immune responses. We identified TGFβ1 production and MHC-II presentation by leukemic cells as key molecular mechanisms that allowed conversion of BAp:I-Ab-specific T cells into the Treg lineage. We uncovered leukemic cells and BAp:I-Ab-specific T cells as two prominent sources of TGFβ1. Genetic ablation of MHC-II or antibody-based depletion of TGFβ resulted in decreased conversion of BAp:I-Ab-specific T cells into Tregs and a corresponding increase in proliferation of BAp:I-Ab-specific T cells in response to leukemia. Thus, our data support the concept that TGFβ1 and antigen presentation by leukemic cells are critical components in the extensive conversion of BAp:I-Ab-specific T cells into the Treg lineage (Fig 7G,H).
Temporal regulation of TGFβ1 production by leukemic cells and BAp:I-Ab-specific T cells remains to be ascertained in this model. One likely model is that initial TGFβ1 production by the leukemic cells induces BAp:I-Ab-specific Treg conversion, which then produce TGFβ1 as well. In this model, leukemic cell-derived antigen presentation and TGFβ1 initiate Treg conversion, and BAp:I-Ab-specific T cell-derived TGFβ1 maintains Treg polarization. Importantly, such a mechanism would allow BAp:I-Ab-specific Tregs to suppress proliferation of other anti-leukemia T cells via TGFβ1 (such as GFP:I-Ab-specific T cells in our model).
Regardless of the temporal expression patterns of TGFβ1, the data presented herein point to the importance of the leukemia microenvironment. Specifically, the leukemia microenvironment has copious amounts of TGFβ, as well as antigen presentation by the leukemic cells, which protects the leukemia from the host immune response. This model requires leukemic cells to present antigen on MHC-II as well as produce TGFβ1, thus suggesting that these two mechanisms may synergize to induce immune suppression.
Our findings provide evidence that Treg depletion can modestly extend survival of leukemic mice and that this correlates with increased priming and Th1-skewing of BAp:I-Ab-specific T cells. Finally, even though current immunotherapy is largely focused on tumor neo-antigens, our studies support the notion that cross-reactive T cells can play a role in the immune response to leukemia. Therefore our work may inform future therapeutic options which are aimed at reducing the number or function of Treg cells present in cancer.
Acknowledgments
Funding
NIH P30CA77598 supports the University of Minnesota Flow Cytometry Resource. LSM is supported by a NIH fellowship F31CA183226. KEV was supported by T32CA009138. KEP is supported by the Robertson Foundation/Cancer Research Institute Irvington Fellowship. MKJ is supported by PO1AI35296, and MAF is supported by R01CA151845, R01CA154998, R56AI113138, and R01CA185062 from the National Institutes of Health, the University of Minnesota Masonic Cancer Center, and by a Leukemia & Lymphoma Scholar Award.
We thank Gregory Hubbard, Alyssa Kne, Christopher Reis, Amy Mack and Emilea Sykes for assistance with mouse husbandry, Justin Taylor for experimental design, Kristin Hogquist for Bim−/− mice, and Dan Mueller for Foxp3DTR/DTR mice, Markus Muschen for BCR-ABL-IRES-GFP retrovirus, Christine Henzler for statistical advice, and Lynn Heltemes-Harris, Shawn Mahmud, and Dan Kaplan for commentary on the manuscript.
References
- 1.Snyder A, Makarov V, Merghoub T, Yuan J, Zaretsky MJ, Desrichard A, Walsh LA, Postow MA, Wong P, Ho TS, Hollmann TJ, Bruggeman C, Kannan K, Li Y, Elipenahli C, Liu C, Harbison CT, Wang L, Ribas A, Wolchok JD, Chan TA. Genetic basis for clinical response to CTLA-4 blockade in melanoma. The New England journal of medicine. 2014;371:2189–2199. doi: 10.1056/NEJMoa1406498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, Lee W, Yuan J, Wong P, Ho TS, Miller ML, Rekhtman N, Moreira AL, Ibrahim F, Bruggeman C, Gasmi B, Zappasodi R, Maeda Y, Sander C, Garon EB, Merghoub T, Wolchok JD, Schumacher TN, Chan TA. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science (New York, NY) 2015 doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Segal NH, Parsons DW, Peggs KS, Velculescu V, Kinzler KW, Vogelstein B, Allison JP. Epitope landscape in breast and colorectal cancer. Cancer research. 2008;68:889–892. doi: 10.1158/0008-5472.CAN-07-3095. [DOI] [PubMed] [Google Scholar]
- 4.Robbins PF, Lu YC, El-Gamil M, Li YF, Gross C, Gartner J, Lin JC, Teer JK, Cliften P, Tycksen E, Samuels Y, Rosenberg SA. Mining exomic sequencing data to identify mutated antigens recognized by adoptively transferred tumor-reactive T cells. Nature medicine. 2013;19:747–752. doi: 10.1038/nm.3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tran E, Turcotte S, Gros A, Robbins PF, Lu YC, Dudley ME, Wunderlich JR, Somerville RP, Hogan K, Hinrichs CS, Parkhurst MR, Yang JC, Rosenberg SA. Cancer immunotherapy based on mutation-specific CD4+ T cells in a patient with epithelial cancer. Science (New York, NY) 2014;344:641–645. doi: 10.1126/science.1251102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matsushita H, Vesely MD, Koboldt DC, Rickert CG, Uppaluri R, Magrini VJ, Arthur CD, White JM, Chen YS, Shea LK, Hundal J, Wendl MC, Demeter R, Wylie T, Allison JP, Smyth MJ, Old LJ, Mardis ER, Schreiber RD. Cancer exome analysis reveals a T-cell-dependent mechanism of cancer immunoediting. Nature. 2012;482:400–404. doi: 10.1038/nature10755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gubin MM, Zhang X, Schuster H, Caron E, Ward JP, Noguchi T, Ivanova Y, Hundal J, Arthur CD, Krebber WJ, Mulder GE, Toebes M, Vesely MD, Lam SS, Korman AJ, Allison JP, Freeman GJ, Sharpe AH, Pearce EL, Schumacher TN, Aebersold R, Rammensee HG, Melief CJ, Mardis ER, Gillanders WE, Artyomov MN, Schreiber RD. Checkpoint blockade cancer immunotherapy targets tumour-specific mutant antigens. Nature. 2014;515:577–581. doi: 10.1038/nature13988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Linnemann C, van Buuren MM, Bies L, Verdegaal EM, Schotte R, Calis JJ, Behjati S, Velds A, Hilkmann H, Atmioui DE, Visser M, Stratton MR, Haanen JB, Spits H, van der Burg SH, Schumacher TN. High-throughput epitope discovery reveals frequent recognition of neo-antigens by CD4+ T cells in human melanoma. Nature medicine. 2015;21:81–85. doi: 10.1038/nm.3773. [DOI] [PubMed] [Google Scholar]
- 9.Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Borresen-Dale AL, Boyault S, Burkhardt B, Butler AP, Caldas C, Davies HR, Desmedt C, Eils R, Eyfjord JE, Foekens JA, Greaves M, Hosoda F, Hutter B, Ilicic T, Imbeaud S, Imielinski M, Jager N, Jones DT, Jones D, Knappskog S, Kool M, Lakhani SR, Lopez-Otin C, Martin S, Munshi NC, Nakamura H, Northcott PA, Pajic M, Papaemmanuil E, Paradiso A, Pearson JV, Puente XS, Raine K, Ramakrishna M, Richardson AL, Richter J, Rosenstiel P, Schlesner M, Schumacher TN, Span PN, Teague JW, Totoki Y, Tutt AN, Valdes-Mas R, van Buuren MM, van ‘t Veer L, Vincent-Salomon A, Waddell N, Yates LR, Zucman-Rossi J, Futreal PA, McDermott U, Lichter P, Meyerson M, Grimmond SM, Siebert R, Campo E, Shibata T, Pfister SM, Campbell PJ, Stratton MR. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rowley JD. Chromosome translocations: dangerous liaisons revisited. Nature reviews Cancer. 2001;1:245–250. doi: 10.1038/35106108. [DOI] [PubMed] [Google Scholar]
- 11.Bocchia M, Korontsvit T, Xu Q, Mackinnon S, Yang SY, Sette A, Scheinberg DA. Specific human cellular immunity to bcr-abl oncogene-derived peptides. Blood. 1996;87:3587–3592. [PubMed] [Google Scholar]
- 12.Riva G, Luppi M, Barozzi P, Quadrelli C, Basso S, Vallerini D, Zanetti E, Morselli M, Forghieri F, Maccaferri M, Volzone F, Del Giovane C, D’Amico R, Locatelli F, Torelli G, Comoli P, Potenza L. Emergence of BCR-ABL-specific cytotoxic T cells in the bone marrow of patients with Ph+ acute lymphoblastic leukemia during long-term imatinib mesylate treatment. Blood. 2010;115:1512–1518. doi: 10.1182/blood-2009-06-230391. [DOI] [PubMed] [Google Scholar]
- 13.Boulos N, Mulder HL, Calabrese CR, Morrison JB, Rehg JE, Relling MV, Sherr CJ, Williams RT. Chemotherapeutic agents circumvent emergence of dasatinib-resistant BCR-ABL kinase mutations in a precise mouse model of Philadelphia chromosome-positive acute lymphoblastic leukemia. Blood. 2011;117:3585–3595. doi: 10.1182/blood-2010-08-301267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kamijo T, Zindy F, Roussel MF, Quelle DE, Downing JR, Ashmun RA, Grosveld G, Sherr CJ. Tumor suppression at the mouse INK4a locus mediated by the alternative reading frame product p19ARF. Cell. 1997;91:649–659. doi: 10.1016/s0092-8674(00)80452-3. [DOI] [PubMed] [Google Scholar]
- 15.Barnden MJ, Allison J, Heath WR, Carbone FR. Defective TCR expression in transgenic mice constructed using cDNA-based alpha- and beta-chain genes under the control of heterologous regulatory elements. Immunology and cell biology. 1998;76:34–40. doi: 10.1046/j.1440-1711.1998.00709.x. [DOI] [PubMed] [Google Scholar]
- 16.McSorley SJ, Asch S, Costalonga M, Reinhardt RL, Jenkins MK. Tracking salmonella-specific CD4 T cells in vivo reveals a local mucosal response to a disseminated infection. Immunity. 2002;16:365–377. doi: 10.1016/s1074-7613(02)00289-3. [DOI] [PubMed] [Google Scholar]
- 17.Kim JM, Rasmussen JP, Rudensky AY. Regulatory T cells prevent catastrophic autoimmunity throughout the lifespan of mice. Nature immunology. 2007;8:191–197. doi: 10.1038/ni1428. [DOI] [PubMed] [Google Scholar]
- 18.Bouillet P, Metcalf D, Huang DC, Tarlinton DM, Kay TW, Kontgen F, Adams JM, Strasser A. Proapoptotic Bcl-2 relative Bim required for certain apoptotic responses, leukocyte homeostasis, and to preclude autoimmunity. Science (New York, NY) 1999;286:1735–1738. doi: 10.1126/science.286.5445.1735. [DOI] [PubMed] [Google Scholar]
- 19.Hogquist KA, Jameson SC, Heath WR, Howard JL, Bevan MJ, Carbone FR. T cell receptor antagonist peptides induce positive selection. Cell. 1994;76:17–27. doi: 10.1016/0092-8674(94)90169-4. [DOI] [PubMed] [Google Scholar]
- 20.Williams RT, Roussel MF, Sherr CJ. Arf gene loss enhances oncogenicity and limits imatinib response in mouse models of Bcr-Abl-induced acute lymphoblastic leukemia. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:6688–6693. doi: 10.1073/pnas.0602030103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pear WS, Miller JP, Xu L, Pui JC, Soffer B, Quackenbush RC, Pendergast AM, Bronson R, Aster JC, Scott ML, Baltimore D. Efficient and rapid induction of a chronic myelogenous leukemia-like myeloproliferative disease in mice receiving P210 bcr/abl-transduced bone marrow. Blood. 1998;92:3780–3792. [PubMed] [Google Scholar]
- 22.Heltemes-Harris LM, Willette MJ, Ramsey LB, Qiu YH, Neeley ES, Zhang N, Thomas DA, Koeuth T, Baechler EC, Kornblau SM, Farrar MA. Ebf1 or Pax5 haploinsufficiency synergizes with STAT5 activation to initiate acute lymphoblastic leukemia. The Journal of experimental medicine. 2011;208:1135–1149. doi: 10.1084/jem.20101947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moon JJ, Chu HH, Hataye J, Pagan AJ, Pepper M, McLachlan JB, Zell T, Jenkins MK. Tracking epitope-specific T cells. Nature protocols. 2009;4:565–581. doi: 10.1038/nprot.2009.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pauken KE, Linehan JL, Spanier JA, Sahli NL, Kalekar LA, Binstadt BA, Moon JJ, Mueller DL, Jenkins MK, Fife BT. Cutting Edge: Type 1 Diabetes Occurs despite Robust Anergy among Endogenous Insulin-Specific CD4 T Cells in NOD Mice. Journal of immunology (Baltimore, Md : 1950) 2013;191:4913–4917. doi: 10.4049/jimmunol.1301927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schenkel JM, Fraser KA, Vezys V, Masopust D. Sensing and alarm function of resident memory CD8(+) T cells. Nature immunology. 2013;14:509–513. doi: 10.1038/ni.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mullighan CG, Williams RT, Downing JR, Sherr CJ. Failure of CDKN2A/B (INK4A/B-ARF)-mediated tumor suppression and resistance to targeted therapy in acute lymphoblastic leukemia induced by BCR-ABL. Genes & development. 2008;22:1411–1415. doi: 10.1101/gad.1673908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nelson RW, Beisang D, Tubo NJ, Dileepan T, Wiesner DL, Nielsen K, Wuthrich M, Klein BS, Kotov DI, Spanier JA, Fife BT, Moon JJ, Jenkins MK. T cell receptor cross-reactivity between similar foreign and self peptides influences naive cell population size and autoimmunity. Immunity. 2015;42:95–107. doi: 10.1016/j.immuni.2014.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davey GM, Kurts C, Miller JF, Bouillet P, Strasser A, Brooks AG, Carbone FR, Heath WR. Peripheral deletion of autoreactive CD8 T cells by cross presentation of self-antigen occurs by a Bcl-2-inhibitable pathway mediated by Bim. The Journal of experimental medicine. 2002;196:947–955. doi: 10.1084/jem.20020827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stritesky GL, Xing Y, Erickson JR, Kalekar LA, Wang X, Mueller DL, Jameson SC, Hogquist KA. Murine thymic selection quantified using a unique method to capture deleted T cells. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:4679–4684. doi: 10.1073/pnas.1217532110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.King C, Garza EN, Mazor R, Linehan JL, Pastan I, Pepper M, Baker D. Removing T-cell epitopes with computational protein design. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:8577–8582. doi: 10.1073/pnas.1321126111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kurotaki D, Kon S, Bae K, Ito K, Matsui Y, Nakayama Y, Kanayama M, Kimura C, Narita Y, Nishimura T, Iwabuchi K, Mack M, van Rooijen N, Sakaguchi S, Uede T, Morimoto J. CSF-1-dependent red pulp macrophages regulate CD4 T cell responses. Journal of immunology (Baltimore, Md : 1950) 2011;186:2229–2237. doi: 10.4049/jimmunol.1001345. [DOI] [PubMed] [Google Scholar]
- 32.Yamanouchi S, Kuwahara K, Sakata A, Ezaki T, Matsuoka S, Miyazaki J, Hirose S, Tamura T, Nariuchi H, Sakaguchi N. A T cell activation antigen, Ly6C, induced on CD4+ Th1 cells mediates an inhibitory signal for secretion of IL-2 and proliferation in peripheral immune responses. European journal of immunology. 1998;28:696–707. doi: 10.1002/(SICI)1521-4141(199802)28:02<696::AID-IMMU696>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 33.Marshall HD, Chandele A, Jung YW, Meng H, Poholek AC, Parish IA, Rutishauser R, Cui W, Kleinstein SH, Craft J, Kaech SM. Differential expression of Ly6C and T-bet distinguish effector and memory Th1 CD4(+) cell properties during viral infection. Immunity. 2011;35:633–646. doi: 10.1016/j.immuni.2011.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hale JS, Youngblood B, Latner DR, Mohammed AU, Ye L, Akondy RS, Wu T, Iyer SS, Ahmed R. Distinct memory CD4+ T cells with commitment to T follicular helper- and T helper 1-cell lineages are generated after acute viral infection. Immunity. 2013;38:805–817. doi: 10.1016/j.immuni.2013.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mahmud SA, Manlove LS, Farrar MA. Interleukin-2 and STAT5 in regulatory T cell development and function. Jak-Stat. 2013;2:e23154. doi: 10.4161/jkst.23154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huber S, Schramm C, Lehr HA, Mann A, Schmitt S, Becker C, Protschka M, Galle PR, Neurath MF, Blessing M. Cutting edge: TGF-beta signaling is required for the in vivo expansion and immunosuppressive capacity of regulatory CD4+CD25+ T cells. Journal of immunology (Baltimore, Md : 1950) 2004;173:6526–6531. doi: 10.4049/jimmunol.173.11.6526. [DOI] [PubMed] [Google Scholar]
- 37.Maude SL, Frey N, Shaw PA, Aplenc R, Barrett DM, Bunin NJ, Chew A, Gonzalez VE, Zheng Z, Lacey SF, Mahnke YD, Melenhorst JJ, Rheingold SR, Shen A, Teachey DT, Levine BL, June CH, Porter DL, Grupp SA. Chimeric antigen receptor T cells for sustained remissions in leukemia. The New England journal of medicine. 2014;371:1507–1517. doi: 10.1056/NEJMoa1407222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wolchok JD, Kluger H, Callahan MK, Postow MA, Rizvi NA, Lesokhin AM, Segal NH, Ariyan CE, Gordon RA, Reed K, Burke MM, Caldwell A, Kronenberg SA, Agunwamba BU, Zhang X, Lowy I, Inzunza HD, Feely W, Horak CE, Hong Q, Korman AJ, Wigginton JM, Gupta A, Sznol M. Nivolumab plus ipilimumab in advanced melanoma. The New England journal of medicine. 2013;369:122–133. doi: 10.1056/NEJMoa1302369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ma J, Li M, Mei L, Zhou Q, Liu L, Yu X, Che G. Double suicide genes driven by kinase domain insert containing receptor promoter selectively kill human lung cancer cells. Genetic vaccines and therapy. 2011;9:6. doi: 10.1186/1479-0556-9-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schreiber RD, Old LJ, Smyth MJ. Cancer immunoediting: integrating immunity’s roles in cancer suppression and promotion. Science (New York, NY) 2011;331:1565–1570. doi: 10.1126/science.1203486. [DOI] [PubMed] [Google Scholar]
- 41.Braumuller H, Wieder T, Brenner E, Assmann S, Hahn M, Alkhaled M, Schilbach K, Essmann F, Kneilling M, Griessinger C, Ranta F, Ullrich S, Mocikat R, Braungart K, Mehra T, Fehrenbacher B, Berdel J, Niessner H, Meier F, van den Broek M, Haring HU, Handgretinger R, Quintanilla-Martinez L, Fend F, Pesic M, Bauer J, Zender L, Schaller M, Schulze-Osthoff K, Rocken M. T-helper-1-cell cytokines drive cancer into senescence. Nature. 2013;494:361–365. doi: 10.1038/nature11824. [DOI] [PubMed] [Google Scholar]