Abstract
Earlier studies reported that a cell membrane protein Annexin A2 (AnxA2) plays multiple roles in the development, invasion and metastasis of cancer. Recent studies have demonstrated that AnxA2 also functions in immunity against infection, but the underlying mechanism remains largely elusive. Using a mouse infection model, we now reveal a crucial role of AnxA2 in host defense against Pseudomonas aeruginosa (Pa), as anxa2−/− mice manifested severe lung injury, systemic dissemination, and increased mortality compared to wild-type (WT) littermates. In addition, anxa2−/− mice exhibited elevated inflammatory cytokines (TNF-α, IL-6, IL-1β and IFN-γ), decreased bacterial clearance by macrophages, and increased superoxide release in the lung. We further identified an unexpected molecular interaction between AnxA2 and Fam13A (Family with sequence similarity 13, member A), which activated Rho GTPase. P. aeruginosa infection induced autophagosome formation by inhibiting Akt1 and mTOR. Our results indicate that AnxA2 regulates autophagy and thereby contributing to host immunity against bacteria through Akt1-mTOR-ULK1/2 signaling pathway.
Keywords: AnxA2, Pseudomonas aeruginosa, Autophagy, Fam13A, Akt1-mTOR-ULK1/2 signaling pathway
Introductions
Annexin A2 (AnxA2), a member of the Annexins family, is expressed in various human cells, such as endothelial cells, mononuclear cells, macrophages, marrow cells and some tumor cells (1). Accumulating evidence indicates that AnxA2 plays multiple roles in signal transduction, cell proliferation, differentiation, apoptosis, endocytosis and exocytosis, and inflammation (1, 2). AnxA2 down-regulation is associated with the occurrence, invasion and metastasis of cancer, while its up-regulation is related to the development, invasion, metastasis and drug resistance of hepatocellular carcinoma, colorectal cancer, breast cancer, acute promyelocytic leukemia, and renal cell carcinoma (3). In addition, AnxA2 has been shown to play diverse roles in pulmonary diseases, such as non-small cell lung cancer, adenocarcinoma, chronic obstructive pulmonary disease (COPD) and chronic inflammatory diseases (4), thus AnxA2 may be a novel diagnostic serum biomarker for lung cancer and other diseases (5). For instance, AnxA2 is involved in p53-induced apoptosis in lung cancer (6). Increasing secretion of collagen VI and pulmonary elasticity of bronchial epithelial cells (7) along with increased AnxA2 on lung epithelial cell surface were recognized by severe acute respiratory syndrome-associated coronavirus spike domain 2 antibodies (8). An epithelial cell surface AnxA2/p11 complex is also required for efficient invasion of Salmonella typhimurium (9). Evidence also suggests that AnxA2 directly interacts with Rab14 in alveolar type II epithelial cells (10). In addition, AnxA2 inducer or inhibitor dramatically modulates AnxA2-related cell functions. Investigators have demonstrated that cationic lipid guided AnxA2 shRNA attenuates tumor growth and metastasis in a mouse lung cancer stem cell model (11), while IFN-γ stimulates AnxA2 expression in lung epithelial cells to enhance apoptosis (8).
Pseudomonas aeruginosa (Pa) is an opportunistic bacterial pathogen causing acute and chronic pulmonary infection in immunocompromised people (12), such as patients with cystic fibrosis (CF) and COPD; and most of the morbidity and pathophysiology associated with these diseases is due to innate hyper-susceptibility to bacterial infection (13). Innate immunity, primarily through inflammatory cytokine production, cellular recruitment, and phagocytic clearance by neutrophils and macrophages, is key to host control of Pa infection (14). Recently, macroautophagy (hereafter referred to as autophagy) in macrophages has been involved in host defense, and directly impacts immunity and inflammatory response (15). Autophagy participates in the elimination of invasive bacteria through autolysosome by functioning as downstream factors of pattern recognition receptors (PPRs, such as TLRs) (16) and pathogen-associated molecular patterns (PAMPs) (17).
Our previous work indicated that autophagy plays a role in immune defense against P. aeruginosa invasion (15). However, roles of AnxA2 in host defense of Pa infection have not been defined. Investigation indicated that cell surface AnxA2 serves as a receptor for Pa in mammalian epithelial cells for bacterial internalization into host cells (18). Evidence also suggests that AnxA2 participates in endocytosis and EGFR-mediated signal transduction in macrophages (19), because suppression of AnxA2 impairs phagocytic ability of peritoneal macrophages (20). In this study, we hypothesized that AnxA2 may play roles in host defense against Pa infection. To this end, we used wild-type (WT) and anxa2−/− mice to investigate the role and underlying mechanism of AnxA2 in Pa infection, particularly the participation of various autophagy related factors. We demonstrate that AnxA2 regulates autophagosome formation after PAO1 infection through Akt1-mTOR-ULK1/2 signaling pathway.
Material and methods
Mice
C57BL/6 female mice (6–8 weeks) were obtained from the Jackson Laboratory (21), and anxa2−/− mice that are constructed based on C57BL/6J mice were kindly provided by Dr. K. Hajjar of Cornell University (22). Exon 3 and 4 of anxa2 were disrupted with a cassette containing neomycin phosphotransferase driven by the phosphoglucokinase promoter, to generate anxa2−/− mice (22). Animals were kept in a specific pathogen-free facility of University of North Dakota (23). All animal studies were approved by the UND IACUC committee and performed in accordance with the animal care and institutional guidelines (IACUC approval #1204-4). The animal experimental procedures including treatment, care, and endpoint choice were followed the ARRIVE reporting guidelines.
Primary cells and cell lines
Mice were sacrificed and the thoracic cavity and trachea were dissected. A small incision was made in the trachea via 1-mL syringe with an angiocath (BD Biosciences, Franklin Lakes, NJ), and recovered to a sterile tube. The lungs were lavaged three times with 1 mL of phosphate-buffered saline (PBS) containing 1% fetal bovine serum (FBS, Life Technologies, Grand Island, NY). The retained bronchoalveolar lavage (BAL) fluid was centrifuged at 600 g for 5 min at 4 °C. The cell pellets were resuspended in RPMI 1640 medium (Life Technologies) supplemented with 10% FBS and incubated on culture plate for 1 h at 37 °C/5% CO2 incubator to allow attachment of macrophages. Non-adherent cells were removed by washing with normal saline. Murine MLE-12 lung type II epithelial cells and MH-S alveolar macrophage cells were obtained from American Type Culture Collection (Manassas, VA) and cultured following the manufacturer’s instructions (24).
Bacteria preparation and infection experiments
The Pa WT strain, PAO1, was kindly provided by Dr. S. Lory (Harvard University) (25). PAK and PAO1-EGFP were obtained from Dr. G. Pier (Harvard University) (26). PAO1 Xen-41 was obtained from PerkinElmer-Caliper (Waltham, MA). K. pneumonia was provided from Dr. V. Miller (University of North Carolina) (27). E. coli DH5-α was obtained from New England Biolabs (Ipswich, MA). Bacteria were grown for about 16 h in lysogeny broth (LB) at 37 °C with 220 rpm shaking. The bacteria were pelleted by centrifugation at 5000 g. Various mammalian cells were changed to antibiotic-free medium and infected by bacteria in an MOI of 20:1 bacteria-cell ratio. To control for LC3 degradation, 5 mM NH4Cl was added into the medium for 2 h of bacterial infection in indicated in vitro LC3 immunoblotting experiments (28). This short time culture with low NH4Cl concentration did not cause significant cellular alterations. Mouse was anesthetized with 45 mg/kg ketamine and intranasally instilled 0.5 × 107 clonal formation units (CFU) of PAO1 in 50 μL PBS (six mice/group) (29). Mice were monitored for symptoms and euthanized when they were moribund.
In vivo imaging
At various time points of post-infection, the whole body of Pa Xen-41 infected mice was imaged under an IVIS XRII system (PerkinElmer, Waltham, MA) following the user guides provided by PerkinElmer-Caliper (30).
RNA isolation and qRT-PCR
RNA was isolated from indicated primary alveolar macrophages (AM). A 50-ng DNA-free RNA was used for the first strand of cDNA synthesis using a SuperScript III first-strand synthesis system (Life Technologies). qRT-PCR was performed using the iTag Universal SYBR Green Supermix (Bio-Rad, Hercules, CA) and gene-specific primers (SI Table S1, synthesized in Integrated DNA Technologies, Coralville, IA), in a CFX Connect™ Real-time PCR Detection System (Bio-Rad). Relative transcript levels were first normalized CT values to GAPDH, and then normalized to the indicated control (2−ΔΔCT) (31).
Plasmid construction
Fam13A gene was amplified from MH-S genome DNA with specific primers (SI Table S1, synthesized in Integrated DNA Technologies) by PCR and cloned into the BamH I and Hind III sites of pcDNA3.1 vector (Addgene, Cambridge, MA). Constructed plasmids were electroporated into DH5α using an Electroporator 2510 system (settings: 25 μF, 200 Ω, 2.5 kV; Eppendorf, Hauppauge, NY). Transformants were selected and maintained in an LB medium containing 100 mM Ampicillin (Sigma-Aldrich, St. Louis, MO). All of the nuclease, polymerase and ligase used in molecular cloning were bought from New England Biolabs Inc.
Transfection of siRNA, plasmids, activators and inhibitors
AnxA2, Fam13A, Atg5, Atg7, Beclin1 and siNC siRNAs were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). MH-S cells were transfected with siRNA (5 pM), LC3-RFP G120A, pcDNA3.1 and pcDNA3.1-Fam13A (100 ng) plasmids using LipofectAmine 2000 (Life Technologies) for 24 h following the manufacture’s instruction. In indicated case, anxa2−/− AM cells were treated with 5 μM autophagy activator Trehalose (Tre, Sigma-Aldrich), while WT AM cells were treated with 5 μM 3-Methyladenine autophagy inhibitor (3-MA, Sigma-Aldrich) for 2 h before and during PAO1 infection as indicated.
Bacterial burden assay
AM cells from BAL fluid and ground lung, spleen, liver and kidney tissues, were homogenized with PBS and spread on LB dishes to enumerate bacterial numbers. The dishes were cultured in a 37 °C incubator overnight, and colonies were counted. Duplicates were done for each sample and control (32).
Nitroblue tetrazolium assay (NBT)
This assay is based the color change of NBT dye upon reduction by released superoxide. AM cells from BAL were grown in a 96-well plate in serum-containing medium at 37 °C for 4 hours and NBT (1 μg/mL) dye (Sigma-Aldrich) to each well. Cells were incubate for another 1 hour or until color developed. The dye is yellow and gives a blue color formazan product upon reduction by superoxide. The reduction was terminated by adding 100 μL of stop solution (10% DMSO; 10% SDS in 50 mM HEPES buffer). The plate was kept at room temperature overnight for complete dissolution of formazan and absorbance at 560 nm was recorded using a multiscan plate reader to quantify the concentration of superoxide anion (24). Triplicates were done for each sample and control.
Dihydrodichlorofluorescein diacetate (H2DCF) assay
Dihydrodichlorofluorescein diacetate dye (Life Technologies) does not normally fluoresce but emits green fluorescence upon reaction with superoxide inside cells. AM cells were treated as above and an equal amount of dye was added. After 10-min incubation, fluorescence was measured using a fluorometer, using 485 nm excitation and a 528 nm emission filter (33).
3-(4,5-Dimethylthiazol-2-yl)-2,5-dimethyltetrazolium bromide (MTT) assay
This assay measures the color change of 3-(4,5-Dimethylthiazol-2-yl)-2,5-dimethyltetrazolium bromide (MTT, Sigma-Aldrich) upon reduction by enzymes to assess the viability of cells. Cells were treated as above, and equal amount of dye was added. After 1 hour incubation, reaction was stopped by stop solution and left at room temperature overnight for complete dissolution of formazan and absorbance at 560 nm was recorded using a multiscan plate reader to quantify the concentration of superoxide anion (34).
Mitochondrial potential assay
JC-1 mitochondrial membrane potential assay kit (Cayman chemical, Ann Arbor, MI) was used for this assay, following the manufacture’s instruction. The cytofluorimetric, lipophilic cationic dye, 5,5′,6,6′-tetrachloro-1,1′3,3-tetracthylbenzimidazolylcarbocyanine idodide (JC-1), can selectively enter into mitochondria and reversibly change color from green to red as the membrane potential increases. AM cells were treated as above and an equal amount of dye was added. After 30-min incubation, fluorescence was measured using a fluorometer, using 560 nm excitation and a 595 nm emission filter for detecting healthy cells, and 485 nm excitation and a 535 nm emission filter for detecting dead cells (35).
Histological analysis
Lung tissues of three independent mice were fixed in 10% formalin (Sigma-Aldrich) for 24 h and then embedded in paraffin using a routine histologic procedure. Four-micrometer sections were cut, stained by standard H&E, and examined for differences in morphology postinfection (36).
Inflammatory cytokine profiling
Cytokine concentrations of TNF-α, IL-6, IL-1β and IFN-γ were measured by ELISA kits stained from eBioscience Co. (San Diego, CA), in samples of BAL fluid collected at the indicated times after infection. BAL fluids were collected and 100 μL aliquots of samples were added to the coated microtiter wells. The cytokine concentrations were determined with corresponding detection HRP-conjugated antibodies. The values were read at 450 nm (36).
CFU count assay
Phagocytosis and intracellular killing were assessed by a CFU assay as previously described (37). Indicated AM cells were challenged with indicated bacteria at a ratio of 20:1. The numbers of internalized and killed bacteria were assessed after 1 or 2 hours of incubation. After 1 hour, gentamicin was added to the medium at 300 μg/mL for 30 minutes to kill extracellular bacteria. Cells were first counted by hemocytometer (Sigma-Aldrich). Then cells were lysed, and bacterial CFU were determined. Phagocytosed bacteria was calculated as CFU (1h)/Cell number. A second series of internalization assays was run in parallel to determine the number of viable bacteria following 2 hours of incubation. After the same treatment to remove extracellular bacteria, cells were incubated for another 1 hour and lysed for analysis of intracellular bacterial CFU. The killing efficiency was calculated as [CFU (1 h) − CFU (2 h)]/CFU (1 h) and normalized to the control group (37).
Phagocytosis assay
Indicated AMs were challenged with PAO1 EGFP at an MOI of 20:1 for 1 hour and then stained with Cholera toxin B chain (CTB, Sigma-Aldrich) to track membrane sphingolipid-rich lipid rafts for 30 minutes. Cells were observed under an LSM 510 Meta Confocal Microscope (Carl Zeiss Micro Imaging, Thornwood, NY). After observation, cells were washed three times with PBS and centrifuged to remove extracellular bacteria, and then lysed. Intracellular bacteria were incubated on LB dishes and analyzed using an IVIS XRII system (30).
Autophagy-based PCR Primers assay
RNA was isolated from AnxA2-silenced or negative MH-S cells after 4 h PAO1 postinfection. DNA-free RNA (100-ng) was used for the first strand of cDNA synthesis using a SuperScript III first-strand synthesis system (Life Technologies). PCR primers assay was performed using the iTag Universal SYBR Green Supermix and gene specific primers that attached on the bottom of the Autophagy M384 predesigned 384 well panel (Bio-Rad), in a CFX Connect™ Real-time PCR Detection System (Bio-Rad). PCR primers assay data were analyzed using PrimePCR analysis software (Bio-Rad).
Immunoblotting
Mouse monoclonal Abs against LC3, β-Actin, Akt1, and rabbit polyclonal Abs against AnxA2, mTOR, ULK1, p-Akt1, and p-mTOR were obtained from Santa Cruz Biotechnology. Rabbit polyclonal Abs against Fam13A was obtained from Proteintech Group (Chicago, IL), while p-ULK1 (ser317) was obtained from Cell Signaling Technology (Danvers, MA). The samples derived from cells and lung homogenates were lysed in RIPA buffer, separated by electrophoresis on 12% SDS-PAGE gels and transferred to nitrocellulose transfer membranes (GE Amersham Biosciences, Pittsburgh, PA). Proteins were detected by western blotting using primary Abs at a concentration of 1/200 (Santa Cruz Biotechnology) or 1/1000 (Cell Signaling Technology) and were incubated overnight. Labelling of the first Abs was detected using relevant secondary Abs conjugated to HRP (Santa Cruz Biotechnology), detected using ECL regents (Santa Cruz Biotechnology) (38). Gel bands were quantified by Quantity One software (Bio Rad) and error bar represents for three independent immunoblotting assays (38). Phosphorylated and total protein levels were determined and quantified by three independent successive immunoblotting membranes.
Immunoprecipitation
To obtain whole-cell lysates, indicated AM cells and MH-S cells were homogenized in lysis buffer containing phosphatase inhibitor (1:10000) and protease inhibitors (1:50, ThermoFisher Scientific, Waltham, MA). Then total cell lysate were mixed with indicated IP antibody, which were coupled to agarose beads (A/G, 50:50, ThermoFisher Scientific). Immunoprecipitates were separated by SDS-PAGE and transferred to nitrocellulose transfer membranes. Proteins were detected using detective Abs and were incubated overnight. Labelling of the first Abs was detected using relevant secondary Abs conjugated to HRP and detected using ECL regents (23).
LC3 puncta observation
Indicated AM cells and MH-S cells were re-transfected with LC3-RFP G120A plasmids for 24 h. Cells were infected with PAO1-EGFP at a MOI of 20:1 for 2 h. Cells were observed under an LSM 510 Meta Confocal Microscope. LC3 puncta values are derived from 100 cells/sample (33).
Immunostaining
AM cells isolated from WT mice after 24 h post PAO1 infection and control mice. Cells were individually incubated with both primary anti-AnxA2 Ab and anti-Fam13A Ab, and then the second FITC-conjugated antibodies as described (31). Colocalization were observed under an LSM 510 Meta Confocal Microscope.
Rho GTPase activity assay
Rho GTPase activity in indicated AM cells and MH-S cells was detected by using Pierce™ active Rho GTPase pull-down and detection kit (Life Technologies), following the manufacture’s instruction. Briefly, whole cell lysate are prepared from AM cells using the lysis buffer provided in the kit. Lysate (containing active and inactive Rho GTPase) is incubated with the GST-protein binding domain fusion protein from the respective downstream effector protein and glutathione resin. Unbound lysate proteins, including inactive or GDP-bound GTPase, are removed using the spin columns. The active GTPase population is recovered from the glutathione resin using SDS-PAGE loading buffer and analyzed by immunoblotting (39).
Statistical analysis
Most experiments were conducted in triplicate. Differences between 2 groups were compared by one-way ANOVA (Tukey’s post hoc) using GraphPad Prism 5 software, while mice survival rates were calculated using Kaplan-Meier curve (40).
Results
anxa2−/− mice are highly susceptible to PAO1 infection
To investigate the role of AnxA2 in Pa infection process, we intranasally instilled PAO1 (0.5 × 107 CFU/mouse) to anxa2−/− and WT mice (with otherwise similar genetic backgrounds) to establish an acute pneumonia model and compared the survival rates of these two groups of mice (6 mice per group). As shown in Fig. 1A, anxa2−/− mice exhibited increased lethality (50% of anxa2−/− mice died within 38 h postinfection). At 68 h, all anxa2−/− mice died, whereas 80% of WT mice remained alive. This result was shown in Kaplan-Meier survival curves (p = 0.021, log-rank test). To directly examine the in vivo infection process, we also instilled mice with PAO1 Xen-41 (an engineered bacterium emitting bioluminescence for imaging) at the same dose per mouse and observed the pattern of bacterial dissemination. We found that anxa2−/− mice displayed much broader distribution of bioluminescence in the area of thoracic cavity after 2 h postinfection and continuously expanding compared to WT mice (Fig. 1B–C). These results of dynamic studies suggest that AnxA2 contributes to host defense against Pa in pneumonia models by slowing down bacterial dissemination.
Figure 1. anxa2−/− mice showed increased susceptibility and mortality against P. aeruginosa infection.
(A) anxa2−/− and WT mice were intranasally challenged with 0.5 × 107 CFU PAO1/mouse, and then maintained up to 96 h. Kaplan-Meier survival curves were obtained (p = 0.021; 95% confidence interval: 16.9–40.8, log-rank test). (B–C) Whole animal imaging of bioluminescence were obtained using IVIS XRII system at different time points, after anxa2−/− and WT mice challenged with 0.5 × 107 CFU PAO1 Xen-41/mouse (arrows indicating PAO1 spread regions). Data are representative of three independent experiments expressed as means ± SEM (one-way ANOVA with Tukey’s post hoc; *p ≤ 0.05; **p ≤ 0.005).
AnxA2 loss results in increased oxidation, lung injury and inflammatory response
To further analyze the cause of infection lethality in anxa2−/− mice, we examined bacterial burdens in the lung, liver, spleen and kidney, as well as BAL fluid and blood isolated from PAO1-infected mice. Bacterial CFUs significantly increased in the indicated organs of anxa2−/− mice compared to the same organs of WT mice (Fig. 2A–B and SI Fig. 1A–D), demonstrating severe lung injury and pneumonia in AnxA2 deficiency. Increased recruitment of polymorphonuclear neutrophil (PMN) cells into the lung, which is necessary for bacterial clearance, may also contribute to severe lung injury and systemic bacterial infection (41). To test this idea, we quantified PMN infiltration and observed increased PMN in the BAL fluid of anxa2−/− mice when compared to that of WT mice (Fig. 2C).
Figure 2. Increased bacterial burdens, PMN, oxidation injury and intense inflammatory response in lung of anxa2−/− mice.
The anxa2−/− and WT mice were infected with 0.5 × 107 CFU PAO1/mouse for 24 h. (A) Bacterial burdens in the lungs after homogenization in PBS. (B) Bacterial loads in BAL derived from infected mice. (C) PMN cell percentages evaluated in BAL versus total nuclear cells using HEMA-3 staining. (D) and (E) Superoxide production in AM cells was detected by NBT assay at a wavelength of 560 nm, and H2DCF assay at a wavelength of 488 nm, respectively. (F) Mitochondrial potential of AM cells measured by JC-1 fluorescence assay at a wavelength of 532 nm. (G) Lung injury and inflammation assessed by histology. Lungs from indicated mice were embedded in formalin. Sections were analyzed by H&E staining. (H) Inflammatory cytokines in BAL fluids were assessed by ELISA. Data are representative of three independent experiments expressed as means ± SEM (one-way ANOVA with Tukey’s post hoc; *p ≤ 0.05; **p ≤ 0.005).
Phagocyte-released reactive oxygen species (ROS) are crucial for host defense against bacterial infection by either augmenting antibiotic activity or directly participating in bacterial killing in lysosomes (42). However, increased levels of ROS may damage cellular organelles and thereby cause tissue injury (43). To measure oxidative stress, anxa2−/− and WT mice were infected with PAO1, and AM cells were isolated for analysis of oxidation levels. As determined by NBT assay (Fig. 2D), AM cells of anxa2−/− mice showed an approximately 2-fold increase in oxidative stress at 24 h postinfection compared to those of WT mice. This result was further confirmed using H2DCF assay (Fig. 2E), a sensitive fluorescence method for quantifying superoxide (44). To determine the viability of cells after infection, we determined the mitochondrial membrane potential and data showed a decreased mitochondrial membrane potential in AM cells of anxa2−/− mice using JC-1 fluorescence assay (Fig. 2F). Thus our data indicate that increased oxidation resulted in apoptosis of alveolar macrophages.
As a direct indicator of lung injury, lung histology was examined 24 h post-PAO1 infection. Although both anxa2−/− and WT mice showed signs of pneumonia, significant histological alterations were only observed in the lungs of anxa2−/− mice, indicating more severe lung injury in these animals (Fig. 2G) (insets). The insets demonstrated the typical regions with serious tissue damage with broken structure and inflammatory response with PMN penetration. We further measured cytokine concentrations of BAL fluid 24 h post-PAO1 infection for gauging the inflammatory response. In BAL fluid of anxa2−/− mice, TNF-α, IL-6, IL-1β and IFN-γ levels were significantly elevated compared to those in WT mice (Fig. 2H), indicating that lung macrophages might be the main source of increased cytokines. Collectively, these data suggest that the severe lung injury of anxa2−/− mice is caused by excessive ROS accumulation and overly active inflammatory response.
AnxA2 loss results in impaired PAO1-induced autophagy in AM cells
Bacteria burdens of BAL fluid in anxa2−/− mice were much higher than those in WT mice (Fig. 2B), indicating that the ability for bacterial clearance in anxa2−/− mice has been substantially dampened. To confirm this, we first determined the survival of AM cells, an essential condition for maintaining the bacterial clearance function, after 24 h post-PAO1 infection. The result showed a significant decrease in the viability of AM cells in anxa2−/− mice (Fig. 3A) and consistent with this reduced survival rate, the ability for bacterial clearance as quantified by CFU assays was also impaired (Fig. 3B). We then transfected the model murine lung macrophage MH-S cells and the lung epithelia cell MLE-12 cells with small interfering RNA (siRNA) against AnxA2 (siAnxA2) to inhibit AnxA2 expression, and with scrambled siRNA (siNC) as a negative control. After 2 h PAO1 postinfection, similar decreases of both survival and bacterial clearance ability were detected in AnxA2-silenced MH-S cells compared to negative controls (Fig. 3C–D). Note that the survival of AnxA2-silenced MLE-12 cells showed no significant difference from controls (Fig. 3C), indicating that AnxA2 may play more dominant roles in macrophages vs. epithelial cells. To determine whether the role of AnxA2 is limited to Pa or it plays a general role in bacterial defense, other bacterial strains and species were also investigated. When infected with Pa strains PAK and PA14, as well as K. pneumoniae and E. coli for 2 h, both the survival and bacterial clearance ability were significantly decreased in AnxA2-silenced MH-S cells (SI Fig. 1), suggesting that the role of AnxA2 may extend to a broad range of species of Gram-negative bacteria.
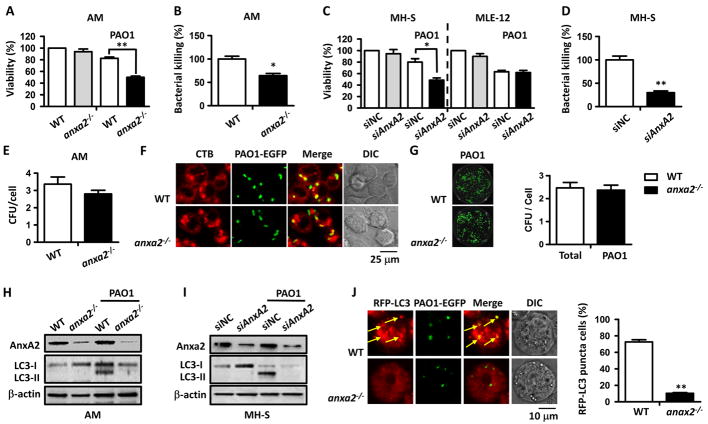
Figure 3. AnxA2 deficiency blunted PAO1-induced autophagy in AM cells.
The anxa2−/− and WT mice were infected with 0.5 × 107 CFU PAO1/mouse for 24 h. (A) Viability of indicated AM cells was determined by MTT assay at a wavelength of 570 nm. (B) Bacterial killing by AM cells was tested by CFU assay. (C) AnxA2-silenced MH-S and MLE-12 cells were infected with PAO1 at an MOI of 20:1 for 2 h. Viability of indicated MH-S and MLE-12 cells was determined by MTT assay at a wavelength of 570 nm. (D) Bacterial killing by MH-S cells was detected in by CFU assay. (E) Phagocytosis of anxa2−/− and WT AMs infected with PAO1 measured by CFU count. (F) CTB-rhodamine stained anxa2−/− and WT AMs (red) were challenged with PAO1 EGFP, at an MOI of 20:1 for 1 h. Uptake of these bacteria was observed by immunofluorescence confocal microscope. (G) Phagocytized PAO1 EGFP were cultured on LB dishes and detected using an IVIS XRII imaging system. (H) PAO1 infected anxa2−/− and WT AM cells were lysed for measuring LC3 levels by immunoblotting. (I) LC3 levels in AnxA2-silenced and MH-S control cells determined by immunoblotting. During PAO1 infection, 5 mM NH4Cl was added to exclude LC3 degradation. (J) Purified anxa2−/− and WT AM cells were transfected with RFP-LC3 G120A plasmids for 24 h, and then infected with PAO1-EGFP for 1 h (MOI = 20:1). Cells with apparent puncta (at least 10 per cell) were considered as LC3-RFP positive cells (arrows indicating colocalization of LC3 puncta and PAO1 EGFP). Values are derived from 100 cells/sample. Data are representative of three independent experiments expressed as means ± SEM (one-way ANOVA with Tukey’s post hoc; *p ≤ 0.05; **p ≤ 0.005).
A previous study reported that human epithelial cell surface AnxA2 could recognize PAO1, indicating that AnxA2 may be required to allow internalization of Pa (18). However, whether AnxA2 facilitates phagocytosis in AMs remains unclear. Here, CFU assay showed that the number of PAO1 phagocytized by anxa2−/− AM cells is similar with that of WT AM cells, after 2 h post-PAO1 infection (Fig. 3E). Confocal laser scanning microscopy (CLSM) also showed a close colocalization of PAO1 EGFP with CTB-stained both from WT and anxa2−/− AM cells (Fig. 3F). In addition, we lysed these infected AM cells to measure phagocytized PAO1 on LB dishes using an IVIS imaging system. Phagocytosis of PAO1 EGFP by anxa2−/− AM cells was similar with that of WT AM cells (Fig. 3G). These data collectively suggest that AnxA2 is not required for bacterial phagocytosis in AM cells.
In the other hand, autophagy plays an essential role in the clearance of PAO1 by macrophages (15). We speculated that AnxA2 was involved in autophagy in AM cells for facilitating bacterial clearance. To test this hypothesis, we determined the expression patterns of LC-3, which is a marker microtubule-associated protein in the autophagosomal membrane, in mouse AM cells by immunoblotting. We found that endogenous LC3-I conversion to LC3-II was dramatically increased in AM cells of WT mice after 24 h PAO1 infection (Fig. 3H). However, this conversion was not detected in AM cells of anxa2−/− mice (Fig. 3H), indicating that loss of AnxA2 inhibited PAO1-induced autophagy in AM cells. Similar results were observed in vitro using AnxA2-silenced MH-S cells after 2 h PAO1 infection (Fig. 3I). A significant increase of LC3-II was detected in MH-S control cells, while the level of LC3-II remained unchanged in AnxA2-silenced MH-S cells (Fig. 3I). We also transfected an RFP-LC3 plasmid into anxa2−/− and WT AM cells, and then infected with PAO1 EGFP. In AM cells from WT mice, PAO1 infection induced significant LC3 puncta, but AnxA2 knockdown resulted in approximately 80% decrease in RFP-LC3 puncta (Fig. 3J). In addition, CLSM showed a colocalization of PAO1 GFP and LC3 (Fig. 3J), indicating that PAO1 clearance is really due to autophagy.
AnxA2 loss results in impaired PAO1-induced autophagy through Akt1-mTOR-ULK1/2 pathway
To further investigate the role of AnxA2 in autophagy, we assessed expression patterns of autophagy related factors in AnxA2-silenced MH-S cells and negative controls, using an autophagy array-based Real-time PCR primer assay. After infection of PAO1, the qPCR primer array assay revealed that three genes were significantly (over 2-fold) upregulated, while 73 genes were down-regulated (Fig. 4A, SI Table S2), indicating that AnxA2 repression may profoundly impair expression of autophagy related factors. Among the upregulated factors, NF-κB is a key transcription regulator that controls diverse gene expression related to host defense against PAO1 infection (45), while Akt1 and mTOR are interconnected junctions within the autophagy regulation pathway (46). Among the downregulated factors, ULK1 and ULK2 were the most significantly decreased factors that directly participate in autophagosome formation (47). Previous studies have investigated a role that mTOR phosphorylates Atg13 and ULK1/2, and associates with the ULK1/2-Atg13-FIP2000 complex to suppress autophagosome formation (46). According to the data of autophagy array-based qPCR assay, we hypothesized that AnxA2 functions in PAO1-induced autophagy through an Akt1-mTOR-ULK1/2 signaling pathway.
Figure 4. AnxA2 deficiency impaired PAO1-induced autophagy through Akt1-mTOR-ULK1/2 pathway.

(A) qPCR primer array assay of autophagy related factors expression in AnxA2-silenced MH-S cells versus MH-S control cells, after PAO1 infection for 4 h. Genes over 2-fold changes among subsets were displayed (n = 2 biological replicates). (B) Autophagy related factors and cell signaling protein levels of anxa2−/− and WT AM cells were determined by immunoblotting, after 24 h postinfection of PAO1. Gel data were quantified from three independent experiments using densitometry with Quantity One. (C) Association of mTOR and ULK1 in AM cells from anxa2−/− and WT mice was determined by immunoblotting with indicated antibody for IP and detection. (D) Purified anxa2−/− AM cells were treated with 5 μM Tre and WT AM cells were treated with 5 μM 3-MA for 4 h before and during PAO1 infection. During PAO1 infection, 5 mM NH4Cl was added to exclude LC3 degradation. LC3 levels of indicated AM cells were measured by immunoblotting. (E) Viability of indicated AM cells was determined by MTT assay at a wavelength of 570 nm. (B) Bacterial killing by indicated AM cells was tested by CFU assay. Data are representative of three independent experiments expressed as means ± SEM (one-way ANOVA with Tukey’s post hoc; *p ≤ 0.05; **p ≤ 0.005).
To validate the results of autophagy array-based qPCR assay and confirm our proposed roles of AnxA2 in PAO1-induced autophagy, we examined the expression levels of predicted factors in AM cells of PAO1-infected anxa2−/− and WT mice by immunoblotting using related antibodies. The result showed that in anxa2−/− AM cells, protein expression of Akt1 and mTOR was highly increased; on the other hand, UKL1 expression was significantly decreased compared to WT AM cells (Fig. 4B). We also observed that phosphorylation of Akt1 and mTOR was increased in anxa2−/− AM cells (Fig. 4B), indicating that in addition to increased protein levels, AnxA2 deficiency also led to enhanced Akt1 and mTOR activation. In WT AM cells, phosphorylation of Akt1 remained at the normal level compared to AM cells without PAO1 infection, while phosphorylation of mTOR seemed to be somewhat inhibited (Fig. 4B). Phosphorylation of the Ser residue at position 317 (Ser317) of ULK1 is an independent marker for ULK1/2-Atg13-FIP200 and mTOR (48). We found that after PAO1 infection, ULK1 Ser317 was dramatically phosphorylated in WT AM cells, but was not phosphorylated in anxa2−/− AM cells (Fig. 4B), indicating that AnxA2 deficiency impaired ULK1/2-Atg13-FIP2000 complex separation from mTOR and autophagosome formation.
To further confirm this result, we prepared total cell lysates of anxa2−/− and WT AM cells to perform Co-IP analysis. After 24 h PAO1 of infection, we used anti-mTOR or anti-ULK1 antibodies to pull down the target protein, respectively, and determined ULK1 and mTOR interaction by immunoblotting using the appropriate antibody. Our results showed that without PAO1 infection, mTOR and ULK1 showed a stable association in anxa2−/− and WT AM cells (Fig. 4C). However, after PAO1 infection, this association was disrupted in WT AM cells, but not in anxa2−/− AM cells (Fig. 4C). This result strongly suggests that AnxA2 plays roles in mTOR-ULK1 association which impacts the autophagy initiation.
To demonstrate that AnxA2 specifically acts by regulating Akt1-mTOR-ULK1/2 signaling pathway, we further treated WT AM cells with 3-MA to inhibit Akt1-mTOR signaling pathway, and simultaneously treated anxa2−/− AM cells with Tre to activate Akt1-mTOR signaling pathway, to mediate autophagy (Fig. 4D). After 2 h PAO1 infection, the viability of 3-MA treated WT AM cells reduced to a similar level of anxa2−/− AM cells, while the survival of Tre treated anxa2−/− AM cells increased to equal to WT AM cells (Fig. 4E). Similar results were observed in of bacterial clearance ability detection of these indicated AM cells (Fig. 4F).
Fam13A is required in PAO1-induced autophagy
Previous studies revealed several new factors involved in diverse pulmonary diseases, including COPD and lung cancer, but it remains unknown whether they are required for host defense against bacterial infection in macrophages. We randomly selected 15 of such factors and measured their transcripts levels of AM cells from WT and anxa2−/− mice after 24 h PAO1 postinfection, by qRT-PCR using specific primers (SI Table S1). After PAO1 infection, transcripts of APOB, Brd4, Cd1d, Fam13A, Malt1, NSF, ScgB, Sirt1 and Tob1 were significantly increased, while transcript of Sirga was reduced (Fig. 5A). Among these 10 proteins, only transcripts of Fam13A and Malt1 showed significant differences between anxa2−/− and control AM cells (Fig. 5A), indicating that they played roles associated with AnxA2 in AMs during PAO1 infection. We further focused on Fam13A because change of Fam13A expression in anxa2−/− AM cells was more significant than Malt1 (Fig. 5A).
Figure 5. Fam13A is in association with AnxA2 in PAO1-induced autophagy.
(A) Transcripts of 15 indicated factors in 24 h post PAO1-infected anxa2−/− and WT mice AM cells, compared to those of control AM cells. (B) Fam13A expression levels in AM cells of PAO1-infected anxa2−/− and WT mice determined by immunoblotting. (C) Fam13A levels in Fam13A-silenced and siNC treated MH-S control cells were determined by immunoblotting. (D) Viability and (E) bacterial killing of Fam13A-silenced MH-S cells determined by an MTT assay at a wavelength of 570 nm and by CFU assay, respectively. (F) Fam13A-silenced and MH-S control cells were transfected with RFP-LC3 G120A plasmids for 24 h, and then infected with PAO1 EGFP for 1 h (MOI = 20:1). Cells with apparent puncta (at least 10 per cell) were considered as LC3-RFP positive cells (arrows indicating colocalization of LC3 puncta and PAO1 EGFP). Values are derived from 100 cells/sample. (G) qPCR primer array assay of autophagy related factors expression in Fam13A-silenced MH-S cells versus MH-S control cells, after PAO1 infection for 4 h. Genes over 2-fold changes among subsets were displayed (n = 2 biological replicates). (H) p-Akt1, p-mTOR and p-ULK1 (Ser317) of Fam13A-silenced and MH-S control cells were determined by immunoblotting, after 2 h postinfection of PAO1. Gel data were quantified from 3 independent experiments using densitometry with Quantity One. Data are representative of three independent experiments expressed as means ± SEM (one-way ANOVA with Tukey’s post hoc; *p ≤ 0.05; **p ≤ 0.005).
As reported, Fam13A is involved in COPD and lung cancer, but little is known about its role in CF and bacterial defense (49). We next detected Fam13A protein levels in indicated AM cells by immunoblotting. We observed a significant induction of Fam13A in PAO1 infected WT AM cells, but not in anxa2−/− AM cells (Fig. 5B), again indicating that Fam13A played roles in AM cells during PAO1 infection. To further confirm our hypothesis, we introduced Fam13A interfering RNA into MH-S cells to block Fam13A expression, while using siNC as a negative control (Fig. 5C). Our results showed that the survival and bacterial clearance ability of Fam13A-silenced MH-S cells were decreased after PAO1 infection (MOI of 20:1 for 2 h) compared to MH-S control cells (Fig. 5D–E), indicating that Fam13A is also required for bacterial clearance in MH-S cells, similar to AnxA2.
Since Fam13A expression was consistently reduced in anxa2−/− AM cells after PAO1 infection (Fig 5A–B), we postulate that Fam13A was also involved in PAO1-induced autophagy. Therefore, we compared autophagosome formation between Fam13A-silenced and MH-S control cells using RFP-LC3 plasmid transfection. The result showed that PAO1 infection induced significant LC3 puncta in MH-S control cells, but this LC3 puncta were diminished in Fam13A-silenced MH-S cells (Fig. 5F), suggesting that Fam13A was required for PAO1-induced autophagy. We again analyzed expression of autophagy related factors of Fam13A-silenced MH-S cells after PAO1 infection, using the autophagy-based qPCR primer assay. The results showed that in Fam13A-silenced MH-S cells, Akt1, mTOR and NFκB were significantly induced and many factors, including ULK1 and ULK2, were downregulated (Fig. 5G and SI Table S2). To validate this array data, we also examined expression levels of predicted factors in Fam13A-silenced and MH-S control cells and found that phosphorylation of Akt1 and mTOR was significantly increased in Fam13A-silenced MH-S cells, while phosphorylation of Ser317 in ULK1 was dramatically inhibited, compared to MH-S control cells (Fig. 5H). This pattern was almost a phenocopy of AnxA2-silenced MH-S cells (Fig. 4A–B, SI Table S2), indicating an extremely critical function for Fam13A in macrophage autophagy during PAO1 infection.
Fam13A directly binds to AnxA2 and activates Rho GTPase to facilitate autophagosome formation during PAO1 infection
To test the potential association between Fam13A and AnxA2 during PAO1 infection, we lysed primary AM cells isolated from WT mice at 24 h PAO1 postinfection, and performed a Co-IP assay with anti-AnxA2 and anti-Fam13A Abs. A stable interaction between AnxA2 and Fam13A was detected (Fig. 6A). CLSM also showed a significant colocalization of AnxA2 and Fam13A in AM cells isolated from post 24 h PAO1 infection WT mice (Fig. 6B), strongly indicating that AnxA2 and Fam13A are associated in AM cells and likely act together in host defense as a single complex. This protein-protein interaction was also detected in siNC-transfected MH-S cells after PAO1 infection (Fig. 6C), but was abolished when either AnxA2 or Fam13A was repressed by RNA interference (Fig. 6C), and could not be rescued with Tre treatment in AnxA2-inhibited MH-S cells (Fig. 6C). In addition, we saw a reduced macrophage viability and bacterial killing in autophagy inhibited MH-S cells by directly silencing some key autophagy factors, including Atg5, Atg7 and Beclin1 (Fig. 6D and SI Fig. 2A–B), again indicating that autophagy plays key roles in bacterial defense in macrophage. The association of AnxA2-Fam13A was stable in all these autophagy inhibited MH-S cells after PAO1 infection (Fig. 6E). Our results collectively suggested the AnxA2-Fam13A complex, rather than canonical autophagy pathways, may be a critical regulator of autophagy.
Figure 6. Fam13A binds AnxA2 and activates Rho GTPase to facilitate autophagy during PAO1 infection.

(A) Association of AnxA2 and Fam13A in AM cells from WT mice were determined by immunoblotting with indicated antibody for IP and detection, after 24 h postinfection of PAO1. (B) AM cells were purified from 24 h post PAO1 infected and control WT mice. Confocal laser scanning microscopy observation showed the colocalization of AnxA2 and Fam13A after PAO1 infection by immune staining. (C) Association of AnxA2 and Fam13A in indicated MH-S cells was determined by immunoblotting with AnxA2 antibody for IP and detection after 2 h postinfection of PAO1. Before infection, MH-S cells were transfected with siNC, siAnxA2 (with and without Tre), siFam13A for 24 h. (D) MH-S cells were transfected with siAtg5, siAtg7, siBeclin1 and pcDNA3.1-Fam13A for 24h. Expressions of Atg5, Atg7, Beclin1 and Fam13A in indicated MH-S cells were determined respectively by immunoblotting. (E) Association of AnxA2 and Fam13A in siNC-, siAtg5-, siAtg7- and siBeclin1-transfected MH-S cells was determined by immunoblotting with AnxA2 antibody for IP and detection after 2 h postinfection of PAO1. (F–H) Total Rho GTPases and activated Rho GTPases in Fam13A-silenced MH-S cells (F), Fam13A overexpressed MH-S cells (G), anxa2−/− AM cells (H) and control cells were determined by immunoblotting, with or without 2 h PAO1 infection. (I) Diagram delineating a pathway in AnxA2 deficient macrophages against PAO1 infection. After PAO1 infection, AnxA2 binds Fam13A to activate Rho GTPase, and subsequently inactivates Akt1-mTOR-ULK1/2 signaling pathway to induce autophagosome formation. Data are representative of three independent experiments.
Protein function prediction analysis suggests that Fam13A contains two conserved Rho GTPase-activating domains (www.ebi.ac.uk/interpro/protein/O94988). We suspected that Fam13A regulates Rho GTPase activity in defense against Pa. To confirm this, we analyzed Rho GTPase activities in WT and Fam13A-silenced MH-S cells, with or without PAO1 infection. We found that Fam13A deficiency directly reduced Rho GTPase activation in MH-S cells, even without PAO1 infection (Fig. 6F and SI Fig. 2E). After PAO1 infection, MH-S control samples exhibited dramatically increased levels of activated GTPase compared to control cells (Fig. 6F and SI Fig. 2E), but the level of activated Rho GTPase in Fam13A-silenced cells is still low (Fig. 6F and SI Fig. 2E). We further cloned Fam13A gene into vector pcDNA3.1 and transfected this plasmid into MH-S cells to overexpress Fam13A, while MH-S transfected with empty pcDNA3.1 vector was used as negative control (Fig. 6D). High expression of Fam13A did not change macrophage viability and bacterial killing in MH-S cells (SI Fig. 2C–D) after PAO1 infection, but directly induced Rho GTPase activation even without PAO1 infection (Fig. 6G and SI Fig. 2F). These data together indicates that Fam13A regulates PAO1-induced Rho GTPase activation, and contributes to host immunity in bacterial defense. The impaired GTPase activation was also detected in anxa2−/− AM cells after PAO1 infection, compared to WT AM cells (Fig. 6H and SI Fig. 2G), suggesting that AnxA2 plays a role in the activation of GTPase via Fam13A.
Discussion
In this study, we reveal a critical role by AnxA2 in host defense against Pa infection. Severe disease phenotype in anxa2−/− mice is observed in our experiments, including decreased survival, increased inflammatory response, and more severe lung injury compared to WT mice (Fig. 1–2 and SI Fig. 1A–D). The increase of bacterial burdens in the lung and other organs of anxa2−/− mice indicates that AnxA2 is a critical contributor to bacterial clearance. Importantly, further data demonstrate that AnxA2 plays an essential role in PAO1-induced autophagy in alveolar macrophages (Fig. 6I). Furthermore, we have identified the underlying mechanism by which AnxA2 impacted autophagy during infection, namely, by directly binding to Fam13A and activating Rho GTPase to regulate the Akt1-mTOR-ULK1/2 signaling axis. Similar phenotypes (increased cell death and decreased bacterial clearance) have been observed in AnxA2-silenced MH-S cells infected with different bacterial strains or species, such as Pa strains PA14, PAK, K. pneumoniae and E. coli (SI Fig. 1E–F). Thus, AnxA2 may render a critical immune defense against a wide range of microorganisms.
Our study established that AnxA2 is involved in host inflammatory response regulation during Pa infection. As reported previously, AnxA2 plays multiple roles in host defense against bacteria. For instance, AnxA2 is predominantly an anti-inflammatory agent largely because of its similarities to Annexin A1, which has been universally demonstrated to play such a role (50). Existing data support a role for AnxA2 in the proinflammatory process: AnxA2 elicits activation of the MAP kinase and nuclear translocation of NF-κB, resulting in inflammatory cytokine production as well as chemokine production. This function was reflected in enhanced macrophage effector function, as AnxA2 dramatically facilitated phagocytosis of E. coli bacteria (51). Other studies have also illustrated the role of AnxA2 in regulating inflammatory response (52). Our results are also in line with this observation as significantly enhanced inflammatory response, lung injury, ROS accumulation and cytokine production were found in anxa2−/− mice following PAO1 infection (Fig 2), and that autophagy array-based qPCR assay showed that AnxA2 negatively regulates host inflammatory response by NF-κB (Fig. 4A).
AnxA2 was previously reported to serve as a receptor for Pa in mammalian epithelial cells and is involved in Pa internalization (18). Whether AnxA2 is required for phagocytosis of PAO1 by AM cells is unknown. Our data showed that phagocytized PAO1 by anxa2−/− AM cells is similar with that of WT AM cells, after PAO1 infection, indicating AnxA2 does not play critical roles in bacterial uptake by AM cells (Fig. 3E–G). Instead, we observed that inhibition of AnxA2 resulted in suppression of autophagosome formation in AM cells after PAO1 infection (Fig 3H–J). Up to date, no literature has reported a mechanistic role of AnxA2 in regulating autophagy and bacterial clearance. The only previous paper demonstrated that AnxA2 expression increased along with autophagy-regulated genes, such as GABARAP, LC3B and ATG3, in human umbilical vein endothelial cells by repeated treatments with resveratrol (53). Here, we found that the reduction in survival only occurred in AnxA2-silenced MH-S cells, but not in MLE-12 cells (Fig 3C), indicating that AnxA2 might play important roles specifically in macrophages rather than epithelial cells. Autophagy array-based qPCR assay and immunoblotting showed increased expression and activation of Akt1 and mTOR in AnxA2-silenced MH-S cells (Fig. 4A), suggesting that AnxA2 regulates PAO1-induced autophagy via an Akt1-mTOR pathway. Autophagy is normally inhibited in metabolically repressed cells by the metabolic checkpoint kinase mTOR (54), but how the Akt1-mTOR signaling pathway induces autophagy upon bacterial infection remains unclear. A previous study determined that infection of epithelial cells with Shigella and Salmonella triggers acute intracellular amino acid starvation due to host membrane damage, and pathogen-induced amino acid starvation was shown to cause downregulation of mTOR activity, resulting in the induction of autophagy (55). Our data provide evidence that AnxA2 regulates pathogen-induced host autophagy through Akt1-mTOR signaling pathway. Generally, PAO1 infection stimulates AnxA2 to interact with free Fam13A in the cytoplasm and activating Rho GTPase. Activated Rho GTPase then inactivates Akt1 and mTOR thus ULK1/2 is released to form the autophagosome (46). On the other hand, AnxA2 deficiency impaired Rho GTPase activation, hence activated mTOR strongly binds ULK1/2 and inhibiting the formation of the autophagosome (Fig. 6I). However, AnxA2 overexpression was strongly associated with rapid recurrence after gemcitabine-adjuvant chemotherapy in patients with resected pancreatic cancer. In GEM-MIA PaCa-2 cells, in which AnxA2 is highly expressed, the levels of p-Akt and p-mTOR were significantly increased (56). Thus, AnxA2 acts as a dual regulator in Akt-mTOR signaling pathway which may be independent of cell nutrition.
Another important finding in this work is that Fam13A is involved in regulation of PAO1-induced autophagy, by activating Rho GTPase. Previously, variants in Fam13A have been found in genome-wide association studies to be associated with lung functions in the general population as well as in several common chronic lung diseases such as COPD, asthma, and idiopathic interstitial pneumonias (57). Sequence analysis has indicated the presence of a Rho GTPase-activating protein domain on exons 2–5 (58). Here, we observed an induction of Fam13A and consistent induction of activated Rho GTPase in AM cells after PAO1 infection, suggesting that Fam13A may mediate Rho GTPase activation, instead of inactivation (Fig. 5B, Fig. 6H and SI Fig. 2G). Further data determined that overexpress of Fam13A could directly induce Rho GTPase activation, and in the contrary, Fam13A deficiency inhibit Rho GTPase activation in AM cells, both with or without PAO1 infection (Fig. 6F–G and SI Fig. 2E–F), indicating that Fam13A directly mediates Rho GTPase activation. In addition, we observed a stable association between AnxA2 and Fam13A after PAO1 infection (Fig. 6A–B), and expression of activated Rho GTPase did not increase in AnxA2-depleted AM cells (Fig. 6H). These results indicated that Rho GTPase regulator function of Fam13A may be strictly limited within AnxA2.
Rho GTPases are normally identified as key regulators of cytoskeletal dynamics (59) and affect several vascular processes, such as endothelial permeability, cell motility, angiogenesis, nitric oxide production, smooth muscle contractility, cell proliferation, differentiation and apoptosis (60, 61). Previous studies also indicated that Rho GTPases play roles in the defense against bacterial pathogen invasion. For example, Rho GTPases are involved in the regulation of pulmonary vascular barrier function (62), in the regulation of macropinosome formation and maturation (63), and act as regulators of immunity in plants and animals (64). However, roles of Rho GTPases in regulation of autophagy, especially in bacteria induced autophagy are still unclear. The only report described that Rho GTPase RhoA positively regulates starvation-induced autophagy through a mechanism that is likely dependent on actin, whereas Rac1 has the opposing effect (65). Here, we determined that Rho GTPases are involved in regulation of PAO1-induced autophagy, as reduced Rho GTPase activation is correlated with dampened autophagosome formation (Fig. 6C and 6F). In the process of autophagosome formation induced by starvation, previous studies have established that RhoA, one of Rho GTPase, triggers a series of signals, leading to actin polymerization (65). However, our data demonstrate that as potential upstream signals, Rho GTPases regulate autophagosome formation via the Akt-mTOR signaling pathway. Gordon et al. support a model in which RhoA GTPase represses mTOR signaling upstream of Akt1 (66), which is consistent with our observation.
In conclusion, we have demonstrated a typical phenotype of Pa infection in anxa2−/− mice, suggesting an important role for AnxA2 in bacterial defense in mice. AnxA2 deficiency impaired autophagosome formation and the bacterial clearance ability as well as intensified inflammatory responses. Importantly, we revealed a previously unidentified mechanistic role for AnxA2 in association with Fam13A to activate Rho GTPases. These activated Rho GTPases in turn inactivated Akt1 and mTOR to release ULK1/2 for forming autophagosome during Pa infection. We supposed that AnxA2 not only affects autophagy mediated bacterial clearances during Pa infection, but also some other pathways, because autophagy plays diverse other important roles, such as inflammasome activation (67, 68). Overall, our findings provide insights into the role of AnxA2 in the regulation of autophagy against Pa and may indicate a novel therapeutic approach to combat bacterial infection.
Supplementary Material
Acknowledgments
We thank S. Rolling of UND imaging core for help with confocal imaging. We also thank Dr. K. Hajjar of Cornell University for kindly providing anxa2−/− mice and thank Dr. Xiao-min Yin of Indiana University for providing LC3-RFP G120A plasmids.
Financial support. This work was supported by the Flight Attendant Medical Research Institute (FAMRI, Grant #103007), National Institute of Health (AI109317-01A1, AI101973-01, and AI097532-01A1).
Abbreviations used in this study
- AnxA2
Annexin A2
- anxa2−/−
AnxA2-deficient
- AM
alveolar macrophages
- BAL
bronchoalveolar lavage
- CF
cystic fibrosis
- CLSM
confocal laser scanning microscopy
- COPD
chronic obstructive pulmonary disease
- Fam13A
Family with sequence similarity 13, member A
- Pa
P. aeruginosa
- H2DCF
dihydrodichlorofluorescein diacetate
- MOI
multiple of infection
- PAMP
pathogen-associated molecular pattern
- PMN
polymorphonuclear neutrophil
- PPR
pattern recognition receptor
- ROS
reactive oxygen species
- siRNA
small interference RNA
- WT
wild-type
References
- 1.Gerke V, Creutz CE, Moss SE. Annexins: linking Ca2+ signalling to membrane dynamics. Nat Rev Mol Cell Biol. 2005;6:449–461. doi: 10.1038/nrm1661. [DOI] [PubMed] [Google Scholar]
- 2.Moss SE, Morgan RO. The annexins. Genome Biol. 2004;5:219. doi: 10.1186/gb-2004-5-4-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang X, Liu S, Guo C, Zong J, Sun MZ. The association of annexin A2 and cancers. Clin Transl Oncol. 2012;14:634–640. doi: 10.1007/s12094-012-0855-6. [DOI] [PubMed] [Google Scholar]
- 4.Cui JW, Wang YL. Expression and function of Annexin II in lung cancer tissue. Asian Pac J Trop Med. 2013;6:150–152. doi: 10.1016/S1995-7645(13)60012-7. [DOI] [PubMed] [Google Scholar]
- 5.Yang J, Yang F, Nie J, Zou X, Tian H, Qin Y, Liu C. Evaluation of Annexin A2 as a novel diagnostic serum biomarker for lung cancer. Cancer Biomarkers. 2014 doi: 10.3233/CBM-140455. [DOI] [PubMed] [Google Scholar]
- 6.Huang Y, Jin Y, Yan CH, Yu Y, Bai J, Chen F, Zhao YZ, Fu SB. Involvement of Annexin A2 in p53 induced apoptosis in lung cancer. Mol Cell Biochem. 2008;309:117–123. doi: 10.1007/s11010-007-9649-5. [DOI] [PubMed] [Google Scholar]
- 7.Dassah M, Almeida D, Hahn R, Bonaldo P, Worgall S, Hajjar KA. Annexin A2 mediates secretion of collagen VI, pulmonary elasticity and apoptosis of bronchial epithelial cells. J Cell Sci. 2014;127:828–844. doi: 10.1242/jcs.137802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fang Y, Lin C, Liao P, Kuo Y, Wang S, Yeh T, Shieh C, Su I, Lei H, Lin Y. Annexin A2 on lung epithelial cell surface is recognized by severe acute respiratory syndrome-associated coronavirus spike domain 2 antibodies. Mol Immunol. 2010;47:1000–1009. doi: 10.1016/j.molimm.2009.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jolly C, Winfree S, Hansen B, Steele-Mortimer O. The Annexin A2/p11 complex is required for efficient invasion of Salmonella Typhimurium in epithelial cells. Cell Microbiol. 2014;16:64–77. doi: 10.1111/cmi.12180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gou D, Mishra A, Weng T, Su L, Chintagari NR, Wang Z, Zhang H, Gao L, Wang P, Stricker HM, Liu L. Annexin A2 interactions with Rab14 in alveolar type II cells. The Journal of biological chemistry. 2008;283:13156–13164. doi: 10.1074/jbc.M801532200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Andey T, Marepally S, Patel A, Jackson T, Sarkar S, O’Connell M, Reddy RC, Chellappan S, Singh P, Singh M. Cationic lipid guided short-hairpin RNA interference of annexin A2 attenuates tumor growth and metastasis in a mouse lung cancer stem cell model. J Control Release. 2014;184:67–78. doi: 10.1016/j.jconrel.2014.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lyczak J, Cannon C, Pier G. Establishment of Pseudomonas aeruginosa infection: lessons from a versatile opportunist. Microbes Infect. 2000;2:1051–1060. doi: 10.1016/s1286-4579(00)01259-4. [DOI] [PubMed] [Google Scholar]
- 13.Sousa AM, Pereira MO. Pseudomonas aeruginosa Diversification during Infection Development in Cystic Fibrosis Lungs-A Review. Pathogens. 2014;3:680–703. doi: 10.3390/pathogens3030680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lovewell RR, Patankar YR, Berwin B. Mechanisms of phagocytosis and host clearance of Pseudomonas aeruginosa. American journal of physiology Lung cellular and molecular physiology. 2014;306:L591–603. doi: 10.1152/ajplung.00335.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yuan K, Huang C, Fox J, Laturnus D, Carlson E, Zhang B, Yin Q, Gao H, Wu M. Autophagy plays an essential role in the clearance of Pseudomonas aeruginosa by alveolar macrophages. J Cell Sci. 2012;125:507–515. doi: 10.1242/jcs.094573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saitoh T, Fujita N, Jang MH, Uematsu S, Yang BG, Satoh T, Omori H, Noda T, Yamamoto N, Komatsu M, Tanaka K, Kawai T, Tsujimura T, Takeuchi O, Yoshimori T, Akira S. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature. 2008;456:264–268. doi: 10.1038/nature07383. [DOI] [PubMed] [Google Scholar]
- 17.Delgado MA, Elmaoued RA, Davis AS, Kyei G, Deretic V. Toll-like receptors control autophagy. EMBO J. 2008;27:1110–1121. doi: 10.1038/emboj.2008.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kirschnek S, Adams C, Culbins E. Annexin II is a novel receptor for Pseudomonas aeruginosa. Biochemical and Biophysical Research Communications. 2004;327:900–906. doi: 10.1016/j.bbrc.2004.12.089. [DOI] [PubMed] [Google Scholar]
- 19.Wos M, Bandorowicz-Pikula J. Participation of annexins in endocytosis and EGFR-mediated signal transduction. Postepy Biochem. 2014;60:55–61. [PubMed] [Google Scholar]
- 20.Wu MH, Chuang PC, Lin YJ, Tsai SJ. Suppression of annexin A2 by prostaglandin E(2) impairs phagocytic ability of peritoneal macrophages in women with endometriosis. Hum Reprod. 2013;28:1045–1053. doi: 10.1093/humrep/det003. [DOI] [PubMed] [Google Scholar]
- 21.Drab M, Verkade P, Elger M, Kasper M, Lohn M, Lauterbach B, Menne J, Lindschau C, Mende F, Luft FC, Schedl A, Haller H, Kurzchalia TV. Loss of caveolae, vascular dysfunction, and pulmonary defects in caveolin-1 gene-disrupted mice. Science. 2001;293:2449–2452. doi: 10.1126/science.1062688. [DOI] [PubMed] [Google Scholar]
- 22.Ling Q, Jacovina AT, Deora A, Febbraio M, Simantov R, Silverstein RL, Hempstead B, Mark WH, Hajjar KA. Annexin II regulates fibrin homeostasis and neoangiogenesis in vivo. The Journal of clinical investigation. 2004;113:38–48. doi: 10.1172/JCI200419684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tan SR, Gan C, Li RP, Ye Y, Zhang S, Wu X, Yang YY, Fan W, Wu M. A novel chemosynthetic peptide with beta-sheet motif efficiently kills Klebsiella pneumoniae in a mouse model. Int J Nanomedicine. 2015;10:1045–1059. doi: 10.2147/IJN.S73303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kannan S, Huang H, Seeger D, Audet A, Chen Y, Huang C, Gao H, Li S, Wu M. Alveolar epithelial type II cells activate alveolar macrophages and mitigate P. Aeruginosa infection. PLoS One. 2009;4:e4891. doi: 10.1371/journal.pone.0004891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kulasakara H, Lee V, Brencic A, Liberati N, Urbach J, Miyata S, Lee DG, Neely AN, Hyodo M, Hayakawa Y, Ausubel FM, Lory S. Analysis of Pseudomonas aeruginosa diguanylate cyclases and phosphodiesterases reveals a role for bis-(3′-5′)-cyclic-GMP in virulence. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:2839–2844. doi: 10.1073/pnas.0511090103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Priebe GP, Brinig MM, Hatano K, Grout M, Coleman FT, Pier GB, Goldberg JB. Construction and characterization of a live, attenuated aroA deletion mutant of Pseudomonas aeruginosa as a candidate intranasal vaccine. Infection and immunity. 2002;70:1507–1517. doi: 10.1128/IAI.70.3.1507-1517.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lawlor MS, Hsu J, Rick PD, Miller VL. Identification of Klebsiella pneumoniae virulence determinants using an intranasal infection model. Mol Microbiol. 2005;58:1054–1073. doi: 10.1111/j.1365-2958.2005.04918.x. [DOI] [PubMed] [Google Scholar]
- 28.Hart PD, Young MR. Ammonium chloride, an inhibitor of phagosome-lysosome fusion in macrophages, concurrently induces phagosome-endosome fusion, and opens a novel pathway: studies of a pathogenic mycobacterium and a nonpathogenic yeast. The Journal of experimental medicine. 1991;174:881–889. doi: 10.1084/jem.174.4.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kannan S, Audet A, Huang H, Chen LJ, Wu M. Cholesterol-rich membrane rafts and Lyn are involved in phagocytosis during Pseudomonas aeruginosa infection. Journal of immunology. 2008;180:2396–2408. doi: 10.4049/jimmunol.180.4.2396. [DOI] [PubMed] [Google Scholar]
- 30.Li X, Ye Y, Zhou X, Huang C, Wu M. Atg7 enhances host defense against infection via downregulation of superoxide but upregulation of nitric oxide. Journal of immunology. 2015;194:1112–1121. doi: 10.4049/jimmunol.1401958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou X, Li X, Ye Y, Zhao K, Zhuang Y, Li Y, Wei Y, Wu M. MicroRNA-302b augments host defense to bacteria by regulating inflammatory responses via feedback to TLR/IRAK4 circuits. Nature communications. 2014;5:3619. doi: 10.1038/ncomms4619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liang H, Deng X, Li X, Ye Y, Wu M. Molecular mechanisms of master regulator VqsM mediating quorum-sensing and antibiotic resistance in Pseudomonas aeruginosa. Nucleic acids research. 2014;42:10307–10320. doi: 10.1093/nar/gku586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu M, Huang H, Zhang W, Kannan S, Weaver A, McKibben M, Herington D, Zeng H, Gao H. Host DNA repair proteins in response to Pseudomonas aeruginosa in lung epithelial cells and in mice. Infection and immunity. 2011;79:75–87. doi: 10.1128/IAI.00815-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu M, Tang SL, Zang RJ, Yu H. Selective killing of tumor cells in vitro by an immunotoxin composed of ricin and monoclonal antibody against Ia antigen. International journal of immunopharmacology. 1990;12:235–239. doi: 10.1016/0192-0561(90)90058-u. [DOI] [PubMed] [Google Scholar]
- 35.Li G, Yuan K, Yan C, Fox J, 3rd, Gaid M, Breitwieser W, Bansal AK, Zeng H, Gao H, Wu M. 8-Oxoguanine-DNA glycosylase 1 deficiency modifies allergic airway inflammation by regulating STAT6 and IL-4 in cells and in mice. Free radical biology & medicine. 2012;52:392–401. doi: 10.1016/j.freeradbiomed.2011.10.490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yan J, Liu X, Wang Y, Jiang X, Liu H, Wang M, Zhu X, Wu M, Tien P. Enhancing the potency of HBV DNA vaccines using fusion genes of HBV-specific antigens and the N-terminal fragment of gp96. The journal of gene medicine. 2007;9:107–121. doi: 10.1002/jgm.998. [DOI] [PubMed] [Google Scholar]
- 37.Yuan K, Huang C, Fox J, Gaid M, Weaver A, Li G, Singh BB, Gao H, Wu M. Elevated inflammatory response in caveolin-1-deficient mice with Pseudomonas aeruginosa infection is mediated by STAT3 protein and nuclear factor kappaB (NF-kappaB) The Journal of biological chemistry. 2011;286:21814–21825. doi: 10.1074/jbc.M111.237628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li G, Fox J, 3rd, Liu Z, Liu J, Gao GF, Jin Y, Gao H, Wu M. Lyn mitigates mouse airway remodeling by downregulating the TGF-beta3 isoform in house dust mite models. Journal of immunology. 2013;191:5359–5370. doi: 10.4049/jimmunol.1301596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Williams DA, Zheng Y, Cancelas JA. Rho GTPases and regulation of hematopoietic stem cell localization. Methods in enzymology. 2008;439:365–393. doi: 10.1016/S0076-6879(07)00427-2. [DOI] [PubMed] [Google Scholar]
- 40.Kannan S, Audet A, Knittel J, Mullegama S, Gao GF, Min W. Src kinase Lyn is crucial for Pseudomonas aeruginosa internalization into lung cells. Euro J Immuno. 2006;36:1739–1752. doi: 10.1002/eji.200635973. [DOI] [PubMed] [Google Scholar]
- 41.Langereis JD. Neutrophil integrin affinity regulation in adhesion, migration, and bacterial clearance. Cell Adh Migr. 2013;7:476–481. doi: 10.4161/cam.27293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Heid ME, Keyel PA, Kamga C, Shiva S, Watkins SC, Salter RD. Mitochondrial reactive oxygen species induces NLRP3-dependent lysosomal damage and inflammasome activation. Journal of immunology. 2013;191:5230–5238. doi: 10.4049/jimmunol.1301490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sunday ME. Oxygen, gastrin-releasing Peptide, and pediatric lung disease: life in the balance. Front Pediatr. 2014;2:72. doi: 10.3389/fped.2014.00072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roychowdhury S, Luthe A, Keilhoff G, Wolf G, Horn T. Oxidative stress in glial cultures: detection by DAF-2 fluorescence used as a tool to measure peroxynitrite rather than nitric oxide. Glia. 2002;38:103–114. doi: 10.1002/glia.10024. [DOI] [PubMed] [Google Scholar]
- 45.Chai W, Zhang J, Duan Y, Pan D, Liu W, Li Y, Yan X, Chen B. Pseudomonas pyocyanin stimulates IL-8 expression through MAPK and NF-kB pathway in differentiated U937 cells. BMC Microbiology. 2014;14:26. doi: 10.1186/1471-2180-14-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dunlop EA, Tee AR. mTOR and autophagy: a dynamic relationship governed by nutrients and energy. Semin Cell Dev Biol. 2014;36:121–129. doi: 10.1016/j.semcdb.2014.08.006. [DOI] [PubMed] [Google Scholar]
- 47.Wong PM, Puente C, Ganley IG, Jiang X. The ULK1 complex: sensing nutrient signals for autophagy activation. Autophagy. 2013;9:124–137. doi: 10.4161/auto.23323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jung CH, Jun CB, Ro SH, Kim YM, Otto NM, Cao J, Kundu M, Kim DH. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell. 2009;20:1992–2003. doi: 10.1091/mbc.E08-12-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Young R, Hopkins R, Hay B, Whittington C, Epton M, Gamble G. Fam13a locus in COPD is independently associated with lung cancer - evidence of a molecular genetic link between COPD and lung cancer. Dovepress. 2010;2011:1–10. doi: 10.2147/TACG.S15758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gerke V, Moss SE. Annexins: from structure to function. Physiol Rev. 2002;82:331–371. doi: 10.1152/physrev.00030.2001. [DOI] [PubMed] [Google Scholar]
- 51.Swisher JF, Khatri U, Feldman GM. Annexin A2 is a soluble mediator of macrophage activation. J Leukoc Biol. 2007;82:1174–1184. doi: 10.1189/jlb.0307154. [DOI] [PubMed] [Google Scholar]
- 52.Woodham AW, Raff AB, Raff LM, Da Silva DM, Yan L, Skeate JG, Wong MK, Lin YG, Kast WM. Inhibition of Langerhans cell maturation by human papillomavirus type 16: a novel role for the annexin A2 heterotetramer in immune suppression. Journal of immunology. 2014;192:4748–4757. doi: 10.4049/jimmunol.1303190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Takizawa Y, Kosuge Y, Awaji H, Tamura E, Takai A, Yanai T, Yamamoto R, Kokame K, Miyata T, Nakata R, Inoue H. Up-regulation of endothelial nitric oxide synthase (eNOS), silent mating type information regulation 2 homologue 1 (SIRT1) and autophagy-related genes by repeated treatments with resveratrol in human umbilical vein endothelial cells. Br J Nutr. 2013;110:2150–2155. doi: 10.1017/S0007114513001670. [DOI] [PubMed] [Google Scholar]
- 54.Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 55.Tattoli I, Sorbara MT, Vuckovic D, Ling A, Soares F, Carneiro LA, Yang C, Emili A, Philpott DJ, Girardin SE. Amino acid starvation induced by invasive bacterial pathogens triggers an innate host defense program. Cell host & microbe. 2012;11:563–575. doi: 10.1016/j.chom.2012.04.012. [DOI] [PubMed] [Google Scholar]
- 56.Kagawa S, Takano S, Yoshitomi H, Kimura F, Satoh M, Shimizu H, Yoshidome H, Ohtsuka M, Kato A, Furukawa K, Matsushita K, Nomura F, Miyazaki M. Akt/mTOR signaling pathway is crucial for gemcitabine resistance induced by Annexin II in pacreatic cancer cells. J Surg Res. 2012;178:758–767. doi: 10.1016/j.jss.2012.05.065. [DOI] [PubMed] [Google Scholar]
- 57.Li X, Hawkins GA, Ampleford EJ, Moore WC, Li H, Hastie AT, Howard TD, Boushey HA, Busse WW, Calhoun WJ, Castro M, Erzurum SC, Israel E, Lemanske RF, Jr, Szefler SJ, Wasserman SI, Wenzel SE, Peters SP, Meyers DA, Bleecker ER. Genome-wide association study identifies TH1 pathway genes associated with lung function in asthmatic patients. J Allergy Clin Immunol. 2013;132:313–320 e315. doi: 10.1016/j.jaci.2013.01.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cohen M, Reichenstein M, Everts-van der Wind A, Heon-Lee J, Shani M, Lewin HA, Weller JI, Ron M, Seroussi E. Cloning and characterization of FAM13A1--a gene near a milk protein QTL on BTA6: evidence for population-wide linkage disequilibrium in Israeli Holsteins. Genomics. 2004;84:374–383. doi: 10.1016/j.ygeno.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 59.Hall A, Lalli G. Rho and Ras GTPases in axon growth, guidance, and branching. Cold Spring Harb Perspect Biol. 2010;2:a001818. doi: 10.1101/cshperspect.a001818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vega FM, Ridley AJ. Rho GTPases in cancer cell biology. FEBS Lett. 2008;582:2093–2101. doi: 10.1016/j.febslet.2008.04.039. [DOI] [PubMed] [Google Scholar]
- 61.Pedersen E, Brakebusch C. Rho GTPase function in development: how in vivo models change our view. Exp Cell Res. 2012;318:1779–1787. doi: 10.1016/j.yexcr.2012.05.004. [DOI] [PubMed] [Google Scholar]
- 62.Duluc L, Wojciak-Stothard B. Rho GTPases in the regulation of pulmonary vascular barrier function. Cell Tissue Res. 2014;355:675–685. doi: 10.1007/s00441-014-1805-0. [DOI] [PubMed] [Google Scholar]
- 63.Egami Y, Taguchi T, Maekawa M, Arai H, Araki N. Small GTPases and phosphoinositides in the regulatory mechanisms of macropinosome formation and maturation. Front Physiol. 2014;5:374. doi: 10.3389/fphys.2014.00374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kawano Y, Kaneko-Kawano T, Shimamoto K. Rho family GTPase-dependent immunity in plants and animals. Front Plant Sci. 2014;5:522. doi: 10.3389/fpls.2014.00522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Aguilera MO, Beron W, Colombo MI. The actin cytoskeleton participates in the early events of autophagosome formation upon starvation induced autophagy. Autophagy. 2012;8:1590–1603. doi: 10.4161/auto.21459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gordon BS, Kazi AA, Coleman CS, Dennis MD, Chau V, Jefferson LS, Kimball SR. RhoA modulates signaling through the mechanistic target of rapamycin complex 1 (mTORC1) in mammalian cells. Cell Signal. 2014;26:461–467. doi: 10.1016/j.cellsig.2013.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jabir MS, Ritchie ND, Li D, Bayes HK, Tourlomousis P, Puleston D, Lupton A, Hopkins L, Simon AK, Bryant C, Evans TJ. Caspase-1 cleavage of the TLR adaptor TRIF inhibits autophagy and beta-interferon production during Pseudomonas aeruginosa infection. Cell host & microbe. 2014;15:214–227. doi: 10.1016/j.chom.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jabir MS, Hopkins L, Ritchie ND, Ullah I, Bayes HK, Li D, Tourlomousis P, Lupton A, Puleston D, Simon AK, Bryant C, Evans TJ. Mitochondrial damage contributes to Pseudomonas aeruginosa activation of the inflammasome and is downregulated by autophagy. Autophagy. 2015;11:166–182. doi: 10.4161/15548627.2014.981915. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.