Abstract
Knowledge of how microorganisms respond and adapt to low-pressure (LP) environments is limited. Previously, Bacillus subtilis strain WN624 was grown at the near-inhibitory LP of 5 kPa for 1,000 generations and strain WN1106, which exhibited increased relative fitness at 5 kPa, was isolated. Genomic sequence differences between ancestral strain WN624 and LP-evolved strain WN1106 were identified using whole-genome sequencing. LP-evolved strain WN1106 carried amino acid-altering mutations in the coding sequences of only seven genes (fliI, parC, ytoI, bacD, resD, walK, and yvlD) and a single 9-nucleotide in-frame deletion in the rnjB gene that encodes RNase J2, a component of the RNA degradosome. By using a collection of frozen stocks of the LP-evolved culture taken at 50-generation intervals, it was determined that (i) the fitness increase at LP occurred rapidly, while (ii) mutation acquisition exhibited complex kinetics. A knockout mutant of rnjB was shown to increase the competitive fitness of B. subtilis at both LP and standard atmospheric pressure.
INTRODUCTION
Microorganisms exhibit an ability to survive and even thrive in a wide range of harsh environments on Earth, which feature extremes of fundamental physical parameters such as temperature and pressure (1). Organisms able to grow at extremely high temperatures (i.e., thermophiles) or low temperatures (i.e., psychrophiles) have been studied extensively (1), as well as organisms capable of growth at high pressure (HP) (i.e., piezophiles) (1, 2). In sharp contrast, our understanding is extremely limited concerning microbial survival, adaptation, and growth in low-pressure (LP) environments. In part, this reflects a relative scarcity of LP environments on the Earth's surface; the atmospheric pressure at sea level averages ∼101.3 kPa, and the lowest average terrestrial barometric pressure is ∼34 kPa, atop Mount Everest. However, there has been a recent upsurge of interest in studying the response of microbes to LP exposure. First, Earth's upper atmosphere is a global LP environment that poses unique challenges to microbial survival and growth; for example, 5 kPa of pressure corresponds to an altitude of ∼19 km, in the lower stratosphere (3–5). Second, man-made LP environments (e.g., hypobaric chambers at pressures of ∼2 kPa) have proven useful for the long-term storage of high-value agricultural commodities, partly due to LP inhibition of the growth of spoilage microorganisms (6). Third, considerable effort is currently being devoted to the study of the biology of the extraterrestrial environment of Mars, which features an LP atmosphere ranging from ∼0.1 kPa to ∼1 kPa; in this context, LP microbiology is important both for life detection and for planetary protection purposes (7).
To understand how microbes respond and adapt to LP, we previously reported an evolution experiment in which Bacillus subtilis ancestral strain WN624 was propagated in liquid LB medium for 1,000 generations at an LP of 5 kPa; during this experiment, an enhanced growth capability evolved in the culture at 5 kPa (8). After 1,000 generations of evolution, strain WN1106 was isolated from the 5-kPa population and was shown to exhibit higher competitive fitness than the ancestral strain at 5 kPa (8).
In order to elucidate the underlying genomic change(s) leading to increased fitness at LP, we describe in this communication the whole-genome sequencing (WGS) of ancestral strain WN624 and LP-evolved strain WN1106. We found that LP-evolved strain WN1106 contained amino acid-changing mutations in only eight protein-coding genes, which are considered candidates for testing their potential importance in conferring an LP growth phenotype on this strain.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and growth conditions.
All B. subtilis strains and plasmids used in this study are listed in Table 1. Strain GP45, with a complete deletion of the rnjB gene and replacement of it with a spectinomycin resistance cassette (ΔrnjB::spc), was a generous gift from Jörg Stülke. Preparation of chromosomal DNA and B. subtilis competent cells and DNA-mediated transformation were performed as described previously (9, 10). Miller LB liquid or agar medium (11) was used throughout and supplemented when necessary with the appropriate antibiotic (final concentration), as follows: chloramphenicol (Cm, 5 μg/ml), neomycin (Neo, 5 μg/ml), or spectinomycin (Spc, 100 μg/ml). Cells were grown under LP (5 ± 0.2 kPa) as described in detail previously (8, 12). Briefly, liquid cultures were propagated in 125-ml sidearm (Klett) flasks in 10 ml of liquid LB medium containing the appropriate selective antibiotic at 27°C in a temperature-controlled rotary shaker bath with moderate shaking (∼150 rpm). Low pressure was supplied by a programmable pumping system (KNF Neuberger, Trenton, NJ) fitted with 0.2-μm in-line air filters. Under these conditions, evaporation within a 24-h period was negligible. Growth was measured by optical density (OD) using a Klett-Summerson colorimeter fitted with a no. 66 (red, 660-nm) filter (note that for purposes of comparison, 100 Klett units = 1 OD660 = ∼1 × 108 cells per ml). At daily intervals, the OD of each population was determined, each culture was diluted 1:100 into fresh selective medium, and incubation was continued. Under this regimen, each population progressed through ∼6.6 generations per day. At ∼50-generation intervals, an aliquot of each culture was stored in 25% (vol/vol) glycerol at −70°C.
TABLE 1.
B. subtilis strains and plasmids used in this study
| Strain or plasmid | Genotype, phenotype, or description | Source or referencea |
|---|---|---|
| Strains | ||
| GP45 | trpC2 rnjB::spc Spcr | J. Stülke |
| WN624 | trpC2 amyE::spc Spcr, ancestral strain | 17 |
| WN628 | trpC2 amyE::cat Cmr, ancestral strain | 17 |
| WN1106 | trpC2 amyE::spc Spcr, evolved to enhanced growth at 5 kPa | 8 |
| WN1261 | trpC2 amyE::neo, Neor in WN624 background | pECE141→WN624 (tf); this study |
| WN1278 | trpC2, amyE::neo, Neor in WN1106 background | pECE141→WN1106 (tf); this study |
| WN1279 | trpC2 amyE::cat, Cmr in WN1106 background | pDAG32→WN1106 (tf); this study |
| WN1518 | trpC2 amyE::neo rnjB::spc, Neor Spcr in WN624 background | GP45→WN1261 (tf); this study |
| WN1519 | trpC2 amyE::neo rnjB::spc, Neor Spcr in WN1106 background | GP45→WN1278 (tf); this study |
| 50–1,000 generations | amyE::spc Spcr; 20 frozen stock populations representing generations 50 to 1,000 of the 5-kPa evolution expt | 8 |
| Plasmids | ||
| pECE73 | pCm::Neo antibiotic switching cassette | BGSC (16) |
| pECE141 | pSpc::Neo antibiotic switching cassette | BGSC (16) |
| pDAG32 | pSpc::Cm antibiotic switching cassette | BGSC (16) |
Abbreviations: tf, transformation; BGSC, Bacillus Genetic Stock Center.
DNA extraction and quality control for WGS.
Overnight cultures of strains WN624 and WN1106 were prepared in LB medium containing the appropriate antibiotic at 37°C. Cells were harvested by centrifugation, and RNA-free genomic DNA (gDNA) was purified as previously described (10). DNA concentrations were measured using a Qubit fluorometer and the Quant-iT double-stranded DNA (dsDNA) broad-range assay kit (Life Technologies, Grand Island, NY) according to the manufacturer's guidelines. The 260/280 nm absorbance ratio for DNA purity was determined by UV spectrophotometry to be >1.8 for all samples.
Oligonucleotide primers.
The identities and sequences of all oligonucleotide primers used in this study are presented and explained in Table 2.
TABLE 2.
Oligonucleotide primers used in this study
| Primer type | Olignucleotide | Sequence (5′→3′) |
|---|---|---|
| WN624a | CITZF230 | GGGGAAATCACAGAATGGAA |
| CITZR1225 | ATGTACGGCTCAGCGTTTTC | |
| COMPF1673 | TGAACGAGGCGGTAGTTCTT | |
| COMPR2285 | GTGCTTGACCGCATTAGACA | |
| EPSCF686 | TGACGACCAAACGAAGCATA | |
| EPSCR1311 | TGTTTTTGACCGCCTCTTCT | |
| GERAF855 | TCACCATTTCCGCAGATACA | |
| GERAR1584 | TTGGGAGACGGATAATGGAG | |
| GLTAF2805 | ATGTCATTCGGGTCCTTGAG | |
| GLTAR3774 | TTGCTTTCCAGTGCTCCTTT | |
| ILVCF106 | ACAGGTACAACGGCTTTTGC | |
| ILVCF366 | GCCCAAGAAGACGGACATAA | |
| ILVCR1136 | TTGATGTTCGTTCTCGCTTG | |
| ILVCR1186 | TGTTTCACAAACGGCATCAT | |
| OPPDF446 | GCTGTCCGAAAAAGAAATGC | |
| OPPDR1409 | GATTCACCAACCAGCCCTAA | |
| PGDSF206 | CACACTGGCAAACTGGAAGA | |
| PGDSR1164 | CAAATCCCGTCTCAGGTGTT | |
| RECAF183 | AGCCTGGGCATGTGTTTATC | |
| RECAR1106 | CCCGAACATAACACCGACTT | |
| RLUBF608 | GAAAGGAATCCCGCCTAAAG | |
| RLUBR1198 | GCTCGATACGTTTCCCATTC | |
| RLUDF173 | AGCGGGAAAAAGAAAAAGGA | |
| RLUDR1150 | TTCAGCTTCGTTTATTATTTCAGA | |
| MSWCF372 | GGAGAGCATTCCTGTTTTCG | |
| MSWCR1079 | CGCCATTTCCGTATCAGAAT | |
| SACAF1339 | CGGAGTCTGGCTTTTCAATC | |
| SACAR1993 | CGGCAGCCTGTTTATTTGAT | |
| SCOCF82 | AAAAACCCTTTCGTCGCAAT | |
| SCOCR1037 | GAGCAAGGAAGAAGGGGAAG | |
| SEPFF133 | GCTGAGAGAACTCCGTGACC | |
| SEPFR940 | TCGGAGGGATGATTTTTCTG | |
| SFTAF480 | AGAGAGCGTCCCAGAAATGA | |
| SFTAR1454 | AGACGATTTGGAACCCTGCT | |
| SIGIF137 | TTCTCCTTTGCGAATCCCTA | |
| SIGIR1004 | TCAGCAACGTGACGAATTTT | |
| SSRBF268 | GAATCGCACTCGGCTTAGAC | |
| SSRBR1008 | TGTCCGACAGTTGAAAGCAG | |
| TRMDF262 | TTCTTCAAAAAGCCCAGGAA | |
| TRMDR1147 | AAGTGTCACCAGGACGGAAC | |
| UVRXF275 | GGCTGTAACAATGGGGCTTA | |
| UVRXR951 | GCATCAACGGAGCACCTATT | |
| VEGF160 | GCTTTACGCCGTTTATGGAA | |
| VEGR804 | AAAGGGCAAAACAAAAAGCA | |
| YOJAR937 | TGTTGCCATTGCCATAAAGA | |
| YOQAF81 | CGGAGTGACAAGTGAAATGC | |
| YOZTF187 | TTGGATGTCCCGATGAAAAT | |
| YOZTR842 | CTTGAGAGCGGGATGGTAAT | |
| YPIBF97 | ATTTTGCGAATCGAGGATGA | |
| YPIBR806 | TCTCCACCTCATCGTTCTCC | |
| YQEZF354 | CGTTCGTTACAAGACGCAAA | |
| YQEZR1217 | AGCTGAGGCGATCAAGAGAG | |
| YUTEF14 | TTGTGGGAGACTGGGGATAC | |
| YUTER749 | ACAAATTCACACGCCTCCTC | |
| YXBDF121 | TGACCCCAGAGCAAATCTTC | |
| YXBDR712 | TTTCGGGCTTCATCTTTCTG | |
| YXJMF412 | TCCTGACCGATTTCTTTTGG | |
| YXJMR919 | TCTGAATCCTCCTTGCTGCT | |
| WN1106b | bacD-F3871584 | AGAGCAGCACGGAAATCTTCA |
| bacD-R3872064 | TCATCGCTGATCTTGGAGGC | |
| fliI-F1695745 | TGCTGATGAAAAAGCTCAAAAAGG | |
| fliI-R1696207 | GCGGTTCTCCAAAAGCATCG | |
| parC-F1935866 | ACACGGTTGAATTTGTGCCG | |
| parC-R1936322 | TCACGAACCTCTGAGATGCC | |
| resD-F2417342 | GAAGCTTTTCCCGATCATACACC | |
| resD-R2417791 | ATGGTGATGAAGCCATTGCC | |
| rnjB+261F | AACAAGCTGTCCGTTCCAGT | |
| rnjB+771R | TTCCGGCTACGGCAATCTTT | |
| walK-F4152887 | CAGAGGCTAGCTTTCTGCGT | |
| walK-R4153352 | GTGGCTGGGAAACAAACGAC | |
| yto-F2998178 | TCATCCGTGTTAAAGCCCCC | |
| yto-R2998587 | ATCGATTCACTGCCTGTCGG | |
| yvlD-F3606521 | GCACAACAGAGGGAGTGCAA | |
| yvlD-R3607016 | GCCAGCCTCATTTTATCGATCTT |
Primers used for PCR amplification and confirmation by Sanger sequencing of the SNP-containing regions found in strain WN624, which differed from the strain 168 sequence (see Table S1 in the supplemental material).
Primers used to verify SNPs identified in strain WN1106 (see Table 3).
WGS and mutation identification.
Samples of DNA from each strain were submitted to Vanderbilt Technologies for Advanced Genomics, Vanderbilt University, Nashville, TN (http://vantage.vanderbilt.edu/). WGS was performed using the Illumina HiSeq 2000 system (Illumina Inc., San Diego, CA). Details of the processing of the raw data and the identification of mutations can be found in the Materials and Methods in the supplemental material. The sequencing reads were mapped to the Bacillus subtilis strain 168 reference genome (GenBank accession number AL009126.3). Mapping statistics for each strain are listed in Table S1 in the supplemental material. Single-nucleotide polymorphisms (SNPs) and insertion/deletions identified by WGS are listed for strains WN624 (see Table S2 in the supplemental material) and WN1106 (Table 3). Mutational calls were analyzed based on (i) presence in the evolved strain but not the ancestor, (ii) high mapping quality as determined by the Unified Genotyper tool, and (iii) visual confirmation from BamView, a mapping visualization tool.
TABLE 3.
SNPs and deletion present in LP-evolved strain WN1106 and absent in ancestral strain WN624
| Gene | Position on genome | Annotated function(s) | Mutation | Amino acid change |
|---|---|---|---|---|
| fliI | 1695979 | Flagellum-specific ATP synthase; motility and chemotaxis | C→A | P35T |
| rnjB | 1749958 | RNase J2; RNA processing and degradation | ΔAGATCGCCA | Δ183-AKI-185 |
| parC | 1936060 | Subunit of DNA topoisomerase IV; chromosome segregation and compaction | G→C | D205H |
| resD | 2417575 | Two-component response regulator; regulation of anaerobic respiration | G→A | P110Q |
| ytoI | 2998392 | Unknown | C→A | V77F |
| yvlD | 3606764 | Unknown | A→T | Stop120K |
| bacD | 3871798 | Alanine-anticapsin ligase; bacilysin biosynthesis | C→T | E97N |
| walK | 4153105 | Two-component sensor kinase; control of cell wall metabolism | G→A | T195M |
| rnjBa | 1749949 | RNase J2 | C→A | C177Stop |
Point mutation found in rnjB, generations 600 to 950.
Mutation verification.
SNPs in strain WN624 found to differ from the published strain 168 sequence (see Table S2 in the supplemental material) and the seven SNPs and the single insertion/deletion found in WN1106 (Table 3) were all PCR amplified using the primer pairs listed in Table 2 and verified by Sanger sequencing either at the Plant-Microbe Genomics Facility at The Ohio State University or at the University of Florida Interdisciplinary Center for Biotechnology Research (UF-ICBR).
Kinetics of mutation sweeps during LP evolution.
To determine at which time each mutation occurred during the 5-kPa evolution experiment that resulted in strain WN1106 (8), aliquots of 20 frozen glycerol stock cultures that had been stored at 50-generation intervals during the 1,000-generation experiment (8) (Table 1) were thawed and grown overnight in liquid LB medium plus Spc. Genomic DNA was prepared from each population and PCR amplified with the primer pairs indicated in Table 2. The sequence chromatographs from the amplified regions were inspected to identify the location of each SNP, which was present as a double peak in heterogeneous populations. Each peak height was measured at this position and converted into a ratio of the mutant nucleotide fluorescence signal to the total fluorescence signal at that position. The proportion of the mutant allele peak height to the total peak height versus the generation was then plotted for each mutation.
In silico analysis of mutations.
Secondary-structure predictions were performed using the online CFSSP tool at www.biogem.org (13, 14). Tertiary-structure predictions were conducted using Swiss Model Workspace for RnjB, BacD, ResD, ParC, and FliI mutant protein sequences with the PDB structure available used as a reference, i.e., 3ZQ4, 3VMM, 1B00, 1ZVU, and 2DPY, respectively. Structural alignments were conducted using Pymol. ResD was aligned and its structure was determined based on two transcriptional response regulators whose structures have been determined, Escherichia coli PhoB and Mycobacterium tuberculosis MtrA. Alignments were performed using the online Clustal Omega tool (http://www.ebi.ac.uk/Tools/msa/clustalo/). Detailed discussion of in silico analyses is presented in Results in the supplemental material.
Competition assays.
Relative fitness gains or losses that occurred during the 5-kPa evolution experiment were determined by competition assays as described previously (8). From the 20 frozen stocks from LP-evolved populations taken at 50-generation intervals, individual overnight cultures were prepared in liquid LB medium plus Spc. Each culture was competed against either strain WN628 (a congenic Cmr version of ancestral strain WN624) or WN1279 (a congenic Cmr version of LP-evolved strain WN1106). Fresh overnight cultures of each strain to be competed were grown in LB medium containing the appropriate antibiotic, and both strains to be competed were diluted 1:200 into the same flask containing LB medium without antibiotic. The mixed cultures were propagated as described above for three to four passages of 1:100 dilutions. At daily intervals, aliquots were removed from the culture and the relative numbers of each strain were determined by serial dilution and viable counts on the respective selective medium. Competition assays were performed in duplicate at 27°C, 5 kPa (8, 12), and relative fitness values were calculated as follows. A selection coefficient, S, was calculated from each competition by the formula S = [lnR(t)/R(t − 1)]/t, where R is the ratio of the number of bacteria of the test strain to the number of bacteria of the reference strain, and t is the number of generations (15). Relative fitness is defined as 1 + S.
Statistical analyses.
Basic statistical parameters and analyses of variance (ANOVA) were performed using commercial statistical software (Kaleidagraph, version 4.5.2; Synergy Software, Reading, PA). Differences with a P of ≤0.05 were considered statistically significant.
RESULTS AND DISCUSSION
WGS was used to determine the genomic changes that occurred in strain WN1106 after 1,000 generations of experimental evolution from ancestral strain WN624 at 5 kPa.
Genome of ancestral strain WN624.
Ancestral strains WN624 and WN628 were originally constructed by transformation of an amyE::spc or an amyE::cat cassette, respectively (16) into the version of laboratory strain B. subtilis 168 housed in W. L. Nicholson's strain collection (17). Comparison of the strain WN624 genome sequence with the published sequence of strain 168 (18) resulted in identification of 30 SNPs differing between the two strains, and the presence of these SNPs was verified by Sanger sequencing (see Table S2 in the supplemental material). Interestingly, 22 of these SNPs were also found to occur in the published genome sequence of B. subtilis strain QB928, one of the original standard kit strains constructed in the Dedonder lab in France during the 1970s to facilitate genetic mapping experiments (19). The construction of QB928 was somewhat complex, but its genetic markers were transferred to a strain 168 genetic background by transformation. The sequence polymorphisms that were identified strongly suggest that WN624, WN628, and QB928 share an ancestral strain derived from the original strain 168 of Burkholder and Giles (20).
Genome of LP-evolved strain WN1106.
The genomic sequence of LP-evolved strain WN1106 differed from the ancestral WN624 sequence by only seven SNPs and a single 9-bp deletion (Table 3). SNPs were located within the coding regions of five known genes, fliI, parC, resD, bacD, and walK, and in two genes of unknown function, ytoI and yvlD (Table 3); no mutations were found in noncoding or intergenic regions. In addition, a 9-bp deletion was identified in the rnjB gene, which we designated rnjB(Δ9). This deletion was predicted to cause an in-frame, 3-amino-acid deletion in the rnjB gene product, RNase J2 (Table 3). Inspection of the DNA sequence surrounding the rnjB(Δ9) mutation revealed that this deletion was likely caused by a recombination event occurring between a pair of 6-bp direct repeats separated by 3 bp (Fig. 1).
FIG 1.
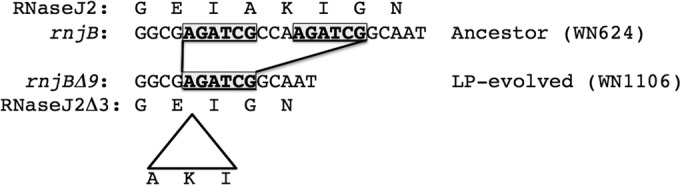
DNA sequence of the rnjB gene and deduced amino acid sequence of RNase J2 in ancestral strain WN624 (top) and the DNA sequence of the rnjB(Δ9) mutant and deduced amino acid sequence of the resulting RNase J2Δ3 in LP-evolved strain WN1106 (bottom). Direct repeats (boxed, boldface type) are indicated. A triangle indicates deletion of 3 amino acids (AKI) from the deduced RNase J2 sequence in strain WN1106.
Kinetics of mutational sweeps during LP evolution.
To determine when each of the mutations occurring in strain WN1106 arose during the 5-kPa evolution experiment, the DNA sequence surrounding each mutation was PCR amplified from each of the 20 frozen stock cultures preserved at ∼50-generation intervals (Table 1), using the PCR primer pairs described in Table 2. The proportion of mutant alleles in each population, estimated from the sequencing chromatographs as described in Materials and Methods, was plotted against the generation of the experiment (Fig. 2A). From inspection of the data, it could be seen that the mutations appeared and swept the culture with distinct kinetic patterns. The patterns could roughly be grouped into (i) mutations occurring early (generations 200 to 400) during the evolution experiment [e.g., the SNPs in fliI, parC, ytoI, and the rnjB(Δ9) deletion] and (ii) mutations detected toward the end (generations 800 to 1000) of the evolution experiment (e.g., the SNPs in walK, yvlD, resD, and bacD) (Fig. 2A).
FIG 2.
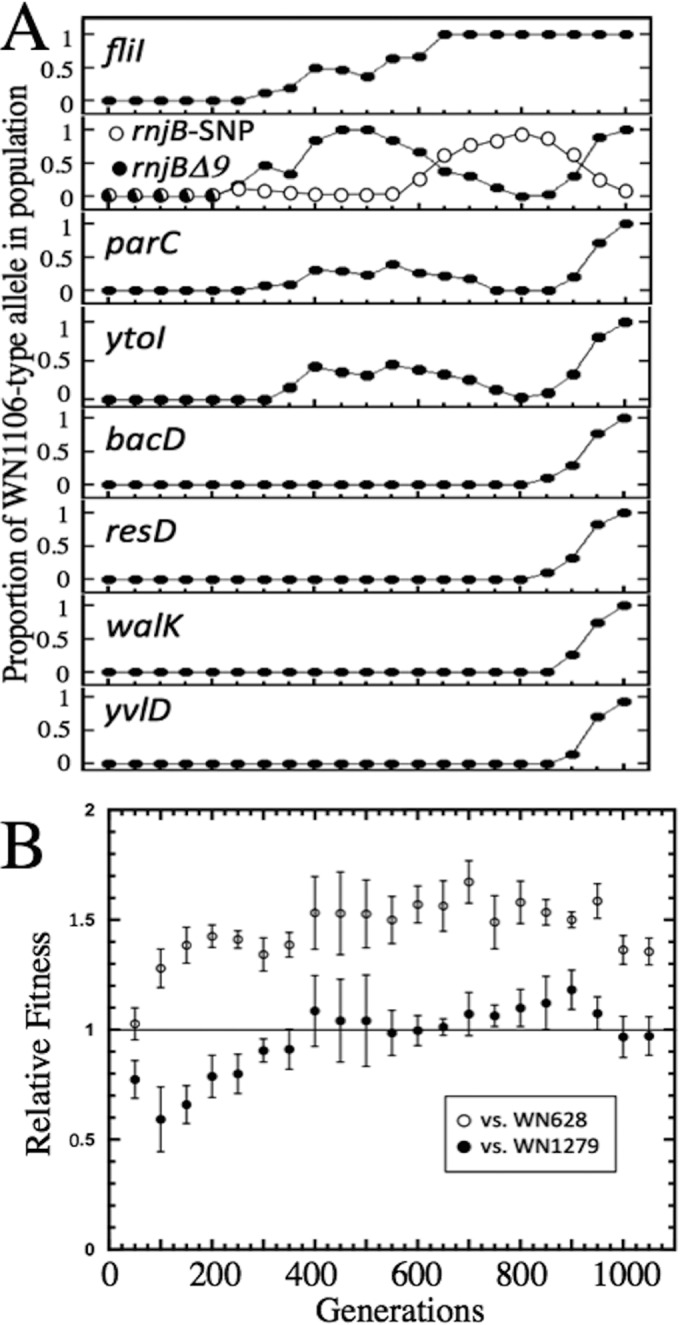
(A) Kinetics of mutational sweeps during LP evolution experiment. Data are depicted as the proportion of the mutant allele divided by the total (mutant + ancestral) allele (filled circles). For rnjB, a second allele was discovered during this experiment, i.e., an SNP resulting in a nonsense mutation at codon 177 (Table 3), which is depicted by white circles. (B) Results from competition experiments in which each frozen stock of the LP-evolving populations (Spcr), taken at 50-generation intervals, was competed at 5 kPa against either ancestral strain WN628 (Cmr; open circles) or LP-evolved strain WN1279 (Cmr; filled circles). Competition at the zero-generation mark used ancestor strain WN624. Data are shown as averages ± standard deviations of results of duplicate experiments. See the text for details.
Details of the dynamics of mutations detected during LP evolution. (i) SNP in fliI.
The most straightforward example of an early mutation was the SNP in fliI that was first detected at generation 300 and then swept through and apparently became fixed in the population by generation 650 (Fig. 2A). In support of previous reports that fliI mutants are deficient in flagella and hence nonmotile, we confirmed by phase-contrast microscopy that overnight cultures of LP-evolved strain WN1106 were not motile, whereas overnight cultures of ancestral strain WN624 were highly motile (data not shown).
(ii) Deletion and SNP in rnjB.
Inspection of the sequence surrounding rnjB revealed that the rnjB(Δ9) deletion was first detected at generation 250 and had apparently swept the population by generation 450. However, the deletion did not become fixed in the population, but rather its frequency declined from generations 550 to 800. The decline of rnjB(Δ9) was soon followed at generation 600 by the appearance of a new subpopulation carrying an SNP in rnjB that resulted in the replacement of codon C177 with a premature stop codon in the deduced RNase J2 amino acid sequence (Table 3). By generation 800, this new SNP in rnjB had come to dominate the population; however, from generations 800 to 1000, it was again replaced by the original rnjB(Δ9) deletion (Fig. 2A).
(iii) SNPs in parC and ytoI.
The appearance of the SNPs detected in parC and ytoI followed a general pattern similar to that seen with the rnjB(Δ9) deletion: both SNPs appeared early at generations 200 to 300, rose in frequency to a maximum at generation 500, decreased in frequency until generation 800, and then rapidly swept the population by generation 1000 (Fig. 2A).
(iv) SNPs in bacD, resD, walK, and yvlD.
In all cases, the SNPs identified in bacD, resD, walK, and yvlD did not appear until generations 850 to 900 and thereafter rapidly increased in the population (Fig. 2A).
Taken together, the data presented in Fig. 2A illustrate the complex dynamics that the population experienced during its evolution to enhanced growth at 5 kPa. It is particularly interesting to note that the population carrying the particular set of mutations identified in strain WN1106 did not actually arise until that last 150 to 200 generations of the experiment, suggesting that a strong selective sweep occurred of a strain which had acquired this particular collection of mutations. At present, it is not possible to distinguish which mutation(s) was responsible for the sweep from generations 850 to 1000 and which were genetic “hitchhikers” (21). Of course, the approach leading to the results illustrated in Fig. 2A yields information about the history of only the eight mutations identified in strain WN1106 (Table 3); a more comprehensive picture of the dynamics of this LP evolution experiment would require WGS of each of the frozen stock populations.
Gains in competitive fitness during LP evolution.
To determine the fitness gains that occurred during evolution at 5 kPa, each frozen stock population stored at 50-generation intervals (Spcr) was competed at 5 kPa in duplicate against congenic ancestral strain WN628 (Cmr) or LP-evolved strain WN1279 (Cmr; congenic to LP-evolved strain WN1106) (Fig. 2B). Interestingly, this competition experiment revealed that the LP-evolved population gained a significant increase in relative fitness over ancestral strain WN628 after only 100 generations of exposure to 5 kPa, and fitness rapidly increased until at 200 generations its value was 1.43 ± 0.05 (Fig. 2B). Thereafter, the relative fitness of the LP-evolving population rose slowly to a maximum value of 1.67 ± 0.1 at generation 650 and then fell to a final value of 1.36 ± 0.06 by generation 1000 (Fig. 2B). This final relative fitness value compares well with that of strain WN1106 (1.26 ± 0.2), which had been originally isolated from the 1,000-generation culture (8, 12).
Examination of the data from the competition experiments of the frozen cultures versus LP-evolved strain WN1279 indicated that at generation 0, the population exhibited a lower relative fitness of 0.77 ± 0.09 (Fig. 2B). Because the population at generation 0 is a clonal population of ancestral strain WN624, this value was expected and corresponds well with the previously determined relative fitness of ancestral strain WN624 versus LP-evolved strain WN1106 (0.79 ± 0.2) (8, 12). A steady rise in relative fitness of the LP-evolving population was noted until generation 350, by which time the relative fitness of the LP-evolving population versus strain WN1279 had stabilized at essentially ∼1 for the remainder of the 1,000-generation experiment (Fig. 2B).
Examination of the data (Fig. 2B) indicated that increased fitness at 5 kPa occurred very early during the LP evolution experiment, reaching a level commensurate with LP-evolved strain WN1106 by the first 150 generations. Interestingly, none of the SNPs identified by WGS of strain WN1106 were found at significant levels by 150 generations (Fig. 2A). Furthermore, it was observed that LP fitness of the evolving population actually was highest between generations 400 to 700 and even declined by generation 1000 (Fig. 2B). Taken together, the data strongly suggest that the mutation(s) which increased LP fitness arose early in the population but was subsequently lost. Such a putative mutation(s) could be identified by WGS of the evolving populations from earlier time points in the experiment and recovered for further testing.
Knockout mutation ΔrnjB::spc results in increased fitness.
Examination of the data in Fig. 2 indicated that the earliest-appearing and most persistent mutation(s) in a gene of global regulatory significance during LP evolution appeared to reside within the rnjB gene–in both the rnjB(Δ9) deletion and the rnjB nonsense mutation, respectively. We thus reasoned that the rnjB gene product may be involved in LP growth and that further testing of rnjB mutations was warranted. We were curious to determine how the rnjB mutations in strain WN1106 affected the activity of RNaseJ2, if at all. We therefore obtained from the Stülke laboratory strain GP45 carrying a deletion-insertion mutation which removed the entire rnjB gene and replaced it with a Spcr cassette (ΔrnjB::spc). Strains WN1518 (ΔrnjB::spc amyE::neo in a WN624 background) and WN1519 (ΔrnjB::spc amyE::neo in a WN1106 background) were constructed (Table 1) in order to directly test the effect of the ΔrnjB::spc knockout mutation in competition experiments versus their congenic counterparts, ancestral strain WN628 (amyE::cat) and LP-evolved strain WN1106 (rnjBΔ9 amyE::spc). (Note that strains WN1519 and WN1106 also both contain the full contingent of SNPs at the seven other loci described in Table 3.) Competition experiments were performed in liquid LB medium at 27°C and at either ∼101 kPa or 5 kPa.
Several interesting observations came from this experiment (Fig. 3). First, the ΔrnjB::spc knockout strain WN1518 demonstrated a higher relative fitness (1.42 ± 0.09) than the congenic ancestral strain WN628 at 5 kPa (Fig. 3), indeed even greater than that previously measured for strain WN1106 at 5 kPa (1.26 ± 0.2) (8, 12). Thus, the ΔrnjB::spc knockout strain appeared to confer higher relative fitness at LP than did the rnjB(Δ9) in-frame deletion.
FIG 3.
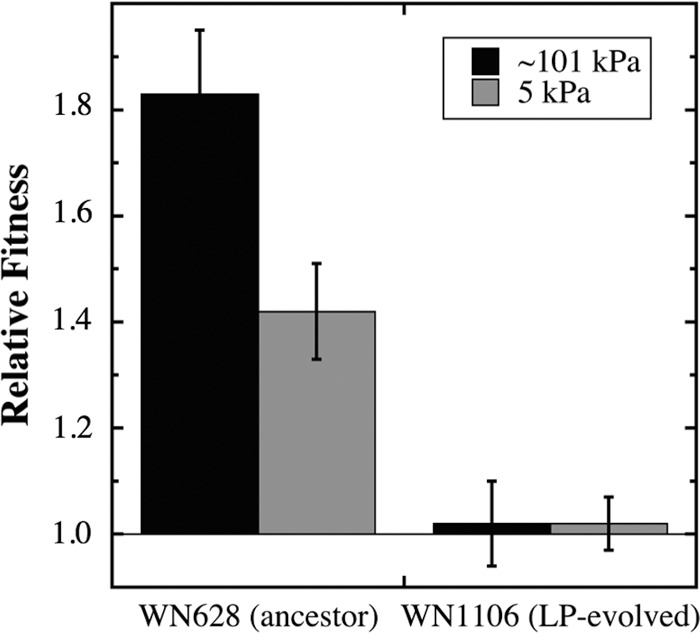
Competition experiments to test the effect of the rnjB::spc knockout mutation on relative fitness. (Left) Strain WN1518 (ΔrnjB::spc amyE::neo in WN624 background) was competed against congenic ancestral strain WN628 (amyE::cat). (Right) Strain WN1519 (ΔrnjB::spc amyE::neo in WN1106 background) was competed against congenic strain WN1106 [rnjB(Δ9) amyE::spc]. Competitions were performed in liquid LB medium at 27°C and either ∼101 kPa (black bars) or 5 kPa (gray bars). Data are averages ± standard deviations of results of duplicate experiments.
Second, surprising to us was the observation that the ΔrnjB::spc knockout strain WN1518 also exhibited a dramatically higher relative fitness than the congenic ancestral strain WN628 (1.83 ± 0.12) at Earth's normal atmospheric pressure of ∼101 kPa (Fig. 3). This result is in stark contrast to the previous observation that ancestral strains WN628 and LP-evolved strain WN1106 showed no difference in relative fitness when competed at ∼101 kPa (8). The observation suggests that deletion of rnjB somehow allows B. subtilis to grow more efficiently in LB medium at 27°C, an observation we are at present investigating in further detail.
Third, strain WN1519 carrying the ΔrnjB::spc knockout mutation exhibited no relative fitness advantage compared to congenic strain WN1106 carrying the rnjB(Δ9) mutation, when the two strains were competed at either ∼101 kPa or 5 kPa (Fig. 3). This observation indicated that the effect of the rnjB(Δ9) in-frame deletion in WN1106 is essentially the same as that of the complete loss of the rnjB gene in strain WN1519, suggesting that the 3-codon in-frame deletion in WN1106 might render RNase J2 inactive. While this third observation apparently stands at odds with the first two, it is possible that the relative fitnesses measured in the competitions of strain WN1519 against WN1106 are altered somehow by the presence of the additional seven SNPs in these two strains. A more definitive answer to this discrepancy can be obtained by construction of a strain carrying only the rnjB(Δ9) mutation in a clean genetic background; this work is currently in progress.
In summary, this communication describes the use of WGS to investigate the experimental evolution of B. subtilis to enhanced growth at the near-inhibitory LP of 5 kPa. Genome sequencing of LP-evolved strain WN1106 revealed mutations in only eight protein-coding genes after 1,000 generations of evolution at LP (Table 3). Each gene will be discussed below in order of appearance in the evolving culture. In addition, a detailed in silico analysis of each gene product can be found in Results in the supplemental material.
rnjB.
Two separate mutations in the rnjB gene, rnjB(Δ9) and rnjB nonsense, were detected as early as generation 250, and their relative proportions fluctuated until rnjB(Δ9) swept the population at generations 850 to 1000 (Fig. 2A). The rnjB gene encodes RNase J2 (also known as RnjB), which together with RNase J1 (also known as RnjA) form an RNase J1/J2 complex comprising part of the RNA degradosome, one of the major mRNA global processing systems in Bacillus subtilis, reviewed previously (22, 23). The known primary function of RnjB is that of an endoribonuclease (24, 25), whereas RnjA, the essential component of the complex, exerts the main exoribonuclease function as well as acting as an endoribonuclease (22, 25, 26). (It should be noted here that the actual function and activity of RNase J2 are currently unclear and a subject of debate.) To date, no phenotype has ever been ascribed to an rnjB mutant; however, our results reported here have uncovered a role for rnjB in regulating the growth of B. subtilis at both LP and standard atmospheric pressure and 27°C (Fig. 3).
It should be noted that short deletions at repeat regions which alter enzyme activity and cell fitness under stressful conditions are not unprecedented. A well-studied example in B. subtilis is the activation of a second glutamate dehydrogenase (GDH) encoded by the cryptic gudB gene via an in-frame 9-bp deletion event; this deletion occurs at high frequency in response to glutamate starvation stress in mutants lacking the major GDH (27–29). A second example concerns in vivo development of daptomycin resistance in a clinical isolate of Enterococcus faecalis, which was shown to result from in-frame deletions in three genes involved in the response to antibiotics causing cell envelope stress (30).
fliI.
A mutation in fliI, the gene encoding an ATPase needed for export of flagellar proteins (31), was detected at generation 300 and swept the population by generation 650. Mutations that result in a decrease in or lack of motility have been reported previously in laboratory evolution experiments in Myxococcus xanthus (32), Campylobacter jejuni (33), Escherichia coli (34), and B. subtilis (35, 36). It is thought that during long-term growth in shake flasks, where nutrient gradients cannot form, motility and chemotaxis functions serve no selective advantage and are energetically expensive; thus, nonmotile mutants enjoy a growth advantage (37). It may be that for long-duration growth in flask cultures, motility loss is an early step toward fitness increase regardless of any externally imposed selective pressure.
ytoI.
Although the SNP in ytoI appeared at generation 300 during LP evolution, it failed to sweep the population until generations 850 to 1000 and may be a genetic hitchhiker. Although widely conserved among the genus Bacillus, the function of the ytoI gene product is currently unknown. It has been predicted to be a transcription factor belonging to the GntR family (38), a prediction supported by its localization to the nucleoid region (39). Protein BLAST at NCBI revealed that the putative 48-kDa ytoI gene product contains a number of highly conserved protein domains (winged helix-turn-helix, DRTGG, CBS pair, and hot-dog superfamily) and is classified to COG (Clusters of Orthologous Groups) 4109, “Predicted transcriptional regulator containing CBS domains.”
parC.
The parC gene encodes ParC, topoisomerase IV subunit A, which is an essential protein involved in chromosome decatenation and replication fork movement (40, 41). It was noted that the SNPs in both ytoI and parC both arose at generation 300, soon after the appearance of the rnjB(Δ9) mutation, and all three mutations followed essentially similar trajectories (Fig. 2A), suggesting that these three mutations may share a lineage. Because ParC is an essential gene product, it might be predicted that the SNP identified would alter, rather than abolish, its activity.
bacD.
The SNP detected in bacD arose late in the LP evolution experiment at around generation 850 and rapidly swept through the population. The bacD gene encodes BacD, an l-amino acid ligase which in vivo is responsible for the ATP-dependent ligation of l-alanine and l-anticapsin and the production of the extracellular dipeptide antibiotic bacilysin (42). It is difficult to envision how a bacD mutation would confer a selective advantage in liquid culture, and thus it seems rational to presume that the SNP in bacD may be a genetic hitchhiker.
resD.
The SNP detected in resD also appeared late in the LP evolution experiment at generation 800. The resDE operon encodes the ResD and ResE proteins, which comprise a well-characterized two-component system in B. subtilis that regulates anaerobic gene expression; ResE serves as the sensor kinase, and ResD is the response regulator (43). Whether the SNP in ResD is a genetic hitchhiker or might actually exert an effect on LP growth is unclear at present. Previous experiments showed that LP-evolved strain WN1106 actually exhibited decreased fitness compared to ancestral strain WN624 when competed at low-oxygen (1% O2) and standard pressure (∼101 kPa) (12). However, this observation does not rule out the possibility that the SNP in resD might be beneficial under low-oxygen conditions coupled with LP.
walK.
The SNP identified in walK also appeared late in LP evolution at generation 900. The walK gene is located downstream from walR, and their gene products form an essential two-component system which controls cell wall metabolism, WalK being the sensor kinase of this system. First discovered in B. subtilis, the WalK-WalR two-component system has since been found to be widespread among the low-G+C Gram-positive bacteria (44, 45). The activity of WalK has been reported to respond to fluctuations in membrane fluidity, and depletion of WalK in the cell activates the two-component system DesK-DesR, resulting in increased des transcription (44). It was reported previously that WN1106 had increased transcription of des at 5 kPa, but it was unclear what the underlying reason for this increase was, as localized sequencing of the des and desKR genes did not reveal any mutations (12). It may be that the underlying genomic explanation for upregulation of des at 5 kPa in WN1106 is due to the mutation in walK. Because WalK is an essential gene product, it might be predicted that the SNP identified would alter, rather than abolish, its activity.
yvlD.
The SNP in yvlD appeared late in LP evolution (generations 850 to 1000) and may be a genetic “hitchhiker.” The yvlD gene is known to be part of the SigW regulon and is induced by cell wall stress (46, 47). This gene is highly conserved among the family Bacillaceae, and protein BLAST at NCBI revealed that the putative 12-kDa ytoI gene product is likely an integral membrane protein of unknown function. It is classified under COG1950 “Predicted membrane protein” and belongs to the domain of unknown function (DUF) 360 superfamily.
Aside from mutations in rnjB, at present the putative roles, if any, of the additional seven SNPs identified in LP-evolved strain WN1106 are unclear. However, it is interesting to note that both the walK and yvlD gene products are related to maintenance of cell wall integrity. Experiments are currently in progress to further elucidate the role(s) played by the gene products identified here in their putative enhancement of LP growth in B. subtilis.
Supplementary Material
ACKNOWLEDGMENTS
We thank: Jörg Stülke for the generous donation of strain GP45, Babraham Bioinformatics, developers of the FASTQC tool for visualization of read qualities, and the anonymous reviewers for their excellent suggestions for improvement of the manuscript.
This research was supported by grants from the NASA Astrobiology: Exobiology and Evolutionary Biology program (NNX08AO15G) to W.L.N. and the NASA Earth and Space Science Fellowship (NESSF) Program (NNX13AP74H) to S.M.W. D.R.Z. is supported by the National Science Foundation under award no. 1349029.
Opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the National Science Foundation.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01690-15.
REFERENCES
- 1.Horikoshi K, Antranikian G, Bull AT, Robb FT, Stetter KO (ed). 2011. Extremophiles handbook. Springer, Tokyo, Japan. [Google Scholar]
- 2.Michiels C, Bartlett DH, Aertsen A (ed). 2008. High-pressure microbiology. ASM Press, Washington, DC. [Google Scholar]
- 3.Smith DJ, Timonen HJ, Jaffe DA, Griffin DW, Birmele MN, Perry KD, Ward PD, Roberts MS. 2013. Intercontinental dispersal of bacteria and archaea by transpacific winds. Appl Environ Microbiol 79:1134–1139. doi: 10.1128/AEM.03029-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith DJ, Griffin DW, McPeters RD, Ward PD, Schuerger AC. 2011. Microbial survival in the stratosphere and implications for global dispersal. Aerobiologia 27:319–332. doi: 10.1007/s10453-011-9203-5. [DOI] [Google Scholar]
- 5.Smith DJ, Griffin DW, Schuerger AC. 2010. Stratospheric microbiology at 20 km over the Pacific Ocean. Aerobiologia 26:35–46. doi: 10.1007/s10453-009-9141-7. [DOI] [Google Scholar]
- 6.Burg SP. 2004. Postharvest physiology and hypobaric storage of fresh produce. CABI Publishing, Cambridge, MA. [Google Scholar]
- 7.Rummel JD, Beaty DW, Jones MA, Bakermans C, Barlow NC, Boston P, Chevrier V, Clark B, de Vera J-P, Gough RV, Hallsworth JE, Head JW, Hipkin VJ, Kieft TL, McEwen AS, Mellon MT, Mikucki J, Nicholson WL, Omelon CR, Peterson R, Roden E, Sherwood Lollar B, Tanaka KL, Viola D, Wray JJ. 2014. A new analysis of Mars ‘Special Regions’: findings of the second MEPAG Special Regions Science Analysis Group (SR-SAG2). Astrobiology 14:887–968. doi: 10.1089/ast.2014.1227. [DOI] [PubMed] [Google Scholar]
- 8.Nicholson WL, Fajardo-Cavazos P, Fedenko J, Ortiz-Lugo JL, Rivas-Castillo A, Waters SM, Schuerger AC. 2010. Exploring the low-pressure growth limit: evolution of Bacillus subtilis in the laboratory to enhanced growth at 5 kilopascals. Appl Environ Microbiol 76:7559–7565. doi: 10.1128/AEM.01126-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boylan RJ, Mendelson NH, Brooks D, Young FE. 1972. Regulation of the bacterial cell wall: analysis of a mutant of Bacillus subtilis defective in biosynthesis of teichoic acid. J Bacteriol 110:281–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cutting SM, Vander Horn PB. 1990. Genetic analysis, p 27–74. In Harwood CR, Cutting SM (ed), Molecular biological methods for Bacillus. John Wiley and Sons, Sussex, United Kingdom. [Google Scholar]
- 11.Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 12.Fajardo-Cavazos P, Waters SM, Schuerger AC, George S, Marois JJ, Nicholson WL. 2012. Evolution of Bacillus subtilis to enhanced growth at low pressure: up-regulated transcription of des-desKR, encoding the fatty acid desaturase system. Astrobiology 12:258–270. doi: 10.1089/ast.2011.0728. [DOI] [PubMed] [Google Scholar]
- 13.Chou PY, Fasman GD. 1974. Conformational parameters for amino acids in helical, beta-sheet, and random coil regions calculated from proteins. Biochemistry 13:211–222. doi: 10.1021/bi00699a001. [DOI] [PubMed] [Google Scholar]
- 14.Chou PY, Fasman GD. 1974. Prediction of protein conformation. Biochemistry 13:222–245. doi: 10.1021/bi00699a002. [DOI] [PubMed] [Google Scholar]
- 15.Dykhuizen D. 1990. Experimental studies of natural selection in bacteria. Annu Rev Ecol Syst 21:373–398. doi: 10.1146/annurev.es.21.110190.002105. [DOI] [Google Scholar]
- 16.Steinmetz M, Richter R. 1994. Plasmids designed to alter the antibiotic resistance expressed by insertion mutations in Bacillus subtilis, through in vivo recombination. Gene 142:79–83. doi: 10.1016/0378-1119(94)90358-1. [DOI] [PubMed] [Google Scholar]
- 17.Maughan H, Callicotte V, Hancock A, Birky CW, Nicholson WL, Masel J. 2006. The population genetics of phenotypic deterioration in experimental populations of Bacillus subtilis. Evolution 60:686–695. doi: 10.1111/j.0014-3820.2006.tb01148.x. [DOI] [PubMed] [Google Scholar]
- 18.Kunst F, Ogasawara N, Moszer I, Albertini AM, Alloni G, Azevedo V, Bertero MG, Bessieres P, Bolotin A, Borchert S, Borriss R, Boursier L, Brans A, Braun M, Brignell SC, Bron S, Brouillet S, Bruschi CV, Caldwell B, Capuano V, Carter NM, Choi SK, Codani JJ, Connerton IF, Cummings NJ, Daniel RA, Denziot F, Devine KM, Düsterhöft A, Ehrlich SD, Emmerson PT, Entian KD, Errington J, Fabret C, Ferrari E, Foulger D, Fritz C, Fujita M, Fujita Y, Fuma S, Galizzi A, Galleron N, Ghim SY, Glaser P, Goffeau A, Golightly EJ, Grandi G, Guiseppi G, Guy BJ, Haga K, et al. 1997. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 19.Dedonder RA, Lepesant JA, Lepesant-Kejzlarová J, Billault A, Steinmetz M, Kunst F. 1977. Construction of a kit of reference strains for rapid genetic mapping in Bacillus subtilis 168. Appl Environ Microbiol 33:989–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burkholder PR, Giles NHJ. 1947. Induced biochemical mutations in Bacillus subtilis. Am J Bot 34:345–348. doi: 10.2307/2437147. [DOI] [PubMed] [Google Scholar]
- 21.Burke MK. 2012. How does adaptation sweep through the genome? Insights from long-term selection experiments. Proc Soc Biol Sci 279:5029–5038. doi: 10.1098/rspb.2012.0799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Condon C. 2010. What is the role of RNase J in mRNA turnover? RNA Biol 7:316–321. doi: 10.4161/rna.7.3.11913. [DOI] [PubMed] [Google Scholar]
- 23.Lehnik-Habrink M, Lewis RJ, Mäder U, Stülke J. 2012. RNA degradation in Bacillus subtilis: an interplay of essential endo- and exoribonucleases. Mol Microbiol 84:1005–1017. doi: 10.1111/j.1365-2958.2012.08072.x. [DOI] [PubMed] [Google Scholar]
- 24.Even S, Pellegrini O, Zig L, Labas V, Vinh J, Bréchemmier-Baey D, Putzer H. 2005. Ribonucleases J1 and J2: two novel endoribonucleases in B. subtilis with functional homology to E. coli RNase E. Nucleic Acids Res 33:2141–2152. doi: 10.1093/nar/gki505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mathy N, Hébert A, Mervelet P, Bénard L, Dorléans A, Li de la Sierra-Gallay I, Noirot P, Putzer H, Condon C. 2010. Bacillus subtilis ribonucleases J1 and J2 form a complex with altered enzyme behaviour. Mol Microbiol 75:489–498. doi: 10.1111/j.1365-2958.2009.07004.x. [DOI] [PubMed] [Google Scholar]
- 26.Mäder U, Zig L, Kretschmer J, Homuth G, Putzer H. 2008. mRNA processing by RNases J1 and J2 affects Bacillus subtilis gene expression on a global scale. Mol Microbiol 70:183–196. doi: 10.1111/j.1365-2958.2008.06400.x. [DOI] [PubMed] [Google Scholar]
- 27.Gunka K, Tholen S, Gerwig J, Herzberg C, Stülke J, Commichau FM. 2012. A high-frequency mutation in Bacillus subtilis: requirements for the decryptification of the gudB glutamate dehydrogenase gene. J Bacteriol 194:1036–1044. doi: 10.1128/JB.06470-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gunka K, Stannek L, Care RA, Commichau FM. 2013. Selection-driven accumulation of suppressor mutants in Bacillus subtilis: the apparent high mutation frequency of the cryptic gudB gene and the rapid clonal expansion of gudB(+) suppressors are due to growth under selection. PLoS One 8:e66120. doi: 10.1371/journal.pone.0066120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Belitsky BR, Sonenshein AL. 1998. Role and regulation of Bacillus subtilis glutamate dehydrogenase genes. J Bacteriol 180:6298–6305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arias CA, Panesso D, McGrath DM, Qin X, Mojica MF, Miller C, Diaz L, Tran TT, Rincon S, Barbu EM, Reyes J, Roh JH, Lobos E, Sodergren E, Pasqualini R, Arap W, Quinn JP, Shamoo Y, Murray BE, Weinstock GM. 2011. Genetic basis for in vivo daptomycin resistance in enterococci. N Engl J Med 365:892–900. doi: 10.1056/NEJMoa1011138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Claret L, Calder SR, Higgins M, Hughes C. 2003. Oligomerization and activation of the FliI ATPase central to bacterial flagellum assembly. Mol Microbiol 48:1349–1355. doi: 10.1046/j.1365-2958.2003.03506.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Velicer GJ, Kroos L, Lenski RE. 1998. Loss of social behaviors by Myxococcus xanthus during evolution in an unstructured habitat. Proc Natl Acad Sci U S A 95:12376–12380. doi: 10.1073/pnas.95.21.12376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jerome JP. 2012. Genome evolution of Campylobacter jejuni during experimental adaptation. PhD thesis Michigan State University, East Lansing, MI. [Google Scholar]
- 34.Conrad TM, Frazier M, Joyce AR, Cho BK, Knight EM, Lewis NE, Landick R, Palsson B. 2010. RNA polymerase mutants found through adaptive evolution reprogram Escherichia coli for optimal growth in minimal media. Proc Natl Acad Sci U S A 107:20500–20505. doi: 10.1073/pnas.0911253107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maughan H, Nicholson WL. 2011. Increased fitness and alteration of metabolic pathways during Bacillus subtilis evolution in the laboratory. Appl Environ Microbiol 77:4105–4118. doi: 10.1128/AEM.00374-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brown CT, Fishwick LK, Chokshi BM, Cuff MA, Jackson JM, Oglesby T, Rioux AT, Rodriguez E, Stupp GS, Trupp AH, Woollcombe-Clarke JS, Wright TN, Zaragoza WJ, Drew JC, Triplett EW, Nicholson WL. 2011. Whole-genome sequencing and phenotypic analysis of Bacillus subtilis mutants following evolution under conditions of relaxed selection for sporulation. Appl Environ Microbiol 77:6867–6877. doi: 10.1128/AEM.05272-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mitchell JG. 2002. The energetics and scaling of search strategies in bacteria. Am Nat 160:727–740. doi: 10.1086/343874. [DOI] [PubMed] [Google Scholar]
- 38.Leyn SA, Kazanov MD, Sernova NV, Ermakova EO, Novichkov PS, Rodionov DA. 2013. Genomic reconstruction of the transcriptional regulatory network in Bacillus subtilis. J Bacteriol 195:2463–2473. doi: 10.1128/JB.00140-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meile JC, Wu LJ, Ehrlich SD, Errington J, Noirot P. 2006. Systematic localisation of proteins fused to the green fluorescent protein in Bacillus subtilis: identification of new proteins at the DNA replication factory. Proteomics 6:2135–2146. doi: 10.1002/pmic.200500512. [DOI] [PubMed] [Google Scholar]
- 40.Barnes MH, LaMarr WA, Foster KA. 2003. DNA gyrase and DNA topoisomerase of Bacillus subtilis: expression and characterization of recombinant enzymes encoded by the gyrA, gyrB and parC, parE genes. Protein Expr Purif 29:259–264. doi: 10.1016/S1046-5928(03)00068-8. [DOI] [PubMed] [Google Scholar]
- 41.Khodursky AB, Peter BJ, Schmid MB, DeRisi J, Botstein D, Brown PO, Cozzarelli NR. 2000. Analysis of topoisomerase function in bacterial replication fork movement: use of DNA microarrays. Proc Natl Acad Sci U S A 97:9419–9424. doi: 10.1073/pnas.97.17.9419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shomura Y, Hinokuchi E, Ikeda H, Senoo A, Takahashi Y, Saito J, Komori H, Shibata N, Yonetani Y, Higuchi Y. 2012. Structural and enzymatic characterization of BacD, an l-amino acid dipeptide ligase from Bacillus subtilis. Protein Sci 21:707–716. doi: 10.1002/pro.2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nakano M, Zuber P. 2002. Anaerobiosis, p 393–404. In Sonenshein AL, Hoch JA, Losick R (ed), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, DC. [Google Scholar]
- 44.Bisicchia P, Noone D, Lioliou E, Howell A, Quigley S, Jensen T, Jarmer H, Devine KM. 2007. The essential YycFG two-component system controls cell wall metabolism in Bacillus subtilis. Mol Microbiol 65:180–200. doi: 10.1111/j.1365-2958.2007.05782.x. [DOI] [PubMed] [Google Scholar]
- 45.Dubrac S, Bisicchia P, Devine KM, Msadek T. 2008. A matter of life and death: cell wall homeostasis and the WalKR (YycGF) essential signal transduction pathway. Mol Microbiol 70:1307–1322. doi: 10.1111/j.1365-2958.2008.06483.x. [DOI] [PubMed] [Google Scholar]
- 46.Cao M, Wang T, Ye R, Helmann JD. 2002. Antibiotics that inhibit cell wall biosynthesis induce expression of the Bacillus subtilis sigma(W) and sigma(M) regulons. Mol Microbiol 45:1267–1276. doi: 10.1046/j.1365-2958.2002.03050.x. [DOI] [PubMed] [Google Scholar]
- 47.Huang X, Gaballa A, Cao M, Helmann JD. 1999. Identification of target promoters for the Bacillus subtilis extracytoplasmic function sigma factor, sigma W. Mol Microbiol 31:361–371. doi: 10.1046/j.1365-2958.1999.01180.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


