Abstract
Corynebacterium glutamicum, a model organism in microbial biotechnology, is known to metabolize glucose under oxygen-deprived conditions to l-lactate, succinate, and acetate without significant growth. This property is exploited for efficient production of lactate and succinate. Our detailed analysis revealed that marginal growth takes place under anaerobic conditions with glucose, fructose, sucrose, or ribose as a carbon and energy source but not with gluconate, pyruvate, lactate, propionate, or acetate. Supplementation of glucose minimal medium with tryptone strongly enhanced growth up to a final optical density at 600 nm (OD600) of 12, whereas tryptone alone did not allow growth. Amino acids with a high ATP demand for biosynthesis and amino acids of the glutamate family were particularly important for growth stimulation, indicating ATP limitation and a restricted carbon flux into the oxidative tricarboxylic acid cycle toward 2-oxoglutarate. Anaerobic cultivation in a bioreactor with constant nitrogen flushing disclosed that CO2 is required to achieve maximal growth and that the pH tolerance is reduced compared to that under aerobic conditions, reflecting a decreased capability for pH homeostasis. Continued growth under anaerobic conditions indicated the absence of an oxygen-requiring reaction that is essential for biomass formation. The results provide an improved understanding of the physiology of C. glutamicum under anaerobic conditions.
INTRODUCTION
Corynebacterium glutamicum is a Gram-positive soil bacterium belonging to the order Corynebacteriales within the Actinobacteria (1, 2). It is used for the industrial production of the bulk products l-glutamate and l-lysine (3.0 and 2.2 million tons per year, respectively) (3) and of several other amino acids with a lower market volume. C. glutamicum has become a model organism in microbial biotechnology, and it was shown that a variety of other products besides amino acids can be efficiently synthesized with strains of this species, such as organic acids, diamines, or biofuels (4–7). Therefore, C. glutamicum has become a platform organism for white biotechnology (8–11).
C. glutamicum is nowadays described as a facultative anaerobic organism, based on studies showing that the type strain ATCC 13032 as well as strain R can grow under anoxic conditions when nitrate is present as terminal electron acceptor (12, 13). Negligible growth was observed in the absence of nitrate. Nitrate is reduced by the membrane-bound nitrate reductase NarGHIJ to nitrite, which accumulates in the medium under strictly anoxic conditions, as C. glutamicum does not possess a nitrite reductase (14). Due to the toxicity of nitrite (15), anaerobic growth of C. glutamicum by nitrate respiration in axenic culture is poor (12, 13). Nitrate is also reduced to nitrite under oxygen-limited conditions, but in this case nitrite can be partially metabolized again (16).
Under oxygen-limited conditions in the absence of nitrate, C. glutamicum was found to convert glucose to lactate, succinate, and acetate, which are excreted into the medium (17, 18). This type of metabolism was reported not to be associated with growth and was used to develop efficient strains for the production of, e.g., succinate (19, 20), d-lactate (21), isobutanol (22), or l-valine (23). For the corresponding production processes, aerobically grown cells are packed at high cell density under oxygen-deprived conditions, where they remain metabolically active and produce and excrete the products mentioned above. Analysis of the carbon flux under anaerobic conditions in a 100% argon atmosphere by 13C nuclear magnetic resonance (NMR) showed that 95% of the glucose is metabolized in glycolysis and 5% in the pentose phosphate pathway (24); 97% of the succinate was formed in the reductive tricarboxylic acid (TCA) cycle and only 3% in the oxidative TCA cycle, with the latter providing reducing equivalents required for the redox balance (24). Replacement of the argon atmosphere with a 100% CO2 atmosphere increased the succinate yield about 3-fold to ∼0.9 mol/mol glucose and the acetate yield about 2-fold to ∼0.3 mol/mol, while the lactate yield was decreased about 3-fold to ∼0.4 mol/mol (24). Gene expression studies revealed that several genes involved in glycolysis, lactate formation, C3 carboxylation, and the reductive TCA cycle were upregulated under oxygen deprivation, such as tpi, gapA, pgk, ldhA, ppc, and mdh, whereas genes of the oxidative TCA cycle were downregulated (25). In support of these data, the activities of glyceraldehyde 3-phosphate dehydrogenase (GAPDH), NAD+-dependent lactate dehydrogenase, and NAD+-dependent malate dehydrogenase (MDH) were found to be increased under oxygen-deprived conditions compared to aerobic conditions (26).
Recently, we described a mutant of C. glutamicum in which the branched aerobic respiratory chain is nonfunctional due to the deletion of the qcrCAB genes for the cytochrome bc1 complex and of the cydAB genes for cytochrome bd oxidase (27). This DOOR strain (mnemonic for “devoid of oxygen respiration”) showed very poor growth under aerobic conditions in glucose minimal medium, which could be stimulated by supplementation with peptone, however. Under these conditions, the DOOR strain displayed a fermentative type of catabolism with l-lactate as the major product and acetate and succinate as minor products (27). Based on these results, we reexamined the potential of C. glutamicum to grow anaerobically by fermentation.
MATERIALS AND METHODS
Bacterial strains, media, and culture conditions.
The bacterial strains used or constructed in the course of this work are listed in Table 1. Escherichia coli DH5α was grown aerobically at 37°C on LB agar plates (LB medium with 1.5% [wt/vol] agar) or in 5 ml LB medium at 170 rpm (28). When appropriate, kanamycin was added to a final concentration of 50 μg ml−1. C. glutamicum was routinely cultivated at 30°C. For the construction of C. glutamicum strain AtpG-S273P, BHIS agar plates (brain heart infusion [BHI] agar [Difco, Detroit, MI, USA] with 1.0 M sorbitol) and BHIS medium were used. When appropriate, kanamycin was added to a final concentration of 25 μg ml−1. For anaerobic growth experiments, C. glutamicum strains were aerobically precultivated at 30°C in 5 ml BHIS medium for 8 h (170 rpm). A second precultivation was performed in 100-ml baffled shake flasks at 30°C and 130 rpm overnight in CGXII minimal medium (29) with 100 mM glucose, fructose, sucrose, ribose, sodium gluconate, sodium pyruvate, sodium acetate, sodium lactate, or sodium propionate or 222 mM glucose (4%, wt/vol) as a carbon and energy source. The cells of these precultures were harvested by centrifugation (5,000 × g, 20°C, 10 min). For anaerobic cultivations, 50 ml of CGXII minimal medium with the indicated carbon sources and different concentrations of supplements was prepared in 60-ml serum bottles closed air-tight with a butyl rubber stopper, followed by flushing of the medium with sterile nitrogen gas for 30 min. A 0.3-ml portion of a sterile l-cysteine solution (60 g liter−1) was added with a syringe immediately before inoculation of the medium to an optical density at 600 nm (OD600) of 1 with aerobically grown cells washed twice with 0.9% (wt/vol) sodium chloride solution. Cells were grown at 30°C with gentle stirring (50 rpm). Sampling was carried out aseptically under constant nitrogen flushing.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Strains | ||
| C. glutamicum | ||
| ATCC 13032 | Biotin-auxotrophic wild-type strain | 75 |
| AtpG-S273P | ATCC 13032 derivative with a point mutation in the atpG gene (γ-subunit of the F1Fo-ATP synthase) leading to a Ser273Pro exchange | This work, based on reference 73 |
| ΔF1Fo | ATCC 13032 derivative with an in-frame deletion of the atpBEFHAGDC genes | 31 |
| E. coli DH5α | F− ϕ80dlacΔ(lacZ)M15 Δ(lacZYA-argF)U169 endA1 recA1 hsdR17 (rK− mK+) deoR thi-1 phoA supE44 gyrA96 relA1 | Invitrogen (Karlsruhe, Germany) |
| Plasmids | ||
| pGA1-AtpG-S273P | Ampr; pUC derivative containing a 1,006-bp fragment which covers the flanking regions of the C. glutamicum atpG gene and a mutation leading to an AtpG-S273P exchange | GeneArt (Regensburg, Germany) |
| pK19mobsacB | Kanr; vector for allelic exchange in C. glutamicum (pK18 oriVE.c., sacB, lacZα) | 76 |
| pK19mobsacB-AtpG-S273P | Kanr; pK19mobsacB derivative containing a 984-bp SmaI fragment containing the atpG-S273P gene and its flanking regions | This work |
For cultivation under controlled conditions, a 1.4-liter bioreactor (Multifors Multi-Fermenter system; Infors, Einsbach, Germany) containing 600 ml morpholinepropanesulfonic acid (MOPS)-free CGXII minimal medium with 4% (wt/vol) glucose was used. The medium was flushed for 60 min with sterile N2 (0.9 liter min−1) before inoculation with a syringe to an OD600 of 1. During cultivation, the medium was flushed with sterile N2 at a flow rate of 0.3 liter min−1 and agitated with a paddle mixer at 200 rpm. The pH was monitored using a pH controller and maintained at pH 5.5, 6.0, 6.5, 7.0, 7.5, 8.0, and 8.5 by addition of 3 M KOH or 3 M HCl. The nitrogen gas was mixed with CO2 gas as indicated. The serial inoculation experiments were performed in MOPS-free CGXII minimal medium with 4% (wt/vol) glucose and 15 g liter−1 tryptone as a carbon and energy source. The pH was kept at 7.5, and the bioreactor was flushed with 90% N2 and 10% CO2 at 0.3 liter min−1. At the indicated time points, 60 ml of culture was withdrawn using a syringe and transferred immediately to another bioreactor with 540 ml of fresh anaerobic medium. When the medium of the main culture was supplemented with tryptone or peptone (BD, Heidelberg, Germany), the preculture was supplemented as well.
Determination of growth parameters and quantification of sugars and organic acids in the culture supernatant.
Cell growth was monitored by measuring the OD600 using an Ultrospec 500pro spectrophotometer (Amersham Biosciences, Freiburg, Germany) for cultivations in serum bottles or an Ultrospec 3100pro spectrophotometer for cultivations in bioreactors. An OD600 of 1 corresponds to 0.25 g cell dry weight (CDW) per liter (30). The culture samples were centrifuged twice (10,000 × g, 4°C, 5 min) to remove the cells, and the resulting supernatants were analyzed for the presence of sugars and organic acids by high-pressure liquid chromatography (HPLC) (Agilent Technologies, Waldbronn, Germany) as described previously (31). Sucrose was measured with the Enzytec kit (Biopharm, Darmstadt, Germany) following the manufacturer's manual.
DNA techniques.
All enzymes used for restriction and ligation of DNA were purchased from Roche Diagnostics (Mannheim, Germany) or New England BioLabs (Frankfurt am Main, Germany). All oligonucleotides were synthesized by Eurofins MWG Operon. Routine methods such as PCR, restriction, and ligation were carried out according to standard protocols (28). Plasmid DNA was isolated with the QIAprep Spin Miniprep kit from Qiagen (Hilden, Germany). E. coli was transformed by the RbCl method (32). Transformation of C. glutamicum was performed as described previously (33). All plasmid constructs described below were controlled by DNA sequencing (LGC Genomics, Berlin, Germany).
RESULTS
Fermentative growth of C. glutamicum under anaerobic conditions.
Motivated by the capacity of C. glutamicum for anaerobic growth by nitrate respiration (12, 13) and for aerobic fermentative growth of the oxygen respiration-deficient DOOR mutant (27), we analyzed in more detail the potential of this species for anaerobic growth by fermentation. For this purpose, the wild type was cultured under anoxic conditions in serum bottles containing CGXII minimal medium with a 100 mM concentration of either glucose, fructose, sucrose, ribose, gluconate, pyruvate, acetate, lactate, or propionate as the sole carbon and energy source. All these compounds can serve as sole carbon and energy sources for aerobic growth of C. glutamicum (34–41). During cultivation, the growth, pH, carbon source consumption, and formation of organic acids were followed. On glucose, fructose, sucrose, and ribose, C. glutamicum grew linearly from an OD600 of 1 to values of between 1.9 and 2.7 (Fig. 1A and Table 2) and excreted lactate, succinate, and acetate, causing an acidification of the medium. This is shown exemplarily for glucose in Fig. 1B. Calculation of the carbon balance showed that 85 to 100% of the consumed carbon sources are metabolized to lactate, succinate, and acetate (Table 2). With gluconate, pyruvate, lactate, propionate, and acetate as carbon sources, no growth was observed, as shown in Fig. 1A exemplarily for gluconate. The absence of growth with these carbon sources makes it very unlikely that the slight growth observed with glucose, fructose, sucrose, and ribose was due to residual amounts of oxygen.
FIG 1.
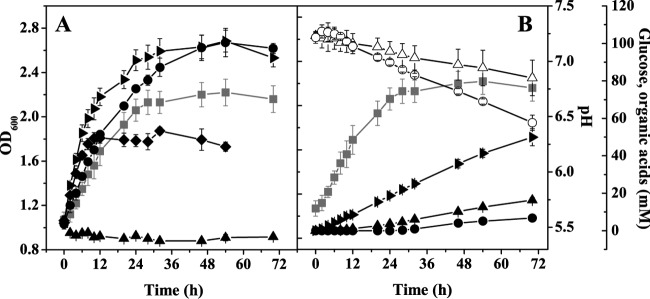
(A) Anaerobic growth of C. glutamicum with different carbon sources. The wild type was cultivated in serum bottles containing CGXII minimal medium with 100 mM glucose (■), fructose (▶), sucrose (◆), ribose (●), or gluconate (▲) as the sole carbon and energy source. When pyruvate, lactate, propionate, or acetate was used, the same negative result as with gluconate was obtained. Average values and standard deviations for at least three independent replicates are shown. (B) Biomass formation (■), glucose consumption (○), lactate formation (▶), succinate formation (▲), acetate formation (●), and pH changes (△) during anaerobic growth of the wild type in serum bottles with 100 mM glucose. Average values and standard deviations for four independent replicates are shown.
TABLE 2.
Biomass formation, growth rate, carbon source consumption, and organic acid formation for wild-type C. glutamicum after 46 h of anaerobic cultivation in serum bottles in CGXII minimal medium with the indicated carbon sources or with glucose in the presence or absence of tryptonea
| Carbon source | Biomass formed (gCDW liter−1) | Growth rate (h−1) | C source consumed (mM) | C source consumption rate (nmol min−1 mgCDW−1)b | Organic acid formation (mM)c |
Carbon balance (%)d | ||
|---|---|---|---|---|---|---|---|---|
| Lactate | Succinate | Acetate | ||||||
| Glucose (100 mM) | 0.55 ± 0.03 | NEGe | 29 ± 2 | 19 ± 3 | 36 ± 2 (0.81) | 10 ± 1 (0.34) | 13 ± 9 (0.45) | 100 |
| Fructose (100 mM) | 0.66 ± 0.03 | NEG | 43 ± 3 | 24 ± 2 | 48 ± 5 (1.12) | 14 ± 3 (0.33) | 9 ± 4 (0.21) | 85 |
| Sucrose (100 mM) | 0.45 ± 0.02 | NEG | 37 ± 1 | 30 ± 2 | 53 ± 3 (1.43) | 10 ± 0 (0.27) | 3 ± 0 (0.08) | 88 |
| Ribose (100 mM) | 0.66 ± 0.03 | NEG | 46 ± 4 | 25 ± 2 | 48 ± 4 (1.04) | 11 ± 1 (0.24) | 6 ± 1 (0.13) | 87 |
| Glucose (222 mM) | ||||||||
| Without tryptone | 0.52 ± 0.01 | NEG | 25 ± 5 | 17 ± 2 | 27 ± 10 (1.08) | 9 ± 1 (0.36) | 10 ± 8 (0.40) | 91 |
| With 5 g/liter tryptone | 2.06 ± 0.03 | 0.11 ± 0.01 | 80 ± 6 | 14 ± 2 | 138 ± 2 (1.73) | 23 ± 0 (0.29) | 9 ± 0 (0.11) | 119 |
| With 10 g/liter tryptone | 2.76 ± 0.02 | 0.11 ± 0.01 | 101 ± 7 | 13 ± 1 | 159 ± 4 (1.57) | 26 ± 1 (0.26) | 10 ± 0 (0.10) | 99 |
| With 15 g/liter tryptone | 2.93 ± 0.04 | 0.10 ± 0.01 | 103 ± 9 | 13 ± 2 | 175 ± 3 (1.70) | 30 ± 0 (0.29) | 7 ± 5 (0.07) | 107 |
Mean values and standard deviations from three independent experiments are shown.
The carbon source consumption rate was calculated for the 46-h period using the final cell dry weight (CDW).
The values in parentheses show the yield of the corresponding fermentation product in moles/mole of carbon source.
The carbon balance was calculated by considering sugars consumed and lactate, succinate, and acetate formed but not biomass formation and consumption of tryptone constituents.
NEG, no exponential growth observed.
The fact that gluconate did not allow fermentative growth whereas ribose did was puzzling. Both carbon sources are phosphorylated after uptake and then catabolized in the pentose phosphate pathway. The difference is that 6-phosphogluconate first has to be oxidized to ribulose 5-phosphate by 6-phosphogluconate dehydrogenase, with concomitant reduction of NADP+. This enzyme is inhibited by NADPH, which is a key mechanism for controlling cellular NADPH synthesis (42). If NADPH reoxidation was not fast enough under anaerobic fermentative conditions, NADPH could accumulate and cause a complete inhibition of 6-phosphogluconate dehydrogenase activity, which in turn would prevent growth on gluconate. Attempts to achieve anaerobic growth with gluconate by expressing the E. coli genes pntAB (43) or udhA (sthA), coding for a membrane-bound and a soluble transhydrogenase, respectively, were not successful (data not shown). Remarkably, anaerobic growth was possible when a combination of gluconate and pyruvate was used, although each substrate alone did not allow growth. When using a combination of 100 mM gluconate and 25 mM pyruvate, 11 mM gluconate and all of the pyruvate was consumed and converted to succinate (13 mM), lactate (7 mM), acetate (14 mM), and in addition l-alanine (2 mM). l-Alanine is synthesized in C. glutamicum from pyruvate by the aminotransferases AlaT and AvtA using l-glutamate and l-valine as amino donors, respectively (44). The regeneration of the amino donors from the corresponding 2-oxoacids requires NADPH. Alanine formation is therefore an NADPH-consuming process, which proved to be essential for anaerobic growth with gluconate and pyruvate. An alanine-auxotrophic ΔalaT ΔavtA mutant (44) was unable to grow anaerobically on this substrate combination in the presence of 3 mM l-alanine but showed growth in l-alanine-supplemented glucose medium (data not shown). Besides l-alanine formation, the high succinate/lactate ratio of 1.9 was a peculiar characteristic of fermentative growth on the mixture of gluconate and pyruvate. Succinate formation requires C3 carboxylation to C4. Carboxylation of pyruvate can proceed not only via the ATP-dependent pyruvate carboxylase (45) but also via malic enzyme, which catalyzes the reversible NADPH-dependent conversion of pyruvate to malate (40). As the latter reaction provides another possibility for NADPH oxidation, we also analyzed a ΔmalE mutant and found that it was unable to grow anaerobically with gluconate and pyruvate but showed growth with glucose (data not shown). This suggests that both conversion of pyruvate to alanine and reductive carboxylation of pyruvate to malate are necessary for reoxidation of NADPH formed by 6-phosphogluconate dehydrogenase.
Stimulation of anaerobic fermentative growth by peptides and amino acids.
With the aim to improve anaerobic growth of C. glutamicum, cultivations in serum bottles were performed using CGXII minimal medium with 4% (wt/vol) glucose and various concentrations of BHI, nutrient broth, yeast extract, Casamino Acids, peptone, or tryptone. All supplements improved growth, with tryptone being the most effective one (data not shown). Supplementation with 5, 10, and 15 g liter−1 tryptone increased the final OD600 from 2 to about 8, 11, and 12, respectively (Fig. 2A). Higher tryptone concentrations did not stimulate growth any further. In the presence of tryptone, cells grew exponentially with a rate of 0.11 h−1 (Table 2). A control culture containing 15 g liter−1 tryptone but no glucose did not grow at all (Fig. 2A), showing the strict requirement for sugar. The rate of glucose consumption in the presence of tryptone (24.6 ± 3.5 nmol mgCDW−1 min−1) was comparable to that in the absence of tryptone (25.8 ± 4.0 nmol mgCDW−1 min−1) within the first 30 h of cultivation, with lactate, succinate, and acetate as products (Fig. 2B and Table 2). Under aerobic conditions, a minimal medium containing 15 g liter−1 tryptone as the sole carbon source enabled growth from an OD600 of 1 to an OD600 of 4 within 24 h, accompanied by an alkalinization of the medium from pH 7.1 to pH 8.2 (data not shown).
FIG 2.
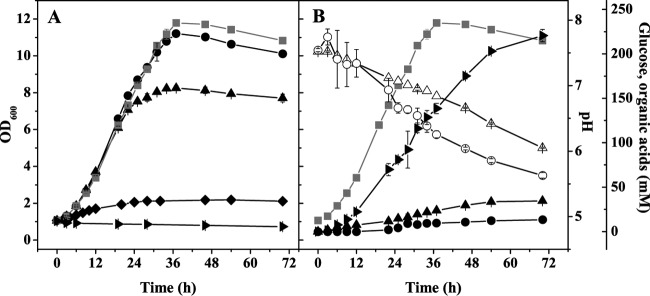
(A) Influence of different tryptone concentrations (◆, 0 g liter−1; ▲, 5 g liter−1; ●, 10 g liter−1; ■, 15 g liter−1) on anaerobic growth of C. glutamicum in serum bottles containing CGXII minimal medium with 4% (wt/vol) glucose. In addition, growth in a medium containing only 15 g liter−1 tryptone (▶) but no glucose is shown. (B) Biomass formation (■), glucose consumption (○), lactate formation (▶), succinate formation (▲), acetate formation (●), and pH changes (△) during anaerobic growth of wild-type C. glutamicum in serum bottles containing CGXII minimal medium with 4% (wt/vol) glucose and 15 g liter−1 tryptone. Average values and standard deviations for three independent replicates are shown.
We subsequently tested the influence of different groups of amino acids on anaerobic growth with glucose. In a first series of experiments, three amino acid mixtures were formed based on the ATP demand of their synthesis (46), i.e., mix I (high ATP demand; R, M, H, W, and C), mix II (intermediate ATP demand; I, K, T, Q, F, P, and Y), and mix III (no ATP demand; A, D, E, G, L, S, and V) (Table 3; see Table S1 in the supplemental material). As shown in Fig. S1 in the supplemental material, the simultaneous addition of all three mixtures (each amino acid in a final concentration of 0.8 mM, corresponding to a total amino acid concentration of about 2 g/liter) allowed a growth rate similar to that with 15 g/liter tryptone (Table 3) and a final OD600 of 6 (50% of the value obtained with 15 g/liter tryptone). Supplementation with mix I and either mix II or mix III still allowed reasonable growth rates and final OD values of 5.2 and 4.6, respectively. In contrast, supplementation with either mix II plus mix III or with each mix alone improved growth much less than the above-mentioned combinations and led to OD values of between 2.7 and 3.3 (Table 3). According to these results, the best growth was achieved with combinations including mix I (containing the most ATP-demanding amino acids), but surprisingly, mix I alone stimulated growth only weakly, suggesting another limitation. We therefore performed a second series of experiments in which we added all amino acids except one of the five amino acid families (aspartate family, glutamate family, serine family, pyruvate family, and aromatic amino acid family) or l-histidine (Table 3). Only the lack of the amino acids of the glutamate family led to a significant reduction of growth and of the final OD600 (by about 50%), whereas the lack of amino acids of the pyruvate family had practically no impact on biomass formation (Table 3). Thus, besides the ATP-demanding amino acids, amino acids of the glutamate family also limit anaerobic growth, which presumably is due to an insufficient carbon flux into the oxidative TCA cycle to form 2-oxoglutarate (24).
TABLE 3.
Growth rate and biomass formed after 46 h of cultivation of wild-type C. glutamicum under anaerobic conditions in serum bottles with CGXII minimal medium containing 4% (wt/vol) glucose and different amino acid supplementsa
| Amino acid supplement (0.8 mM each) | μ (h−1) | Biomass formed (gCDW liter−1) | % biomassb |
|---|---|---|---|
| None | NEGc | 0.52 ± 0.01 | 35 |
| Mix I (high ATP demand; R, M, H, W, C, N) | 0.03 ± <0.01 | 0.71 ± 0.01 | 47 |
| Mix II (intermediate ATP demand; I, K, T, Q, F, P, Y) | 0.04 ± <0.01 | 0.67 ± 0.09 | 45 |
| Mix III (no ATP demand; A, D, E, G, L, S, V) | 0.05 ± <0.01 | 0.83 ± 0.10 | 55 |
| Mix I + II | 0.05 ± 0.01 | 1.31 ± 0.01 | 87 |
| Mix I + III | 0.06 ± <0.01 | 1.14 ± 0.08 | 76 |
| Mix II + III | 0.06 ± <0.01 | 0.67 ± 0.04 | 45 |
| Mix I + II + III | 0.10 ± <0.01 | 1.50 ± 0.07 | 100 |
| Glutamate family (E, Q, R, P) | NEG | 0.69 | 46 |
| Serine family (S, C, G) | NEG | 0.56 | 37 |
| Histidine | NEG | 0.56 | 37 |
| Aspartate family (D, M, N, T, I, K) | NEG | 0.52 | 35 |
| Pyruvate family (A, V, L) | NEG | 0.53 | 35 |
| Aromatic amino acid family (F, Y, W) | NEG | 0.52 | 35 |
| All amino acids except: | |||
| Pyruvate family | NEG | 1.43 | 95 |
| Histidine | NEG | 1.29 | 86 |
| Aromatic amino acid family | NEG | 1.25 | 83 |
| Aspartate family | NEG | 1.17 | 78 |
| Serine family | NEG | 1.13 | 75 |
| Glutamate family | NEG | 0.79 | 53 |
Mean values and standard deviations are shown for experiments with three replicates, and mean values are shown for experiments with two replicates.
Biomass obtained with supplementation of all amino acids set as 100%.
NEG, no exponential growth observed.
pH tolerance range during anaerobic fermentative growth.
Under aerobic conditions, C. glutamicum shows maximal growth rates at between pH 7.0 and 8.5 and effective pH homeostasis in the pH range between 6.0 and 9.0, in which the internal pH is kept at 7.5 ± 0.5 (47). At pH 6.0, C. glutamicum regulates its internal pH by proton extrusion via the respiratory chain and potassium influx via the potassium channel CglK (48, 49). During anaerobic fermentative growth with 100 mM glucose in the absence of tryptone, the pH dropped to 6.9 (Fig. 1B), and during anaerobic growth with 222 mM glucose in the presence of 15 g liter−1 tryptone, it dropped to 6.1 (Fig. 2B). pH homeostasis might become a problem for C. glutamicum during anaerobic fermentative growth, because organic acids are produced and not consumed later on and because protons cannot be pumped out of the cell by the respiratory chain. We therefore analyzed the pH tolerance of C. glutamicum under anaerobic conditions in more detail. For this purpose, cells were cultivated in a bioreactor using MOPS-free CGXII minimal medium with 4% (wt/vol) glucose and, when indicated, 15 g liter−1 tryptone. The external pH was kept constant at 6.0, 6.5, 7.0, 7.5, and 8.0 in the absence of tryptone and at 5.5, 6.0, 6.5, 7.0, 7.5, 8.0, and 8.5 in the presence of tryptone. Under both conditions, the highest final optical densities were observed at pH 7.5, which was previously shown to be the preferred internal pH under aerobic conditions (Fig. 3). Higher or lower pH values led to reduced growth. In the absence of tryptone, no growth was observed at pH 6.0 (Fig. 3A), and only weak growth occurred at pH 8.0 after a lag phase of 12 to 24 h (Fig. 3B). In the presence of tryptone, C. glutamicum showed weak growth even at pH 5.5 (Fig. 3C) and 8.5 (Fig. 3D). At all tested pH values, growth with tryptone was better than that without. Thus, effective pH homeostasis was possible in the range from pH 6.5 to 8.0 in the absence of tryptone and from pH 5.5 to 8.5 in the presence of tryptone, showing that control of the cytoplasmic pH was still possible and more efficient in the presence of tryptone.
FIG 3.
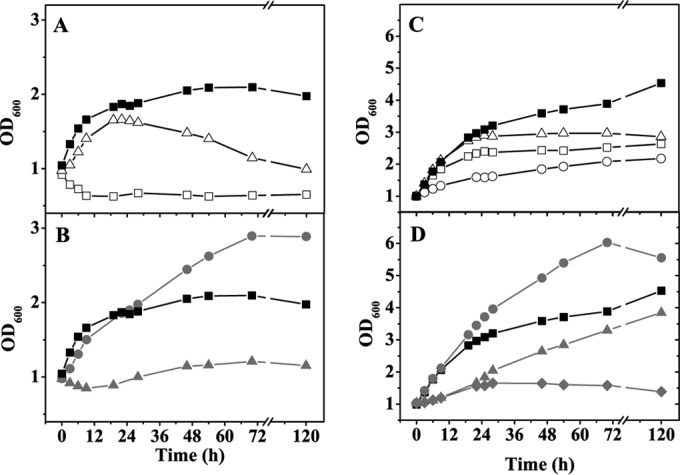
Influence of pH on anaerobic growth of C. glutamicum in a bioreactor containing MOPS-free CGXII minimal medium with 4% (wt/vol) glucose and either no tryptone (A and B) or 15 g liter−1 tryptone (C and D). The following pH values were tested: 5.5 (○), 6.0 (□), 6.5 (△), 7.0 (■), 7.5 (●), 8.0 (▲), and 8.5 (◆). The bioreactor was flushed with pure N2 at 0.3 liter min−1. The pH was kept constant by addition of 3 M KOH or 3 M HCl. Average values for at least two independent replicates are shown.
Carbon dioxide demand of anoxic C. glutamicum cells.
In the course of our experiments we noticed that the final OD600 of wild-type C. glutamicum in CGXII minimal medium with 4% (wt/vol) glucose and 15 g liter−1 tryptone under anaerobic conditions in serum bottles was much higher than that in the bioreactor cultivations (Fig. 2 and 4). We assumed that the bioreactor cultures could be limited in carbon dioxide for oxaloacetate biosynthesis via phosphoenolpyruvate (PEP) carboxylase and pyruvate carboxylase and for fatty and mycolic acid synthesis. In contrast to the case in the closed serum bottles, the carbon dioxide formed by the cells in the bioreactor is removed to a large extent by the constant flushing with nitrogen gas. To test this assumption, cultivations were performed in which the bioreactor was flushed with nitrogen gas containing 1%, 2.5%, 5%, 7.5%, or 10% (vol/vol) carbon dioxide. As shown in Fig. 4, both the growth rate and the maximal OD600 were significantly improved with increasing CO2 concentrations. At 10% (vol/vol) CO2, a maximal OD600 of 12.5 and a growth rate of 0.14 h−1 was achieved, the latter being 27% higher than in serum bottles. The molar yields of the lactate, succinate, and acetate were not significantly changed at 10% (vol/vol) CO2 compared to cultivation without CO2 supplementation (see Table S2 in the supplemental material). Higher CO2 concentrations did not stimulate growth any further. The observation that anaerobic cultivation in bioreactors with continuous flushing led to a OD600 comparable to that for growth in serum bottles argues against the possibility of residual oxygen in the gas flow, as this should have allowed higher cell densities.
FIG 4.
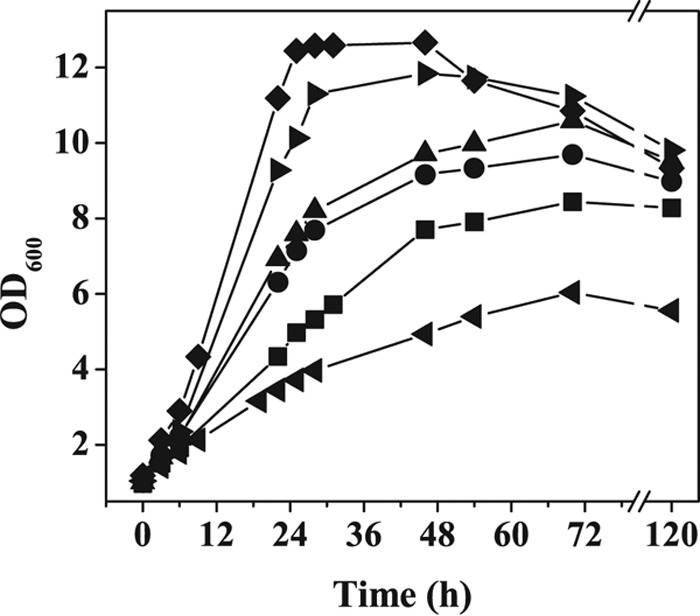
Influence of CO2 on anaerobic growth of C. glutamicum in a bioreactor using MOPS-free CGXII minimal medium with 4% (wt/vol) glucose and 15 g liter−1 tryptone at a constant pH of 7.5. The bioreactor was flushed with a gas mixture composed of N2 and the indicated CO2 concentrations: 10% (◆), 7.5% (▶), 5% (▲), 2.5% (●), 1% (■), or 0% (◀). Average values for at least two independent replicates are shown.
Serial growth under anaerobic conditions.
In order to test whether anaerobic fermentative growth of C. glutamicum is temporally limited due to the existence of an oxygen-dependent reaction that is essential for biomass formation in the long term, we performed serial inoculation experiments in MOPS-free CGXII minimal medium containing 4% (wt/vol) glucose and 15 g liter−1 tryptone (Fig. 5). We transferred 60 ml of the first culture at an OD600 of 5 into another bioreactor containing 540 ml of fresh anoxic medium, resulting in a starting OD600 of the new culture of about 0.5. This procedure was repeated seven times. During these seven cultivations the cells stayed vital and grew with a growth rate of between 0.09 and 0.13 h−1, suggesting the absence of an oxygen-dependent reaction that is essential for continued biomass formation by C. glutamicum.
FIG 5.
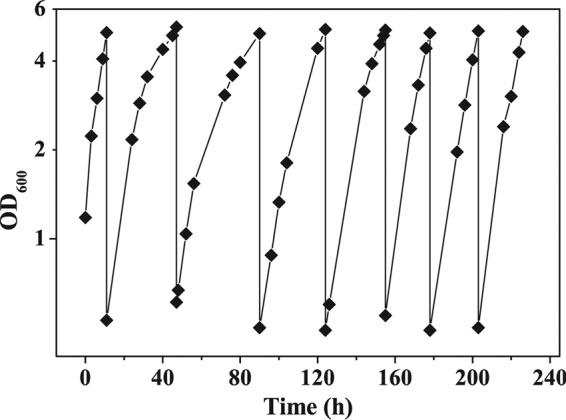
Serial inoculation experiment with wild-type C. glutamicum. The cells were cultivated anaerobically in bioreactors containing MOPS-free CGXII minimal medium with 4% (wt/vol) glucose and 15 g liter−1 tryptone at a constant pH of 7.5. The bioreactors were flushed with 90% N2 and 10% CO2 at 0.3 liter min−1. When the cultures reached an OD600 of about 5, 60 ml of the culture was transferred to another bioreactor with 540 ml fresh anoxic medium.
DISCUSSION
C. glutamicum is currently described as a facultative anaerobe that can grow anaerobically only by nitrate respiration. Here we demonstrate that C. glutamicum is also able to grow anaerobically in the absence of nitrate on glucose, fructose, sucrose, and ribose by mixed-acid fermentation with lactate, succinate, and acetate as products. The mentioned carbon sources are catabolized via glycolysis or the pentose phosphate pathway (Fig. 6) and allow ATP synthesis by substrate-level phosphorylation. No growth was observed on gluconate, pyruvate, lactate, acetate, and propionate. Fermentative growth with acetate and propionate was not expected, as they do not allow net ATP synthesis by substrate-level phosphorylation. Anaerobic growth by lactate fermentation occurs in nature, as shown by propionibacteria that gain net ATP by conversion of lactate to propionate, acetate, and CO2 either via the methylmalonyl coenzyme A (methylmalonyl-CoA) pathway or via the acrylyl-CoA pathway. However, the key enzymes of these pathways, transcarboxylase, lactyl-CoA dehydratase, and acrylyl-CoA reductase, are not present in C. glutamicum. In the case of pyruvate, an exergonic fermentation pathway can be formulated, in which one molecule of pyruvate is converted by pyruvate dehydrogenase, phosphotransacetylase, and acetate kinase to acetate and CO2 along with formation of NADH and ATP. The NADH could be reoxidized by reduction of a second molecule of pyruvate to l-lactate by the NAD+-dependent lactate dehydrogenase, resulting in the following overall reaction: 2 pyruvate− + H2O → acetate− + CO2 + lactate− (ΔG° = −95.1 kJ/mol acetate). In contrast to acetate, propionate, and lactate, pyruvate as the sole carbon source was partially metabolized under anaerobic conditions to lactate and acetate, but this conversion was not associated with growth, possibly because the energy budget of the cells did not allow for gluconeogenesis.
FIG 6.
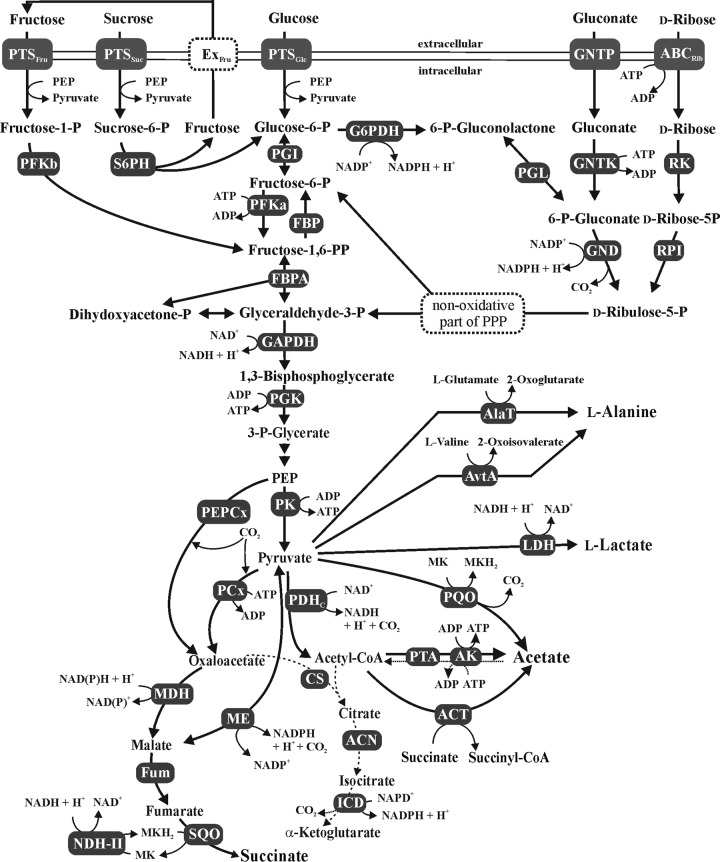
Catabolism of glucose, fructose, sucrose, gluconate, and ribose by C. glutamicum and anaerobic central metabolism. Abbreviations: ABCRib, ABC-type transporter for ribose; ACN, aconitase; AK, acetate kinase; CoA, coenzyme A; ACT, acetyl-CoA:CoA transferase; AlaT, alanine aminotransferase; AvtA, valine-pyruvate transaminase; CS, citrate synthase; ExFru, exporter for fructose; FBPA, fructose-bisphosphate aldolase; FUM, fumarase; Fru, fructose; GAP, glyceraldehyde-3-phosphate; GAPDH, GAP dehydrogenase; Glu, glucose; G6PDH, glucose-6-phosphate dehydrogenase; GND, 6-phosphate-gluconate dehydrogenase; GNTK, gluconate kinase; ICD, isocitrate dehydrogenase; LDH, lactate dehydrogenase; MDH, malate dehydrogenase; ME, malic enzyme; NDH-II, NADH dehydrogenase; P, phosphate; PP, bisphosphate; PPP, pentose phosphate pathway; PCx, pyruvate carboxylase; PDHC, pyruvate dehydrogenase complex; PEP, phosphoenolpyruvate; PEPCx, PEP carboxylase; PFKa, 6-phosphofructokinase; PFKb, 1-phosphofructokinase; PGI, glucose-6-phosphate isomerase; PGK, phosphoglycerate kinase; PGL, 6-phosphogluconolactonase; PK, pyruvate kinase; PQO, pyruvate:quinone oxidoreductase; PTA, phosphotransacetylase; PTS, phosphotransferase system; RK, ribokinase; RPI, ribose 5-phosphate isomerase; S6PH, sucrose 6-phosphate hydrolase; Suc, sucrose; SQO, succinate:menaquinone oxidoreductase. Note that SQO and NADH dehydrogenase are membrane-integral or membrane-associated enzymes, respectively, which is not shown in the figure.
Strong evidence was obtained that the inability for to grow anaerobically with gluconate was due to an insufficient capacity for reoxidation of NADPH generated by 6-phosphogluconate dehydrogenase. Supplementation with pyruvate could overcome this problem, as pyruvate allows NADPH reoxidation both by conversion to l-alanine and by reductive carboxylation to l-malate. The responsible enzymes, the transaminases AlaT and AvtA and malic enzyme MalE, were shown to be essential for fermentative growth with the gluconate-pyruvate mixture.
Addition of tryptone or other peptide sources or of amino acids strongly enhanced growth of C. glutamicum under anaerobic fermentative conditions with the sugars mentioned above, whereas tryptone alone did not allow growth. C. glutamicum is known to have efficient peptide uptake systems, although they have not yet been characterized in detail (50–54). At least a fraction of the peptides present in tryptone are taken up and then hydrolyzed by peptidases to amino acids, which then are used for protein synthesis. In agreement, supplementation with amino acids rather than tryptone also improved anaerobic growth. A mixture of all amino acids was required to achieve maximal growth (Table 3). In Table 4, an overview of proposed or experimentally verified peptide and amino acid transport systems of C. glutamicum is given. It also includes amino acid exporters, which allow export of amino acids that cannot be degraded and accumulate in the cell when their uptake, e.g., as peptides, is more rapid than their incorporation into proteins, which can lead to toxic effects (55). As proteins account for about 50% of the dry weight of cells (56) and de novo amino acid biosynthesis requires a significant fraction of the cellular energy demand, the supplementation of the medium with amino acids or peptides will save a lot of energy. This energy can be used for the synthesis of other building blocks, leading to improved growth, or for maintenance and homeostatic processes.
TABLE 4.
Overview of putative or verified peptide and amino acid transport systems of C. glutamicum based on the genome annotation and available literature
| Category | Cg no. | NCgl no. | Gene name | Annotation | Reference(s) |
|---|---|---|---|---|---|
| Putative secondary peptide transporters | Cg2884 | NCgl2514 | Dipeptide/tripeptide permease | 77 | |
| Cg3382 | NCgl2949 | Dipeptide/tripeptide permease | 77 | ||
| Putative peptide ABC transporters | Cg2181 | NCgl1915 | ABC-type peptide transport system, secreted component | 77 | |
| Cg2182 | NCgl1916 | ABC-type peptide transport system, permease component | 77 | ||
| Cg2183 | NCgl1917 | ABC-type peptide transport system, permease component | 77 | ||
| Cg2184 | NCgl1918 | ATPase component of peptide ABC-type transport system, contains duplicated ATPase domains | 77 | ||
| Cg2549 | NCgl2238 | ABC-type dipeptide/oligopeptide/nickel transport system, secreted component | 77 | ||
| Cg2550 | NCgl2239 | ABC-type dipeptide/oligopeptide/nickel transport systems, permease components | 77 | ||
| Cg2551 | NCgl2240 | ABC-type dipeptide/oligopeptide/nickel transport system, permease component | 77 | ||
| Cg2552 | NCgl2241 | ATPase component of ABC-type transport system, contains duplicated ATPase domain | 77 | ||
| Cg2675 | NCgl2350 | ATPase component of ABC-type transport system, contains duplicated ATPase domains | 77 | ||
| Cg2676 | NCgl2351 | ABC-type dipeptide/oligopeptide/nickel transport systems, permease component | 77 | ||
| Cg2677 | NCgl2352 | ABC-type dipeptide/oligopeptide/nickel transport system, permease component | 77 | ||
| Cg2678 | NCgl2353 | ABC-type dipeptide/oligopeptide/nickel transport systems, secreted component | 77 | ||
| Putative orphan periplasmic peptide binding proteins | Cg1249 | NCgl1054 | ABC-type dipeptide/oligopeptide/nickel transport system, secreted component | 77 | |
| Cg1864 | NCgl1592 | dciAE | ABC-type dipeptide/oligopeptide/nickel transport systems, secreted component | 77 | |
| Cg2610 | NCgl2294 | ABC-type dipeptide/oligopeptide/nickel transport system, secreted component | 77 | ||
| Putative or verified secondary amino acid importers | Cg0254 | NCgl0203 | Putative Na+/alanine symporter (possibly d-alanine) | 77 | |
| Cg0555 | NCgl0453 | Putative d-serine/d-alanine/glycine transporter | 77 | ||
| Cg0568 | NCgl0464 | gapP | γ-Aminobutyric acid transporter weakly inhibited by asparagine and glutamine | 78 | |
| Cg0953 | NCgl0799 | Putative Na+/proline, Na+/pantothenate symporter | 77 | ||
| Cg1016 | NCgl0856 | betP | Glycine betaine transporter | 79 | |
| Cg1105 | NCgl0929 | lysI | l-Lysine permease | 80 | |
| Cg1167 | NCgl0985 | metS | Na+/methionine/alanine transporter, small subunit | 81 | |
| Cg1169 | NCgl0986 | metP | Na+/methionine/alanine transporter, large subunit | 81 | |
| Cg1257 | NCgl1062 | aroP | Aromatic amino acid and histidine transporter | 82, 83 | |
| Cg1305 | NCgl1108 | pheP | Phenylalanine transport system | 84 | |
| Cg1314 | NCgl1116 | putP | Na+/proline symporter | 85 | |
| Cg2537 | NCgl2228 | brnQ | Branched-chain amino acid uptake carrier | 86 | |
| Cg2539 | NCgl2230 | ectP | Ectoine/proline/glycine betaine carrier | 87 | |
| Cg2563 | NCgl2251 | lcoP | Ectoine/betaine transporter | 88 | |
| Cg3080 | NCgl2683 | gltS | Na+/glutamate symporter | 89 | |
| Cg3357 | NCgl2925 | trpP | Putative tryptophan-specific permease, 5-methyltryptophan resistance | 90 | |
| Cg3395 | NCgl2961 | proP | Proline/ectoine uptake system | 87 | |
| Putative or verified amino acid ABC transporters | Cg0737 | NCgl0610 | metQ | ABC-type methionine transport system, secreted lipoprotein component | 81 |
| Cg0736 | NCgl0609 | metN | ABC-type methionine transport system, ATPase component | 81 | |
| Cg0735 | NCgl0608 | metI | ABC-type methionine transport system, permease component | 81 | |
| Cg2136 | Cgl1875 | gluA | ABC transporter for glutamate uptake, ATPase component | 91 | |
| Cg2137 | NCgl1876 | gluB | ABC transporter for glutamate uptake, secreted glutamate binding protein | 91 | |
| Cg2138 | NCgl1877 | gluC | ABC transporter for glutamate uptake, permease component | 91 | |
| Cg2139 | NCgl1878 | gluD | ABC transporter for glutamate uptake, permease component | 91 | |
| Cg2467 | NCgl2168 | ABC transporter, ATP-binding protein | 77 | ||
| Cg2468 | NCgl2169 | Putative branched-chain amino acid ABC-type transport system, permease component | 77 | ||
| Cg2470 | NCgl2170 | ABC transporter, secreted substrate-binding protein | 77 | ||
| Putative or verified amino acid exporters | Cg0183 | NCgl0143 | Putative threonine efflux protein | 77 | |
| Cg2574 | NCgl2262 | Putative threonine efflux protein | 77 | ||
| Cg2905 | NCgl2533 | thrE | Threonine export carrier | 92 | |
| Cg2941 | NCgl2566 | Putative threonine efflux protein | 77 | ||
| Cg0314 | NCgl0254 | brnF | Exporter for branched-chain amino acids and methionine, large subunit | 93, 94 | |
| Cg0315 | NCgl0255 | brnE | Exporter for branched-chain amino acids and methionine, small subunit | 93, 94 | |
| Cg3412 | NCgl2976 | azlD | Predicted branched-chain amino acid permease (azaleucine resistance) | 77 | |
| Cg3413 | NCgl2977 | azlC | Predicted branched-chain amino acid permease (azaleucine resistance) | 77 | |
| Cg1424 | NCgl1214 | lysE | Lysine/arginine efflux permease | 55 | |
| Cg1434 | NCgl1221 | yggB, mscCG | Glutamate exporter | 95 |
pH homeostasis is a crucial point for anaerobic fermentative growth of C. glutamicum, as acids are formed as end products, causing an acidification of the medium. At the same time, the major components of the respiratory chain involved in proton export, in particular the cytochrome bc1-aa3 supercomplex, are inactive. Under aerobic respiratory conditions, the growth optimum of C. glutamicum in minimal medium was reported to be between pH 7 and 8.5, and the cells grew at between pH 4 and 10 (47). Under anaerobic fermentative conditions, the growth optimum in minimal medium without tryptone was pH 7.5, and cells grew at between pH 6.5 and 8.0. Thus, the optimal pH range and the pH range allowing growth were drastically reduced under anaerobic conditions, indicating that pH homeostasis is a severe problem. Various possibilities to cope with acid stress have been described in bacteria, such as amino acid decarboxylases like glutamate decarboxylase or lysine decarboxylase (57–59). The protein encoded by cg1261 (NCgl1065) in the genome of C. glutamicum was annotated to be a member of the lysine decarboxylase protein family, but it has not been characterized functionally, and no evidence has been found for its involvement in acid stress adaptation in genome-wide transcriptome studies (47, 60). An alternative way to cope with acid stress is the hydrolysis of urea via urease, as found, e.g., in Helicobacter pylori (61). A urease is present in C. glutamicum and is induced under nitrogen starvation, whereas there is no evidence for an involvement in the acid stress response (62, 63). A limited number of reactions are assumed to be relevant for proton extrusion under fermentative conditions in C. glutamicum. (i) The export of lactate (pKa = 3.90) and succinate (pKa1 = 4.16, pKa2 = 5.61) by nongrowing C. glutamicum cells at an extracellular pH of 5.7 and an intracellular pH of 6.3 was reported to be coupled to the export of 1.4 and 2.3 to 2.7 protons, respectively (24), thereby contributing to the establishment of a proton motive force. The driving force for this proton export is the 10-fold-higher intracellular compared to extracellular concentration of lactate and succinate, as measured by 13C NMR (24). For succinate, the exporter SucE has been identified in C. glutamicum (64, 65) but still awaits a detailed biochemical characterization, including the stoichiometry of cotransported ions. For lactate, an exporter has not yet been described for C. glutamicum. Uptake of succinate and other C4-dicarboxylates is catalyzed by the secondary transporters DccT and DctA in C. glutamicum; however, their expression levels are very low in the wild type (66, 67). Uptake of l-lactate presumably involves a secondary transporter encoded by cg3226 (NCgl2816); however, as the absence of this gene does not prevent growth on lactate, further lactate uptake systems probably exist (68). As in the case of SucE, the putative lactate permease Cg3226 has not yet been characterized biochemically. (ii) The reduction of endogenously formed fumarate to succinate by succinate:menaquinone oxidoreductase (Fig. 6) is presumably coupled with the export of two protons (14, 69) and thus contributes to proton motive force generation. (iii) Proton export driven by ATP hydrolysis via F1Fo-ATP synthase (4 H+/ATP) was reported for several Gram-positive bacteria (59, 70, 71), including Mycobacterium smegmatis (72). We tested the role of F1Fo-ATP synthase in the anaerobic fermentative growth of C. glutamicum by analyzing AtpG-S273P and ΔF1Fo mutants, which have a strongly reduced (73, 74) or no (31) F1-ATPase activity. As shown in Fig. S2 in the supplemental material, fermentative growth of both mutants was improved in glucose medium without tryptone and impaired in glucose medium supplemented with tryptone. A straightforward explanation of these opposing effects cannot be given, but the results demonstrate that F1Fo-ATP synthase plays a role in anaerobic fermentative growth, either for proton export at the cost of ATP hydrolysis or for ATP synthesis at the cost of proton import. In conclusion, all of the three processes can be assumed to contribute to pH homeostasis under anaerobic conditions.
During anaerobic fermentative growth in bioreactors which were constantly flushed with nitrogen gas, supplementation with 1 to 10% (vol/vol) CO2 strongly improved growth. CO2 is required on one hand in the pathway of succinate formation via the reductive branch of the TCA cycle, where oxaloacetate has to be synthesized by carboxylation of PEP or pyruvate, and on the other hand for biosynthetic reactions, in particular fatty and mycolic acid synthesis. In the previous study by Rados et al. (24), the presence of 20% CO2 reduced the lactate yield from 1.22 to 0.73 mol/mol glucose and increased the succinate yield from 0.29 to 0.59 mol/mol and the acetate yield from 0.13 to 0.19 mol/mol. In contrast, the yields of lactate, succinate, and acetate were changed only moderately in our experiments with 10% CO2 (see Table S2 in the supplemental material). This difference can be explained by the different conditions used in the experiments. Rados et al. (24) used nongrowing cells resuspended to an OD600 of 100 in a buffer without nitrogen, sulfur, or phosphorus sources, whereas we analyzed growing cells at a starting OD600 of 1 in a minimal medium supplemented with 15 g/liter tryptone. Therefore, it has to be assumed that in our experiments, CO2 was used to enhance CO2-requiring biosynthetic reactions, in particular acetyl-CoA carboxylation for fatty and mycolic acid synthesis, rather than for increasing pyruvate and PEP carboxylation.
In summary, we demonstrate that C. glutamicum is able to grow under anaerobic conditions without nitrate by mixed-acid fermentation. In the presence of tryptone or amino acid supplements, cell densities which correspond to 20% of those reached under aerobic conditions can be obtained. Fermentative growth was possible over many generations, supporting the view that none of the reactions involved in biomass formation is strictly dependent on oxygen.
Supplementary Material
ACKNOWLEDGMENT
This work was financially supported by the German Federal Ministry of Education and Research (BMBF) within the GenoMik-Transfer project “Corynebacterium: improving flexibility and fitness for industrial production (FlexFit)” by grant 0315589A.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02413-15.
REFERENCES
- 1.Liebl W. 2006. Corynebacterium—nonmedical. Prokaryotes 3:796–818. [Google Scholar]
- 2.Gao B, Gupta RS. 2012. Phylogenetic framework and molecular signatures for the main clades of the phylum Actinobacteria. Microbiol Mol Biol Rev 76:66–112. doi: 10.1128/MMBR.05011-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ajinomoto Co., Inc. 31 July 2014, posting date First quarter-FY2014. Market and other information. Ajinomoto Co., Inc., Tokyo, Japan: http://www.ajinomoto.com/en/ir/pdf/Q1-FY14_data_E.pdf. [Google Scholar]
- 4.Wieschalka S, Blombach B, Bott M, Eikmanns BJ. 2013. Bio-based production of organic acids with Corynebacterium glutamicum. Microb Biotechnol 6:87–102. doi: 10.1111/1751-7915.12013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schneider J, Wendisch VF. 2011. Biotechnological production of polyamines by bacteria: recent achievements and future perspectives. Appl Microbiol Biotechnol 91:17–30. doi: 10.1007/s00253-011-3252-0. [DOI] [PubMed] [Google Scholar]
- 6.Becker J, Wittmann C. 2012. Bio-based production of chemicals, materials and fuels—Corynebacterium glutamicum as versatile cell factory. Curr Opin Biotechnol 23:631–640. doi: 10.1016/j.copbio.2011.11.012. [DOI] [PubMed] [Google Scholar]
- 7.Blombach B, Eikmanns BJ. 2011. Current knowledge on isobutanol production with Escherichia coli, Bacillus subtilis and Corynebacterium glutamicum. Bioeng Bugs 2:346–350. doi: 10.4161/bbug.2.6.17845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eggeling L, Bott M. 2005. Handbook of Corynebacterium glutamicum. CRC Press, Taylor & Francis Group, Boca Raton, FL, USA. [Google Scholar]
- 9.Burkovski A. 2008. Corynebacteria: genomics and molecular biology. Caister Academic Press, Norfolk, United Kingdom. [Google Scholar]
- 10.Yukawa H, Inui M (ed). 2013. Corynebacterium glutamicum: biology and biotechnology. Microbiology monographs, vol 23 Springer, Heidelberg, Germany. [Google Scholar]
- 11.Burkovski A. 2015. Corynebacterium glutamicum—from systems biology to biotechnological applications, p 198 Caister Academic Press, Norfolk, United Kingdom. [Google Scholar]
- 12.Takeno S, Ohnishi J, Komatsu T, Masaki T, Sen K, Ikeda M. 2007. Anaerobic growth and potential for amino acid production by nitrate respiration in Corynebacterium glutamicum. Appl Microbiol Biotechnol 75:1173–1182. doi: 10.1007/s00253-007-0926-8. [DOI] [PubMed] [Google Scholar]
- 13.Nishimura T, Vertes AA, Shinoda Y, Inui M, Yukawa H. 2007. Anaerobic growth of Corynebacterium glutamicum using nitrate as a terminal electron acceptor. Appl Microbiol Biotechnol 75:889–897. doi: 10.1007/s00253-007-0879-y. [DOI] [PubMed] [Google Scholar]
- 14.Bott M, Niebisch A. 2003. The respiratory chain of Corynebacterium glutamicum. J Biotechnol 104:129–153. doi: 10.1016/S0168-1656(03)00144-5. [DOI] [PubMed] [Google Scholar]
- 15.Bowman LAH, McLean S, Poole RK, Fukuto JM. 2011. The diversity of microbial responses to nitric oxide and agents of nitrosative stress: close cousins but not identical twins. Adv Microb Physiol 59:135–219. doi: 10.1016/B978-0-12-387661-4.00006-9. [DOI] [PubMed] [Google Scholar]
- 16.Platzen L, Koch-Koerfges A, Weil B, Brocker M, Bott M. 2014. Role of flavohaemoprotein Hmp and nitrate reductase NarGHJI of Corynebacterium glutamicum for coping with nitrite and nitrosative stress. FEMS Microbiol Lett 350:239–248. doi: 10.1111/1574-6968.12318. [DOI] [PubMed] [Google Scholar]
- 17.Dominguez H, Nezondet C, Lindley ND, Cocaign M. 1993. Modified carbon flux during oxygen limited growth of Corynebacterium glutamicum and the consequences for amino acid overproduction. Biotechnol Lett 15:449–454. doi: 10.1007/BF00129316. [DOI] [Google Scholar]
- 18.Inui M, Murakami S, Okino S, Kawaguchi H, Vertes AA, Yukawa H. 2004. Metabolic analysis of Corynebacterium glutamicum during lactate and succinate productions under oxygen deprivation conditions. J Mol Microbiol Biotechnol 7:182–196. doi: 10.1159/000079827. [DOI] [PubMed] [Google Scholar]
- 19.Okino S, Noburyu R, Suda M, Jojima T, Inui M, Yukawa H. 2008. An efficient succinic acid production process in a metabolically engineered Corynebacterium glutamicum strain. Appl Microbiol Biotechnol 81:459–464. doi: 10.1007/s00253-008-1668-y. [DOI] [PubMed] [Google Scholar]
- 20.Litsanov B, Brocker M, Bott M. 2012. Toward homosuccinate fermentation: metabolic engineering of Corynebacterium glutamicum for anaerobic production of succinate from glucose and formate. Appl Environ Microbiol 78:3325–3337. doi: 10.1128/AEM.07790-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Okino S, Suda M, Fujikura K, Inui M, Yukawa H. 2008. Production of d-lactic acid by Corynebacterium glutamicum under oxygen deprivation. Appl Microbiol Biotechnol 78:449–454. doi: 10.1007/s00253-007-1336-7. [DOI] [PubMed] [Google Scholar]
- 22.Blombach B, Riester T, Wieschalka S, Ziert C, Youn JW, Wendisch VF, Eikmanns BJ. 2011. Corynebacterium glutamicum tailored for efficient isobutanol production. Appl Environ Microbiol 77:3300–3310. doi: 10.1128/AEM.02972-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hasegawa S, Suda M, Uematsu K, Natsuma Y, Hiraga K, Jojima T, Inui M, Yukawa H. 2013. Engineering of Corynebacterium glutamicum for high-yield l-valine production under oxygen deprivation conditions. Appl Environ Microbiol 79:1250–1257. doi: 10.1128/AEM.02806-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rados D, Turner DL, Fonseca LL, Carvalho AL, Blombach B, Eikmanns BJ, Neves AR, Santos H. 2014. Carbon flux analysis by 13C nuclear magnetic resonance to determine the effect of CO2 on anaerobic succinate production by Corynebacterium glutamicum. Appl Environ Microbiol 80:3015–3024. doi: 10.1128/AEM.04189-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Inui M, Suda M, Okino S, Nonaka H, Puskas LG, Vertes AA, Yukawa H. 2007. Transcriptional profiling of Corynebacterium glutamicum metabolism during organic acid production under oxygen deprivation conditions. Microbiology 153:2491–2504. doi: 10.1099/mic.0.2006/005587-0. [DOI] [PubMed] [Google Scholar]
- 26.Yamamoto S, Sakai M, Inui M, Yukawa H. 2011. Diversity of metabolic shift in response to oxygen deprivation in Corynebacterium glutamicum and its close relatives. Appl Microbiol Biotechnol 90:1051–1061. doi: 10.1007/s00253-011-3144-3. [DOI] [PubMed] [Google Scholar]
- 27.Koch-Koerfges A, Pfelzer N, Platzen L, Oldiges M, Bott M. 2013. Conversion of Corynebacterium glutamicum from an aerobic respiring to an aerobic fermenting bacterium by inactivation of the respiratory chain. Biochim Biophys Acta 1827:699–708. doi: 10.1016/j.bbabio.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook J, Russell D. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 29.Keilhauer C, Eggeling L, Sahm H. 1993. Isoleucine synthesis in Corynebacterium glutamicum: molecular analysis of the ilvB-ilvN-ilvC operon. J Bacteriol 175:5595–5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kabus A, Niebisch A, Bott M. 2007. Role of cytochrome bd oxidase from Corynebacterium glutamicum in growth and lysine production. Appl Environ Microbiol 73:861–868. doi: 10.1128/AEM.01818-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koch-Koerfges A, Kabus A, Ochrombel I, Marin K, Bott M. 2012. Physiology and global gene expression of a Corynebacterium glutamicum ΔF1Fo-ATP synthase mutant devoid of oxidative phosphorylation. Biochim Biophys Acta 1817:370–380. doi: 10.1016/j.bbabio.2011.10.006. [DOI] [PubMed] [Google Scholar]
- 32.Hanahan D. 1985. Techniques for transformation of E. coli, p 109–135. In Glover DM. (ed), DNA cloning, vol 1 IRL Press, Oxford, United Kingdom. [Google Scholar]
- 33.van der Rest ME, Lange C, Molenaar D. 1999. A heat shock following electroporation induces highly efficient transformation of Corynebacterium glutamicum with xenogeneic plasmid DNA. Appl Microbiol Biotechnol 52:541–545. doi: 10.1007/s002530051557. [DOI] [PubMed] [Google Scholar]
- 34.Blombach B, Seibold GM. 2010. Carbohydrate metabolism in Corynebacterium glutamicum and applications for the metabolic engineering of l-lysine production strains. Appl Microbiol Biotechnol 86:1313–1322. doi: 10.1007/s00253-010-2537-z. [DOI] [PubMed] [Google Scholar]
- 35.Wendisch VF. 2003. Genome-wide expression analysis in Corynebacterium glutamicum using DNA microarrays. J Biotechnol 104:273–285. doi: 10.1016/S0168-1656(03)00147-0. [DOI] [PubMed] [Google Scholar]
- 36.Frunzke J, Engels V, Hasenbein S, Gätgens C, Bott M. 2008. Co-ordinated regulation of gluconate catabolism and glucose uptake in Corynebacterium glutamicum by two functionally equivalent transcriptional regulators, GntR1 and GntR2. Mol Microbiol 67:305–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Letek M, Valbuena N, Ramos A, Ordonez E, Gil JA, Mateos LM. 2006. Characterization and use of catabolite-repressed promoters from gluconate genes in Corynebacterium glutamicum. J Bacteriol 188:409–423. doi: 10.1128/JB.188.2.409-423.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Netzer R, Krause M, Rittmann D, Peters-Wendisch PG, Eggeling L, Wendisch VF, Sahm H. 2004. Roles of pyruvate kinase and malic enzyme in Corynebacterium glutamicum for growth on carbon sources requiring gluconeogenesis. Arch Microbiol 182:354–363. doi: 10.1007/s00203-004-0710-4. [DOI] [PubMed] [Google Scholar]
- 39.Wendisch VF, Spies M, Reinscheid DJ, Schnicke S, Sahm H, Eikmanns BJ. 1997. Regulation of acetate metabolism in Corynebacterium glutamicum: transcriptional control of the isocitrate lyase and malate synthase genes. Arch Microbiol 168:262–269. doi: 10.1007/s002030050497. [DOI] [PubMed] [Google Scholar]
- 40.Gourdon P, Baucher MF, Lindley ND, Guyonvarch A. 2000. Cloning of the malic enzyme gene from Corynebacterium glutamicum and role of the enzyme in lactate metabolism. Appl Environ Microbiol 66:2981–2987. doi: 10.1128/AEM.66.7.2981-2987.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nentwich SS, Brinkrolf K, Gaigalat L, Hüser AT, Rey DA, Mohrbach T, Marin K, Pühler A, Tauch A, Kalinowski J. 2009. Characterization of the LacL-type transcriptional repressor RbsR controlling ribose transport in Corynebacterium glutamicum ATCC 13032. Microbiology 155:150–164. doi: 10.1099/mic.0.020388-0. [DOI] [PubMed] [Google Scholar]
- 42.Moritz B, Striegel K, De Graaf AA, Sahm H. 2000. Kinetic properties of the glucose-6-phosphate and 6-phosphogluconate dehydrogenases from Corynebacterium glutamicum and their application for predicting pentose phosphate pathway flux in vivo. Eur J Biochem 267:3442–3452. doi: 10.1046/j.1432-1327.2000.01354.x. [DOI] [PubMed] [Google Scholar]
- 43.Kabus A, Georgi T, Wendisch VF, Bott M. 2007. Expression of the Escherichia coli pntAB genes encoding a membrane-bound transhydrogenase in Corynebacterium glutamicum improves l-lysine formation. Appl Microbiol Biotechnol 75:47–53. doi: 10.1007/s00253-006-0804-9. [DOI] [PubMed] [Google Scholar]
- 44.Marienhagen J, Eggeling L. 2008. Metabolic function of Corynebacterium glutamicum aminotransferases AlaT and AvtA and impact on l-valine production. Appl Environ Microbiol 74:7457–7462. doi: 10.1128/AEM.01025-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peters-Wendisch PG, Kreutzer C, Kalinowski J, Patek M, Sahm H, Eikmanns BJ. 1998. Pyruvate carboxylase from Corynebacterium glutamicum: characterization, expression and inactivation of the pyc gene. Microbiology 144:915–927. doi: 10.1099/00221287-144-4-915. [DOI] [PubMed] [Google Scholar]
- 46.Neidhardt FC, Ingraham JL, Schaechter M. 1990. Physiology of the bacterial cell: a molecular approach. Sinauer Associates Inc., Sunderland, MA, USA. [Google Scholar]
- 47.Follmann M, Ochrombel I, Krämer R, Trötschel C, Poetsch A, Rückert C, Hüser A, Persicke M, Seiferling D, Kalinowski J, Marin K. 2009. Functional genomics of pH homeostasis in Corynebacterium glutamicum revealed novel links between pH response, oxidative stress, iron homeostasis and methionine synthesis. BMC Genomics 10:621. doi: 10.1186/1471-2164-10-621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Follmann M, Becker M, Ochrombel I, Ott V, Krämer R, Marin K. 2009. Potassium transport in Corynebacterium glutamicum is facilitated by the putative channel protein CglK, which is essential for pH homeostasis and growth at acidic pH. J Bacteriol 191:2944–2952. doi: 10.1128/JB.00074-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ochrombel I, Ott L, Krämer R, Burkovski A, Marin K. 2011. Impact of improved potassium accumulation on pH homeostasis, membrane potential adjustment and survival of Corynebacterium glutamicum. Biochim Biophys Acta 1807:444–450. doi: 10.1016/j.bbabio.2011.01.008. [DOI] [PubMed] [Google Scholar]
- 50.Erdmann A, Weil B, Krämer R. 1993. Lysine secretion by wild-type Corynebacterium glutamicum triggered by dipeptide uptake. J Gen Microbiol 139:3115–3122. doi: 10.1099/00221287-139-12-3115. [DOI] [Google Scholar]
- 51.Zittrich S, Krämer R. 1994. Quantitative discrimination of carrier-mediated excretion of isoleucine from uptake and diffusion in Corynebacterium glutamicum. J Bacteriol 176:6892–6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hermann T, Krämer R. 1996. Mechanism and regulation of isoleucine excretion in Corynebacterium glutamicum. Appl Environ Microbiol 62:3238–3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bellmann A, Vrljic M, Patek M, Sahm H, Krämer R, Eggeling L. 2001. Expression control and specificity of the basic amino acid exporter LysE of Corynebacterium glutamicum. Microbiology 147:1765–1774. [DOI] [PubMed] [Google Scholar]
- 54.Mustafi N, Grünberger A, Kohlheyer D, Bott M, Frunzke J. 2012. The development and application of a single-cell biosensor for the detection of l-methionine and branched-chain amino acids. Metab Eng 14:449–457. doi: 10.1016/j.ymben.2012.02.002. [DOI] [PubMed] [Google Scholar]
- 55.Vrljic M, Sahm H, Eggeling L. 1996. A new type of transporter with a new type of cellular function: l-lysine export from Corynebacterium glutamicum. Mol Microbiol 22:815–826. doi: 10.1046/j.1365-2958.1996.01527.x. [DOI] [PubMed] [Google Scholar]
- 56.Marx A, de Graaf AA, Wiechert W, Eggeling L, Sahm H. 1996. Determination of the fluxes in the central metabolism of Corynebacterium glutamicum by nuclear magnetic resonance spectroscopy combined with metabolite balancing. Biotechnol Bioeng 49:111–129. [DOI] [PubMed] [Google Scholar]
- 57.Slonczewski JL, Fujisawa M, Dopson M, Krulwich TA. 2009. Cytoplasmic pH measurement and homeostasis in bacteria and archaea. Adv Microb Physiol 55:1–79. doi: 10.1016/S0065-2911(09)05501-5. [DOI] [PubMed] [Google Scholar]
- 58.Krulwich TA, Sachs G, Padan E. 2011. Molecular aspects of bacterial pH sensing and homeostasis. Nat Rev Microbiol 9:330–343. doi: 10.1038/nrmicro2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cotter PD, Hill C. 2003. Surviving the acid test: responses of gram-positive bacteria to low pH. Microbiol Mol Biol Rev 67:429–453. doi: 10.1128/MMBR.67.3.429-453.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jakob K, Satorhelyi P, Lange C, Wendisch VF, Silakowski B, Scherer S, Neuhaus K. 2007. Gene expression analysis of Corynebacterium glutamicum subjected to long-term lactic acid adaptation. J Bacteriol 189:5582–5590. doi: 10.1128/JB.00082-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stingl K, Altendorf K, Bakker EP. 2002. Acid survival of Helicobacter pylori: how does urease activity trigger cytoplasmic pH homeostasis? Trends Microbiol 10:70–74. doi: 10.1016/S0966-842X(01)02287-9. [DOI] [PubMed] [Google Scholar]
- 62.Nolden L, Beckers G, Möckel B, Pfefferle W, Nampoothiri KM, Krämer R, Burkovski A. 2000. Urease of Corynebacterium glutamicum: organization of corresponding genes and investigation of activity. FEMS Microbiol Lett 189:305–310. doi: 10.1111/j.1574-6968.2000.tb09248.x. [DOI] [PubMed] [Google Scholar]
- 63.Beckers G, Bendt AK, Krämer R, Burkovski A. 2004. Molecular identification of the urea uptake system and transcriptional analysis of urea transporter- and urease-encoding genes in Corynebacterium glutamicum. J Bacteriol 186:7645–7652. doi: 10.1128/JB.186.22.7645-7652.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fukui K, Koseki C, Yamamoto Y, Nakamura J, Sasahara A, Yuji R, Hashiguchi K, Usuda Y, Matsui K, Kojima H, Abe K. 2011. Identification of succinate exporter in Corynebacterium glutamicum and its physiological roles under anaerobic conditions. J Biotechnol 154:25–34. doi: 10.1016/j.jbiotec.2011.03.010. [DOI] [PubMed] [Google Scholar]
- 65.Huhn S, Jolkver E, Krämer R, Marin K. 2011. Identification of the membrane protein SucE and its role in succinate transport in Corynebacterium glutamicum. Appl Microbiol Biotechnol 89:327–335. doi: 10.1007/s00253-010-2855-1. [DOI] [PubMed] [Google Scholar]
- 66.Youn JW, Jolkver E, Krämer R, Marin K, Wendisch VF. 2008. Identification and characterization of the dicarboxylate uptake system DccT in Corynebacterium glutamicum. J Bacteriol 190:6458–6466. doi: 10.1128/JB.00780-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Youn JW, Jolkver E, Krämer R, Marin K, Wendisch VF. 2009. Characterization of the dicarboxylate transporter DctA in Corynebacterium glutamicum. J Bacteriol 191:5480–5488. doi: 10.1128/JB.00640-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stansen C, Uy D, Delaunay S, Eggeling L, Goergen JL, Wendisch VF. 2005. Characterization of a Corynebacterium glutamicum lactate utilization operon induced during temperature-triggered glutamate production. Appl Environ Microbiol 71:5920–5928. doi: 10.1128/AEM.71.10.5920-5928.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Matsushita K. 2013. Respiratory chain and energy metabolism of Corynebacterium glutamicum, p 315–334. In Yukawa H, Inui M (ed), Corynebacterium glutamicum: biology and biotechnology. Springer, Heidelberg, Germany. [Google Scholar]
- 70.Sheng J, Marquis RE. 2006. Enhanced acid resistance of oral streptococci at lethal pH values associated with acid-tolerant catabolism and with ATP synthase activity. FEMS Microbiol Lett 262:93–98. doi: 10.1111/j.1574-6968.2006.00374.x. [DOI] [PubMed] [Google Scholar]
- 71.Harold FM. 1986. The vital force: a study of bioenergetics. W. H. Freeman and Company, New York, NY. [Google Scholar]
- 72.Rao M, Streur TL, Aldwell FE, Cook GM. 2001. Intracellular pH regulation by Mycobacterium smegmatis and Mycobacterium bovis BCG. Microbiology 147:1017–1024. [DOI] [PubMed] [Google Scholar]
- 73.Sekine H, Shimada T, Hayashi C, Ishiguro A, Tomita F, Yokota A. 2001. H+-ATPase defect in Corynebacterium glutamicum abolishes glutamic acid production with enhancement of glucose consumption rate. Appl Microbiol Biotechnol 57:534–540. doi: 10.1007/s002530100778. [DOI] [PubMed] [Google Scholar]
- 74.Sawada K, Kato Y, Imai K, Li L, Wada M, Matsushita K, Yokota A. 2012. Mechanism of increased respiration in an H+-ATPase-defective mutant of Corynebacterium glutamicum. J Biosci Bioeng 113:467–473. doi: 10.1016/j.jbiosc.2011.11.021. [DOI] [PubMed] [Google Scholar]
- 75.Abe S, Takayama K, Kinoshita S. 1967. Taxonomical studies on glutamic acid producing bacteria. J Gen Appl Microbiol 13:279–301. doi: 10.2323/jgam.13.279. [DOI] [Google Scholar]
- 76.Schäfer A, Tauch A, Jäger W, Kalinowski J, Thierbach G, Pühler A. 1994. Small mobilizable multipurpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19—selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69–73. doi: 10.1016/0378-1119(94)90324-7. [DOI] [PubMed] [Google Scholar]
- 77.Kalinowski J, Bathe B, Bartels D, Bischoff N, Bott M, Burkovski A, Dusch N, Eggeling L, Eikmanns BJ, Gaigalat L, Goesmann A, Hartmann M, Huthmacher K, Krämer R, Linke B, McHardy AC, Meyer F, Möckel B, Pfefferle W, Pühler A, Rey DA, Rückert C, Rupp O, Sahm H, Wendisch VF, Wiegrabe I, Tauch A. 2003. The complete Corynebacterium glutamicum ATCC 13032 genome sequence and its impact on the production of l-aspartate-derived amino acids and vitamins. J Biotechnol 104:5–25. doi: 10.1016/S0168-1656(03)00154-8. [DOI] [PubMed] [Google Scholar]
- 78.Zhao Z, Ding JY, Ma WH, Zhou NY, Liu SJ. 2012. Identification and characterization of γ-aminobutyric acid uptake system GabPCg (NCgl0464) in Corynebacterium glutamicum. Appl Environ Microbiol 78:2596–2601. doi: 10.1128/AEM.07406-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Peter H, Burkovski A, Krämer R. 1996. Isolation, characterization, and expression of the Corynebacterium glutamicum betP gene, encoding the transport system for the compatible solute glycine betaine. J Bacteriol 178:5229–5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Seep-Feldhaus AH, Kalinowski J, Pühler A. 1991. Molecular analysis of the Corynebacterium glutamicum lysl gene involved in lysine uptake. Mol Microbiol 5:2995–3005. doi: 10.1111/j.1365-2958.1991.tb01859.x. [DOI] [PubMed] [Google Scholar]
- 81.Trötschel C, Follmann M, Nettekoven JA, Mohrbach T, Forrest LR, Burkovski A, Marin K, Krämer R. 2008. Methionine uptake in Corynebacterium glutamicum by MetQNI and by MetPS, a novel methionine and alanine importer of the NSS neurotransmitter transporter family. Biochemistry 47:12698–12709. doi: 10.1021/bi801206t. [DOI] [PubMed] [Google Scholar]
- 82.Wehrmann A, Morakkabati S, Krämer R, Sahm H, Eggeling L. 1995. Functional analysis of sequences adjacent to dapE of Corynebacterium glutamicum reveals the presence of aroP, which encodes the aromatic amino acid transporter. J Bacteriol 177:5991–5993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shang X, Zhang Y, Zhang G, Chai X, Deng A, Liang Y, Wen T. 2013. Characterization and molecular mechanism of AroP as an aromatic amino acid and histidine transporter in Corynebacterium glutamicum. J Bacteriol 195:5334–5342. doi: 10.1128/JB.00971-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhao Z, Ding JY, Li T, Zhou NY, Liu SJ. 2011. The ncgl1108 (PhePCg) gene encodes a new l-Phe transporter in Corynebacterium glutamicum. Appl Microbiol Biotechnol 90:2005–2013. doi: 10.1007/s00253-011-3245-z. [DOI] [PubMed] [Google Scholar]
- 85.Peter H, Bader A, Burkovski A, Lambert C, Krämer R. 1997. Isolation of the putP gene of Corynebacterium glutamicum and characterization of a low-affinity uptake system for compatible solutes. Arch Microbiol 168:143–151. doi: 10.1007/s002030050480. [DOI] [PubMed] [Google Scholar]
- 86.Tauch A, Hermann T, Burkovski A, Krämer R, Pühler A, Kalinowski J. 1998. Isoleucine uptake in Corynebacterium glutamicum ATCC 13032 is directed by the brnQ gene product. Arch Microbiol 169:303–312. doi: 10.1007/s002030050576. [DOI] [PubMed] [Google Scholar]
- 87.Peter H, Weil B, Burkovski A, Krämer R, Morbach S. 1998. Corynebacterium glutamicum is equipped with four secondary carriers for compatible solutes: identification, sequencing, and characterization of the proline/ectoine uptake system, ProP, and the ectoine/proline/glycine betaine carrier, EctP. J Bacteriol 180:6005–6012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Steger R, Weinand M, Krämer R, Morbach S. 2004. LcoP, an osmoregulated betaine/ectoine uptake system from Corynebacterium glutamicum. FEBS Lett 573:155–160. doi: 10.1016/j.febslet.2004.07.067. [DOI] [PubMed] [Google Scholar]
- 89.Trötschel C, Kandirali S, Diaz-Achirica P, Meinhardt A, Morbach S, Krämer R, Burkovski A. 2003. GltS, the sodium-coupled l-glutamate uptake system of Corynebacterium glutamicum: identification of the corresponding gene and impact on l-glutamate production. Appl Microbiol Biotechnol 60:738–742. doi: 10.1007/s00253-002-1170-x. [DOI] [PubMed] [Google Scholar]
- 90.Heery DM, Fitzpatrick R, Dunican LK. 1994. A sequence from a tryptophan-hyperproducing strain of Corynebacterium glutamicum encoding resistance to 5-methyltryptophan. Biochem Biophys Res Commun 201:1255–1262. doi: 10.1006/bbrc.1994.1840. [DOI] [PubMed] [Google Scholar]
- 91.Kronemeyer W, Peekhaus N, Krämer R, Sahm H, Eggeling L. 1995. Structure of the gluABCD cluster encoding the glutamate uptake system of Corynebacterium glutamicum. J Bacteriol 177:1152–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Simic P, Sahm H, Eggeling L. 2001. l-Threonine export: use of peptides to identify a new translocator from Corynebacterium glutamicum. J Bacteriol 183:5317–5324. doi: 10.1128/JB.183.18.5317-5324.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kennerknecht N, Sahm H, Yen MR, Patek M, Saier MH Jr, Eggeling L. 2002. Export of l-isoleucine from Corynebacterium glutamicum: a two-gene-encoded member of a new translocator family. J Bacteriol 184:3947–3956. doi: 10.1128/JB.184.14.3947-3956.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Trötschel C, Deutenberg D, Bathe B, Burkovski A, Krämer R. 2005. Characterization of methionine export in Corynebacterium glutamicum. J Bacteriol 187:3786–3794. doi: 10.1128/JB.187.11.3786-3794.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Nakamura J, Hirano S, Ito H, Wachi M. 2007. Mutations of the Corynebacterium glutamicum NCgl1221 gene, encoding a mechanosensitive channel homolog, induce l-glutamic acid production. Appl Environ Microbiol 73:4491–4498. doi: 10.1128/AEM.02446-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.