Abstract
Seasonal variation in the phylogenetic composition of Synechococcus assemblages in estuarine and coastal waters of Hong Kong was examined through pyrosequencing of the rpoC1 gene. Sixteen samples were collected in 2009 from two stations representing estuarine and ocean-influenced coastal waters, respectively. Synechococcus abundance in coastal waters gradually increased from 3.6 × 103 cells ml−1 in March, reaching a peak value of 5.7 × 105 cells ml−1 in July, and then gradually decreased to 9.3 × 103 cells ml−1 in December. The changes in Synechococcus abundance in estuarine waters followed a pattern similar to that in coastal waters, whereas its composition shifted from being dominated by phycoerythrin-rich (PE-type) strains in winter to phycocyanin-only (PC-type) strains in summer owing to the increase in freshwater discharge from the Pearl River and higher water temperature. The high abundance of PC-type Synechococcus was composed of subcluster 5.2 marine Synechococcus, freshwater Synechococcus (F-PC), and Cyanobium. The Synechococcus assemblage in the coastal waters, on the other hand, was dominated by marine PE-type Synechococcus, with subcluster 5.1 clades II and VI as the major lineages from April to September, when the summer monsoon prevailed. Besides these two clades, clade III cooccurred with clade V at relatively high abundance in summer. During winter, the Synechococcus assemblage compositions at the two sites were similar and were dominated by subcluster 5.1 clades II and IX and an undescribed clade (represented by Synechococcus sp. strain miyav). Clade IX Synechococcus was a relatively ubiquitous PE-type Synechococcus found at both sites, and our study demonstrates that some strains of the clade have the ability to deal with large variation of salinity in subtropical estuarine environments. Our study suggests that changes in seawater temperature and salinity caused by the seasonal variation of monsoonal forcing are two major determinants of the community composition and abundance of Synechococcus assemblages in Hong Kong waters.
INTRODUCTION
Members of the Synechococcus group of widely distributed and abundant picocyanobacteria are important primary producers in the surface waters of global oceans (1). Strains of Synechococcus are both phenotypically and phylogenetically diverse and dynamic (2, 3). Based on gene markers, like the 16S rRNA gene, marine Synechococcus strains form a well-defined clade termed cluster 5 (4, 5), which is divided into 3 subclusters: 5.1, 5.2, and 5.3. Of these, subcluster 5.1 is the most abundant and diverse subcluster in marine environments and is further divided into at least 9 clades (6). The high genetic diversity of Synechococcus is reflected in the ecogeographic and temporal distribution of different ecotypes. Subcluster 5.1 clade I mainly dominates in temperate mesotrophic ocean waters, while clade II is mainly present in offshore, continental shelf, and oligotrophic warm waters (4, 7). Subcluster 5.2 Synechococcus strains are phycocyanin-enriched (PC-type) euryhaline strains widely distributed in coastal and estuarine waters (6). Chen et al. suggested that members of this subcluster exhibit higher genetic diversity than subcluster 5.1 Synechococcus due to their complex habitats (8). Subcluster 5.3 Synechococcus strains are less studied, phycoerythrin-enriched (PE-type) strains (pigment types 3b and 3d) (http://roscoff-culture-collection.org/strains/shortlists/taxonomic-groups/marine-synechococcus) and include at least six clades (9).
In addition to the inherent genetic diversity, environmental factors also determine the variations in Synechococcus abundance and diversity in a particular environment (10–15). Temperature, for example, is one of the most important parameters determining the abundance of Synechococcus (10, 16, 17). However, to date, only a few studies have reported the temporal changes in the Synechococcus assemblage structure in detail (4, 18, 19).
Hong Kong coastal waters have a unique and highly dynamic hydrographic setting and therefore are ideal sites to study the diversity and spatiotemporal variation in the Synechococcus community composition (20). This region is influenced by several different water masses, including the freshwater discharge from the Pearl River, oceanic water of the South China Sea, and the Fujian-Zhejiang coastal current. The relative strengths and interactions of these water masses are modulated by the annual cycle of the monsoonal wind. A recent study found that estuarine waters of the Pearl River were dominated by PC-type Synechococcus, while the coastal waters on the other side of Hong Kong were dominated by PE-type Synechococcus (21). However, the phylogenetic composition of PE- and PC-type Synechococcus strains in Hong Kong waters remains unknown. Therefore, the present study was conducted to reveal the seasonal variation of the Synechococcus assemblage composition in coastal and estuarine waters of Hong Kong, particularly in response to the changes in the Pearl River plume. PCR-based molecular methods have been successfully employed to examine the community structure of Synechococcus in marine waters (7, 22). In addition to the 16S rRNA gene, gene markers with even higher genetic resolution, such as the rRNA internally transcribed spacer (ITS), narB, ntcA, and rpoC1, are now being used to study the community structure of Synechococcus (22, 23). In fact, rpoC1 was the first gene marker to show the presence of different Synechococcus clades in seawater samples (24), which seemed to be a promising approach.
In this study, we applied pyrosequencing of the rpoC1 gene to investigate Synechococcus assemblage compositions in Hong Kong waters. Because of their sequencing depth, pyrosequencing methods reveal information on both dominant members and rare species (25, 26). We documented high diversity of Synechococcus strains in Hong Kong waters and recorded spatiotemporal variation of Synechococcus assemblage compositions. Both the phenotypic and phylogenetic compositions of Synechococcus assemblages are markedly influenced by river inputs.
MATERIALS AND METHODS
Sample collection.
Field sampling was conducted at two Hong Kong marine stations, NM3 (113°56.7′E, 22°21.3′N) and PM7 (114°17.7′E, 22°20.4′N), with different hydrographic and trophic conditions (Fig. 1). NM3, located in the Pearl River estuary, represented estuarine waters, while PM7, at Port Shelter in the eastern waters of Hong Kong, represented ocean-influenced coastal water (here termed coastal waters). Surface water samples were collected from the two stations every month from March to December 2009, except in August and November.
FIG 1.
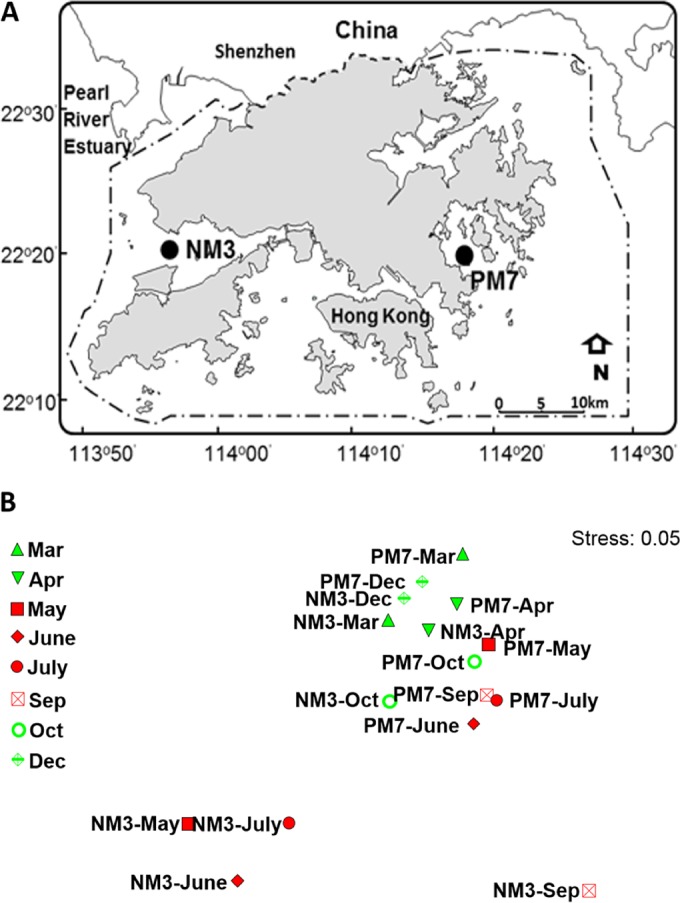
(A) Sampling stations. NM3 is located in the western waters of Hong Kong, which are influenced by freshwater discharge from the Pearl River, whereas PM7 is a typical coastal station located in the eastern waters of Hong Kong. (Adapted from reference 21 with permission of the publisher [copyright 2013 Society for Applied Microbiology and John Wiley & Sons Ltd.].) (B) NMDS plot showing the relationship of hydrographical and trophic conditions at sampling stations.
At each station, 0.5 liter of surface seawater was prefiltered through a 3.0-μm (47-mm) polycarbonate membrane (Pall Corporation) and then filtered onto a 0.22-μm (47-mm) polycarbonate membrane. Samples were frozen at −80°C immediately after filtration. The temperature and salinity of the seawater were measured using a YSI 6600 (YSIs, USA). Samples for nutrient and chlorophyll a (Chl a) analyses were collected from the two stations each month. Total inorganic nitrogen (TIN) and phosphate concentrations were measured using a Skalar Nutrient Analyzer (ThermoFisher, USA). The Chl a concentration was measured using a Turner Designs (Sunnyvale, CA) fluorometer after extraction with 90% acetone (27).
DNA extraction, PCR, and sequencing.
DNA was extracted using the enzyme/phenol-chloroform protocol (28). Extracted DNA was eluted in Tris-EDTA (TE) buffer (10 mM Tris, 1 mM EDTA, pH 8.0) and kept at −20°C until further use. The PCR followed the protocol of Mühling et al. (23). The nested-PCR primers are highly specific for Synechococcus species and therefore are suitable for amplifying the rpoC1 gene from low-DNA-concentration templates (23). The first round of PCR used the primer rpoC1-N5 and the C-terminal primer rpoC1-C, and the PCR products were used as templates for the second-round PCR with the modified primers rpoC1-39F (5′-ccatctcatccctgcgtgtctccgactcagnnnnnnnnnnGGNATYGTYTGYGAGCGYTG) and rpoC1-462R (5′-cctatcccctgtgtgccttggcagtctcagCGYAGRCFCTTGRTCAGCTT) (23). (Sequences in lowercase letters are 454 adapters for binding used in pyrosequencing, and the “n” repeat represents 10-nt barcode sequences used to identify samples. The sequences in uppercase letters are targeted to the rpoC1 gene.) The PCR products were gel purified using a Qiaquick gel purification kit (Qiagen, Hilgen, Germany) as described by the manufacturer. Library quantification was done by fluorometry using the Quant-iT picoGreen double-stranded DNA (dsDNA) assay kit (Invitrogen, USA). The amplicons were mixed in equal amounts and sequenced in a two-region 454 run on a GS PicoTiterPlate using a GS Junior pyrosequencing system (Roche, 454 Life Sciences, Branford, CT, USA) according to the manufacturer's instructions.
Postrun sequence analyses.
Analysis of rpoC1 sequence was conducted using the microbial ecology community software program mothur (http://www.mothur.org/wiki/Download_mothur) (29). The reads were processed by removing tags and primers; only reads with an average quality score above 20 and read lengths between 300 bp and 500 bp were accepted. Sequence denoise was carried out using the command shhh.seqs with sigma value 0.01. Chimeras were analyzed in the mothur software package using the command chimera.uchime (30). After the above-described quality control, 6,247 high-quality sequences were randomly subsampled from each sample and were further analyzed with mothur for alignment and DNA distance calculations.
The richness estimator (ACE), diversity index (Shannon index; H′), coverage, and operational taxonomic units (OTUs) were calculated at 95% similarity using mothur's summary. single routine. Rarefaction curves were calculated using ACE richness index data and the number of observed OTUs.
The representative sequence of each OTU was extracted and identified by local BLAST search using Bio-Edit (31), and the sequences used as references are listed in Table S1 in the supplemental material. Sequences that were less than 90% identical to the reference sequences were assigned as unclassified. The expectation value cutoff used was 0.01. The relative abundance of each lineage was summarized and used to calculate the abundance of Synechococcus lineages in each month (abundance of each lineage = relative abundance × total Synechococcus abundance measured by flow cytometry).
Heat maps showing the relative abundances and relationships of the top 50 most abundant OTUs were generated using HemI (32). The relative abundance of each OTU was square root transformed. Average linkage clustering was performed by using Pearson correlation matrices. The representative sequence of each OTU was classified using the database mentioned above. BLASTn was also applied to sequence identity analysis (http://www.ncbi.nlm.nih.gov/).
Phylogenetic trees of PC-type Synechococcus sequences were constructed in MEGA 5 using the maximum-likelihood method (ML) with a Kimura 2-parameter model (33). The initial tree for ML was based on BIONJ (64). The nearest relatives were retrieved from National Center for Biotechnology Information (NCBI) reference sequences. A heat map showing the relative abundance of each OTU was generated in iTol (34). Nonmetric multidimensional scaling (NMDS) analysis, the analysis of molecular variance (AMOVA) test, and similarity percentage (SIMPER) analysis were applied using Primer 5 (Primer-E-Ltd., United Kingdom) to compare the community compositions based on the relative abundances of lineages identified from Hong Kong waters.
The relationship between environmental parameters and Synechococcus community composition was studied by redundancy analysis (RDA) using CANOCO V4.5 (Microcomputer Power, USA). A matrix was generated using the relative abundance of each lineage transformed by square root transformation. Environmental data were normalized using Z-score transformation. Centering and normalizing options were employed, producing scores centered and standardized to unit variance. The significance of the eigenvalues and species-environment correlations of the first three axes were determined by Monte Carlo tests (500 permutations).
Salinity tolerance of a clade IX Synechococcus strain in Hong Kong waters.
In order to study the salinity tolerance of Synechococcus lineages, we evaluated the growth rates of 6 strains—WH 8012 (clade II), WH 7803 (clade V), PS01 (clade V), MW03 (clade VIII), MW02 (clade IX), and WH 5701 (subcluster 5.2)—at various salinities. MW02 and MW03 were isolated from estuarine waters of Hong Kong. The ITS gene sequence of MW02 shows it is a clade IX Synechococcus strain related to Synechococcus sp. strain RS9901 (BLASTN). PS01 is a low-phycourobilin (PUB) Synechococcus strain, while MW02 and MW03 are PE-only and PC-only Synechococcus strains, respectively. All the cultures were grown in f/2 medium enriched artificial seawater (65) (with 50 μM ammonium as an N source) with different salinities (15, 22, 28, and 34 ppt) for 15 days in plant growth chambers at 25°C under a photon flux density of 2.5 × 1015 quanta s−1 cm−2 in a 12-h/12-h light-dark cycle. The optical density at 440 nm (OD440) of each culture was measured every day. Growth rates (day−1) were calculated as ln (Nt/N0)/dt. N0 and Nt are the OD440 values of strains at the beginning and at the end of the culture period, and dt is the culture period (in days).
In vivo absorption spectra.
The in vivo absorption spectra of the Synechococcus cultures were measured to determine the optical properties of Synechococcus strains (2, 35). An aliquot of the exponentially growing culture was transferred to a cuvette, and the in vivo absorption spectrum was measured from 400 to 700 nm using a spectrophotometer (UH5300; Hitachi, USA). The scan rate was 1 nm s−1. The spectra were normalized at 440 nm.
Accession numbers.
The ITS gene sequence of MW02 has been submitted to NCBI with accession number KP113680. All 200,079 raw sequences obtained from this study have been deposited in the NCBI Sequence Read Archive (SRA) under accession numbers SRR1583674 (PM7-Mar), SRR1106828 (PM7-Apr), SRR1583675 (PM7-May), SRR1583673 (PM7-June), SRR2242694 (PM7-July), SRR1583676 (PM7-Sep), SRR2242696 (PM7-Oct), SRR1107754 (PM7-Dec), SRR1583670 (NM3-Mar), SRR1107750 (NM3-Apr), SRR1583671 (NM3-May), SRR1583669 (NM3-June), SRR2242693 (NM3-July), SRR1583672 (NM3-Sep), SRR2242695 (NM3-Oct), and SRR1107753 (NM3-Dec).
RESULTS
Environmental conditions at the sampling stations.
The estuarine station (NM3) is strongly influenced by freshwater discharge from the Pearl River, particularly during the wet season (April to September; 80% of the discharge of the Pearl River occurs during this period [36]), making it a low-salinity and high-nutrient environment. On the other hand, the coastal station (PM7) located on the east side of Hong Kong did not receive direct river input and developed stratified conditions with nutrient depletion in the upper mixed layer in the summer (37). At PM7, the highest TIN and phosphate concentrations occurred in December because of the nutrient-rich Fujian-Zhejiang coastal current water brought to the region under the northeast monsoon (Table 1). We found that the two stations had very different hydrographic and trophic conditions during the period from May to September (Table 1 and Fig. 1B). The highest dissimilarity of environmental conditions between the two stations (90.9%) was found in May, while the lowest (16.4%) was found in December.
TABLE 1.
Environmental parameters of sample stations
| Sample | Temp (°C) | TIN (μmol/liter) | Phosphate (μmol/liter) | Salinity (ppt) | Chl a (mg/m3) |
|---|---|---|---|---|---|
| PM7-Mar | 18.95 | 1.81 | 0.41 | 34.49 | 4.31 |
| PM7-Apr | 22.01 | 1.29 | 0.39 | 34.78 | 2.88 |
| PM7-May | 27.30 | 3.15 | 0.47 | 35.52 | 0.78 |
| PM7-June | 27.70 | 7.31 | 0.44 | 29.47 | 7.13 |
| PM7-July | 29.70 | 2.63 | 0.45 | 30.84 | 2.27 |
| PM7-Sep | 28.90 | 6.71 | 0.46 | 31.45 | 4.27 |
| PM7-Oct | 27.60 | 3.05 | 0.55 | 33.30 | 0.99 |
| PM7-Dec | 19.80 | 10.45 | 0.61 | 33.87 | 2.74 |
| NM3-Mar | 21.00 | 24.33 | 0.60 | 29.92 | 0.70 |
| NM3-Apr | 22.90 | 16.69 | 0.48 | 31.70 | 2.76 |
| NM3-May | 26.30 | 78.28 | 1.97 | 22.84 | 2.83 |
| NM3-June | 29.00 | 64.37 | 1.84 | 18.41 | 5.17 |
| NM3-July | 28.30 | 35.67 | 1.64 | 19.94 | 3.13 |
| NM3-Sep | 29.40 | 14.27 | 0.63 | 28.51 | 19.60 |
| NM3-Oct | 27.60 | 13.22 | 1.30 | 32.30 | 0.85 |
| NM3-Dec | 20.60 | 9.94 | 0.95 | 33.93 | 2.05 |
Sequencing statistics.
A total 200,079 rpoC1 raw sequences were obtained from 16 samples. After removing the noise, poor-quality reads, and chimera sequences, 130,739 high-quality sequences were obtained. Following random subsampling, 6,247 high-quality sequences from each sample were obtained and used for further analysis (see Table S2 in the supplemental material).
Rarefaction analysis was applied to evaluate whether screening of 6,247 sequences was sufficient to estimate the diversity of Synechococcus communities (see Fig. S1 in the supplemental material). The rarefaction curves of the observed OTUs did not saturate biodiversity, while some ACE index curves were close to stabilization. The coverage value of Synechococcus assemblages ranged from 0.825 to 0.965. The lowest value was detected in the sample PM7-July (see Table S2 in the supplemental material).
Spatiotemporal variation of Synechococcus lineages.
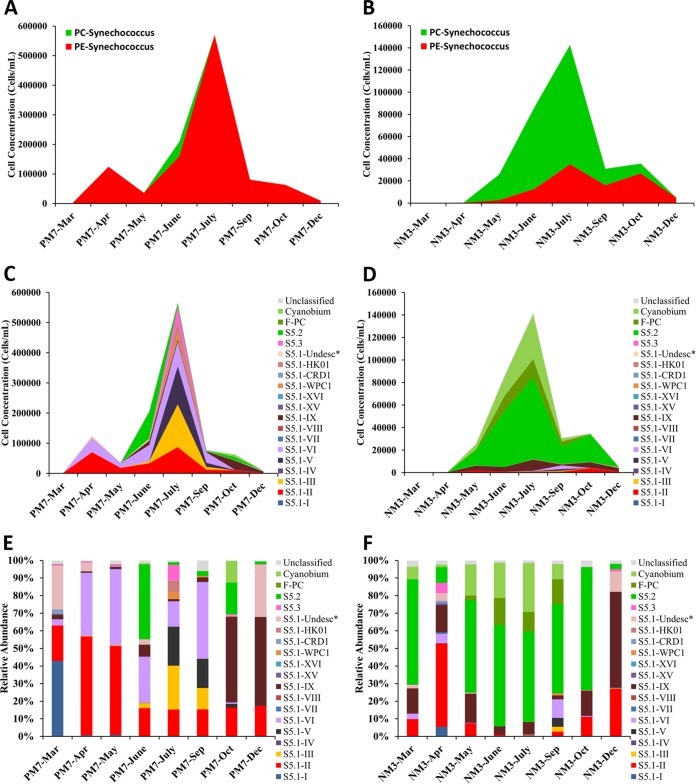
In the coastal waters, Synechococcus abundance gradually increased from 3.6 × 103 cells ml−1 in March to the maximum value of 5.7 × 105 cells ml−1 in July and then gradually decreased to 9.3 × 103 cells ml−1 in December. The Synechococcus assemblage in coastal waters was dominated by PE-type Synechococcus (Fig. 2A). Changes of Synechococcus abundance in estuarine waters followed a pattern similar to that in coastal waters, but the community was dominated by PC-type Synechococcus (Fig. 2B).
FIG 2.
Abundances and relative abundances of Synechococcus lineages in Hong Kong waters. (A and B) Abundances of PC-type and PE-type Synechococcus in Hong Kong waters measured by flow cytometry analysis (replotted from Liu et al. [21]). (C and D) Calculated abundances of Synechococcus lineages in each month (abundance of each lineage = relative abundance × total Synechococcus abundance measured by flow cytometry). (E and F) Relative abundances of Synechococcus lineages in samples collected in each month. *, S5.1-Undesc is the undescribed clade represented by Synechococcus sp. strain miyav.
Based on our database, more than 98% of our sequences were classified. All three marine Synechococcus subclusters (subclusters 5.1, 5.2, and 5.3) were detected at both stations. The most abundant subclusters in the coastal waters and the estuarine waters were subclusters 5.1 and 5.2, respectively (Fig. 2C and D). In total, 15 clades of subcluster 5.1 Synechococcus were detected in Hong Kong waters. Subcluster 5.3 Synechococcus sequences were only a minor component at both stations, accounting for only 1.53% and 0.96% of the total sequences in coastal waters and estuarine waters, respectively. Besides marine Synechococcus, Cyanobium and freshwater PC-type Synechococcus (F-PC) were also detected (Fig. 2C and D).
At the coastal station, the most abundant lineage was clade II, followed by clades VI and IX (Fig. 2E). Clade II Synechococcus was the core lineage that could be detected in all months. Clade VI was the second major lineage during the wet season, while clade IX was the dominant lineage in the dry season. Clade III, which cooccurred with clade V only, had high abundance in the coastal waters in July and September. Clade I was observed only in March. The undescribed clade (miyav) (22), represented by strain Synechococcus sp. strain miyav, mainly occurred in December and March. In July, many Synechococcus lineages contributed to the high abundance of Synechococcus in the coastal waters, including clades II, III, V, VI, WPC1, HK01, and S5.3 (Fig. 2E). In coastal waters, subcluster 5.2 Synechococcus could be found only occasionally, typically in summer (Fig. 2C and E).
Conversely, at the estuarine station (NM3), lineages of subcluster 5.2, F-PC, and Cyanobium were highly abundant during the wet season and peaked in July, likely owing to high freshwater discharge from the Pearl River (Fig. 2D). A phylogenetic tree shows that OTUs belonging to S5.2, F-PC, and Cyanobium are affiliated with Synechococcus sp. strain WH 8007, freshwater Synechococcus group D strains (38), and Synechococcus sp. strain PCC 9005 (Cyanobium), respectively (see Fig. S2 in the supplemental material). In comparison to the coastal waters, the diversity and abundance of subcluster 5.1 Synechococcus in estuarine waters were low (Fig. 2D and F). Subcluster 5.1 Synechococcus strains in estuarine waters were mainly contributed by clade II and clade IX; the former occurred in the dry season, while the latter occurred in all months at relatively high abundance (Fig. 2D and F). The undescribed clade (miyav) was also an important component in December (Fig. 2F). Clades IV, VII, VIII, XV, XVI, and CRD were minor lineages in Hong Kong waters. These lineages mainly occurred at NM3 in April and at PM7 in March and April (Fig. 2E and F).
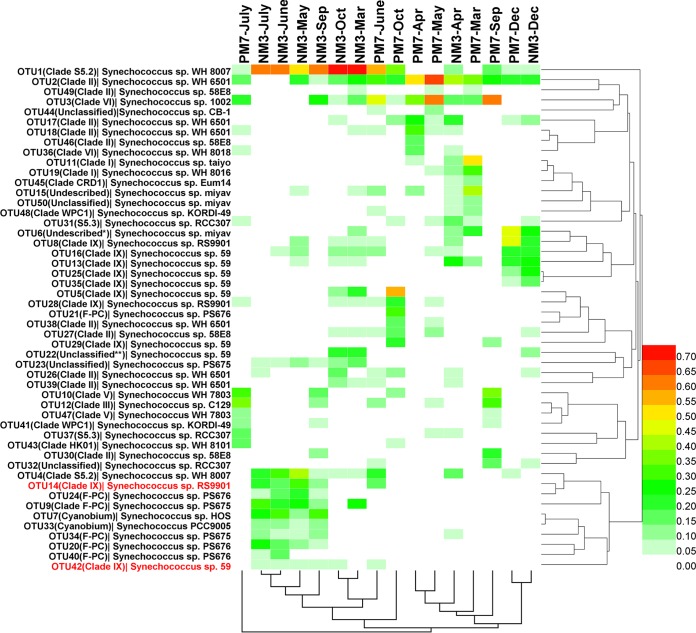
At the OTU level, we found that high abundance of clade II and clade IX Synechococcus were comprised of multiple OTUs (Fig. 3). Clade IX OTUs were affiliated with Synechococcus sp. strain 59 and Synechococcus sp. RS9901, respectively. Unlike other clade IX OTUs, which mainly occurred in winter, OTU14 and OTU42 dominated estuarine waters in summer and clustered together with subcluster 5.2 Synechococcus, Cyanobium, and freshwater Synechococcus (Fig. 3).
FIG 3.
Heat map displaying the relative abundances of the top 50 most abundant OTUs across the samples. The data were transformed by square root transformation. The OTU name, lineage, and most similar strain in the NCBI database are shown on the left. OTU14 and OTU42 (red), affiliated with clade IX Synechococcus sp. RS9901, have a niche similar to that of PC-type Synechococcus. *, undescribed clade represented by Synechococcus sp. strain miyav; **, unclassified OTU (the OTU's representative sequence was less than 90% identical to the reference sequences in this study).
Some OTUs showed similar niche preferences. For example, OTU10 (clade V), OTU12 (clade III), and OTU41 (WPC1) were abundant at PM7 in July and September; OTU11 (clade I), OTU19 (clade I), and OTU45 (clade CRD1) were relatively highly abundant in samples NM3-Apr and PM7-Mar.
Richness and diversity of Synechococcus assemblages in Hong Kong waters.
The Shannon diversity index of Synechococcus communities at the two study sites ranged from 1.83 to 5.22. The highest Synechococcus community diversity and richness in coastal waters and estuarine waters occurred in July and April, respectively (Fig. 4). The diversity index of the Synechococcus community in coastal waters was much higher than that in estuarine waters in July, when the highest Synechococcus abundance was recorded at both stations (Fig. 2 and 4). The richness index, ACE, showed results similar to those of the Shannon index (Fig. 4). Even though the abundance of Synechococcus was low in winter, the diversity of Synechococcus was still high at both stations.
FIG 4.
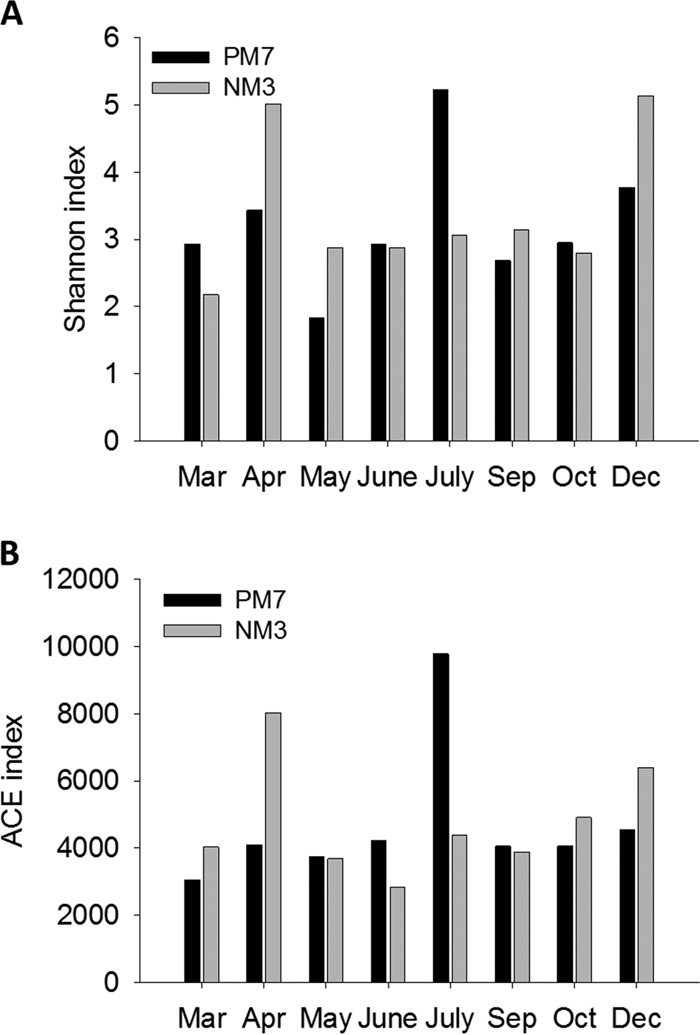
Diversity and richness indices of Synechococcus assemblages in Hong Kong waters.
Similarity among Synechococcus assemblages in Hong Kong waters.
NMDS plots showed that the compositions of Synechococcus assemblages at the two stations were similar in December (Dry) but differed greatly in the wet season (Fig. 5). The assemblages formed three groups, PM7(Summer), NM3(Wet), and Dry. The similarity of assemblages in each group was higher than 60%. The first group, PM7(Summer), was composed of samples collected from stratified water at PM7 when the summer monsoon prevailed. Sample PM7-June, however, deviated from this group, presumably due to the high abundance of subcluster 5.2 Synechococcus, which originated in the estuarine waters and was brought to the coastal waters by freshwater discharge. The second group, NM3(Wet), was composed of samples collected at NM3 from March to October, excluding those collected in April, when freshwater discharge from the Pearl River influenced the station. The last group, Dry, contains samples PM7-Dec, PM7-Oct, and NM3-Dec. The compositions of the three groups were significantly different (AMOVA; P < 0.05). SIMPER analysis was performed to determine each lineage's contribution to dissimilarity between the three groups. The dissimilarities were 88.2% between PM7(Summer) and NM3(Wet), 77.1% between PM7(Summer) and Dry, and 73.1% between NM3(Wet) and Dry. In summer, the difference between the assemblages in NM3 and PM7 was mainly contributed by subcluster 5.2, clade VI, clade II, and Cyanobium. In NM3, the variation of Synechococcus assemblages between summer and winter was mainly caused by an increase of subcluster 5.2 and a decrease of clade IX in summer; while in PM7, it was mainly contributed by clade IX and clade VI (Fig. 6).
FIG 5.
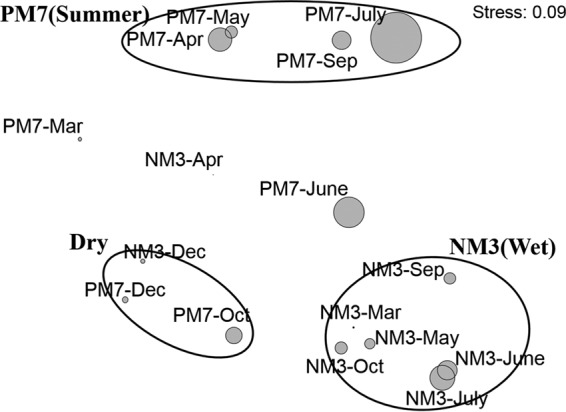
NMDS plot showing the relationship of the Synechococcus communities based on the relative abundance of each Synechococcus lineage. The samples formed three groups, PM7(Summer), NM3(Wet), and Dry. The AMOVA test showed that the compositions of the three groups were significantly different from each other. The sizes of the bubbles indicate the abundance of Synechococcus in each sample (the abundance of each sample was square root transformed).
FIG 6.
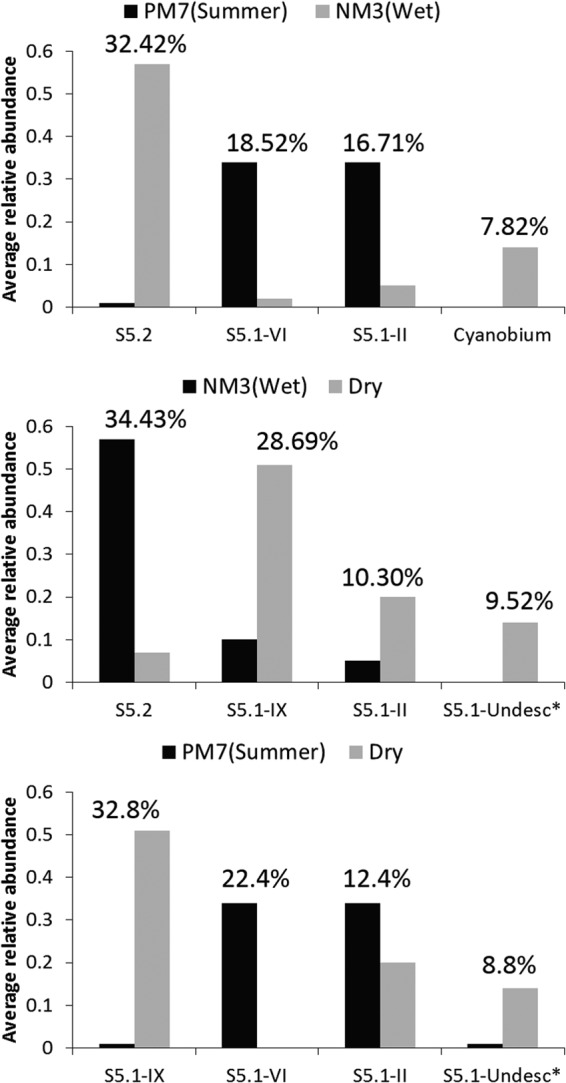
Four lineages that contribute most to the variation among group PM7(Summer), group NM3(Wet), and group Dry using SIMPER analysis. The bars show the average relative abundance of each lineage in each group; the numbers above the bars are the contributions of each lineage to the dissimilarity between groups. The three groups were determined according to an NMDS plot. *, undescribed clade (Undesc).
Impact of environmental parameters on Synechococcus community compositions.
The sum of all canonical eigenvalues showed that 57.0% of the observed assemblage variation could be accounted for by environmental variables. Of all the measured environmental parameters, salinity (Monte Carlo test; P = 0.002; F ratio = 6.57) and temperature (Monte Carlo test; P = 0.012; F ratio = 3.72) had significant impacts and explained 32.0% and 22.0% variance of the Synechococcus assemblages, respectively (Fig. 7).
FIG 7.
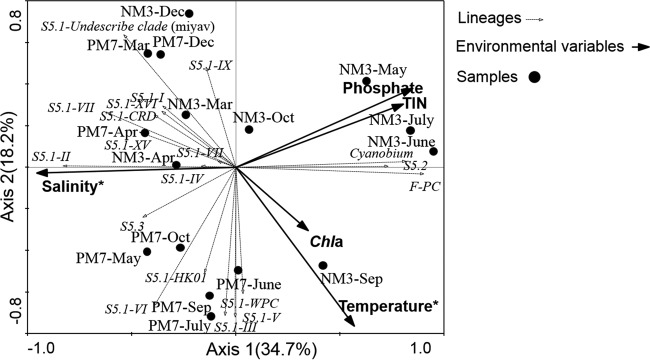
Correlation triplot based on an RDA depicting the relationship between the environmental factors and Synechococcus communities. The relative abundance of each lineage was normalized by square root transformation, and environmental data were Z-score transformed. *, factors significantly correlated with variation in the Synechococcus community (P < 0.05).
The abundances of Synechococcus clades I, IV, VII, CRD1, XV, and XVI were positively related to salinity but negatively related to Chl a and temperature. Clade II abundance was positively related to salinity and negatively related to TIN and phosphate. Subcluster 5.2, Cyanobium, and F-PC Synechococcus abundances were negatively related to salinity but positively related to temperature. Clade IX and the undescribed clade (represented by Synechococcus sp. strain miyav) were dominant in the dry season and were negatively related to temperature (Fig. 7).
Growth of Synechococcus strains in media with different salinities.
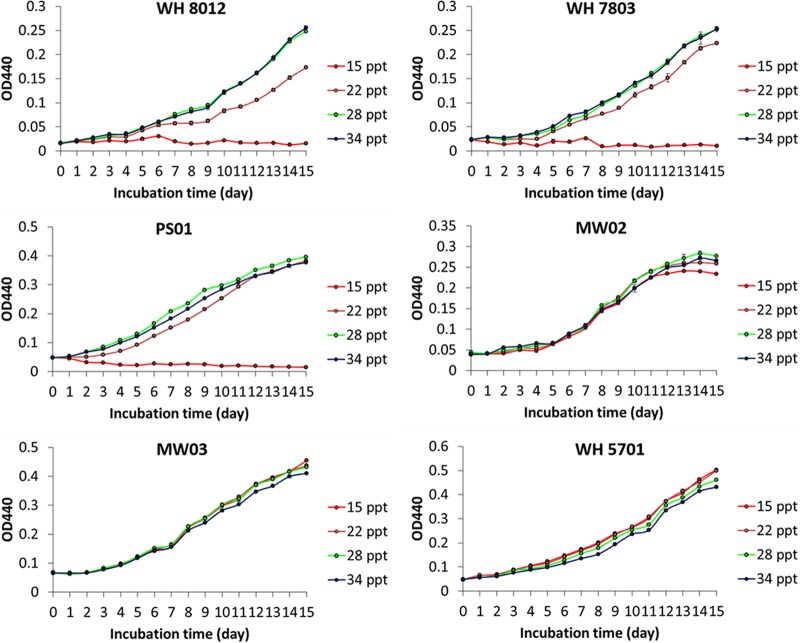
Both PC-type Synechococcus strains (MW03 and WH 5701) showed high tolerance for salinity variations and grew at salinities from 15 ppt to 34 ppt. The PE-type Synechococcus strains WH 7803, PS01, and WH 8012 could not tolerate salinity lower than 15 ppt (Fig. 8). WH 7803 and WH 8012 showed higher growth rates at salinities of 28 ppt and 34 ppt, while PS01 had the highest growth rates (0.14 ± 0.001 day−1) at 28 ppt. In comparison, PE-type Synechococcus sp. strain MW02 (pigment type 2) (see Fig. S3 in the supplemental material), isolated from the estuarine waters of Hong Kong, could tolerate a wide range of salinity, which is consistent with its distribution in Hong Kong waters. MW02 showed the highest growth rates (0.12 ± 0.007 day−1) at 28 ppt.
FIG 8.
Growth curves of Synechococcus strains in f/2 media with different salinities. MW03 and WH 5701 are PC-type Synechococcus. WH 8012, WH 7803, PS01, and MW02 are PE-type Synechococcus.
DISCUSSION
Pyrosequencing the rpoC1 gene from 16 samples from Hong Kong waters showed different patterns of seasonal variation in the Synechococcus assemblage composition in estuarine waters and coastal waters. Synechococcus assemblage diversity was high in high-temperature and stratified coastal waters in summer but relatively low in freshwater-influenced estuarine waters. In summer, the Synechococcus assemblage in estuarine waters was dominated by PC-type Synechococcus strains, which were composed of subcluster 5.2, freshwater Synechococcus, and Cyanobium. In contrast, the Synechococcus assemblage in coastal waters in summer was dominated by PE type subcluster 5.1 Synechococcus lineages. PE-type Synechococcus strains isolated from estuarine waters were highly tolerant of variations in salinity.
Sensitivity of the pyrosequencing application.
Different gene markers have been used to characterize marine Synechococcus diversity (22, 39, 40). Most protein gene markers provide higher resolution of genetic diversity in marine Synechococcus than the 16S rRNA gene (4, 23). The ITS region is a widely used and accurate marker for Synechococcus identification, but it did not seem suitable for pyrosequencing, as the method would not cover the whole ITS gene sequence. In contrast, a 423-bp fragment of the rpoC1 gene used for Synechococcus identification could be covered by the output of the 454 pyrosequencing method and is single copy in Synechococcus cells. In this study, all major Synechococcus clades were detected in Hong Kong waters by pyrosequencing of the rpoC1 gene, which indicates that the method is suitable for evaluating marine Synechococcus assemblage composition and diversity.
As mentioned above, only a few studies have reported about the Synechococcus community composition in Hong Kong waters. The most recent one (21), which used the flow cytometry approach, reported the presence of PC-type Synechococcus in the coastal waters only in June, in contrast to our findings of the presence of the same clade throughout most of the year. These contradictions may have arisen due to the differences in the methods used in the two studies. We also found Cyanobium in estuarine waters of Hong Kong, which had not been reported in a previous study based on clone library analysis (21). Use of next-generation DNA-sequencing methods with high sensitivity essentially yields more insights into the microbial community composition, and we highlight the need for such approaches to fully discover the community composition of Synechococcus lineages.
Abundance and diversity of Synechococcus assemblages in Hong Kong waters.
Temperature is an important factor that determines the abundance of Synechococcus (41–43). Growth of Synechococcus exceeds its grazing mortality in summer, resulting in a high abundance of Synechococcus. In winter, low growth rates caused by low temperature make the bacteria unable to keep pace with grazing, and as a result, Synechococcus abundance becomes low. This is similar to what we found, where Synechococcus abundance in Hong Kong waters is low in winter but high in summer (Fig. 2A and B). In response to increases of temperature during the wet season (April to September), Synechococcus cell density increased from several hundred cells ml−1 to more than 5.5 × 105 cells ml−1 in coastal waters and 1.4 × 105 cells ml−1 in estuarine waters in July. Low salinity, highly turbid, and high-nutrient estuarine waters mainly selected for subcluster 5.2 Synechococcus, while relatively oligotrophic coastal water selected for subcluster 5.1 Synechococcus (Fig. 2D and E). These results agree with the definition of subcluster 5.1 and 5.2 Synechococcus (44) and support the idea that Synechococcus groups can serve as indicator organisms for estuarine hydrodynamics (45).
Based on pyrosequencing of the rpoC1 gene, we found 17 clades representing Synechococcus subclusters 5.1, 5.2, and 5.3 and also freshwater Synechococcus and Cyanobium species (Fig. 2). In comparison, studies of the tropical/subtropical Pacific Ocean (9), Chesapeake Bay (46), East China Sea (19), California current (18), and Sargasso Sea (47) found about 6 to 11 Synechococcus clades. Although 12 or 13 Synechococcus clades were reported in the Gulf of Aqaba by constructing ntcA gene clone libraries and pyrosequencing of the V6 region of the 16S rRNA gene, the data were combined from several different studies conducted in multiple years (48). Our results suggest that Hong Kong coastal waters are one of the regions with the world's highest Synechococcus diversity.
Seasonal variation of Synechococcus assemblages in Hong Kong coastal waters.
Seasonal variation of the Synechococcus assemblage composition in coastal waters was mainly caused by different subcluster 5.1 lineages. Clade II dominated the Synechococcus assemblages throughout the year; clade III occurred in summer, when stratification developed; and clade VI was a major lineage during the transition periods between mixing and stratification, which is in agreement with the study conducted in the Gulf of Aqaba (48). However, that study was unable to distinguish clades V and VI, and therefore, no information on the niche difference between the two clades was obtained. We found that clade V, which occurred only in highly stratified waters from July to September, had a narrower niche than clade VI, even though they cooccur in summer. Clade IX, which was first found in the Gulf of Aqaba (4), has a low abundance in the global ocean (7) and is also even a rare species in the Gulf of Aqaba (49), but it thrived in Hong Kong coastal waters during winter. It is worth noting that the diversity of the Synechococcus assemblage was highest in the coastal waters of Hong Kong in July (Fig. 4). Overall, our results suggest that Hong Kong coastal waters are an ideal environment for the growth of most subcluster 5.1 and 5.3 Synechococcus lineages when it has high temperature and is strongly influenced by the water from the South China Sea driven by the southwest monsoon. In addition to the abundant clades described above, several opportunistic Synechococcus groups with narrow niches were found in Hong Kong coastal waters. For example, clade I Synechococcus strains are typical of temperate cold waters (7) and also occur in upwelling waters (50) and mixing waters (48). They appeared in Hong Kong waters during winter mixing. Clades XV and XVI are capable of chromatic adaptation and were first isolated from the Sargasso Sea (latitude 34° to 35°N) (47). It has been suggested that these two clades are adapted to low-light conditions (50). In Hong Kong waters, they appeared in March, when strong mixing occurred. Clade III has a narrow temperature spectrum (51), and it was found in July, when the temperature was higher than 29°C. The apparently quick appearance (and disappearance) of these clades in response to changes in the environment meets the criteria for a “microbial seed bank,” which suggests that microorganisms are able to enter a reversible state of low metabolic activity under harsh environmental conditions and can be resuscitated when the conditions favor growth (52).
Seasonal variation of Synechococcus assemblages in Hong Kong estuarine waters.
Unlike in coastal waters, the increase in discharge from the Pearl River shifted the Synechococcus community structure in estuarine waters from PE-type subcluster 5.1 lineages to PC-type Synechococcus lineages. PC-type Synechococcus strains have greater absorbance in red light, enabling them to grow well in highly turbid, red-light-dominant estuarine waters (53). Also, they have large genomes (54), potentially to deal with complex hydrographic conditions in the estuarine environment. Moreover, it has been suggested that PC-type Synechococcus strains have a greater ability to deal with salinity variation than marine PE-type Synechococcus (4, 55). Although high abundance of PC-type Synechococcus strains was often found in estuarine waters (56, 57), only a few studies have reported their community composition at the molecular level. In this study, we found that PC-type Synechococcus assemblages in Hong Kong estuarine waters were composed of marine-source Synechococcus (subcluster 5.2, represented by Synechococcus sp. WH 8007), freshwater Synechococcus (F-PC), and Cyanobium (see Fig. S2 in the supplemental material). This result supports the hypothesis, proposed by Liu et al. (21), that the reported Prochlorococcus-like picocyanobacteria in fresh and brackish waters (58, 59) are likely PC-type Synechococcus or Cyanobium. Subcluster 5.2 Synechococcus sp. WH 8007, which was the most abundant PC-type Synechococcus strain in Hong Kong waters, is also the major PC-type Synechococcus in Chesapeake Bay (46). Similar to the result of the study in Chesapeake Bay, we also did not find Synechococcus strains in Hong Kong waters that were closely related to Synechococcus sp. strain WH 5701 (8), which is another representative strain of subcluster 5.2. This suggests that strain clusters represented by WH 5701 and WH 8007 have different niches.
Besides the PC-type subcluster 5.2 Synechococcus strains, we also found that the increase in the freshwater Synechococcus abundance followed the increase in the discharge from the Pearl River. This implies that advection of the allochthonous Synechococcus population from the river inflow can be important for the estuarine ecosystem (60) during the wet season. Chen et al. reported that freshwater Synechococcus strains are rare in the Chesapeake Bay, where high abundance of PC-type Synechococcus was detected (46). In contrast, freshwater Synechococcus was found in Hong Kong waters, based on sequencing of the cpcBA gene (21). Consistently, more than 1.539 × 104 cells ml−1 of freshwater Synechococcus strains were detected in estuarine waters in July in this study (Fig. 2B). We further revealed that freshwater Synechococcus strains in Hong Kong estuarine waters were affiliated with PC-type Synechococcus sp. strain PS676, which was isolated from Lake Teganuma (Japan). The presence of a high abundance of freshwater Synechococcus strains in Hong Kong estuarine waters suggests freshwater Synechococcus strains are abundant in the Pearl River.
Cyanobium mainly occurs in freshwater and brackish environments and was also detected in estuarine waters of Hong Kong during May to September, when the discharge from the Pearl River was high. Many studies have reported that the taxonomic distinction between the Cyanobium and Synechococcus taxa is poorly defined (61, 62). Indeed, phylogenetic analysis shows that the Cyanobium strains found in this study were most closely related to Synechococcus sp. PCC9005 (Cyanobium) and formed a cluster with Synechococcus sp. WH 5701 (see Fig. S2 in the supplemental material).
PE-type Synechococcus in estuarine waters in summer.
Subcluster 5.1 Synechococcus strains have been defined as strictly marine strains that are unable to grow well in low-salinity environments (44). For example, PE-type Synechococcus sp. strain WH 7803 and strain WH 7805 cannot grow well when the salinity is less than 28 ppt (55). However, recently, Chen et al. also showed that PE-type Synechococcus sp. strain CB0205 and strain CB0208, isolated from the Chesapeake Bay, are able to grow in SN medium with a wide range of salinities (8). In this study, we noticed that subcluster 5.1 clade IX Synechococcus sp. MW02, isolated from Hong Kong waters, could also likely survive in low-salinity waters based on pyrosequencing data (Fig. 2), and this was confirmed by a growth experiment (Fig. 8). Either this is a characteristic of all clade IX strains or it might be that some Synechococcus clades have different subclades adapted to different environments (Fig. 3). This result may also support the finding of Junier et al. (63) that some freshwater Synechococcus strains are closely related to clade IX Synechococcus sp. RS9901.
Conclusions.
Using a high-throughput sequencing method, we found an unprecedentedly high phylogenetic diversity of Synechococcus strains, an important unicellular cyanobacterial primary producer, in subtropical waters of Hong Kong. Temperature and salinity are the main factors influencing the Synechococcus community composition in Hong Kong waters. Furthermore, the variation in freshwater discharge from the Pearl River strongly influences the abundance and composition of the Synechococcus community in estuarine waters. Unlike most PE-type Synechococcus strains, which cannot survive in low-salinity estuarine waters, we found that some clade IX Synechococcus strains that are common in our study sites can grow at a range of salinities. Further research on the genome of euryhaline PE-type Synechococcus strains, especially clade IX, may help us to better understand the mechanism by which these Synechococcus cells respond to salinity changes.
Supplementary Material
ACKNOWLEDGMENTS
We thank Chen Xihan for collecting the DNA samples.
This work was funded by the Hong Kong Research Grants Council (GRF 661912 and 661813). Support from the State Key Laboratory in Marine Pollution (SKLMP) Seed Collaborative Research Fund under project SKLMP/SCRF/0006 is also acknowledged.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01895-15.
REFERENCES
- 1.Waterbury JB, Watson SW, Guillard RR, Brand LE. 1979. Widespread occurrence of a unicellular, marine, planktonic, cyanobacterium. Nature 277:293–294. doi: 10.1038/277293a0. [DOI] [Google Scholar]
- 2.Haverkamp TH, Schouten D, Doeleman M, Wollenzien U, Huisman J, Stal LJ. 2009. Colorful microdiversity of Synechococcus strains (picocyanobacteria) isolated from the Baltic Sea. ISME J 3:397–408. doi: 10.1038/ismej.2008.118. [DOI] [PubMed] [Google Scholar]
- 3.Choi DH, Noh JH. 2009. Phylogenetic diversity of Synechococcus strains isolated from the East China Sea and the East Sea. FEMS Microbiol Ecol 69:439–448. doi: 10.1111/j.1574-6941.2009.00729.x. [DOI] [PubMed] [Google Scholar]
- 4.Fuller NJ, Marie D, Partensky F, Vaulot D, Post AF, Scanlan DJ. 2003. Clade-specific 16S ribosomal DNA oligonucleotides reveal the predominance of a single marine Synechococcus clade throughout a stratified water column in the Red Sea. Appl Environ Microbiol 69:2430–2443. doi: 10.1128/AEM.69.5.2430-2443.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Everroad RC, Wood AM. 2012. Phycoerythrin evolution and diversification of spectral phenotype in marine Synechococcus and related picocyanobacteria. Mol Phylogenet Evol 64:381–392. doi: 10.1016/j.ympev.2012.04.013. [DOI] [PubMed] [Google Scholar]
- 6.Scanlan DJ, Ostrowski M, Mazard S, Dufresne A, Garczarek L, Hess WR, Post AF, Hagemann M, Paulsen I, Partensky F. 2009. Ecological genomics of marine picocyanobacteria. Microbiol Mol Biol Rev 73:249–299. doi: 10.1128/MMBR.00035-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zwirglmaier K, Jardillier L, Ostrowski M, Mazard S, Garczarek L, Vaulot D, Not F, Massana R, Ulloa O, Scanlan DJ. 2008. Global phylogeography of marine Synechococcus and Prochlorococcus reveals a distinct partitioning of lineages among oceanic biomes. Environ Microbiol 10:147–161. [DOI] [PubMed] [Google Scholar]
- 8.Chen F, Wang K, Kan J, Bachoon DS, Lu J, Lau S, Campbell L. 2004. Phylogenetic diversity of Synechococcus in the Chesapeake Bay revealed by ribulose-1,5-bisphosphate carboxylase-oxygenase (RuBisCO) large subunit gene (rbcL) sequences. Aquat Microb Ecol 36:153–164. doi: 10.3354/ame036153. [DOI] [Google Scholar]
- 9.Huang S, Wilhelm SW, Harvey HR, Taylor K, Jiao N, Chen F. 2012. Novel lineages of Prochlorococcus and Synechococcus in the global oceans. ISME J 6:285–297. doi: 10.1038/ismej.2011.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chiang K-P, Kuo M-C, Chang J, Wang R-H, Gong G-C. 2002. Spatial and temporal variation of the Synechococcus population in the East China Sea and its contribution to phytoplankton biomass. Cont Shelf Res 22:3–13. doi: 10.1016/S0278-4343(01)00067-X. [DOI] [Google Scholar]
- 11.Tsai A-Y, Gong G-C, Sanders RW, Chiang K-P. 2013. Relationship of Synechococcus abundance to seasonal ocean temperature ranges. Terr Atmos Ocean Sci 24:925–932. doi: 10.3319/TAO.2013.06.17.01(Oc). [DOI] [Google Scholar]
- 12.Campbell L, Liu H, Nolla HA, Vaulot D. 1997. Annual variability of phytoplankton and bacteria in the subtropical North Pacific Ocean at Station ALOHA during the 1991-1994 ENSO event. Deep Sea Res 44:167–192. doi: 10.1016/S0967-0637(96)00102-1. [DOI] [Google Scholar]
- 13.Marston MF, Sallee JL. 2003. Genetic diversity and temporal variation in the cyanophage community infecting marine Synechococcus species in Rhode Island's coastal waters. Appl Environ Microbiol 69:4639–4647. doi: 10.1128/AEM.69.8.4639-4647.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Campbell L, Vaulot D. 1993. Photosynthetic picoplankton community structure in the subtropical North Pacific Ocean near Hawaii (station ALOHA). Deep Sea Res 40:2043–2060. doi: 10.1016/0967-0637(93)90044-4. [DOI] [Google Scholar]
- 15.Rabouille S, Edwards CA, Zehr JP. 2007. Modelling the vertical distribution of Prochlorococcus and Synechococcus in the North Pacific subtropical ocean. Environ Microbiol 9:2588–2602. doi: 10.1111/j.1462-2920.2007.01376.x. [DOI] [PubMed] [Google Scholar]
- 16.Flombaum P, Gallegos JL, Gordillo RA, Rincón J, Zabala LL, Jiao N, Karl DM, Li WK, Lomas MW, Veneziano D. 2013. Present and future global distributions of the marine Cyanobacteria Prochlorococcus and Synechococcus. Proc Natl Acad Sci U S A 110:9824–9829. doi: 10.1073/pnas.1307701110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sandaa R-A, Larsen A. 2006. Seasonal variations in virus-host populations in Norwegian coastal waters: focusing on the cyanophage community infecting marine Synechococcus spp. Appl Environ Microbiol 72:4610–4618. doi: 10.1128/AEM.00168-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tai V, Palenik B. 2009. Temporal variation of Synechococcus clades at a coastal Pacific Ocean monitoring site. ISME J 3:903–915. doi: 10.1038/ismej.2009.35. [DOI] [PubMed] [Google Scholar]
- 19.Choi DH, Noh JH, Shim J. 2013. Seasonal changes in picocyanobacterial diversity as revealed by pyrosequencing in temperate waters of the East China Sea and the East Sea. Aquat Microb Ecol 71:75–90. doi: 10.3354/ame01669. [DOI] [Google Scholar]
- 20.Lee JH, Harrison PJ, Kuang C, Yin K. 2006. Eutrophication dynamics in Hong Kong coastal waters: physical and biological interactions, p 187–206. The environment in Asia Pacific harbours. Springer, Berlin, Germany. [Google Scholar]
- 21.Liu H, Jing H, Wong TH, Chen B. 2014. Co-occurrence of phycocyanin- and phycoerythrin-rich Synechococcus in subtropical estuarine and coastal waters of Hong Kong. Environ Microbiol Rep 6:90–99. doi: 10.1111/1758-2229.12111. [DOI] [PubMed] [Google Scholar]
- 22.Ahlgren NA, Rocap G. 2012. Diversity and distribution of marine Synechococcus: multiple gene phylogenies for consensus classification and development of qPCR assays for sensitive measurement of clades in the ocean. Front Microbiol 3:213. doi: 10.3389/fmicb.2012.00213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mühling M, Fuller NJ, Somerfield PJ, Post AF, Wilson WH, Scanlan DJ, Joint I, Mann NH. 2006. High resolution genetic diversity studies of marine Synechococcus isolates using rpoC1-based restriction fragment length polymorphism. Aquat Microb Ecol 45:263–275. doi: 10.3354/ame045263. [DOI] [Google Scholar]
- 24.Ferris MJ, Palenik B. 1998. Niche adaptation in ocean cyanobacteria. Nature 396:226–228. doi: 10.1038/24297. [DOI] [Google Scholar]
- 25.Sogin ML, Morrison HG, Huber JA, Welch DM, Huse SM, Neal PR, Arrieta JM, Herndl GJ. 2006. Microbial diversity in the deep sea and the underexplored “rare biosphere”. Proc Natl Acad Sci U S A 103:12115–12120. doi: 10.1073/pnas.0605127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kunin V, Engelbrektson A, Ochman H, Hugenholtz P. 2010. Wrinkles in the rare biosphere: pyrosequencing errors can lead to artificial inflation of diversity estimates. Environ Microbiol 12:118–123. doi: 10.1111/j.1462-2920.2009.02051.x. [DOI] [PubMed] [Google Scholar]
- 27.Chen B, Liu H, Landry MR, Dai M, Huang B, Sune J. 2009. Close coupling between phytoplankton growth and microzooplankton grazing in the western South China Sea. Limnol Oceanogr 54:1084–1097. doi: 10.4319/lo.2009.54.4.1084. [DOI] [Google Scholar]
- 28.Riemann L, Leitet C, Pommier T, Simu K, Holmfeldt K, Larsson U, Hagström Å. 2008. The native bacterioplankton community in the central Baltic Sea is influenced by freshwater bacterial species. Appl Environ Microbiol 74:503–515. doi: 10.1128/AEM.01983-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schloss PD, Gevers D, Westcott SL. 2011. Reducing the effects of PCR amplification and sequencing artifacts on 16S rRNA-based studies. PLoS One 6:e27310. doi: 10.1371/journal.pone.0027310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98. [Google Scholar]
- 32.Deng W, Wang Y, Liu Z, Cheng H, Xue Y. 2014. Hemi: a toolkit for illustrating heatmaps. PLoS One 9:e111988. doi: 10.1371/journal.pone.0111988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Letunic I, Bork P. 2007. Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics 23:127–128. doi: 10.1093/bioinformatics/btl529. [DOI] [PubMed] [Google Scholar]
- 35.Six C, Thomas J-C, Garczarek L, Ostrowski M, Dufresne A, Blot N, Scanlan DJ, Partensky F. 2007. Diversity and evolution of phycobilisomes in marine Synechococcus spp.: a comparative genomics study. Genome Biol 8:R259. doi: 10.1186/gb-2007-8-12-r259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao H. 1990. Evolution of the Pearl River estuary. China Ocean Press, Beijing, China. [Google Scholar]
- 37.Yung Y-K, Wong C, Yau K, Qian P. 2001. Long-term changes in water quality and phytoplankton characteristics in Port Shelter, Hong Kong, from 1988-1998. Mar Pollut Bull 42:981–992. doi: 10.1016/S0025-326X(01)00066-2. [DOI] [PubMed] [Google Scholar]
- 38.Robertson BR, Tezuka N, Watanabe MM. 2001. Phylogenetic analyses of Synechococcus strains (cyanobacteria) using sequences of 16S rDNA and part of the phycocyanin operon reveal multiple evolutionary lines and reflect phycobilin content. Int J Syst Evol Microbiol 51:861–871. doi: 10.1099/00207713-51-3-861. [DOI] [PubMed] [Google Scholar]
- 39.Rocap G, Distel DL, Waterbury JB, Chisholm SW. 2002. Resolution of Prochlorococcus and Synechococcus ecotypes by using 16S-23S ribosomal DNA internal transcribed spacer sequences. Appl Environ Microbiol 68:1180–1191. doi: 10.1128/AEM.68.3.1180-1191.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dall'Agnol LT, Ghilardi R Jr, Mcculloch JA, Schneider H, Schneider MP, Silva A. 2012. Phylogenetic and gene trees of Synechococcus: choice of the right marker to evaluate the population diversity in the Tucuruí Hydroelectric Power Station Reservoir in Brazilian Amazonia. J Plankton Res 34:245–257. doi: 10.1093/plankt/fbr109. [DOI] [Google Scholar]
- 41.Agawin NS, Duarte CM, Agusti S. 1998. Growth and abundance of Synechococcus sp. in a Mediterranean bay: seasonality and relationship with temperature. Mar Ecol Prog Ser 170:45–53. doi: 10.3354/meps170045. [DOI] [Google Scholar]
- 42.Li WK. 1998. Annual average abundance of heterotrophic bacteria and Synechococcus in surface ocean waters. Limnol Oceanogr 43:1746–1753. doi: 10.4319/lo.1998.43.7.1746. [DOI] [Google Scholar]
- 43.Moisan TA, Blattner KL, Makinen CP. 2010. Influences of temperature and nutrients on Synechococcus abundance and biomass in the southern Mid-Atlantic Bight. Cont Shelf Res 30:1275–1282. doi: 10.1016/j.csr.2010.04.005. [DOI] [Google Scholar]
- 44.Herdman M, Castenholz R, Waterbury J, Rippka R. 2001. Form-genus XIII. Synechococcus, p 508–512. In Boone DR, Castenholz RW (ed), Bergey’s manual of systematic bacteriology, 2nd ed, vol 1 The Archaea and deeply branching and phototrophic Bacteria. Springer, New York, NY. [Google Scholar]
- 45.Rajaneesh K, Mitbavkar S. 2013. Factors controlling the temporal and spatial variations in Synechococcus abundance in a monsoonal estuary. Mar Environ Res 92:133–143. doi: 10.1016/j.marenvres.2013.09.010. [DOI] [PubMed] [Google Scholar]
- 46.Chen F, Wang K, Kan J, Suzuki MT, Wommack KE. 2006. Diverse and unique picocyanobacteria in Chesapeake Bay, revealed by 16S-23S rRNA internal transcribed spacer sequences. Appl Environ Microbiol 72:2239–2243. doi: 10.1128/AEM.72.3.2239-2243.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ahlgren NA, Rocap G. 2006. Culture isolation and culture-independent clone libraries reveal new marine Synechococcus ecotypes with distinctive light and N physiologies. Appl Environ Microbiol 72:7193–7204. doi: 10.1128/AEM.00358-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Post AF, Penno S, Zandbank K, Paytan A, Huse SM, Welch DM. 2011. Long term seasonal dynamics of Synechococcus population structure in the Gulf of Aqaba, Northern Red Sea. Front Microbiol 2:131. doi: 10.3389/fmicb.2011.00131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Penno S, Lindell D, Post AF. 2006. Diversity of Synechococcus and Prochlorococcus populations determined from DNA sequences of the N-regulatory gene ntcA. Environ Microbiol 8:1200–1211. doi: 10.1111/j.1462-2920.2006.01010.x. [DOI] [PubMed] [Google Scholar]
- 50.Ahlgren NA, Noble A, Patton AP, Roache-Johnson K, Jackson L, Robinson D, McKay C, Moore LR, Saito MA, Rocap G. 2014. The unique trace metal and mixed layer conditions of the Costa Rica upwelling dome support a distinct and dense community of Synechococcus. Limnol Oceanogr 59:2166–2184. doi: 10.4319/lo.2014.59.6.2166. [DOI] [Google Scholar]
- 51.Pittera J, Humily F, Thorel M, Grulois D, Garczarek L, Six C. 2014. Connecting thermal physiology and latitudinal niche partitioning in marine Synechococcus. ISME J 8:1221–1236. doi: 10.1038/ismej.2013.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lennon JT, Jones SE. 2011. Microbial seed banks: the ecological and evolutionary implications of dormancy. Nat Rev Microbiol 9:119–130. doi: 10.1038/nrmicro2504. [DOI] [PubMed] [Google Scholar]
- 53.Stomp M, Huisman J, de Jongh F, Veraart AJ, Gerla D, Rijkeboer M, Ibelings BW, Wollenzien UI, Stal LJ. 2004. Adaptive divergence in pigment composition promotes phytoplankton biodiversity. Nature 432:104–107. doi: 10.1038/nature03044. [DOI] [PubMed] [Google Scholar]
- 54.Dufresne A, Ostrowski M, Scanlan DJ, Garczarek L, Mazard S, Palenik BP, Paulsen IT, de Marsac NT, Wincker P, Dossat C. 2008. Unraveling the genomic mosaic of a ubiquitous genus of marine cyanobacteria. Genome Biol 9:R90. doi: 10.1186/gb-2008-9-5-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lu J, Chen F, Hodson RE. 2001. Distribution, isolation, host specificity, and diversity of cyanophages infecting marine Synechococcus spp. in river estuaries. Appl Environ Microbiol 67:3285–3290. doi: 10.1128/AEM.67.7.3285-3290.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Katano T, Nakano S-I, Mitamura O, Yoshida H, Azumi H, Matsuura Y, Tanaka Y, Maezono H, Satoh Y, Satoh T. 2008. Abundance and pigment type composition of picocyanobacteria in Barguzin Bay, Lake Baikal. Limnol Oceanogr 9:105–114. [Google Scholar]
- 57.Wang K, Wommack KE, Chen F. 2011. Abundance and distribution of Synechococcus spp. and cyanophages in the Chesapeake Bay. Appl Environ Microbiol 77:7459–7468. doi: 10.1128/AEM.00267-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Corzo A, Jiménez-Gómez F, Gordillo F, Garcia-Ruiz R, Niell F. 1999. Synechococcus and Prochlorococcus-like populations detected by flow cytometry in a eutrophic reservoir in summer. J Plankton Res 21:1575–1581. doi: 10.1093/plankt/21.8.1575. [DOI] [Google Scholar]
- 59.Shang X, Zhang L, Zhang J. 2007. Prochlorococcus-like populations detected by flow cytometry in the fresh and brackish waters of the Changjiang Estuary. J Mar Biol Assoc UK 87:643–648. doi: 10.1017/S0025315407055191. [DOI] [Google Scholar]
- 60.Waleron M, Waleron K, Vincent WF, Wilmotte A. 2007. Allochthonous inputs of riverine picocyanobacteria to coastal waters in the Arctic Ocean. FEMS Microbiol Ecol 59:356–365. doi: 10.1111/j.1574-6941.2006.00236.x. [DOI] [PubMed] [Google Scholar]
- 61.Lopes VR, Ramos V, Martins A, Sousa M, Welker M, Antunes A, Vasconcelos VM. 2012. Phylogenetic, chemical and morphological diversity of cyanobacteria from Portuguese temperate estuaries. Mar Environ Res 73:7–16. doi: 10.1016/j.marenvres.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 62.Honda D, Yokota A, Sugiyama J. 1999. Detection of seven major evolutionary lineages in cyanobacteria based on the 16S rRNA gene sequence analysis with new sequences of five marine Synechococcus strains. J Mol Evol 48:723–739. doi: 10.1007/PL00006517. [DOI] [PubMed] [Google Scholar]
- 63.Junier P, Witzel K-P, Hadas O. 2007. Genetic diversity of cyanobacterial communities in Lake Kinneret (Israel) using 16S rRNA gene, psbA and ntcA sequence analyses. Aquat Microb Ecol 49:233–241. doi: 10.3354/ame01161. [DOI] [Google Scholar]
- 64.Gascuel O. 1997. BIONJ: an improved version of the NJ algorithm based on a simple model of sequence data. Mol Biol Evol 14:685–695. [DOI] [PubMed] [Google Scholar]
- 65.Guillard RR. 1975. Culture of phytoplankton for feeding marine invertebrates, p 29–60. In Smith WL, Chanley MH (ed), . Culture of marine invertebrate animals. Plenum Press, New York, NY. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.