Abstract
Motility plays an essential role in bacterial fitness and colonization in the plant environment, since it favors nutrient acquisition and avoidance of toxic substances, successful competition with other microorganisms, the ability to locate the preferred hosts, access to optimal sites within them, and dispersal in the environment during the course of transmission. In this work, we have observed that the mutation of the flagellar master regulatory gene, fleQ, alters bacterial surface motility and biosurfactant production, uncovering a new type of motility for Pseudomonas syringae pv. tomato DC3000 on semisolid surfaces. We present evidence that P. syringae pv. tomato DC3000 moves over semisolid surfaces by using at least two different types of motility, namely, swarming, which depends on the presence of flagella and syringafactin, a biosurfactant produced by this strain, and a flagellum-independent surface spreading or sliding, which also requires syringafactin. We also show that FleQ activates flagellum synthesis and negatively regulates syringafactin production in P. syringae pv. tomato DC3000. Finally, it was surprising to observe that mutants lacking flagella or syringafactin were as virulent as the wild type, and only the simultaneous loss of both flagella and syringafactin impairs the ability of P. syringae pv. tomato DC3000 to colonize tomato host plants and cause disease.
INTRODUCTION
Motility plays a pivotal role in the spreading of bacteria across surfaces and colonization, contributing to the formation of structured communities called biofilms (1). Efficient bacterial motility under diverse environmental conditions, from liquid to semisolid and solid surfaces, is achieved by flagellum-dependent swimming and swarming or flagellum-independent twitching, gliding, nonsocial gliding, and sliding (2, 3, 4). Swimming is a flagellum-driven motility observed in bacteria moving through liquids or semisolid media, such as low-percentage agar (0.2% to 0.4%). Twitching is a slow cell movement on surfaces mediated by the extension and retraction of type IV pili (5). Gliding, a surface movement extensively studied in myxobacteria, does not require flagella or pili but involves focal adhesion complexes, cell surface-associated complexes that anchor the bacterium to a substrate and might act as a motor (6). Sliding is a passive form of surface spreading by expansion that is powered by the outward pressure of bacterial growth and facilitated by compounds that reduce friction between cells and surfaces (3). Sliding or spreading by expansion has been observed in a diverse group of bacteria, such as mycobacteria, Bacillus subtilis, Vibrio cholerae, Serratia marcescens, Pseudomonas aeruginosa, or Legionella pneumophila (7–12), in which a strong correlation between sliding and production of surfactants has been established. Furthermore, sliding is easily mistaken for swarming motility and can occur when flagella are disrupted in bacteria that normally would swarm (7, 8, 13, 14). Swarming is a rapid and coordinated movement of bacterial populations over a surface like semisolid agar (0.5% to 1%) and depends on flagella, pili, and the presence of a water film and/or surfactants to enable motility (2–4). Bacterial biosurfactants, in addition to having a role in multiple motility mechanisms, are involved in biofilm structure and maintenance, as well as in the delivery of insoluble signals (15, 16). Additionally, some biosurfactants have been shown to exhibit membrane-disrupting and, thus, zoosporicidal or antimicrobial activities (17). Although biosurfactants include many types of molecules, the lipopeptides, composed of an oligopeptide and a lipid tail, are particularly important and well studied in Pseudomonas and Bacillus species (17).
Bacteria belonging to the genus Pseudomonas are ubiquitous bacteria that are able to colonize a wide range of niches, including the soil, the plant rhizosphere and phyllosphere, and animal tissues. Motility is an important trait for these processes (18–21), biofilm formation (22–24), and pathogenesis in plants (25, 26) and animals (27). Therefore, mutations affecting flagellar regulation, biogenesis, and/or modification affect the bacterial ability to move through the environment (28), display chemotaxis toward attractants (21), and form biofilms (29, 30). In particular, motility is crucial in plant-interacting bacteria; thus, nonmotile mutants of different Pseudomonas fluorescens strains are severely affected in root colonization (20). Flagellum-mediated motility is also an important trait for both epiphytic and pathogenic lifestyles of Pseudomonas syringae (31–33). Nonmotile P. syringae mutants are more sensitive to desiccation and UV exposure than their motile counterparts, presumably because they cannot escape those environmental stresses (31). Additionally, their ability to invade the leaf apoplast and cause disease is severely reduced (26, 31–33).
Biogenesis and assembly of the bacterial flagellum involves a combination of transcriptional, translational, and posttranslational mechanisms that have been elucidated in P. aeruginosa (34, 35). In this strain, flagellar assembly is regulated in a hierarchical way, with the transcriptional activator FleQ on the top of the regulatory cascade, which involves three sigma factors, the housekeeping σ70 and two alternative sigma factors, σ54 (RpoN) and σ28 (FliA). In this four-tiered transcription regulation hierarchy, σ70 directs the expression of class I genes encoding σ54, producing FliA and FleQ, an NtrC family activator protein and a cognate activator of σ54 (34). FleQ and σ54 are required for the transcriptional activation of the class II genes, like fleSR, and more than 20 flagellar proteins (27, 36). Class III genes, including the basal body-related genes, are positively regulated by the activated response regulator FleR in concert with σ54 (34, 37). The transcription of class IV genes, including fliC, which encodes the flagellin, is dependent on the availability of free FliA following the export of the FliA-specific anti-sigma factor FlgM through the hook-basal body apparatus (34, 38). The basic details underlying flagellar assembly and motility in a broad range of bacteria are well understood, but occasionally similar genes exert different functions depending on the strain, like the fliT and fleP genes in P. fluorescens F113 and P. aeruginosa PAO1 (18).
Aside from activating motility, FleQ also regulates genes involved in attachment to surfaces and biofilm formation (39–41). In P. fluorescens Pf0-1, FleQ regulates genes encoding various enzymes, putative lipoproteins, regulators, and hypothetical proteins (40). Recently it has been shown that FleQ is a negative regulator of exopolysaccharide biosynthesis in P. aeruginosa and P. fluorescens SBW25, and its activity is modulated by c-di-GMP (42–45). Mutation of fleQ in P. fluorescens SBW25 is linked to the loss of flagella and swimming motility but also promotes sliding motility, which is dependent on the biosurfactant viscosin (13). P. aeruginosa exhibited similar behavior on swarming plates when both pili and flagella were deleted, and biosurfactant expression was suggested as a major factor regulating this sliding motility (8).
Pseudomonas syringae pv. tomato DC3000 (P. syringae pv. tomato DC3000) causes bacterial speck on tomato and Arabidopsis and represents an important model in molecular plant pathology, since it carries a large repertoire of potential virulence factors, including proteinaceous effectors that are secreted through the type III secretion system and a polyketide phytotoxin called coronatine, which structurally mimics the plant hormone jasmonate. P. syringae pv. tomato DC3000 produces one to five polar flagella (46, 47), but little is known about their expression and regulation. This bacterium also produces the biosurfactant syringafactin, a peptide composed of eight amino acids with six structural variants that differ in a single amino acid and the length of the carbon chain attached (3-hydroxydecanoyl or 3-hydroxydodecanoyl). It is synthesized by a nonribosomal peptide synthetase encoded by an operon composed of two genes, syfA and syfB (48). Syringafactin is essential for swarming in P. syringae pv. tomato DC3000 but not in P. syringae pv. syringae B728a, where the loss of syringafactin severely reduced swarming but did not abolish it completely (48–50).
By using mutants fleQ and fliC, lacking flagella (46, 51), and syfA, impaired in the production of syringafactin (48), we have observed that the fleQ mutation alters bacterial surface motility and biosurfactant production, uncovering a new type of motility for P. syringae pv. tomato DC3000 on semisolid surfaces. Consequently, we aimed to determine the role of FleQ in the regulation of flagellar and syringafactin production genes and how both flagella and biosurfactant act on P. syringae pv. tomato DC3000 motility and virulence in tomato plants.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture media.
The bacterial strains used in this study are listed in Table 1. P. syringae pv. tomato DC3000 and mutants were routinely grown in Luria-Bertani (LB) medium (56) at 28°C. When required, the following antibiotics were added to the cultures/plates: ampicillin (100 μg/ml), chloramphenicol (30 μg/ml), gentamicin (5 μg/ml), kanamycin (50 μg/ml), rifampin (10 μg/ml), and streptomycin (25 μg/ml).
TABLE 1.
Bacterial strains and plasmids used
| Strain or plasmid | Relevant characteristicsa | Reference or source |
|---|---|---|
| P. syringae pv. tomato strains | ||
| DC3000 | Wild type, Rifr | 52 |
| fleQ | fleQ::ΩKm; Rifr Kmr | 46 |
| fliC | fliC (flaA)::miniTn5Cm; Rifr Cmr | 51 |
| pilA | pilA::ΩSm/Sp; Rifr Smr Spr | 53 |
| syfA | ΔsyfA; Rifr | 48 |
| syfA fleQ | ΔsyfA fleQ::ΩKm; Rifr Kmr | This work |
| Plasmids | ||
| pBBR1MCS-5 | Gmr, cloning vector | 54 |
| pBBR1MCS-5-fleQ | Gmr, pBBR1MCS-5 derivative containing the fleQ gene of Pto DC3000 | 55 |
| pHPR-QΩKm | Apr Kmr, pUC18 derivative containing P. syringae pv. tomato DC3000 fleQ gene interrupted by ΩKm | 46 |
Apr, Cmr, Gmr, Kmr, Rifr, Smr, and Spr stand for resistance to ampicillin, chloramphenicol, gentamicin, kanamycin, rifampin, streptomycin, and spectinomycin, respectively.
The syfA fleQ null mutant was constructed by gene replacement. The Apr Kmr suicide plasmid pHPR-QΩKm (46) was electroporated (57) into the P. syringae pv. tomato DC3000 syfA mutant (48). Transformants that acquired the inactivated gene (Kmr) were selected and, among them, we screened for the Ap (100 μg/ml)-sensitive ones, which were expected to be the result of a double-recombination event. One of the Kmr Aps clones was chosen and confirmed by Southern blotting to have the wild-type fleQ gene replaced by the mutant allele fleQ::ΩKm (not shown).
Motility experiments.
For swimming assays, P. syringae pv. tomato DC3000 and mutants were grown on LB plates for 48 h, resuspended in 10 mM MgCl2 and adjusted to an optical density at 660 nm (OD660), of 2.0. Two-microliter aliquots were inoculated in the center of 0.3% agar LB plates and incubated for 48 h at 25°C. The diameters of the swimming halos were measured after 48 h. For surface motility (swarming/sliding) assays, the 2-μl aliquots were inoculated in the center of PAG plates (0.5% protease peptone no. 3 [Difco 212693] and 0.2% glucose with 0.5% Difco Bacto agar) and incubated at 25°C. The surface motility was observed after 24 h and the area measured using ImageJ (58). Three motility plates were used for each strain, and the experiment was repeated with independent cultures.
Microscopy assays.
The Leifson flagellum staining procedure for light microscopy was carried out according to Clark (59), with some modifications. A 1.2% solution of basic fuchsine prepared in 95% ethanol and left overnight at 25°C was mixed with an equal volume of a solution of 0.75% NaCl and 1.5% tannic acid prepared in MilliQ water. The final pH of the dye was adjusted with 1 N NaOH to pH 5.0. Swarmer cells were resuspended in 10 mM MgCl2, adjusted to an OD660 of 1.0, and fixed by adding 1 ml of 4% formaldehyde solution per ml of culture for 20 min. The suspension then was centrifuged, washed with MilliQ water, and resuspended in 1 ml of MilliQ water. The slides were cleaned by soaking them for 24 h at room temperature in acid dichromate solution, rinsed with MilliQ water, and then air dried. A large drop of culture suspension was deposited on the center of the slide and was allowed to run down its length and then air dried. One milliliter of dye was added and kept for 40 min, and the slide was washed with tap water, air dried, mounted with merkoglass, and examined under a Zeiss Axioskop microscope. At least 100 cells were counted per experiment, which was repeated with three independent cultures.
For negative staining, the Formvar-coated nickel grids were applied directly to bacteria grown on the agar surface at the edges of the swarming colonies and left for 5 min to allow bacterial adhesion. The grids then were washed twice in water for 1 min and negatively stained with a 1% solution of potassium phosphotungstic acid for 1 min. The grids were observed under a JEOL JEM-1011 transmission electron microscope at 100 kV at the Microscopy Service of the Estación Experimental del Zaidín, Granada, Spain.
Detection of biosurfactants.
Biosurfactants were detected with the atomized oil assay previously described (49). For a more uniform inoculation of plates, P. syringae pv. tomato DC3000 and mutants were grown on LB plates for 48 h, resuspended in water, and adjusted to an OD660 of 1.0. Ten-microliter aliquots (∼5 × 107 cells) were pipetted onto the surface of LB plates (1.5% agar), which were incubated for 24 h at 20°C and then sprayed with a mist of mineral oil. The diameter of the visible halo of brighter oil drops was measured, and the area of the producing bacterial colony was calculated and subtracted from that of the surfactant halo to yield the adjusted halo area. Four plates were used for each strain, and the experiment was repeated with three independent cultures.
Infection assays.
Seeds of Solanum lycopersicum cultivar Moneymaker (a P. syringae pv. tomato S line) were germinated and grown with 16-h/8-h light/dark cycles at 22°C/16°C day/night and 70% relative humidity in a plant growth chamber. P. syringae pv. tomato DC3000 strains, grown on LB plates for 48 h at 28°C, were suspended in 10 mM MgCl2, and the inoculum was adjusted to 108 CFU/ml. For infections, individual inocula of the strains were sprayed into different plants, and the analysis of the evolution of symptoms and sampling were performed 3 h after inoculation (time zero), when the leaves were dried, and several days after inoculation (3, 6, and 10 days postinfection [dpi]) to determine bacterial populations in plants. Bacteria were recovered from the infected leaves using a 10-mm-diameter cork-borer. Five disks (3.9 cm2) per plant were homogenized by mechanical disruption into 1 ml of 10 mM MgCl2 and counted by plating serial dilutions onto LB plates with the corresponding antibiotics. The severity of symptoms was evaluated as the percentage of necrotic area per leaflet induced by the inoculated strains after 10 days of incubation. The necrotic areas were digitally analyzed using ImageJ (58) on five inoculated leaflets of three different plants.
RNA preparation and assays.
For RNA isolation, cells of P. syringae pv. tomato DC3000 and fleQ strains were grown in PAG plates and incubated at 25°C for 24 h (swarming conditions) or in MMR (7 mM Na-glutamate, 55 mM mannitol, 1.31 mM K2HPO4, 2.2 mM KH2PO4, 0.61 mM MgSO4, 0.34 mM CaCl2, 0.022 mM FeCl3, 0.85 mM NaCl, 0.818 μM biotin, 0.296 μM thiamine, and 0.209 μM calcium pantothenate) minimal medium (60) at 20°C to an OD660 of 1.5. Cells were harvested and washed with 0.1% Sarkosyl, and cell pellets were immediately frozen in liquid nitrogen and conserved at −80°C until RNA isolation. For reverse transcription-quantitative real-time PCR (RT-qPCR) and transcriptome sequencing (RNA-seq), RNA was isolated using TRI Reagent LS (Molecular Research Center, Cincinnati, OH) as described before (61). Residual DNA was removed with the RNase-free DNase I set (Roche).
For RT-qPCR assays, total RNA (1 μg) treated with the RNase-free DNase I set (Qiagen) was retrotranscribed to cDNA with SuperScript II reverse transcriptase (Invitrogen) using random hexamers (Roche) as primers. Quantitative real-time PCR was performed on an iCycler iQ5 (Bio-Rad, Hercules, CA, USA). Control PCRs of the RNA samples not treated with reverse transcriptase also were performed to confirm the absence of contaminating genomic DNA. The specific primer pairs used to amplify cDNA are listed in Table 2, and primer efficiency was optimal (≈100%) for all pairs (62). Relative transcript abundance was calculated using the ΔΔCT method (63). P. syringae pv. tomato transcriptional data were normalized to the housekeeping gene gyrA (61). The expression of a given gene relative to that of gyrA was calculated as the difference in qPCR threshold cycles (ΔCT = CT gene of interest − CT gyrA) The expression of each gene was determined with the control treatment (wild type) as the difference between ΔCT values (ΔΔCT). As one PCR cycle represents a 2-fold difference in template abundance, fold change values were calculated as 2−ΔΔCT.
TABLE 2.
Primers used in this study
| Primer name | Sequence (5′→3′) | Use |
|---|---|---|
| flhA+ | TATTCGCAGCATTGCAGAGG | flhA-flhF intergenic region amplification |
| flhF− | GCATCGAGTCAAAGGCACGC | flhA-flhF intergenic region amplification |
| flhF+ | CAGTATGGACAGTTTCCGCATCGGC | flhF-fleN intergenic region amplification |
| fleN− | TTGTCGCCAATCTCGCTGAACGCC | flhF-fleN intergenic region amplification |
| fleN+RT | AGGCGTTCAGCGAGATTGGCGACA | fleN-fliA intergenic region amplification |
| fliA−RT | ACGCTCGCGCTCTGGCAAATTGG | fleN-fliA intergenic region amplification |
| fleQ-F | GCGAGAGGTTCTGTGCGTGCTGGTC | qRT-PCR expression of fleQ |
| fleQ-R | GCAGCTCGACGGAAGAGTTTTCGCTCA | |
| fleS-F | CGCCAGTCATCTGACCGAGCAGGAA | qRT-PCR expression of fleS |
| fleS-R | GGGCGTTAGGCGGTCCGTCAGC | |
| fleN-F | AGGCGTTCAGCGAGATTGGCGA | qRT-PCR expression of fleN |
| fleN-R | TCGCACACCACCAGCAGCACTTCCT | |
| fliA-F | CGAGGACGGCGCAAGCGGTTC | qRT-PCR expression of fliA |
| fliA-R | ACGCTCGCGCTCTGGCAAATTGG | |
| flgM-F | CACGTACCGCAGCCAGCAAAGACACC | qRT-PCR expression of flgM |
| flgM-R | TAACGGTAGGCAGGTCGGTCAGCTTGTCC | |
| flgF-F | TCGACCAATGGCTTCATGCGTGACCTT | qRT-PCR expression of flgF |
| flgF-R | TCGGGAGTCTGCACGGCAATCCAG | |
| flhA-F | CGTCGTGCTGCTGGTTTGCGTCTAT | qRT-PCR expression of flhA |
| flhA-R | GCATGACCGTCCTGACCGTGAAGC | |
| flhF-F | GCCGCCACGCCCGTTTGC | qRT-PCR expression of flhF |
| flhF-R | GCTGCGGACGGCTGCCTTGT | |
| fliC-F | AAGGCGCACTGCAAGAGTCGACCAAC | qRT-PCR expression of fliC |
| fliC-R | GGTGCTGGCGGAACCGTCAAGC | |
| syfA-F | CGCACGACGCAACGCAAGGAATGG | qRT-PCR expression of syfA |
| syfA-R | ACGCACGGCACTTACCTGGGCAAA | |
| syfR-F | CCGCTGGTCGAACATCACGCACAA | qRT-PCR expression of syfR |
| syfR-R | AGCCACGGTTCCGCCTGTCAA | |
| gyrA-F | GGCAAGGTCACCCGCTTCAAGGAAT | qRT-PCR expression of gyrA |
| gyrA-R | GACCGCCACGCTTGTACTCAGGGAAC |
For RT-PCR assays, total RNA (2 μg) was used for cDNA synthesis with SuperScript II reverse transcriptase (Invitrogen) and random hexamers as primers. Controls yielding the expected amplified fragments from all of the primers pairs were obtained by using chromosomal DNA as the template, and the presence of DNA contamination was checked using the RNA samples without RT treatment.
For comparative transcriptome analysis, total RNA was isolated from P. syringae pv. tomato DC3000 and the fleQ mutant grown under 2 different conditions: from MMR liquid cultures and from swarming plates. Library generation and sequencing was performed by Macrogen Inc. (Seoul, Republic of Korea) using Illumina HiSeq2000 to produce reads of 100 bp. Reads were clipped and trimmed to remove low-quality nucleotides, Illumina adapters, and rRNAs by using SeqTrimNext (http://www.scbi.uma.es). Filtered reads were aligned to the P. syringae pv. tomato DC3000 reference genome (AE016853) with Bowtie (64). SAMtools (65) utilities were used to convert sam files to bam format and to sort, index, and convert them to bedgraph files. Analysis of the data was conducted in the Integrated Genome Browser graphic user interface (http://bioviz.org/igb/).
Immunoblotting.
Total cell extracts were obtained from cultures incubated in LB with shaking at 25°C. Ten milliliters of cells was centrifuged for 10 min at 2,500 × g at 4°C, and the pellets were kept at −80°C until the lysis step. The harvested cells then were resuspended in 200 μl of phosphate-buffered saline (PBS) with protease inhibitor mixture and lysed by sonication. Total cell protein extracts were recovered from the supernatant after centrifuging for 2 min at 15,000 × g and 4°C. Equal amounts of whole-cell lysates were loaded onto 10% polyacrylamide gels, electrophoresed, and transferred to polyvinylidene difluoride (PVDF) membranes. Ponceau S staining of the membranes was performed as a control of loading and transfer of comparable amounts of protein. The membranes were incubated with a 1:10,000 dilution of an anti-FliA antiserum from P. aeruginosa (66) and then with a peroxidase-tagged secondary antibody (anti-rabbit immunoglobulin) for 1 h. The enhanced chemiluminescence (ECL; GE Healthcare) method and Personal FX equipment was used for development. Results were analyzed with Quantity One software (Bio-Rad).
Statistical analysis.
Statistical comparison among different strains or conditions was performed by one-way analysis of variance (ANOVA) with post hoc Tukey's honestly significant difference (HSD) test using R.
Transcriptome accession number.
The RNA-seq analysis was deposited in the public database ArrayExpress at EBI under the accession number E-MTAB-3779.
RESULTS
P. syringae pv. tomato DC3000 employs flagellum-dependent and flagellum-independent syringafactin-dependent surface motilities.
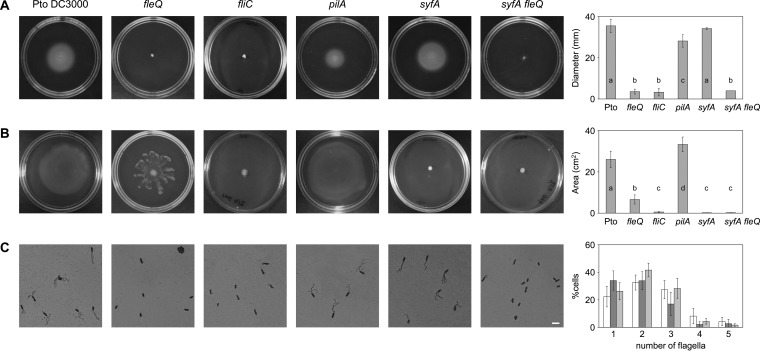
To investigate the role of flagella in P. syringae pv. tomato DC3000 motility, we used an fleQ mutant deficient in the master regulator for flagellar synthesis (46) and an fliC mutant impaired in flagellin production (previously called flaA) (51). Surface (swarming and sliding) motility assays were carried out in PAG (0.5% agar) plates to detect the ability to spread over the agar surface. As a control, we performed swimming assays in LB (0.3% agar) plates and detected the cells moving through the agar (Fig. 1). We observed that the fliC and fleQ mutants remained immobile in swimming plates (Fig. 1A). On PAG, the surface motility of the fliC mutant was severely affected, whereas the fleQ mutant exhibited altered surface spreading on semisolid agar (Fig. 1B). To confirm that the phenotypic changes were due to the loss of FleQ, the mutant was complemented with an intact copy of the fleQ gene in a plasmid. The complemented strain produced flagella, regained the ability to swim through agar, and exhibited a swarming motility phenotype similar to that of the wild type (see Fig. S1 in the supplemental material).
FIG 1.
Motility of P. syringae pv. tomato DC3000 and its mutants. (A) Swimming motility assays were performed with the wild type and the indicated mutants. Pictures were taken 48 h after inoculation and the swimming halo diameter measured. The graphic shows the means and standard deviations from four experiments with three replicates; a, b, and c denote ANOVA categories with significant differences (P < 0.01). (B) Surface motility assays with the wild type and the indicated mutants 24 h after inoculation. The areas of motility were calculated with ImageJ and the means and standard deviations from five experiments with three replicates plotted; a, b, c, and d denote ANOVA categories with significant differences (P < 0.01). (C) Visualization of flagellar abundance in P. syringae pv. tomato DC3000 strains taken from PAG plates. Light microphotographs of cells from PAG plates stained with the Leifson method. The scale bar (white) represents 5 μm. The graphic shows the means and standard deviations of the percentages of P. syringae pv. tomato DC3000 (white), pilA (dark gray), and syfA (light gray) cells with 1 to 5 flagella. The fleQ, fliC, and syfA fleQ mutants did not produce any flagella. At least 100 cells were counted per experiment, which was repeated with four independent cultures. No significant differences in flagellum numbers were detected by one-way ANOVA between the wild type and the syfA mutant.
Swarming is powered by flagella but also is influenced by other surface structures, like type IV pili (TFP) (2–4). To investigate the role TFP in P. syringae pv. tomato DC3000 motility, a pilA knockout mutant (53) lacking those organelles was tested in motility assays (Fig. 1). We observed that the pilA mutant exhibited a 20% reduction on swimming motility (Fig. 1A), but its swarming motility increased 30% compared to that of the wild type (Fig. 1B). Therefore, TFP are dispensable for swimming (Fig. 1A), and they seem to restrict swarming in the wild-type strain (Fig. 1B), as has been observed in P. aeruginosa (8, 67, 68).
The surface motility exhibited by the P. syringae pv. tomato DC3000 fleQ mutant is similar to that reported for different bacteria after blocking flagellum production (7, 8, 13, 14) and could be considered sliding, a passive form of surface spreading which relies on bacterial growth and biosurfactant production (3). As has been mentioned before, P. syringae pv. tomato DC3000 produces the biosurfactant syringafactin, and a mutant blocked in syringafactin production (syfA) could swarm no longer (48, 49). Also, under our experimental conditions (PAG–0.5% agar plates), the swarming of the P. syringae pv. tomato DC3000 syfA mutant was completely inhibited (Fig. 1). However, its swimming motility (in LB–0.3% agar plates) was similar to that of the wild type (Fig. 1).
To assess whether all of those strains produced flagella, a flagellar stain for light microscopy was used in swarmer cells (59), observing that P. syringae pv. tomato DC3000 produced 2 to 5 polar flagella, more than it did in liquid medium-grown cells (46, 47). The fleQ and fliC mutants were devoid of these organelles, as expected; however, the syfA mutant produced flagella similar in number and morphology to those of the wild type (Fig. 1C). These results confirm that the flagellum is essential for swimming motility in P. syringae pv. tomato DC3000 and plays an important role in its surface motility, whereas TFP seem to have an accessory role. On the other hand, syringafactin is not required for swimming but is absolutely indispensable for swarming, at least under the conditions tested.
FleQ is the master regulator of flagellar genes in P. syringae pv. tomato DC3000.
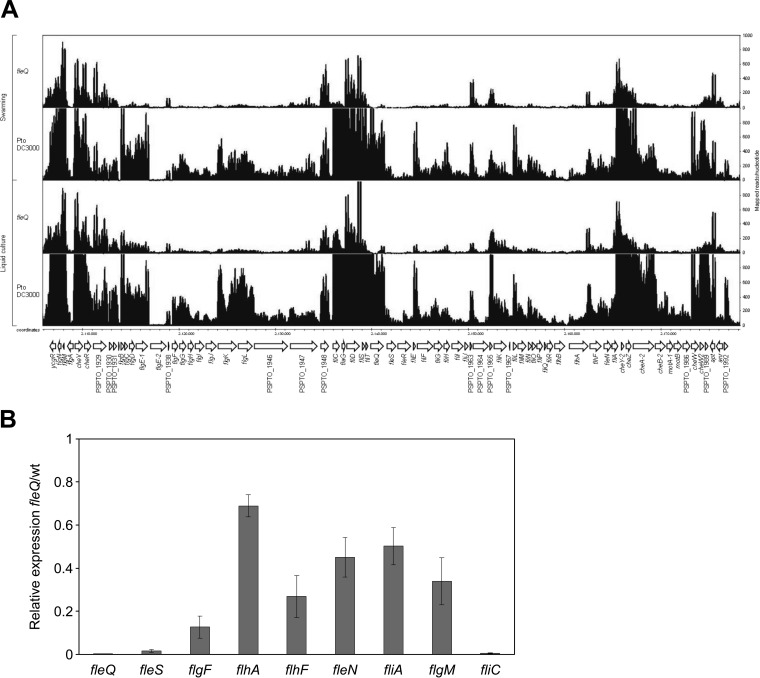
In P. aeruginosa, the master regulator FleQ belongs to the top tier of the flagellar cascade and is required to activate all other flagellar genes, with the exception of fliA (27, 34, 36). To advance our knowledge of the flagellar regulatory cascade in P. syringae pv. tomato DC3000, which has not been characterized so far, and to establish the role of FleQ in this strain, we analyzed the effect of the fleQ deletion on transcript levels by RNA-seq. We compared the wild-type strain and the fleQ mutant in swarming and in liquid culture-grown cells as a control, observing that the majority of the flagellar genes were expressed similarly under both conditions (Fig. 2A). Also, several genes were strongly downregulated in the fleQ mutant, like flgBCDE, flgFGHIJKL, fliC, flaG, fliD, fliS, fliT, fleSR, fliEFGHIJ, cheY-2, cheZ, and cheA-2, among others. These results confirm the role of P. syringae pv. tomato DC3000 FleQ as a master regulator of the flagellar genes, like in other pseudomonads (18, 27, 34, 36, 41). In order to validate the RNA-seq data, we analyzed the expression of several motility genes presumably belonging to different classes of the flagellar gene hierarchy (fleQ, fleS, flhA, flhF, fleN, fliA, flgF, flgM, and fliC) under swarming conditions (Fig. 2B). The RT-qPCR analysis carried out revealed that the expression of fleS and fliC is strictly dependent on FleQ, similar to what occurs in P. aeruginosa. However, transcription of flgF, flhA, flhF, fleN, fliA, and flgM seem to be regulated only to some extent by FleQ, unlike in P. aeruginosa, where fliA is a class I gene which expresses independently of FleQ, flgF, flhA, flhF, and fleN are strictly dependent on the activation by FleQ, and flgM also is positively regulated by FliA (34, 36).
FIG 2.
Expression analysis of flagellar genes in P. syringae pv. tomato DC3000. (A) RNA mapped read distribution of flagellum-encoding genes in P. syringae pv. tomato DC3000 and fleQ. RNA was obtained from P. syringae pv. tomato DC3000 and fleQ grown under swarming conditions and liquid cultures (MMR at 20°C). Drawings are images from IGB software showing reads in several genes belonging to the flagellar cascade. Genomic coordinates denote the position in kilobases of the P. syringae pv. tomato DC3000 genome, and annotated ORFs are shown as arrows. The scale (counts) indicates the number of mapped reads per nucleotide position. (B) Expression of several flagellar genes determined by RT-qPCR. Total RNAs and cell extracts were obtained from bacteria grown in PAG (0.5% agar) plates for 24 h at 25°C. RT-qPCR expression values of the genes in the fleQ mutant were normalized with the housekeeping gene gyrA. The relative expression of flagellar genes was calculated as the fold change between the fleQ mutant and the wild-type strain (which was set to 1.0). Results shown are the means and standard deviations from three independent biological experiments with three technical replicates each. In all cases, differences with respect to the wild type were significant as determined by one-way ANOVA (P < 0.01).
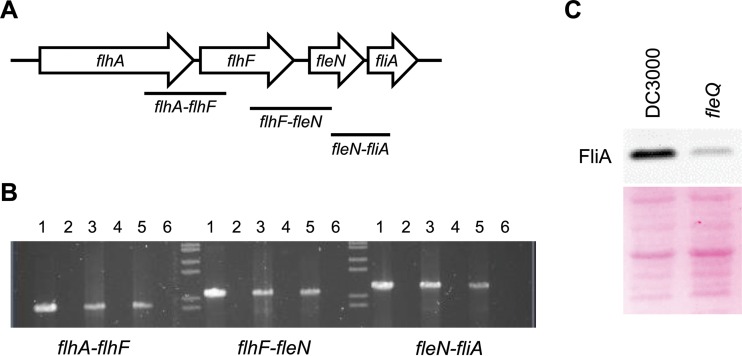
Since the expression of fliA and some adjacent genes (flhA, flhF, and fleN) in P. syringae pv. tomato DC3000 was different from that of P. aeruginosa, we decided to ascertain the transcriptional organization of the flhA-fliA region and its expression in both the wild type and the fleQ mutant. Primers for coamplification of adjacent genes in the flhA-fliA region were designed, and total RNA from cells of P. syringae pv. tomato DC3000 and the fleQ mutant grown on PAG plates (swarming conditions) was subjected to RT-PCR (Fig. 3A). The results show that the flhA-flhF, flhF-fleN, and fleN-fliA primer pairs each yielded an RT-PCR product, indicating that in P. syringae pv. tomato DC3000, flhA-flhF-fleN-fliA is transcribed as a tetracistronic operon (Fig. 3B), which is different from P. aeruginosa, where flhA and fliA are transcribed as monocistronic operons and flhF and fleN are transcribed as a bicistronic operon (34). In fact, the P. syringae pv. tomato DC3000 flhA transcription start site was previously determined (69, 70), and we have identified a putative σ54 promoter upstream of flhA and a FleQ binding site which, similar to that in P. aeruginosa (36, 42), is located within the leader region (+17/+22; see Fig. S2 in the supplemental material). Furthermore, flhA, flhF, fleN, and fliA also were cotranscribed in the P. syringae pv. tomato DC3000 fleQ mutant (Fig. 2A and 3B; also see Fig. S3 for a detailed view). Therefore, flhA-flhF-fleN-fliA genes transcribe as an operon not only in the presence of FleQ but also in its absence, albeit at lower levels, providing basal concentrations of FlhA, FlhF, FleN, and FliA proteins. To corroborate this hypothesis, immunoblots of total cell extracts from the wild type and the fleQ mutant were carried out, showing that the production of FliA was maximal in the wild type and significantly diminished in the fleQ-lacking strain (Fig. 3C). Therefore, not only fliA transcription but also FliA production are reduced in the fleQ mutant, confirming that, in P. syringae pv. tomato DC3000, FliA synthesis is mostly dependent on the master regulator (Fig. 2 and 3). The results presented here corroborate that flagellar biosynthesis is differently regulated in diverse Pseudomonas strains (71).
FIG 3.
Expression of the flhA-fliA region in P. syringae pv. tomato DC3000. (A) Map of the flhA-fliA region. The labeled bars indicate the expected fragments amplified with each primer pair (Table 2), with each of them partially covering two adjacent genes. (B) Results from RT-PCR amplification of the flhA-fliA region. Total RNA was obtained from P. syringae pv. tomato DC3000 and the fleQ mutant grown in swarming conditions. Lanes 1 and 2, positive (chromosomal DNA as the template yielding the expected amplified fragments from all of the primer pairs) and negative (water) controls; lanes 3 and 5, 2 μg of RNA samples from the wild type and the fleQ mutant, respectively, with RT treatment; lanes 4 and 6, the presence of DNA contamination was checked using 2 μg of RNA samples of the wild type and the fleQ mutant, respectively, without RT treatment. (C) Production of FliA by P. syringae pv. tomato DC3000 and fleQ. Protein was detected using an anti-FliA antibody from P. aeruginosa (66). FliA appeared on membranes as a band of approximately 29 kDa in apparent size. Ponceau staining of the membrane was used as a loading and transfer control (bottom).
FleQ negatively regulates syringafactin production in P. syringae pv. tomato DC3000.
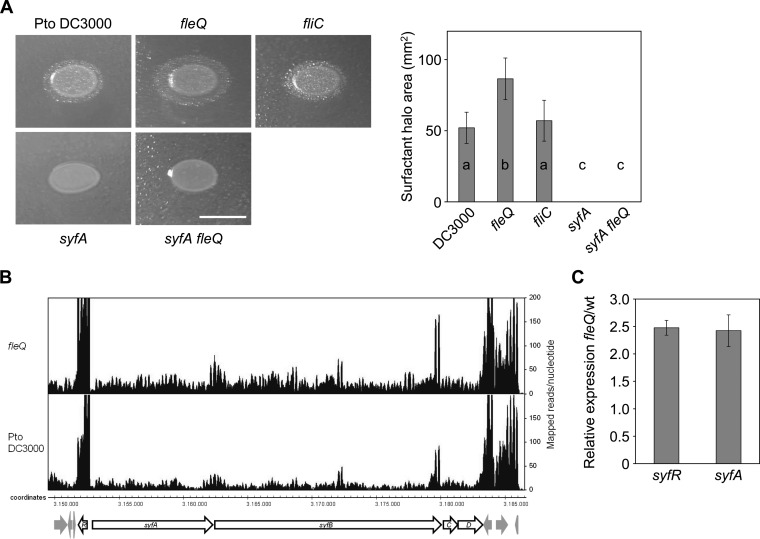
Although the flagellum plays an important role in P. syringae pv. tomato DC3000 surface motility, other mechanisms may facilitate its displacement over surfaces, as suggested by the movement exhibited by the fleQ mutant. In this regard, a P. fluorescens SBW25 fleQ mutant exhibited a spreading surface motility that required the expression of viscB and viscC, which encode a nonribosomal peptide synthetase involved in the production of the lipopeptide surfactant viscosin (13). Likewise, we hypothesized that the inactivation of fleQ in P. syringae pv. tomato DC3000 leads to an increase in the production of surfactant(s). To check this possibility, we assessed the production of surfactants by the atomized oil assay (49). The amount of surfactants produced by the fleQ mutant was increased by 40% compared to that produced by the wild-type strain or the fliC mutant (Fig. 4A), suggesting this is the cause of its altered spreading motility. To test this hypothesis, we constructed an syfA fleQ double mutant, observing that this mutant did not exhibit swimming or surface motility and lacked flagella (Fig. 1). We were unable to generate a complemented mutant for syringafactin production; instead, we carried out RNA-seq to investigate the role of FleQ in syringafactin synthesis at the transcriptional level, noticing that the expression of syfR and syfABCD genes is upregulated in the fleQ mutant (Fig. 4B). To corroborate these results, we carried out RT-qPCR assays and observed that syfR and syfA mRNA levels were increased (2.5- ± 0.1-fold and 2.4- ± 0.3-fold, respectively) in the fleQ mutant (Fig. 4C). syfR (PSPTO_2828) encodes a transcriptional regulator of the LuxR family which seems to function as an activator for the syfABCD genes, since an insertion in syfR resulted in the loss of swarming motility, droplet collapse, and syringafactin accumulation (48). The increased expression of syfR and syfA in the absence of FleQ suggests that the flagellar master regulator is negatively regulating syfR and, consequently, syfAB(CD) expression and syringafactin production in the wild-type strain.
FIG 4.
Syringafactin expression and production in P. syringae pv. tomato DC3000 and its mutants. (A) Comparison of surfactant-induced halos around bacterial colonies grown on LB (1.5% agar) for 24 h at 20°C and visualized by the atomized oil assay. The biosurfactant areas were calculated and the means and standard deviations of three experiments with four replicates plotted; a, b, and c denote ANOVA categories with significant differences (P < 0.01). The scale bar (white) represents 1 cm. (B) P. syringae pv. tomato DC3000 and the fleQ mutant mapped read distribution in the syf region under swarming conditions. Drawings are images from IGB software showing reads in syfR, syfA, syfB, syfC, and syfD. Genomic coordinates denote the position in kilobases of the P. syringae pv. tomato DC3000 genome, and annotated ORFs are shown as arrows. The scale (counts) indicates the number of mapped reads per nucleotide position. (C) Expression analysis of syfA and syfR in P. syringae pv. tomato DC3000 and fleQ determined by RT-qPCR. Total RNAs and cells extracts were obtained from bacteria grown in PAG (0.5% agar) plates for 24 h at 25°C. RT-qPCR expression values of the genes in the fleQ mutant were normalized with the housekeeping gene gyrA. The relative expression of flagellar genes was calculated as the fold change between the fleQ mutant and the wild-type strain (set to 1.0). Results shown are the means and standard deviations from three independent biological experiments with three technical replicates each. In all cases, differences with respect to the wild type were significant as determined by one-way ANOVA (P < 0.01).
Role of P. syringae pv. tomato DC3000 flagellar and nonflagellar motility in virulence.
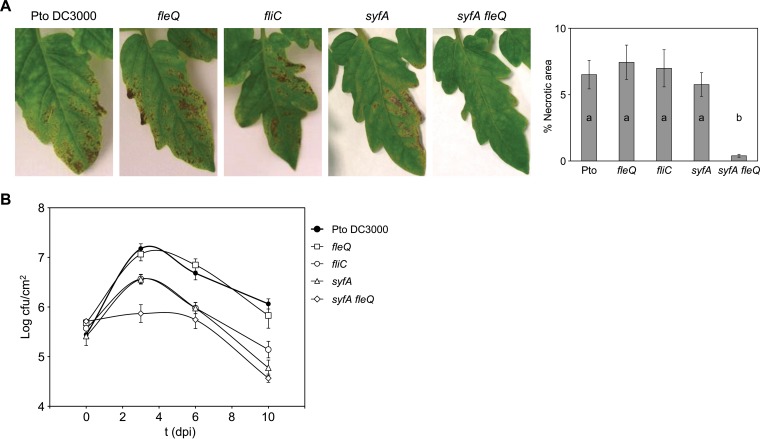
Once we established the role of flagella and syringafactin and their regulation by FleQ in the different movements exhibited by P. syringae pv. tomato DC3000, we next studied whether the flagella and syringafactin contributed to the pathogenic process of P. syringae pv. tomato DC3000. For this, we compared the abilities of the wild type and the fleQ, syfA, and syfA fleQ mutant strains to survive and multiply in tomato leaf tissues by monitoring bacterial populations and development of disease symptoms for 10 days after inoculation by spray. We also used the fliC mutant, since this strain has a functional FleQ but lacks flagella due to the fliC/flagellin deficiency. The disease symptoms caused by the fliC, fleQ, and syfA single mutants were indistinguishable from those caused by the wild-type strain: small water-soaked lesions surrounded by large regions of chlorosis that appeared 2 to 3 days after inoculation. These spots soon turned brown, and the tissue surrounding the spots became yellow. However, the symptoms caused by the syfA fleQ double mutant were far less abundant (Fig. 5A).
FIG 5.
Bacterial growth and symptom development on tomato leaves. (A) Development of symptoms induced on tomato leaves 10 days after inoculation with P. syringae pv. tomato DC3000 and different mutants at 108 CFU/ml by spray. The severity of symptoms was evaluated as the percentage of necrotic area per leaflet induced by the inoculated strains. The graphic shows the means and standard errors from three different experiments (with five leaflets analyzed in each of them); a and b denote ANOVA categories with significant differences (P < 0.01). (B) Time course of growth of P. syringae pv. tomato DC3000 and the fleQ, fliC, syfA, and syfA fleQ mutants in the primary leaves of tomato plants. CFU were quantified at 0, 3, 6, and 10 dpi with approximately 108 CFU/ml by spray. Data represent the averages from three experiments with their standard errors. At 3 days, differences between the wild type and the fleQ mutant and between the fliC and syfA mutant populations were not significant. At 6 and 10 dpi, fliC, syfA, and syfA fleQ mutant populations were significantly different from those of the wild type and the fleQ mutant as determined by one-way ANOVA (P < 0.01); differences between the wild type and the fleQ mutant or among the fliC, syfA, and syfA fleQ mutant populations were not significant.
Before the plant experiments, all of the mutants and the wild type were tested for growth in different laboratory media and showed similar growth curves (not shown). However, their behavior in tomato leaves varied, although the starting populations of the five strains were similar (Fig. 5B). The syfA fleQ mutant was significantly growth impaired in planta, as evidenced by the small population sizes at all time points. The wild type and the fleQ mutant behaved similarly: their populations reached a maximum at 3 dpi (1.8 × 107 to 2.1 × 107 CFU/cm2) and after that decreased to 1.5 × 106 to 1.9 × 106 CFU/cm2 at 10 dpi. The fliC and syfA mutants behaved similarly and exhibited an intermediate phenotype between that of the syfA fleQ mutant and that of the wild type and fleQ: their populations reached a maximum at 3 dpi (4.4 × 106 to 5.4 × 106 CFU/cm2) and after that decreased to 1.0 × 105 to 1.9 × 105 CFU/cm2. These results indicate that only the combined loss of fleQ and syfA causes a significant decrease in the ability of P. syringae pv. tomato DC3000 to colonize tomato plants and cause disease. However, mutants deficient in flagella (fliC) or syringafactin (syfA) were not able to reach population densities as high as those of the wild type or the fleQ mutant but caused similar symptoms. Interestingly, the fleQ mutant, which also lacks flagella, behaved like the wild type in both growth and symptom development.
DISCUSSION
Bacteria can move through their environment in multiple ways. Several genera of bacteria, including pseudomonads, exhibit swarming, a complex form of motility on semisolid surfaces which is absolutely dependent on flagella but also influenced by specific environmental signals, the presence of other surface structures, and the production of surfactants (8, 13, 72). In this work, we present evidence that P. syringae pv. tomato DC3000 is able to display different types of motility. The predominant movement in liquid or semisolid media is swimming, which requires the presence of flagella, whereas the biosurfactant syringafactin is dispensable (Fig. 1A). In contrast, both flagella and syringafactin are required for swarming motility (Fig. 1B). We also show that P. syringae pv. tomato DC3000 displays an alternative form of surface motility that is unmasked upon inactivation of the regulator FleQ. This type of motility is consistent with the notion that bacterial translocation is facilitated by both propulsive forces and those that reduce friction between bacterium and substrate (3, 8, 73, 74). In fact, the absence of flagella is not sufficient to enable sliding motility, and an fliC mutant is not able to spread on plates (Fig. 1B). Furthermore, not only swarming, as it was shown before (48, 49), but also sliding are strictly dependent on the production of the biosurfactant syringafactin, since both an syfA mutant and a double syfA fleQ mutant are not able to spread on plates (Fig. 1B). We should mention that this is a consistent phenotype exhibited under different conditions, KB (0.4 and 0.5% agar) (48, 72) and PAG plates (0.5% agar) (this work), suggesting that both flagellum-dependent motility and flagellum-independent, syringafactin-dependent motility are in operation on the agar plates. The fact that syringafactin production is crucial for both swarming and sliding in P. syringae pv. tomato DC3000 seems to be unique among P. syringae strains, since P. syringae pv. syringae B728a syringafactin mutants exhibited greatly reduced surfactant halos, drop collapse ability, and swarming, but only the lack of 3-(3-hydroxyalkanoyloxy) alkanoic acid (HAA) production in a syringafactin-deficient strain completely abolished surface motility (72).
An fleQ mutant of P. syringae pv. syringae B728a previously was deficient in surfactant (HAA and syringafactin) production (72). However, we have shown the opposite: the P. syringae pv. tomato DC3000 fleQ mutant overproduced syringafactin (Fig. 4A); furthermore, its sliding motility on semisolid surfaces is similar to that of the P. fluorescens SBW25 fleQ mutant, which was shown to require the surfactant viscosin (13). This suggests that the control of biosurfactant production by FleQ is a common feature in pseudomonads. Particularly in P. syringae pv. tomato DC3000, syringafactin production is enhanced under conditions in which fleQ is downregulated, assuring bacterial motility over surfaces. It remains to be determined whether the effect of FleQ on syringafactin production is direct (transcriptional regulation of the syf genes) or, most likely, mediated by additional factors/regulators, since no conserved sites for FleQ binding were detected in the syfR-syfA intergenic region.
We also have established the role of FleQ as the master regulator of flagellar genes in P. syringae pv. tomato DC3000 and spotted some differences in the regulation of flagellar biosynthesis compared to that of other Pseudomonas organisms. We have demonstrated that flhA, flhF, fleN, and fliA transcribe as an operon in the presence of FleQ but also in its absence, although at low levels (Fig. 2 and 3). This situation is different from that of P. aeruginosa, where FleQ regulates the expression of the majority of flagellar genes, with the exception of fliA, grouping fleQ and fliA as class I genes (34). Therefore, we propose that fliA should be considered a class II gene in P. syringae pv. tomato DC3000, like flhA, flhF, and fleN, because its expression is partially dependent on the master regulator. Moreover, the lower levels of FliA detected in the fleQ mutant suggest that FleQ controls expression of fliA not only at the transcriptional level but also posttranscriptionally. In Escherichia coli, FliA degradation increases when its anti-sigma factor FlgM is not available due to secretion or mutation (75). In the P. syringae pv. tomato DC3000 fleQ mutant, flgM expression also is reduced (Fig. 2); therefore, the FliA:FlgM stoichiometry would be altered and FliA degraded. Nevertheless, the low expression of the flhA-flhF-fleN-fliA operon in the absence of FleQ provides basal levels of FliA (Fig. 3) and most likely of FlhA, FlhF, and FleN proteins. Since P. syringae pv. tomato DC3000 produces 1 to 5 polar flagella, whereas P. aeruginosa possesses a single polar flagellum, basal levels of those proteins may be needed for rapid flagellum assembly. Also, those proteins may have additional functions in virulence factor secretion, cytotoxicity, or proper cell division, as has been shown in Bacillus thuringiensis (76), Xanthomonas campestris pv. campestris (77), Yersinia enterocolitica (78), and Campylobacter jejuni (79).
In summary, we have shown that FleQ activates flagellum synthesis and negatively regulates syringafactin production in P. syringae pv. tomato DC3000, thereby controlling not only swimming and swarming but also the sliding motility in this strain.
Bacterial flagella are considered an important virulence factor in P. aeruginosa, C. jejuni, and Salmonella enterica serovar Typhimurium mainly because of their role in motility and chemotaxis, facilitating the pathogen-host interactions and promoting colonization, penetration, and invasion of host tissues (80–82). The importance of flagella in plant colonization is a controversial subject with contradictory results. Some researchers showed that flagella were essential for P. fluorescens root colonization (20) or movement onto the root surface and attachment (83), whereas others suggested that passive movement with water flow played a more important role than motility (84). S. enterica and enterohemorrhagic E. coli require flagella for systemic spread from root to shoot of Arabidopsis thaliana (85), whereas flagella are essential to colonize certain plants only for some Listeria monocytogenes strains (86). Here, we have shown that mutation of fliC in P. syringae pv. tomato DC3000 did not abolish its ability to reach the apoplast and cause infection, although its populations were smaller than those of the wild type (Fig. 5), as was observed before (87). This is different from other Pseudomonas strains (88) and pathovars of P. syringae (30, 33, 89) and suggests that P. syringae pv. tomato DC3000 uses alternative forms of motility that are not flagellum dependent for the colonization of tomato leaves. It has been shown that flagellum-based swarming is insufficient for movement over dry surfaces, where biosurfactants may acquire a more important role by promoting water retention and/or surface spreading (8, 13, 50, 90). In this sense, the P. syringae pv. tomato fliC mutant is able to produce syringafactin at levels comparable to those of the wild type (Fig. 4).
On the other hand, we also have shown that the syfA mutant provoked symptoms similar to those of the wild type, although it was not able to multiply at the same level (Fig. 5). Therefore, the absence of syringafactin did not prevent P. syringae pv. tomato DC3000 from reaching the apoplast and causing infection, which indicates that this surfactant is not phytotoxic per se and is not essential for P. syringae pv. tomato DC3000 colonization of tomato leaves when flagella are present and bacteria can swim. This is different from other phytopathogenic Pseudomonas species, like P. cichorii, whose biosurfactants (cichofactins) significantly contribute to lettuce midrib invasion (91). Moreover, in the fleQ mutant, the overproduction of syringafactin seems to counteract the absence of flagella (compared to the fliC and the syfA fleQ mutants), increasing bacterial survival and/or growth in planta. Therefore, colonization and symptom development in tomato are seriously compromised only when both flagella and syringafactin are missing, but neither flagella nor syringafactin on their own are essential for leaf surface migration, colonization, and virulence in P. syringae pv. tomato DC3000, although they may have a role in leaf invasion and/or survival given that the mutant population levels remain lower than those of the wild type (Fig. 5). Thus, the production of both flagella and syringafactin in P. syringae pv. tomato DC3000 seems to play a role in leaf surface dissemination and also in apoplast colonization, since they probably facilitate access to nutrients and bacterial survival. Since the overproduction of syringafactin compensates for the absence of flagella in P. syringae pv. tomato DC3000 colonization of tomato leaves, we propose that sliding movement and positive chemotaxis toward stomata are the driving forces allowing P. syringae pv. tomato DC3000 to rapidly reach stomata and enter the apoplast, similar to the sliding motility linked to bacterial cell division of rhizobia within legume root infection threads (92, 93). Moreover, P. syringae pv. tomato DC3000 may have developed this alternative form of sliding motility to colonize plant surfaces under conditions in which the production and/or activity of flagella is not optimal or flagellar expression results are counterproductive, since flagellin triggers host immune responses (94) with FleQ as the regulator that coordinates the switching between flagellum-dependent and flagellum-independent, syringafactin-dependent motilities.
Supplementary Material
ACKNOWLEDGMENTS
We thank S. Muñoz and A. Felipe for their technical assistance. We also thank Michael G. Thomas for the syfA mutant and Sheng Yang He for fliC mutants, Reuben Ramphal for the anti-FliA antibody, and Harold Prada for the pBBR1-MCS5::fleQ plasmid.
This research was supported by grants BFU2008-00086 and BIO2011-23032 from the Spanish Ministerio de Ciencia e Innovación/Ministerio de Economía y Competitividad and P08-CVI-3475 and P10-CVI-5800 from the Junta de Andalucía, all of them cofinanced by the European Regional Development Fund. P.V. was supported by a JAE-Pre fellowship from the CSIC, also cofinanced by the European Regional Development Fund. G.A.F. was supported by a Ciência sem Fronteiras fellowship (BEX10043/13-6) from the CAPES-Brazil.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01798-15.
REFERENCES
- 1.Verstraeten N, Braeken K, Debkumari B, Fauvart M, Fransaer J, Vermant J, Michiels J. 2008. Living on a surface: swarming and biofilm formation. Trends Microbiol 16:496–506. doi: 10.1016/j.tim.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 2.Harshey RM. 2003. Bacterial motility on a surface: many ways to a common goal. Annu Rev Microbiol 57:249–273. doi: 10.1146/annurev.micro.57.030502.091014. [DOI] [PubMed] [Google Scholar]
- 3.Henrichsen J. 1972. Bacterial surface translocation: a survey and a classification. Bacteriol Rev 36:478–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kearns DB. 2010. A field guide to bacterial swarming motility. Nat Rev Microbiol 8:634–644. doi: 10.1038/nrmicro2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mattick JS. 2002. Type IV pili and twitching motility. Annu Rev Microbiol 56:289–314. doi: 10.1146/annurev.micro.56.012302.160938. [DOI] [PubMed] [Google Scholar]
- 6.Mignot T, Shaevitz JW, Hartzell PL, Zusman DR. 2007. Evidence that focal adhesion complexes power bacterial gliding motility. Science 315:853–856. doi: 10.1126/science.1137223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Matsuyama T, Bhasin A, Harshey RM. 1995. Mutational analysis of flagellum-independent surface spreading of Serratia marcescens 274 on a low-agar medium. J Bacteriol 177:987–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murray TS, Kazmierczak BI. 2008. Pseudomonas aeruginosa exhibits sliding motility in the absence of type IV pili and flagella. J Bacteriol 190:2700–2708. doi: 10.1128/JB.01620-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Agustí G, Astola O, Rodríguez-Güell E, Julián E, Luquin M. 2008. Surface spreading motility shown by a group of phylogenetically related, rapidly growing pigmented mycobacteria suggests that motility is a common property of mycobacterial species but is restricted to smooth colonies. J Bacteriol 190:6894–6902. doi: 10.1128/JB.00572-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown II, Hase CC. 2001. Flagellum-independent surface migration of Vibrio cholerae and Escherichia coli. J Bacteriol 183:3784–3790. doi: 10.1128/JB.183.12.3784-3790.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fall R, Kearns DB, Nguyen T. 2006. A defined medium to investigate sliding motility in a Bacillus subtilis flagella-less mutant. BMC Microbiol 6:31. doi: 10.1186/1471-2180-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stewart CR, Rossier O, Cianciotto NP. 2009. Surface translocation by Legionella pneumophila: a form of sliding motility that is dependent upon type II protein secretion. J Bacteriol 191:1537–1546. doi: 10.1128/JB.01531-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alsohim AS, Taylor TB, Barrett GA, Gallie J, Zhang XX, Altamirano-Junqueira AE, Johnson LJ, Rainey PB, Jackson RW. 2014. The biosurfactant viscosin produced by Pseudomonas fluorescens SBW25 aids spreading motility and plant growth promotion. Environ Microbiol 16:2267–2281. doi: 10.1111/1462-2920.12469. [DOI] [PubMed] [Google Scholar]
- 14.Matsuyama T, Kaneda K, Nakagawa Y, Isa K, Hara-Hotta H, Yano I. 1992. A novel extracellular cyclic lipopeptide which promotes flagellum-dependent and -independent spreading growth of Serratia marcescens. J Bacteriol 174:1769–1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bais HP, Fall R, Vivanco JM. 2004. Biocontrol of Bacillus subtilis against infection of Arabidopsis roots by Pseudomonas syringae is facilitated by biofilm formation and surfactin production. Plant Physiol 134:307–319. doi: 10.1104/pp.103.028712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuiper I, Lagendijk EL, Pickford R, Derrick JP, Lamers GE, Thomas-Oates JE, Lugtenberg BJ, Bloemberg GV. 2004. Characterization of two Pseudomonas putida lipopeptide biosurfactants, putisolvin I and II, which inhibit biofilm formation and break down existing biofilms. Mol Microbiol 51:97–113. [DOI] [PubMed] [Google Scholar]
- 17.Raaijmakers JM, de Nybroe BIO, Ongena M. 2010. Natural functions of lipopeptides from Bacillus and Pseudomonas: more than surfactants and antibiotics. FEMS Microbiol Rev 34:1037–1062. doi: 10.1111/j.1574-6976.2010.00221.x. [DOI] [PubMed] [Google Scholar]
- 18.Capdevila S, Martinez-Granero FM, Sanchez-Contreras M, Rivilla R, Martin M. 2004. Analysis of Pseudomonas fluorescens F113 genes implicated in flagellar filament synthesis and their role in competitive root colonization. Microbiology 150:3889–3897. doi: 10.1099/mic.0.27362-0. [DOI] [PubMed] [Google Scholar]
- 19.Martínez-Granero F, Rivilla R, Martín M. 2006. Rhizosphere selection of highly motile phenotypic variants of Pseudomonas fluorescens with enhanced competitive colonization ability. Appl Environ Microbiol 72:3429–3434. doi: 10.1128/AEM.72.5.3429-3434.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Weger LA, van der Vlugt CI, Wijfjes AH, Bakker PA, Schippers B, Lugtenberg B. 1987. Flagella of a plant-growth-stimulating Pseudomonas fluorescens strain are required for colonization of potato roots. J Bacteriol 169:2769–2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Weert S, Vermeiren H, Mulders IH, Kuiper I, Hendrickx N, Bloemberg GV, Vanderleyden J, De MR, Lugtenberg BJ. 2002. Flagella-driven chemotaxis towards exudate components is an important trait for tomato root colonization by Pseudomonas fluorescens. Mol Plant Microbe Interact 15:1173–1180. doi: 10.1094/MPMI.2002.15.11.1173. [DOI] [PubMed] [Google Scholar]
- 22.Toutain CM, Caizza NC, Zegans ME, O'Toole GA. 2007. Roles for flagellar stators in biofilm formation by Pseudomonas aeruginosa. Res Microbiol 158:471–477. doi: 10.1016/j.resmic.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 23.O'Toole GA, Kolter R. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol Microbiol 30:295–304. doi: 10.1046/j.1365-2958.1998.01062.x. [DOI] [PubMed] [Google Scholar]
- 24.Choy WK, Zhou L, Syn CK, Zhang LH, Swarup S. 2004. MorA defines a new class of regulators affecting flagellar development and biofilm formation in diverse Pseudomonas species. J Bacteriol 186:7221–7228. doi: 10.1128/JB.186.21.7221-7228.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Czelleng A, Bozso Z, Ott PG, Besenyei E, Varga GJ, Szatmari A, Kiraly L, Klement Z. 2006. Identification of virulence-associated genes of Pseudomonas viridiflava activated during infection by use of a novel IVET promoter probing plasmid. Curr Microbiol 52:282–286. doi: 10.1007/s00284-005-0208-6. [DOI] [PubMed] [Google Scholar]
- 26.Quiñones B, Dulla G, Lindow SE. 2005. Quorum sensing regulates exopolysaccharide production, motility, and virulence in Pseudomonas syringae. Mol Plant Microbe Interact 18:682–693. doi: 10.1094/MPMI-18-0682. [DOI] [PubMed] [Google Scholar]
- 27.Arora SK, Ritchings BW, Almira EC, Lory S, Ramphal R. 1997. A transcriptional activator, FleQ, regulates mucin adhesion and flagellar gene expression in Pseudomonas aeruginosa in a cascade manner. J Bacteriol 179:5574–5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marshall B, Robleto EA, Wetzler R, Kulle P, Casaz P, Levy SB. 2001. The adnA transcriptional factor affects persistence and spread of Pseudomonas fluorescens under natural field conditions. Appl Environ Microbiol 67:852–857. doi: 10.1128/AEM.67.2.852-857.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Casaz P, Happel A, Keithan J, Read DL, Strain SR, Levy SB. 2001. The Pseudomonas fluorescens transcription activator AdnA is required for adhesion and motility. Microbiology 147:355–361. [DOI] [PubMed] [Google Scholar]
- 30.Taguchi F, Takeuchi K, Katoh E, Murata K, Suzuki T, Marutani M, Kawasaki T, Eguchi M, Katoh S, Kaku H, Yasuda C, Inagaki Y, Toyoda K, Shiraishi T, Ichinose Y. 2006. Identification of glycosylation genes and glycosylated amino acids of flagellin in Pseudomonas syringae pv. tabaci. Cell Microbiol 8:923–938. doi: 10.1111/j.1462-5822.2005.00674.x. [DOI] [PubMed] [Google Scholar]
- 31.Haefele DM, Lindow SE. 1987. Flagellar motility confers epiphytic fitness advantages upon Pseudomonas syringae. Appl Environ Microbiol 53:2528–2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schreiber KJ, Desveaux D. 2011. AlgW regulates multiple Pseudomonas syringae virulence strategies. Mol Microbiol 80:364–377. doi: 10.1111/j.1365-2958.2011.07571.x. [DOI] [PubMed] [Google Scholar]
- 33.Taguchi F, Yamamoto M, Ohnishi-Kameyama M, Iwaki M, Yoshida M, Ishii T, Konishi T, Ichinose Y. 2010. Defects in flagellin glycosylation affect the virulence of Pseudomonas syringae pv. tabaci 6605. Microbiology 156:72–80. doi: 10.1099/mic.0.030700-0. [DOI] [PubMed] [Google Scholar]
- 34.Dasgupta N, Wolfgang MC, Goodman AL, Arora SK, Jyot J, Lory S, Ramphal R. 2003. A four-tiered transcriptional regulatory circuit controls flagellar biogenesis in Pseudomonas aeruginosa. Mol Microbiol 50:809–824. doi: 10.1046/j.1365-2958.2003.03740.x. [DOI] [PubMed] [Google Scholar]
- 35.Smith TG, Hoover TR. 2009. Deciphering bacterial flagellar gene regulatory networks in the genomic era. Adv Appl Microbiol 67:257–295. doi: 10.1016/S0065-2164(08)01008-3. [DOI] [PubMed] [Google Scholar]
- 36.Jyot J, Dasgupta N, Ramphal R. 2002. FleQ, the major flagellar gene regulator in Pseudomonas aeruginosa, binds to enhancer sites located either upstream or atypically downstream of the RpoN binding site. J Bacteriol 184:5251–5260. doi: 10.1128/JB.184.19.5251-5260.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ritchings BW, Almira EC, Lory S, Ramphal R. 1995. Cloning and phenotypic characterization of fleS and fleR, new response regulators of Pseudomonas aeruginosa which regulate motility and adhesion to mucin. Infect Immun 63:4868–4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Frisk A, Jyot J, Arora SK, Ramphal R. 2002. Identification and functional characterization of flgM, a gene encoding the anti-sigma 28 factor in Pseudomonas aeruginosa. J Bacteriol 184:1514–1521. doi: 10.1128/JB.184.6.1514-1521.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Deflaun MF, Marshall BM, Kulle EP, Levy SB. 1994. Tn5 insertion mutants of Pseudomonas fluorescens defective in adhesion to soil and seeds. Appl Environ Microbiol 60:2637–2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mastropaolo MD, Silby MW, Nicoll JS, Levy SB. 2012. Novel genes involved in Pseudomonas fluorescens Pf0-1 motility and biofilm formation. Appl Environ Microbiol 78:4318–4329. doi: 10.1128/AEM.07201-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Robleto EA, Lopez-Hernandez I, Silby MW, Levy SB. 2003. Genetic analysis of the AdnA regulon in Pseudomonas fluorescens: nonessential role of flagella in adhesion to sand and biofilm formation. J Bacteriol 185:453–460. doi: 10.1128/JB.185.2.453-460.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baraquet C, Murakami K, Parsek MR, Harwood CS. 2012. The FleQ protein from Pseudomonas aeruginosa functions as both a repressor and an activator to control gene expression from the pel operon promoter in response to c-di-GMP. Nucleic Acids Res 40:7207–7218. doi: 10.1093/nar/gks384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hickman JW, Harwood CS. 2008. Identification of FleQ from Pseudomonas aeruginosa as a c-di-GMP-responsive transcription factor. Mol Microbiol 69:376–389. doi: 10.1111/j.1365-2958.2008.06281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Giddens SR, Jackson RW, Moon CD, Jacobs MA, Zhang XX, Gehrig SM, Rainey PB. 2007. Mutational activation of niche-specific genes provides insight into regulatory networks and bacterial function in a complex environment. Proc Natl Acad Sci U S A 104:18247–18252. doi: 10.1073/pnas.0706739104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baraquet C, Harwood CS. 2013. Cyclic diguanosine monophosphate represses bacterial flagella synthesis by interacting with the Walker A motif of the enhancer-binding protein FleQ. Proc Natl Acad Sci U S A 110:18478–18483. doi: 10.1073/pnas.1318972110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vargas P, Farias GA, Nogales J, Prada H, Carvajal V, Barón M, Rivilla R, Martín M, Olmedilla A, Gallegos MT. 2013. Plant flavonoids target Pseudomonas syringae pv. tomato DC3000 flagella and type III secretion system. Environ Microbiol Rep 5:841–850. doi: 10.1111/1758-2229.12086. [DOI] [PubMed] [Google Scholar]
- 47.Roine E, Wei W, Yuan J, Nurmiaho-Lassila EL, Kalkkinen N, Romantschuk M, He SY. 1997. Hrp pilus: an hrp-dependent bacterial surface appendage produced by Pseudomonas syringae pv. tomato DC3000. Proc Natl Acad Sci U S A 94:3459–3464. doi: 10.1073/pnas.94.7.3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Berti AD, Greve NJ, Christensen QH, Thomas MG. 2007. Identification of a biosynthetic gene cluster and the six associated lipopeptides involved in swarming motility of Pseudomonas syringae pv. tomato DC3000. J Bacteriol 189:6312–6323. doi: 10.1128/JB.00725-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Burch AY, Shimada BK, Browne PJ, Lindow SE. 2010. Novel high-throughput detection method to assess bacterial surfactant production. Appl Environ Microbiol 76:5363–5372. doi: 10.1128/AEM.00592-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Burch AY, Zeisler V, Yokota K, Schreiber L, Lindow SE. 2014. The hygroscopic biosurfactant syringafactin produced by Pseudomonas syringae enhances fitness on leaf surfaces during fluctuating humidity. Environ Microbiol 16:2086–2098. doi: 10.1111/1462-2920.12437. [DOI] [PubMed] [Google Scholar]
- 51.Hu W, Yuan J, Jin QL, Hart P, He SY. 2001. Immunogold labeling of Hrp pili of Pseudomonas syringae pv. tomato assembled in minimal medium and in planta. Mol Plant Microbe Interact 14:234–241. doi: 10.1094/MPMI.2001.14.2.234. [DOI] [PubMed] [Google Scholar]
- 52.Cuppels DA. 1986. Generation and characterization of Tn5 insertion mutations in Pseudomonas syringae pv. tomato. Appl Environ Microbiol 51:323–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roine E, Raineri DM, Romantschuk M, Wilson M, Nunn DN. 1998. Characterization of type IV pilus genes in Pseudomonas syringae pv. tomato DC3000. Mol Plant Microbe Interact 11:1048–1056. doi: 10.1094/MPMI.1998.11.11.1048. [DOI] [PubMed] [Google Scholar]
- 54.Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM, Peterson KM. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
- 55.Prada H. 2014. Papel del segundo mensajero c-di-GMP en Pseudomonas syringae pv. tomato DC3000. Ph.D. thesis University of Granada, Granada, Spain. [Google Scholar]
- 56.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 57.Choi KH, Kumar A, Schweizer HP. 2006. A 10-min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells: application for DNA fragment transfer between chromosomes and plasmid transformation. J Microbiol Methods 64:391–397. doi: 10.1016/j.mimet.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 58.Abràmoff MD, Magalhães PJ, Ram SJ. 2004. Image processing with ImageJ. Biophotonics Int 11:36–42. [Google Scholar]
- 59.Clark WA. 1976. A simplified Leifson flagella stain. J Clin Microbiol 3:632–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Robertsen BK, Aman P, Darvill AG, McNeil M, Albersheim P. 1981. Host-symbiont interactions. V. The structure of acidic extracellular polysaccaharides secreted by Rhizobium leguminosarum and Rhizobium trifolii. Plant Physiol 67:389–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vargas P, Felipe A, Michan C, Gallegos MT. 2011. Induction of Pseudomonas syringae pv. tomato DC3000 MexAB-OprM multidrug efflux pump by flavonoids is mediated by the repressor PmeR. Mol Plant Microbe Interact 24:1207–1219. doi: 10.1094/MPMI-03-11-0077. [DOI] [PubMed] [Google Scholar]
- 62.Michán C, Pueyo C. 2009. Growth phase-dependent variations in transcript profiles for thioredoxin- and glutathione-dependent redox systems followed by budding and hyphal Candida albicans cultures. FEMS Yeast Res 9:1078–1090. doi: 10.1111/j.1567-1364.2009.00558.x. [DOI] [PubMed] [Google Scholar]
- 63.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 64.Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jyot J, Sonawane A, Wu W, Ramphal R. 2007. Genetic mechanisms involved in the repression of flagellar assembly by Pseudomonas aeruginosa in human mucus. Mol Microbiol 63:1026–1038. doi: 10.1111/j.1365-2958.2006.05573.x. [DOI] [PubMed] [Google Scholar]
- 67.Rashid MH, Kornberg A. 2000. Inorganic polyphosphate is needed for swimming, swarming, and twitching motilities of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 97:4885–4890. doi: 10.1073/pnas.060030097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shrout JD, Chopp DL, Just CL, Hentzer M, Givskov M, Parsek MR. 2006. The impact of quorum sensing and swarming motility on Pseudomonas aeruginosa biofilm formation is nutritionally conditional. Mol Microbiol 62:1264–1277. doi: 10.1111/j.1365-2958.2006.05421.x. [DOI] [PubMed] [Google Scholar]
- 69.Filiatrault MJ, Stodghill PV, Bronstein PA, Moll S, Lindeberg M, Grills G, Schweitzer P, Wang W, Schroth GP, Luo S, Khrebtukova I, Yang Y, Thannhauser T, Butcher BG, Cartinhour S, Schneider DJ. 2010. Transcriptome analysis of Pseudomonas syringae identifies new genes, noncoding RNAs, and antisense activity. J Bacteriol 192:2359–2372. doi: 10.1128/JB.01445-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Filiatrault MJ, Stodghill PV, Myers CR, Bronstein PA, Butcher BG, Lam H, Grills G, Schweitzer P, Wang W, Schneider DJ, Cartinhour SW. 2011. Genome-wide identification of transcriptional start sites in the plant pathogen Pseudomonas syringae pv. tomato str. DC3000. PLoS One 6:e29335. doi: 10.1371/journal.pone.0029335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Redondo-Nieto M, Lloret J, Larenas J, Barahona E, Navazo A, Martinez-Granero F, Capdevila S, Rivilla R, Martin M. 2008. Transcriptional organization of the region encoding the synthesis of the flagellar filament in Pseudomonas fluorescens. J Bacteriol 190:4106–4109. doi: 10.1128/JB.00178-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Burch AY, Shimada BK, Mullin SW, Dunlap CA, Bowman MJ, Lindow SE. 2012. Pseudomonas syringae coordinates production of a motility-enabling surfactant with flagellar assembly. J Bacteriol 194:1287–1298. doi: 10.1128/JB.06058-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nogales J, Bernabeu-Roda L, Cuellar V, Soto MJ. 2012. ExpR is not required for swarming but promotes sliding in Sinorhizobium meliloti. J Bacteriol 194:2027–2035. doi: 10.1128/JB.06524-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kinsinger RF, Shirk MC, Fall R. 2003. Rapid surface motility in Bacillus subtilis is dependent on extracellular surfactin and potassium ion. J Bacteriol 185:5627–5631. doi: 10.1128/JB.185.18.5627-5631.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Barembruch C, Hengge R. 2007. Cellular levels and activity of the flagellar sigma factor FliA of Escherichia coli are controlled by FlgM-modulated proteolysis. Mol Microbiol 65:76–89. doi: 10.1111/j.1365-2958.2007.05770.x. [DOI] [PubMed] [Google Scholar]
- 76.Ghelardi E, Celandroni F, Salvetti S, Beecher DJ, Gominet M, Lereclus D, Wong AC, Senesi S. 2002. Requirement of flhA for swarming differentiation, flagellin export, and secretion of virulence-associated proteins in Bacillus thuringiensis. J Bacteriol 184:6424–6433. doi: 10.1128/JB.184.23.6424-6433.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yang TC, Leu YW, Chang-Chien HC, Hu RM. 2009. Flagellar biogenesis of Xanthomonas campestris requires the alternative sigma factors RpoN2 and FliA and is temporally regulated by FlhA, FlhB, and FlgM. J Bacteriol 191:2266–2275. doi: 10.1128/JB.01152-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Young BM, Young GM. 2002. YplA is exported by the Ysc, Ysa, and flagellar type III secretion systems of Yersinia enterocolitica. J Bacteriol 184:1324–1334. doi: 10.1128/JB.184.5.1324-1334.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Balaban M, Hendrixson DR. 2011. Polar flagellar biosynthesis and a regulator of flagellar number influence spatial parameters of cell division in Campylobacter jejuni. PLoS Pathog 7:e1002420. doi: 10.1371/journal.ppat.1002420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Arora SK, Neely AN, Blair B, Lory S, Ramphal R. 2005. Role of motility and flagellin glycosylation in the pathogenesis of Pseudomonas aeruginosa burn wound infections. Infect Immun 73:4395–4398. doi: 10.1128/IAI.73.7.4395-4398.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nachamkin I, Yang XH, Stern NJ. 1993. Role of Campylobacter jejuni flagella as colonization factors for three-day-old chicks: analysis with flagellar mutants. Appl Environ Microbiol 59:1269–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Marchetti M, Sirard JC, Sansonetti P, Pringault E, Kerneis S. 2004. Interaction of pathogenic bacteria with rabbit appendix M cells: bacterial motility is a key feature in vivo. Microbes Infect 6:521–528. doi: 10.1016/j.micinf.2004.02.009. [DOI] [PubMed] [Google Scholar]
- 83.Turnbull GA, Morgan JA, Whipps JM, Saunders JR. 2001. The role of bacterial motility in the survival and spread of Pseudomonas fluorescens in soil and in the attachment and colonisation of wheat roots. FEMS Microbiol Ecol 36:21–31. doi: 10.1111/j.1574-6941.2001.tb00822.x. [DOI] [PubMed] [Google Scholar]
- 84.Bowers JH, Parke JL. 1993. Colonization of pea (Pisum sativum L.) taproots by Pseudomonas fluorescens: effect of soil temperature and bacterial motility. Soil Biol Biochem 25:1693–1701. doi: 10.1016/0038-0717(93)90172-8. [DOI] [Google Scholar]
- 85.Cooley MB, Miller WG, Mandrell RE. 2003. Colonization of Arabidopsis thaliana with Salmonella enterica and enterohemorrhagic Escherichia coli O157:H7 and competition by Enterobacter asburiae. Appl Environ Microbiol 69:4915–4926. doi: 10.1128/AEM.69.8.4915-4926.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gorski L, Duhe JM, Flaherty D. 2009. The use of flagella and motility for plant colonization and fitness by different strains of the foodborne pathogen Listeria monocytogenes. PLoS One 4:e5142. doi: 10.1371/journal.pone.0005142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Clarke CR, Chinchilla D, Hind SR, Taguchi F, Miki R, Ichinose Y, Martin GB, Leman S, Felix G, Vinatzer BA. 2013. Allelic variation in two distinct Pseudomonas syringae flagellin epitopes modulates the strength of plant immune responses but not bacterial motility. New Phytol 200:847–860. doi: 10.1111/nph.12408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Feldman M, Bryan R, Rajan S, Scheffler L, Brunnert S, Tang H, Prince A. 1998. Role of flagella in pathogenesis of Pseudomonas aeruginosa pulmonary infection. Infect Immun 66:43–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Naito K, Taguchi F, Suzuki T, Inagaki Y, Toyoda K, Shiraishi T, Ichinose Y. 2008. Amino acid sequence of bacterial microbe-associated molecular pattern flg22 is required for virulence. Mol Plant Microbe Interact 21:1165–1174. doi: 10.1094/MPMI-21-9-1165. [DOI] [PubMed] [Google Scholar]
- 90.Tremblay J, Richardson AP, Lepine F, Deziel E. 2007. Self-produced extracellular stimuli modulate the Pseudomonas aeruginosa swarming motility behaviour. Environ Microbiol 9:2622–2630. doi: 10.1111/j.1462-2920.2007.01396.x. [DOI] [PubMed] [Google Scholar]
- 91.Pauwelyn E, Huang CJ, Ongena M, Leclere V, Jacques P, Bleyaert P, Budzikiewicz H, Schafer M, Hofte M. 2013. New linear lipopeptides produced by Pseudomonas cichorii SF1-54 are involved in virulence, swarming motility, and biofilm formation. Mol Plant Microbe Interact 26:585–598. doi: 10.1094/MPMI-11-12-0258-R. [DOI] [PubMed] [Google Scholar]
- 92.Gage DJ, Margolin W. 2000. Hanging by a thread: invasion of legume plants by rhizobia. Curr Opin Microbiol 3:613–617. doi: 10.1016/S1369-5274(00)00149-1. [DOI] [PubMed] [Google Scholar]
- 93.Fournier J, Timmers AC, Sieberer BJ, Jauneau A, Chabaud M, Barker DG. 2008. Mechanism of infection thread elongation in root hairs of Medicago truncatula and dynamic interplay with associated rhizobial colonization. Plant Physiol 148:1985–1995. doi: 10.1104/pp.108.125674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zeng W, He SY. 2010. A prominent role of the flagellin receptor FLAGELLIN-SENSING2 in mediating stomatal response to Pseudomonas syringae pv tomato DC3000 in Arabidopsis. Plant Physiol 153:1188–1198. doi: 10.1104/pp.110.157016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.