Abstract
Microbial sulfide oxidation in aquatic environments is an important ecosystem process, as sulfide is potently toxic to aerobic organisms. Sulfide oxidation in anoxic waters can prevent the efflux of sulfide to aerobic water masses, thus mitigating toxicity. The contribution of phototrophic sulfide-oxidizing bacteria to anaerobic sulfide oxidation in the Chesapeake Bay and the redox chemistry of the stratified water column were investigated in the summers of 2011 to 2014. In 2011 and 2013, phototrophic sulfide-oxidizing bacteria closely related to Prosthecochloris species of the phylum Chlorobi were cultivated from waters sampled at and below the oxic-anoxic interface, where measured light penetration was sufficient to support populations of low-light-adapted photosynthetic bacteria. In 2012, 2013, and 2014, light-dependent sulfide loss was observed in freshly collected water column samples. In these samples, extremely low light levels caused 2- to 10-fold increases in the sulfide uptake rate over the sulfide uptake rate under dark conditions. An enrichment, CB11, dominated by Prosthecochloris species, oxidized sulfide with a Ks value of 11 μM and a Vmax value of 51 μM min−1 (mg protein−1). Using these kinetic values with in situ sulfide concentrations and light fluxes, we calculated that a small population of Chlorobi similar to those in enrichment CB11 can account for the observed anaerobic light-dependent sulfide consumption activity in natural water samples. We conclude that Chlorobi play a far larger role in the Chesapeake Bay than currently appreciated. This result has potential implications for coastal anoxic waters and expanding oxygen-minimum zones as they begin to impinge on the photic zone.
INTRODUCTION
The oxidation of sulfide by oxygen is thermodynamically unfavorable for a one-electron transfer and kinetically inhibited for a two-electron transfer (1). Kinetic limitations on sulfide oxidation can be overcome by catalysts, either abiotic (e.g., transition metals [2]) or biotic (e.g., sulfide:quinone oxidoreductase in various microbes [3]). In many environments, particularly those with low oxygen levels and/or limited free transition metals, sulfide oxidation is expected to be microbially mediated (4). Phototrophic sulfide-oxidizing bacteria (PSOB) have been found in a wide array of aqueous environments in which both light and sulfide are present, including sediments (5), hot springs (6), freshwater lakes (7–10), and marine basins (11). In the water column, these bacteria are typically found at depths of between 2 and 20 m from the surface and at light transmittance values from 10% to 0.015% of surface irradiance (12). Biological rates of sulfide oxidation can be orders of magnitude higher than rates of abiotic sulfide oxidation (4); thus, the presence of PSOB has the potential to significantly impact the sulfur cycle and redox chemistry in the environments in which they occur. The degree to which PSOB influence their environment depends upon environmental factors such as light availability, the sulfide concentration, and the rates of abiotic sulfide oxidation processes (13).
Investigations of the presence and role of phototrophic sulfur bacteria in anoxic environments are important for understanding both modern and ancient systems. In modern systems, the development of sulfidic waters can cause stress or toxicity to aerobic organisms such as fish (14, 15). Currently, the volume of low-oxygen waters in oxygen-minimum zones (OMZs) and coastal hypoxic zones is increasing due to a changing global climate (16), and many of these areas are completely anoxic for part of the year. Sulfide has been directly detected in several OMZs (17, 18), and evidence for the cryptic cycling of sulfur with simultaneous sulfate reduction and sulfide oxidation has also been observed (19). As these areas grow, the depth of anoxic water is expected to shoal, moving toward the photic zone (20) and creating potential new niches for PSOB, which could play an important role in mitigating sulfide fluxes. In ancient oceans, PSOB were likely important biogeochemical mediators during the transition of ocean chemistry from anoxic, with periodic widespread sulfidic, conditions to fully oxic conditions (see reference 21 and references therein). Environments that currently experience periodic anoxia and euxinia provide an analogue to the ancient oceans in which to study the effects of changing redox conditions on the chemistry and microbiology of a system, lending insight into the biogeochemical evolution of modern oceans. Thus, further understanding of the environmental niches occupied by PSOB and their impact on sulfide oxidation has widespread implications.
The Chesapeake Bay is a partially stratified estuary extending from the mouth of the Susquehanna River to the Atlantic Ocean (22). Water transport is a two-layer gravitational circulation where saline water flows up-estuary and freshwater flows down-estuary (23). Salinity and temperature gradients, coupled with an influx of nutrients and subsequent biological productivity and decomposition, lead to the development of suboxic-to-anoxic deep water in the summer. A suboxic zone between the oxic surface and anoxic deep layers in which neither oxygen nor sulfide is detectable may form at the interface created by density stratification. In addition to annual and interannual variation in oxygen concentrations, shorter-term variations occur due to tidal forcing and wind-induced mixing of the water column (24), which result in changes in the depth and extent of the interface. The seasonal development of anoxic and sulfidic waters in the Chesapeake Bay has been correlated with shifts in the bacterial community structure (25), and functional gene surveys indicate that the lower-water-column microbial community switches to anaerobic respiration during anoxia, potentially exacerbating the production of sulfide. It is not clear, however, whether there are significant microbial contributions to sulfide oxidation in this system.
In this study, we present field results that indicate that PSOB are a consistent, recurring, and active component of the Chesapeake Bay sulfur cycle. In addition, laboratory experiments with enrichments of PSOB obtained from the Chesapeake Bay allowed determination of kinetic parameters with respect to light and sulfide concentrations. These parameters allow a preliminary investigation into the potential contribution of PSOB to sulfide oxidation in the Chesapeake Bay given observed sulfide concentrations and light profiles in this system.
MATERIALS AND METHODS
Sample collection.
Field studies took place at Station 858 (38°58.8′N, 76°22′E) in the upper Chesapeake Bay south of the Bay Bridge. This site is a hole off the main channel in the mesohaline portion of the bay that is ∼25 m deep and is one of the first sites to stratify in the spring (24). Fieldwork was conducted from 27 to 30 July 2011, 17 to 19 August 2012, 9 to 13 August 2013, and 18 to 22 August 2014 aboard the R/V Hugh R. Sharp. Photosynthetically active radiation (PAR) measurements were made in the water column by using the PAR sensor of an in situ FIRe (fluorescence induction and relaxation) sensor (26). This sensor has a detection limit of 0.034 μmol photons m−2 s−1 and a spectral range of 400 to 700 nm. In 2012 and 2013, all samples were obtained from Niskin bottles on the ship's conductivity, temperature, and depth (CTD) rosette and were collected in cleaned Nalgene bottles on the ship. In 2011 and 2014, samples were taken from either Niskin bottles or water pumped to the surface by a Rule Industries 1,000-gal/h pump. Care was taken during sampling to avoid aeration of the samples by flushing the sample bottle three times and capping the bottle with no headspace immediately after sample collection. Sample processing was rapid (<10 min) in order to preserve redox speciation in the samples.
Measurements of oxygen and sulfide concentrations.
Measurements of oxygen and sulfide concentrations in the water column were made by using in situ voltammetry. The voltammetric system used a 100-μm gold amalgam working electrode, a Ag/AgCl reference electrode, and a platinum counterelectrode. In 2011, this system was interfaced with a DLK-SUB-II electrochemical analyzer attached to a metal cage that was suspended over the side of the ship. In 2012 to 2014, a laboratory DLK 100A potentiostat was used as water was pumped with a pump profiler and measurements were made onboard the ship using a flowthrough cell (26), which has been demonstrated to preserve in situ speciation (27). Voltage was scanned from −0.1 V to −1.8 V at a scan rate of 2,000 mV/s. Using this method, the detection limits are 3 μM for oxygen and 0.2 μM for sulfide and polysulfide.
Bacterial enrichments.
Samples of phototrophic sulfide-oxidizing bacteria were taken within and below the suboxic zone. One milliliter of sample water was injected into a 20-ml septum vial containing anoxic SL-10 medium specialized for PSOB that contained 17.9 mM HCO3− as the carbon source and 2.5 mM H2S as the sole electron donor (28). The vials were incubated under ambient conditions for the duration of the cruise. Upon return to land, the vials were incubated at 20°C to 22°C at a PAR flux value of 5 μmol m−2 s−1 and were subcultured in fresh medium when growth was visibly apparent. This produced a number of stably transferable enrichments that were all similar in appearance (data not shown). One of these, enrichment CB11, was selected and used for subsequent sulfide oxidation experiments.
Identification of phototrophic bacteria in CB11.
Whole-cell absorption spectra (400 to 1,100 nm) for CB11 and pure cultures of related strains were collected on samples taken from actively growing cultures by using a syringe and transferred to a plastic cuvette. Spectra were recorded on a DU730 spectrophotometer within 1 min of removal from the culture.
For phylogenetic analysis, cells were collected from 1 ml of the enrichment culture by centrifugation at 16,400 × g for 2 min. Cells were resuspended in lysis buffer containing 1% (vol/vol) Triton X-100, 20 mM Tris-HCl, and 0.1 mM EDTA at pH 8.0. Cells were lysed by heating the mixture to 95°C for 5 min. After centrifugation, 2 μl of the supernatant was used in a 20-μl PCR mixture with 0.5 mM each primer 27f (5′-GAGTTTGATYHTGGCTCAG-3′) and 1492r (5′-GGTTACCTTGTTACGACTT-3′) PCR products were inserted into pCR2.1-TOPO after ExoSAP IT treatment, and clones were recovered by electroporation into Escherichia coli strain EC100D. Positive clones were identified by colony PCR with primers M13r (5′-AACAGCTATGACCATG-3′) and T7P (5′-TAATACGACTCACTATAGGG-3′). PCR products of the appropriate size were sent for Sanger sequencing with M13r and T7P after ExoSAP-IT treatment at the University of Delaware Sequencing and Genotyping Center. Insert sequences from three clones were assembled into a contig by using CLC Workbench v.7 (Qiagen Inc.), which was aligned to 73 Chlorobi 16S rRNA sequences and those of Ignavibacterium album JCM 16511 and Melioribacter roseum P3M as outgroups by using MUSCLE in MEGA6 (29). The resulting alignment was used to construct a phylogenetic tree created by maximum likelihood analysis using the Kimura 2-parameter model, allowing for both invariant sites and a gamma distribution with 5 rate categories for variable-rate sites.
Automated ribosomal intergenic spacer analysis (ARISA) (30) was used to compare enrichments obtained from Chesapeake Bay samples. Samples were prepared for PCR as described above, but reaction mixtures contained 0.5 μM each primer ITSF (5′-GTCGTAACAAGGTAGCCGTA-3′) and ITSReub (5′-GCCAAGGCATCCACC-3′, with a 5′-NED fluorophore [Applied Biosystems]). PCR products were mixed with a size standard (Liz-1000) and analyzed on a 3130XL genetic analyzer at the University of Delaware Sequencing and Genotyping Center. Electropherograms were analyzed with the PeakStudio software package (http://fodorlab.uncc.edu/software/peakstudio) to calculate fragment sizes and peak areas. Chlorobaculum tepidum TLS, Chlorobium luteolum DSM 273, Chlorobium limicola DSM 245, and Prosthecochloris aestuarii DSM 271 were analyzed as controls, and they all produced ARISA peaks that matched those predicted from their complete genome sequences (data not shown).
Preparation of enrichments for laboratory experiments.
CB11 bacteria were grown in pressurized 100-ml septum vials in a water bath set at 25°C for 48 h under 20 μmol m−2 s−1 PAR flux from an incandescent bulb with a color temperature of 3,000 K. At the end of the growth period, cells were washed three times with anoxic HEPES buffer (0.1 M; pH 7.4) to remove salts and then stored in anoxic HEPES buffer (3 ml) in a sealed 20-ml septum vial to be used in experiments the same day. Anoxic conditions were maintained during transfer of the cells by using a glove bag purged with ultra-high-purity argon. Protein measurements were made by using the Bradford assay (31).
Analytical methods for sulfide loss experiments.
In situ voltammetry with a solid-state 100-μm gold amalgam electrode was used to measure sulfide concentrations throughout the experiments (4, 21). The methods for construction and calibration of the electrodes were outlined previously by Luther et al. (32). Experiments were conducted in a thermostated electrochemical cell that contained ports for the working electrode, the Ag/AgCl reference electrode, and a platinum counterelectrode, which were controlled by using an Analytical Instrument Systems (AIS) DLK-60 potentiostat. High-purity argon gas in the cell headspace was used for maintenance of anoxic conditions during the experiments. Voltammetric scans of solutions were run from −0.1 to −1.8 V with two conditioning steps: one at −0.9 V for 5 s, which prevents sulfide from accumulating onto the electrode surface, and one at −0.1 V for 2 s.
Procedures for shipboard kinetic experiments.
In 2012, 2013, and 2014, kinetic experiments modeled after those described previously by Luther et al. (33) were conducted shipboard by using anoxic bottom water. Water samples were used unfiltered or after being filtered through either a 0.2-μm or a 0.05-μm Nuclepore filter. Experiments were set up in a glove bag purged with ultra-high-purity argon gas. In 2012, as no sulfide was detectable in the water column, anoxic bottom water was amended with 30 μM sulfide. In 2013 and 2014, the bottom waters were sulfidic, and kinetic experiments were performed to monitor the loss of the initial sulfide present in the sample. During these experiments, sulfide loss in the electrochemical cell was monitored by solid-state voltammetry as detailed above. Light intensity was controlled by using a desk lamp with an incandescent bulb with a color temperature of 3,000 K attached to a Variac instrument, which allowed for fine-tuning, and photosynthetic photon flux was monitored by using a Li-Cor Biosciences LI-1400 Data Logger light meter. Light measurements were taken from the center of the electrochemical cell. The observed attenuation of light within the cell was <0.1 μmol m−2 s−1 PAR flux. Dark conditions were maintained by shrouding the electrochemical cell. DCMU [3-(3,4-dichlorophenyl)-1,1-dimethylurea] was added to the sample water (50 μM) prior to experiments in order to inhibit oxygenic photosynthesis. Abiotic control experiments were also conducted by fixing the samples with 1.6% formaldehyde (final concentration) prior to monitoring sulfide loss.
Procedures for laboratory kinetic experiments.
Experiments were conducted to measure the effect of biomass, sulfide concentration, and light intensity on the sulfide oxidation activity of enrichment CB11. All experiments were run in anoxic HEPES buffer (0.1 M; pH 7.4) at 25°C. Light intensity was monitored and manipulated as described above for the shipboard experiments.
Cells in the mid-logarithmic growth phase (10-h doubling time) were used for all experiments. The duration of individual experiments was <1 h. At 5 μmol m−2 s−1 PAR flux and 30 μM sulfide, biomass-normalized sulfide oxidation activity was linear at protein concentrations of between 19 and 90 μg in the electrochemical cell, and changes in this rate due to various external factors could be easily detected. Therefore, all experiments were carried out with this biomass range. The sulfide concentration varied from 5 to 150 μM with 5 μmol m−2 s−1 PAR flux. Rates of sulfide loss obtained under dark conditions at each sulfide concentration were were subtracted from the experimental rate in order to separate sulfide loss from light-dependent activity by the cells. Experiments with various light intensities, from 0 to 10 μmol m−2 s−1 PAR flux, were run with 75 μM sulfide. Abiotic control experiments were conducted for each set of experimental conditions, and these rates were subtracted from those for the biotic experiments.
Determination of kinetic parameters.
Rate data were modeled by using the Michaelis-Menten equation in order to estimate the maximum rate of sulfide oxidation, Vmax, and the half-saturation constant, Ks ([S] is the sulfide concentration):
| (1) |
The data can be algebraically transformed via equation 2 to create a linear Lineweaver-Burke double-reciprocal plot (not shown) in order to obtain more accurate estimates for Vmax and Ks:
| (2) |
Finally, the substrate inhibition model of Luong (34) was used:
| (3) |
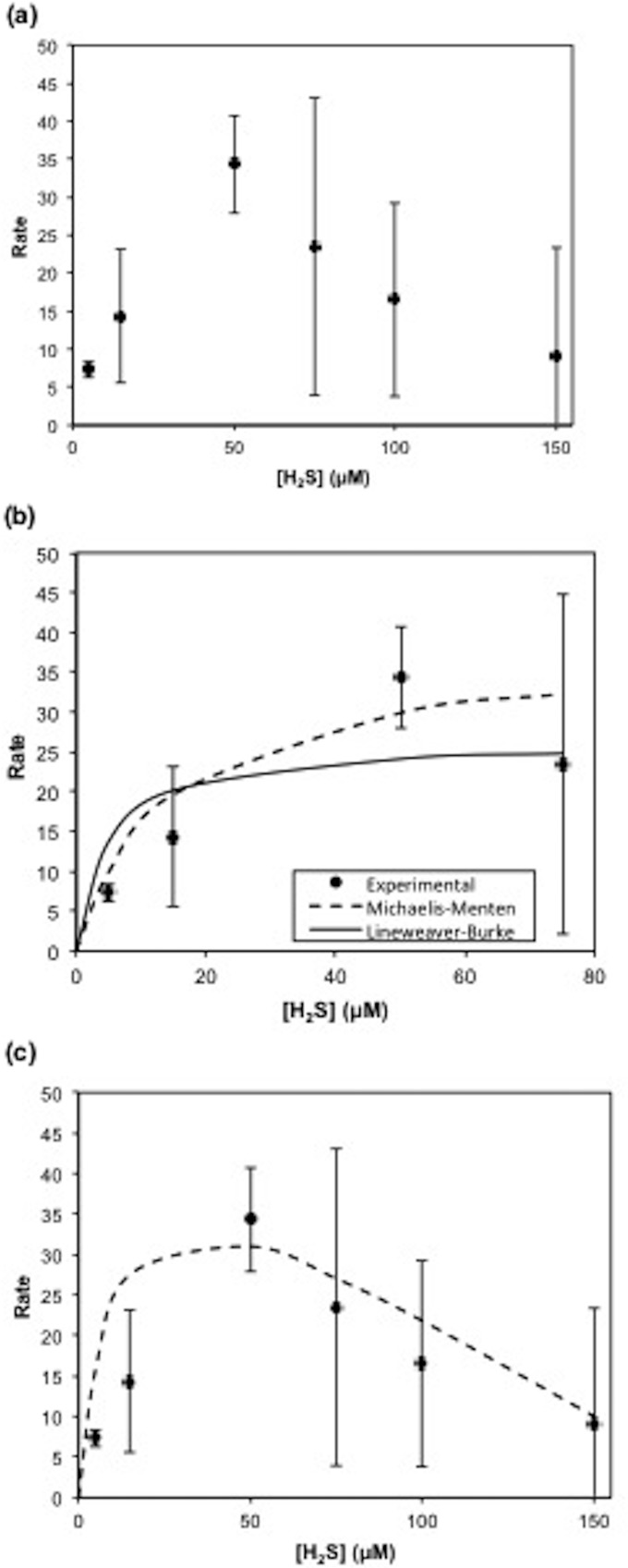
This model includes a term for Smax, the threshold substrate concentration above which activity is inhibited. The exponent n is a fitting term that describes the shape of inhibition. For modeling of the data shown in Fig. 4, n was set equal to 1, describing a linear decrease in the rate of sulfide loss after 50 μM. Kinetic parameters were determined by the best fit of the model to the experimental data using Solver in Microsoft Excel, and standard deviations were calculated by using the Solver Aid Visual Basic subroutine (35). Finally, it should be noted that these parameters are determined based on the specific sulfide oxidation rate in short-term experiments and not on measurements of growth as in other studies (36).
FIG 4.
(a) Effect of sulfide concentration on the rate of sulfide loss at saturating light intensities. (b) Michaelis-Menten and Lineweaver-Burke models. (c) Substrate inhibition model. Rates are in micromolar per minute per milligram protein.
Nucleotide sequence accession numbers.
Sequences derived from enrichments CB11 and CB21 have been deposited in GenBank under accession numbers KR013743 and KT388749, respectively.
RESULTS AND DISCUSSION
Physical and chemical stratification of the Chesapeake Bay water column.
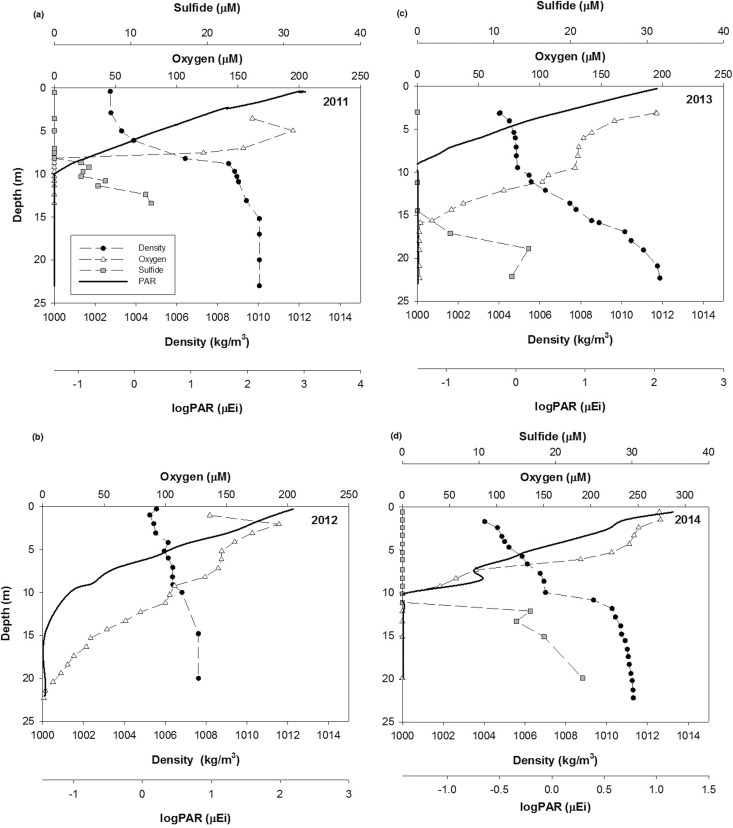
Interannual variation in water column chemistry and the location of the oxic-anoxic interface was evident during the four summers during which this study took place (Fig. 1). In 2011, the water column was strongly stratified with a clear pycnocline. The depth of the oxic-anoxic interface varied between 7 and 10 m, sulfide concentrations were up to 100 μM at depth, and PAR profiles overlapped those of sulfide (Fig. 1a). In 2012 (Fig. 1b), oxygen was present throughout the water column, with only the very deepest waters being suboxic or anoxic, well below the depth of light penetration. Free sulfide was not detected in the water column, although trace quantities of FeS were detected in deepwater samples. In 2013, the water column was stratified, and the oxic-anoxic interface was present at a depth of ∼15 m. Sulfide was measured at concentrations of up to 40 μM and did not overlap detectable PAR (Fig. 1c). In 2014, the water column was stratified, and there was a significant suboxic zone ([O2], <3 μM; [H2S], <0.2 μM) between depths of 10 and 13 m. The concentration of sulfide was up to 70 μM in deep waters. PAR overlapped the suboxic zone but not the sulfidic layer (Fig. 1d).
FIG 1.
Physical and chemical characteristics of the Chesapeake Bay water column over four subsequent summers, 2011 (a), 2012 (b), 2013 (c), and 2014 (d). μEi, microeinsteins.
Cultivation of phototrophic sulfide-oxidizing bacteria.
Phototrophic sulfide-oxidizing bacteria were successfully enriched in samples from the Chesapeake Bay taken at and directly beneath the oxic-anoxic interface in 2011 and 2013 (Table 1).
TABLE 1.
Location and characteristics of successful enrichments of phototrophic sulfide-oxidizing bacteria from the Chesapeake Bay
| Yr | Date (day and mo) | Time | Depth (m) | [H2S] (μM) | PAR (microeinsteins) | pH | Temp (°C) | ARISA peak |
|
|---|---|---|---|---|---|---|---|---|---|
| bp | % of area | ||||||||
| 2011 | 28 July | 07:20 | 11.4 | <0.2 | 0.039 | 7.1 | 24.9 | 578 | 94 |
| 13.5 | 10.4 | 0.038 | 7.03 | 24.4 | 578 | 100 | |||
| 30 July | 08:30 | 6.88 | <0.2 | 0.271 | 7.1 | 26.8 | 578 | 95 | |
| 11.3 | 45.8 | 0.038 | 7.05 | 24.3 | 581 | 56 | |||
| 578 | 40 | ||||||||
| 2013 | 09 August | 07:00 | 18.9 | 14.5 | <0.034 | 7.36 | 25.3 | 578 | 98 |
| 13:30 | 14.9 | 9.4 | 0.038 | 7.39 | 25.2 | 578 | 98 | ||
| 11 August | 07:30 | 13.3 | <0.2 | 0.038 | 7.36 | 25.3 | 578 | 99 | |
| 18.1 | 21.8 | <0.034 | 7.35 | 25.3 | 578 | 97 | |||
| 12 August | 07:40 | 13.1 | <0.2 | 0.038 | 7.44 | 25.4 | 578 | 98 | |
| 17.1 | 13.25 | <0.034 | 7.42 | 25.3 | 578 | 98 | |||
Intact cell absorption spectra for CB11 (data not shown) showed peaks at 469 and 720 nm from antenna bacteriochlorophylls in the chlorosome and a small peak at 810 nm from the baseplate bacteriochlorophylls (37). This spectrum of the enrichment is qualitatively similar to those of other Chlorobi that contain bacteriochlorophyll e (Bchl e) (see reference 35 and references therein). The presence of Bchl e was suggested by spectra of methanol extracts of cells that displayed peaks at 470 and 654 nm (35).
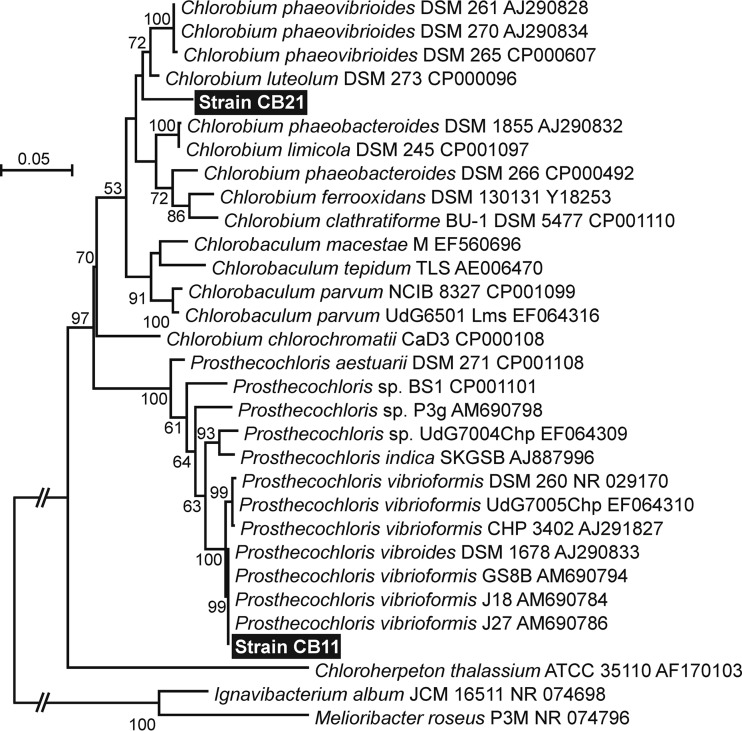
Sequencing of three independently cloned 16S rRNA PCR products from enrichment CB11 produced identical results. Phylogenetic analysis of the sequence shows that the strain dominant in CB11 is very closely related to strains of Prosthecochloris vibrioformis (Fig. 2).
FIG 2.
Maximum likelihood phylogenetic tree of the Chlorobiaceae based on a 1,238-bp alignment of 16S rRNA sequences including the sequences derived from enrichments CB11 and CB21. Sequences collected from databases are identified by organism names as proposed by Imhoff and Thiel (53), followed by strain designations and accession numbers. Numbers at the nodes indicate the percent support observed from 1,000 bootstrap replicates. Only values of >50% are shown. Shortened branch lengths are indicated by a break with two slashes. For all other branches, the bar indicates 0.05 substitutions per site.
Enrichment CB11 was analyzed by ARISA and found to contain one major peak of 578 bp that accounted for 94% of the total peak area detected in the sample (Table 1). This same peak was the dominant ARISA peak observed in all other positive enrichments analyzed except one (Table 1). In the one enrichment where it was not dominant, the CB11 peak still accounted for 40% of the total ARISA peak area. Sequencing of 16S rRNA PCR product clones from this enrichment (CB21) detected both the CB11 sequence and a sequence that is most closely related to C. luteolum DSM 273 (Fig. 2). We conclude that the second strain is the source of the 581-bp ARISA peak dominating this enrichment.
Light-dependent sulfide oxidation in field samples.
For the discussion of both field and laboratory incubation experiments, sulfide loss refers to the sum of two specific processes that will be differentiated when possible. Sulfide uptake indicates the portion of sulfide loss that consists of any sulfide taken into cells whether or not it was oxidized, e.g., sulfide loss observed in the dark in excess of that for the abiotic control. Sulfide oxidation indicates the portion of sulfide loss that is known to have resulted in oxidation, indicated by the detection of oxidation products. CB11 has been shown to produce nanoparticulate sulfur as a product of sulfide oxidation (38).
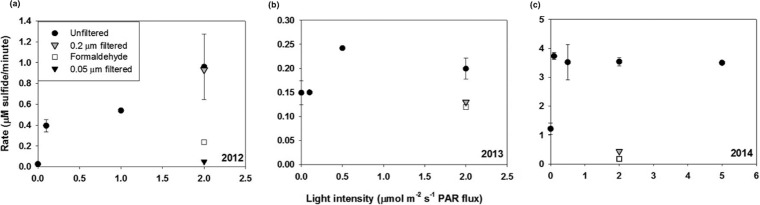
In 2012, free sulfide was not detectable in the water column; however, light-dependent sulfide loss was observed upon the addition of sulfide to bottom-water samples (Fig. 3a). This sulfide loss activity was size dependent: filtration through a 0.2-μm filter had no significant effect on the rate of sulfide loss compared to that in experiments with unfiltered water samples; however, filtration through a 0.05-μm filter effectively stopped sulfide loss. Treatment of the anoxic water sample with formaldehyde also significantly decreased the sulfide loss rate compared to that in untreated water (from 0.96 to 0.34 μM min−1).
FIG 3.
Experiments conducted shipboard demonstrating light-dependent sulfide oxidation in anoxic Chesapeake Bay water samples in 2012 (a), 2013 (b), and 2014 (c).
In 2013, samples were taken from sulfidic waters directly below the oxic-anoxic interface at depths ranging from 14 to 19 m, which contained 10 to 20 μM initial sulfide. Increased light intensity had a significant effect on the rate of sulfide loss again in 2013 (Fig. 3b). Both the addition of formaldehyde and filtration through a 0.2-μm filter resulted in a decrease in the rate of sulfide loss approximately equal to that of the dark rate.
Water samples for the 2014 experiments were collected below the interface, at a depth of ∼15 m, with sulfide concentrations of 50 to 60 μM. In 2014, rates of sulfide loss were an order of magnitude higher than those in 2013 (Fig. 3c). The rate of sulfide loss in the dark was a third of that in the light; however, in contrast to previous years, there were no significant differences between measured rates at various light intensities (0.1 to 5 μmol m−2 s−1 PAR flux). The increased rates of sulfide loss observed in 2014 could be because the redox interface was closer to the surface than in 2013 and thus was exposed to greater light intensities. Given these favorable conditions, the community of PSOB would be active and primed for sulfide oxidation.
Also unique to this year was the observation of polysulfides in kinetic experiments conducted in the light, indicated by a doublet peak at −0.7 V in the voltammetric scan (39, 40). Polysulfides are produced during the metabolism of some green and purple PSOB (41) and are the first product of sulfide oxidation by the enzyme sulfide:quinone oxidoreductase (42), so their presence supports sulfide oxidation activity by PSOB. The dark rate (1.22 μM min−1) in 2014 is an order of magnitude higher than the rate with formaldehyde treatment (0.17 μM min−1), suggesting that sulfide loss in the dark is due to transport into the cell. Sulfide is readily transported through the cell membrane (43), and a similar loss of sulfide in the dark is observed in our laboratory cultures of PSOB as well. Polysulfides were not detected during experiments in the dark, indicating that sulfide was not oxidized.
The data shown in Fig. 3 strongly indicate that the observed sulfide loss is due to PSOB activity, and as such, this is one of the first reports of phototrophic sulfide loss rates in natural samples. First, the addition of formaldehyde significantly inhibited sulfide loss, indicating a biological rather than a chemical process, and the rates of abiotic sulfide oxidation for formaldehyde-treated samples over all 3 years were comparable. Second, the addition of DCMU as an inhibitor of photosystem II did not affect the sulfide loss rate (data not shown), demonstrating that O2 production by cyanobacteria was not the cause of sulfide loss. Finally, the effect of low-intensity light variations on the sulfide loss rate and the successful enrichment of Chlorobi in both 2011 and 2013 indicate that the observed anoxic, light-dependent sulfide loss in anoxic Chesapeake Bay waters is due to the activity of phototrophic sulfur bacteria and not chemotrophic sulfide-oxidizing bacteria. One prior study in the Chesapeake Bay showed rapid sulfide loss (with a half-life for sulfide of ∼15 min) in water samples exposed to light, which did not appear to be due to oxidation by oxygen or trace metals, as this sulfide loss was not observed in the dark or in water samples that had been fixed with formaldehyde (33), similar to the results presented above (Fig. 3). The reproducibility in light-dependent sulfide loss in anoxic water masses in four separate years (1987, 2012, 2013, and 2014) suggests that this activity is a common component of the Chesapeake Bay sulfur cycle despite interannual redox variation.
Sulfide oxidation kinetics in enrichment CB11.
Sulfide oxidation in CB11 follows Michaelis-Menten kinetics at low sulfide concentrations (5 to 75 μM) (Fig. 4a and b); however, this description of the kinetics is not accurate for the entire data set (Fig. 4a). At higher sulfide concentrations (>75 μM) the rate data display characteristics of substrate inhibition and were modeled by using the substrate inhibition model (equation 3) (Fig. 4c) proposed by Luong (34). The kinetic parameters derived from each of these models, the Michaelis-Menten, Lineweaver-Burke, and substrate inhibition models, are summarized in Table 2.
TABLE 2.
Comparison of kinetic parameters derived from enzyme activity models
| Model | Mean Vmax (μM/min/mg protein) ± SD | Mean Ks (μM) ± SD | Mean Smax (μM) ± SD |
|---|---|---|---|
| Michaelis-Menten | 39 ± 14 | 14 ± 13 | |
| Lineweaver-Burke | 26 | 4.5 | |
| Substrate inhibitiona | 51 ± 25 | 11 ± 5 | 190 ± 84 |
See equation 3 (34).
The values for both Vmax and Ks fall within the sulfide range that CB11 would experience in the Chesapeake Bay (≤100 μM) and are comparable to values for the kinetic parameters determined for Chlorobi and other green sulfur bacteria (GSB) (44). Given that the substrate inhibition model is the only model used to fit the entire data set, the maximum rate and half-saturation constant derived from this model are the best description of the kinetic parameters of CB11.
Effect of light intensity on the sulfide oxidation rate.
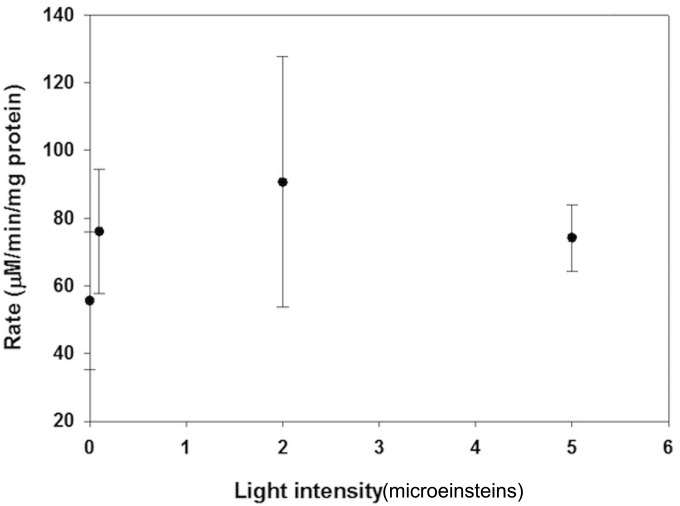
Small increases in light intensity result in significant changes in the sulfide oxidation rate of CB11 over the dark uptake rate (Fig. 5), similar to the results observed in field experiments (Fig. 3a to c). Saturation of the rate is also evident after 2 μmol m−2 s−1 PAR flux, indicating that photoinhibition occurs at very low light intensities.
FIG 5.
Effect of light intensity on the sulfide oxidation rate of laboratory-grown enrichments.
Furthermore, a significant rate of sulfide loss was observed in the dark (57 μM min−1 mg protein−1) (Fig. 5). Photosynthetic sulfide oxidation cannot occur under dark conditions, as light is required for the oxidation of the quinone pool (45); however, uptake of sulfide for later oxidation may occur, as is done with elemental sulfur (13). This dark rate is observed for other phototrophic sulfide-oxidizing bacteria such as Chlorobaculum tepidum and RSC1, a red-colored green sulfur bacterium isolated from a Bahamian sinkhole (A. J. Findlay, D. MacDonald, T. E. Hanson, and G. W. Luther, III, unpublished data). In order to account for all the dark sulfide loss observed, the internal concentration of sulfide in the cells would need to be 1.5 to 2 mM. As the cells are grown in medium containing 2.5 mM sulfide, this uptake is not expected to be inhibitory. The potential for dark uptake is significant, as it suggests that sulfide may be stored inside the cell for future oxidation and that uptake and oxidation of sulfide are independent processes.
A direct comparison between the field and laboratory results is difficult because the biomass in laboratory experiments is likely more concentrated than the in situ biomass; however, sulfide loss rates over different light intensities in both the field and laboratory are within the same order of magnitude and show similar responses to small changes in light intensity. This has wide environmental implications, as small changes in the depth of the chemical interface (and thus in the light reaching it) could greatly impact the sulfide oxidation capacity of the Chesapeake Bay and similar systems where these organisms are present.
Viability of PSOB at in situ light levels.
The light-dependent sulfide loss observed in the Chesapeake Bay samples suggests that the bacteria present are active or have the potential to be active given sufficient light and sulfide. In conventional oceanography and limnology, the photic zone is delineated by light intensities >1% of surface intensity; however, the PSOB found in the Chesapeake Bay are capable of significant sulfide oxidation activity at light intensities as low as 0.0067% of surface intensity (0.1 μmol m−2 s−1 PAR flux) (Fig. 3a to c).
Furthermore, a study by Manske et al. (46) found enrichments of PSOB from the Black Sea to be photosynthetically active at light intensities down to 0.015 μmol m−2 s−1 PAR flux, and field studies have established that short-term variation in the light intensity at the interface can lead to photosynthetic growth, a buildup of green sulfur bacteria, and maintenance of the population (10, 47). The location of the interface in the Chesapeake Bay is known to vary tidally (24), and so this process could be a factor in maintaining PSOB populations. Additionally, field studies of the Black Sea have found that although most cells in the green sulfur bacterial population are photosynthetically inactive and do not grow, the cells are capable of maintaining their cellular ATP pools, and presumably viability, at light intensities as low as 0.0014 μmol m−2 s−1 PAR flux (47). In the Chesapeake Bay, light intensities of 0.015 μmol m−2 s−1 PAR flux are found at depths of 10 to 12.5 m, and light intensities of 0.001 μmol m−2 s−1 PAR flux are found at depths of 12.5 to 15.3 m (Fig. 1).
Potential impact of PSOB on sulfide oxidation in the Chesapeake Bay.
The evidence presented above indicates that phototrophic sulfide oxidation is an important component of the sulfur cycle in the Chesapeake Bay during periods in which the water column is stratified; however, abiotic sulfide oxidation is also expected to occur. Oxidized iron and manganese are the two dominant oxidants for sulfide at the chemical interface:
| (4) |
| (5) |
In order to compare the fractions of biotic and abiotic sulfide oxidation in the Chesapeake Bay, the rates of each process were calculated by using measured parameters for the suboxic zone from 2013 and 2014 (the 2 years in which light-dependent sulfide loss experiments were conducted with field samples and sulfide was detected in the water column).
The highest concentration of iron measured at the chemical interface for both 2013 and 2014 was 1.5 μM. Using the rate constant derived by Yao and Millero (2) and measured concentrations of iron and sulfide, we can determine the rate of sulfide oxidation by iron(III):
| (6) |
where k at 25°C is 148 M−1 min−1.
The highest concentrations of MnO2 at the chemical interface (2013) were 6 μM in 2013 and 2 μM in 2014. The rate law for sulfide oxidation by manganese(IV) oxides was derived by Yao and Millero (48). The value for k in seawater at 25°C is 436 M−1 min−1:
| (7) |
The rate constant and in situ rate of sulfide loss due to phototrophic sulfide-oxidizing bacteria were derived by using data from the laboratory experiments. The rate of sulfide metabolism for phototrophic bacteria is assumed to be dependent on both light and the sulfide concentration (8):
| (8) |
where [H2S] is in molar and [light] is the light intensity in micromoles per square meter per second of PAR flux. A rate order for sulfide (exponent a) of 1 was calculated by the method of initial rates using laboratory experiments with enrichments that isolated the effects of sulfide on sulfide oxidation rates in bacterial enrichments (Fig. 4a). Light is a continuous flux during these measurements and does not become depleted; thus, PSOB activity can be described by a pseudo-first-order rate expression that is dependent solely upon the sulfide concentration at a specific light intensity. The rate equation then becomes
| (9) |
in which the value of k will change with the light intensity and other environmental parameters such as the biomass of PSOB. Using the rates from Fig. 3b and c and the in situ sulfide concentration, a value for k may be derived (for 0.1 μmol m−2 s−1 PAR flux) for 2013 and 2014. The expected rate of biotic sulfide oxidation at the chemical interface can then be calculated from equation 9 (Table 3).
TABLE 3.
Comparison of sulfide oxidation rates for 2013 and 2014
| Yr and pathway for sulfide oxidation | [H2S] (μM) | Concn of oxidant | k | Rate (nM/min) |
|---|---|---|---|---|
| 2013 | ||||
| Iron oxides | 4 | 1.5 μMa | 148 M−1 min−1 | 0.89 |
| Manganese oxide | 4 | 6 μMb | 436 M−1 min−1 | 11 |
| Phototrophic sulfide bacteria | 4 | 0.1 μmol m−2 s−1 PAR fluxc | 0.10 min−1 | 400 |
| 2014 | ||||
| Iron oxides | 10 | 1.5 μMa | 148 M−1 min−1 | 2.2 |
| Manganese oxide | 10 | 2 μMb | 436 M−1 min−1 | 8.72 |
| Phototrophic sulfide bacteria | 10 | 0.1 μmol m−2 s−1 PAR fluxc | 0.075 min−1 | 750 |
Maximum measured concentration of Fe3+ at the interface.
Maximum measured concentration of manganese oxide at the interface.
Lowest light intensity for which measurements of H2S loss rates were made (Fig. 3), which is representative of light intensities measured at the interface in 2011.
Estimate of PSOB biomass in the Chesapeake Bay.
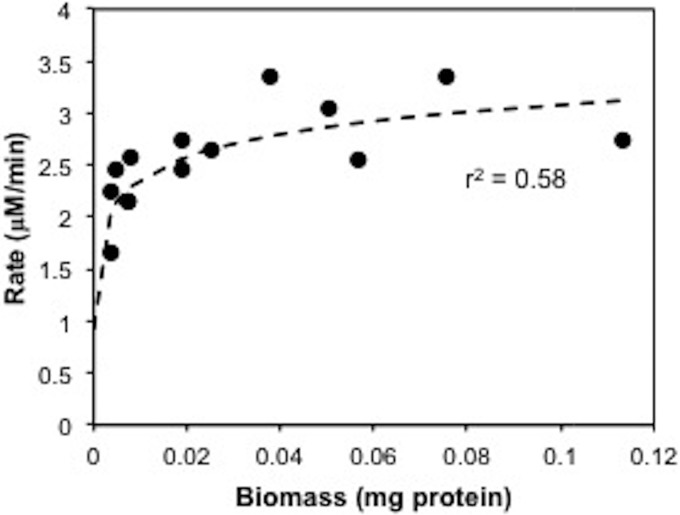
Using the kinetic parameters (Table 2), in situ measurements of both light intensity and sulfide (Fig. 1), and the relationship between biomass (milligram of protein) and the rate of sulfide oxidation (micromolar per minute) for laboratory enrichments (Fig. 6), we are able to estimate the biomass present at the redox interface in 2013 and 2014.
FIG 6.
Relationship between biomass and the rate of sulfide oxidation in CB11.
A rate constant was calculated for 5 μmol m−2 s−1 PAR flux, derived from the experimental data as described above, in order to correlate the in situ rate with the rate measurements obtained under laboratory conditions (30 μM sulfide and 5 μmol m−2 s−1 PAR flux). We thus calculated that the biomasses of phototrophic sulfide oxidizers in the Chesapeake Bay were 11 μg protein liter−1 in 2013 and 20 μg protein liter−1 in 2014. Protein was calibrated with BChl e in order to make a direct comparison with literature values reported as BChl for the Black Sea (46), and as reviewed by VanGemerden and Mas (44). It should be noted that under low-light conditions, phototrophic sulfur bacteria may increase the efficiency of photosynthesis by increasing their pigment content (49), and so BChl is not a direct measure of cell abundance. The specific BChl e content was 45 μg BChl e mg protein−1, which is similar to the values obtained by Overmann et al. (11) for pelagic PSOB from the Black Sea grown at 30 μmol photons m−2 s−2. From this, we estimate that the concentrations of BChl e in the Chesapeake Bay were 0.5 μg liter−1 in 2013 and 0.9 μg liter−1 in 2014. In other stratified environments, the density of phototrophic sulfide-oxidizing bacteria can vary from 0.008 μg liter−1 to 28 mg liter−1 BChl e. Our estimate for the Chesapeake Bay falls in the lower portion of this range, consistent with the low-light conditions in the water column, and is an order of magnitude higher than the maximum BChl e values of 0.054 to 0.068 μg liter−1 observed in the Black Sea (46).
Another way of estimating the population size is to use previously reported values for cellular protein content in marine bacteria (6 × 10−14 to 33 × 10−14 g cell−1 [50]). This leads to Chlorobi population sizes of ∼1 × 108 cells liter−1 in 2013 and 2 × 108 cells liter−1 in 2014. Given a mean bacterial population size of 4 × 109 cells liter−1 for the Chesapeake Bay in the summertime (calculated from data in reference 51), the observed sulfide consumption rates could be explained if CB11-type Chlorobi account for 3 to 5% of the total bacterial community at the interface.
Based upon these calculations, if the populations of PSOB present in 2013 and 2014 were exposed to 0.1 μmol m−2 s−1 PAR flux, they could account for up to 97% of anaerobic sulfide oxidation in the bay in 2013 and 96% in 2014. For comparison, Manske et al. (46) attribute <0.01% of anaerobic sulfide oxidation in the Black Sea to phototrophic sulfide oxidation, based on calculated doubling times of GSB populations.
The capacity of Chlorobi in the Chesapeake Bay for sulfide oxidation will be affected by changes in the interface depth and the corresponding changes in light intensity. Although the interface typically received <0.1 μmol m−2 s−1 PAR flux light in 2013, its location in the water column will vary on time scales of hours, due to tidal currents; days, due to diel light cycles; and months, due to seasonal turnover affecting sulfide and oxygen in the bottom waters (May to August).
It is well established that phototrophic sulfur bacteria are found in most environments in which sulfidic waters or sediments are located within the photic zone (8, 12, 44). More recently, viable phototrophic sulfide-oxidizing bacteria have been found in extremely-low-light environments in the Black Sea (11, 46, 47), a hydrothermal vent at the East Pacific Rise (52), and a stratified lake, Lake Kinneret (10). The consistent recurrence of PSOB related to those in enrichment CB11 in the light-limited water column of the Chesapeake Bay and the observation of light-dependent sulfide loss in field samples add to the growing evidence that phototrophic sulfide-oxidizing bacteria may be a significant component of environments previously considered to be light limited.
ACKNOWLEDGMENTS
This work was supported by National Science Foundation grants to George W. Luther, III (OCE-1131109 and OCE 1155385), and a joint grant to George W. Luther, III, and Thomas E. Hanson (MCB-0919682). This work was also supported by the Delaware Environmental Institute's Environmental Frontiers Grant Program.
We thank Daniel MacDonald for providing PAR data and Bradley Tebo and Matthew Jones for providing MnO2 data. Finally, we thank the captain and crew of the R/V Hugh R. Sharp for successful cruises.
REFERENCES
- 1.Luther GW., III 2010. The role of one- and two-electron transfer reactions in forming thermodynamically unstable intermediates as barriers in multi-electron redox reactions. Aquat Geochem 16:395–420. doi: 10.1007/s10498-009-9082-3. [DOI] [Google Scholar]
- 2.Yao W, Millero FJ. 1996. Oxidation of hydrogen sulfide by hydrous Fe(III) oxides in seawater. Mar Chem 52:1–16. doi: 10.1016/0304-4203(95)00072-0. [DOI] [Google Scholar]
- 3.Gregersen LH, Bryant DA, Frigaard N. 2011. Mechanisms and evolution of oxidative sulfur metabolism in green sulfur bacteria. Front Microbiol 2:116. doi: 10.3389/fmicb.2011.00116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Luther GW, Findlay AJ, MacDonald DJ, Owings SM, Hanson TE, Beinart RA, Girguis PR. 2011. Thermodynamics and kinetics of sulfide oxidation by oxygen: a look at inorganically controlled reactions and biologically mediated processes in the environment. Front Microbiol 2:62. doi: 10.3389/fmicb.2011.00062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Preisler A, de Beer D, Lichtschlag A, Lavik G, Boetius A, Jørgensen BB. 2007. Biological and chemical sulfide oxidation in a Beggiatoa inhabited marine sediment. ISME J 1:341–353. [DOI] [PubMed] [Google Scholar]
- 6.Wahlund TM, Woese CR, Castenholz RW, Madigan MT. 1991. A thermophilic green sulfur bacterium from New Zealand hot springs, Chlorobium tepidum sp nov. Arch Microbiol 156:81–90. [Google Scholar]
- 7.Parkin TB, Brock TD. 1981. The role of phototrophic bacteria in the sulfur cycle of a meromictic lake. Limnol Oceanogr 26:880–890. doi: 10.4319/lo.1981.26.5.0880. [DOI] [Google Scholar]
- 8.Guerrero R, Montesinos E, Pedros-Alio C, Esteve I, Mas J, van Gemerden H, Hofman PAG, Bakker JF. 1985. Phototrophic sulfur bacteria in two Spanish lakes: vertical distribution and limiting factors. Limnol Oceanogr 30:919–931. doi: 10.4319/lo.1985.30.5.0919. [DOI] [Google Scholar]
- 9.Tonolla M, Peduzzi S, Hahn D, Peduzzi R. 2003. Spatio-temporal distribution of phototrophic sulfur bacteria in the chemocline of meromictic Lake Cadagno (Switzerland). Microb Ecol 43:89–98. doi: 10.1111/j.1574-6941.2003.tb01048.x. [DOI] [PubMed] [Google Scholar]
- 10.Rimmer A, Ostrovsky I, Yacobi YZ. 2008. Light availability for Chlorobium phaebacteroides development in Lake Kinneret. J Plankton Res 30:765–776. doi: 10.1093/plankt/fbn037. [DOI] [Google Scholar]
- 11.Overmann J, Cypionka H, Pfennig N. 1992. An extremely low-light-adapted phototrophic sulfur bacterium from the Black Sea. Limnol Oceanogr 37:150–155. doi: 10.4319/lo.1992.37.1.0150. [DOI] [Google Scholar]
- 12.Overmann J. 2008. Ecology of phototrophic sulfur bacteria, p 375–396. In Hell R, Dahl C, Knaff DB, Leustek T (ed), Sulfur metabolism in phototrophic organisms, 1st ed, vol 27 Springer, Dordrecht, The Netherlands. [Google Scholar]
- 13.Overmann J. 1997. Mahoney Lake: a case study of the ecological significance of phototrophic sulfur bacteria, p 251–288. In Jones G. (ed), Advances in microbial ecology, 1st ed, vol 15 Plenum Press, New York, NY. [Google Scholar]
- 14.Eghbal MA, Pennefather PS, O'Brien PJ. 2004. H2S cytotoxicity mechanism involves reactive oxygen species formation and mitochondrial depolarisation. Toxicology 203:69–76. doi: 10.1016/j.tox.2004.05.020. [DOI] [PubMed] [Google Scholar]
- 15.Julian D, April KL, Patel S, Stein JR, Wohlgemuth SE. 2005. Mitochondrial depolarization following hydrogen sulfide exposure in erythrocytes from a sulfide-tolerant marine invertebrate. J Exp Biol 208:4109–4122. doi: 10.1242/jeb.01867. [DOI] [PubMed] [Google Scholar]
- 16.Deutsch C, Brix H, Ito T, Frenzel H, Thompson L. 2011. Climate-forced variability of ocean hypoxia. Science 333:336–339. doi: 10.1126/science.1202422. [DOI] [PubMed] [Google Scholar]
- 17.Lavik G, Stührmann T, Brüchert V, Van der Plas A, Mohrholz V, Lam P, Mussmann M, Fuchs BM, Amann R, Lass U, Kuypers MM. 2009. Detoxification of sulphidic African shelf waters by blooming chemolithotrophs. Nature 457:581–584. doi: 10.1038/nature07588. [DOI] [PubMed] [Google Scholar]
- 18.Schunck H, Lavik G, Desai DK, Großkopf T, Kalvelage T, Löscher CR, Paulmier A, Contreras S, Siegel H, Holtappels M, Rosenstiel P, Schilhabel MB, Graco M, Schmitz RA, Kuypers MM, Laroche J. 2013. Giant hydrogen sulfide plume in the oxygen minimum zone off Peru supports chemolithoautotrophy. PLoS One 8:e68661. doi: 10.1371/journal.pone.0068661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Canfield DE, Stewart FJ, Thamdrup B, De Brabandere L, Dalsgaard T, Delong EF, Revsbech NP, Ulloa O. 2010. A cryptic sulfur cycle in oxygen-minimum-zone waters off the Chilean coast. Science 330:1375–1378. doi: 10.1126/science.1196889. [DOI] [PubMed] [Google Scholar]
- 20.Stramma L, Schmidtko S, Levin LA, Johnson GC. 2010. Ocean oxygen minima expansions and their biological impacts. Deep Sea Res Part I Oceanogr Res Pap 57:587–595. doi: 10.1016/j.dsr.2010.01.005. [DOI] [Google Scholar]
- 21.Hanson TE, Luther GW III, Findlay AJ, MacDonald DJ, Hess D. 2013. Phototrophic sulfide oxidation: environmental insights and methods for kinetic analysis. Front Microbiol 4:382. doi: 10.3389/fmicb.2013.00382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hagy JD, Boynton WR, Keefe CW, Wood KV. 2004. Hypoxia in Chesapeake Bay, 1950-2001: long-term change in relation to nutrient loading and river flow. Estuaries 27:634–658. doi: 10.1007/BF02907650. [DOI] [Google Scholar]
- 23.Pritchard DW. 1952. Salinity distribution and circulation in the Chesapeake Bay estuarine system. J Mar Res 11:106–123. [Google Scholar]
- 24.Lewis BL, Glazer BT, Montbriand PJ, Luther GW III, Nuzzio DB, Deering T, Ma S, Theberge S. 2007. Short-term and interannual variability of redox-sensitive chemical parameters in hypoxic/anoxic bottom waters of the Chesapeake Bay. Mar Chem 105:296–308. doi: 10.1016/j.marchem.2007.03.001. [DOI] [Google Scholar]
- 25.Crump BC, Peranteau C, Beckingham B, Cornwell JC. 2007. Respiratory succession and community succession of bacterioplankton in seasonally anoxic estuarine waters. Appl Environ Microbiol 73:6802–6810. doi: 10.1128/AEM.00648-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.MacDonald DJ, Findlay AJ, Hredzak-Showalter P, Krepski ST, McAllister SM, Cone S, Scott J, Bennett S, Chan CS, Emerson D, Luther GW III. 2014. Using in situ voltammetry as a tool to search for iron oxidizing bacteria: from fresh water wetlands to hydrothermal vent sites. Environ Sci Process Impacts 16:2117–2126. doi: 10.1039/C4EM00073K. [DOI] [PubMed] [Google Scholar]
- 27.Glazer BT, Luther GW III, Konavalov SK, Friederich GE, Nuzzio DB, Trouborst RE, Tebo BM, Clement B, Murray K, Romanov AS. 2006. Documenting the suboxic zone of the Black Sea via high-resolution real-time redox profiling. Deep Sea Res 53:1740–1755. doi: 10.1016/j.dsr2.2006.03.011. [DOI] [Google Scholar]
- 28.Overmann J, Pfennig N. 1989. Pelodictyon phaeoclathratiforme sp. nov., a new brown-colored member of the Chlorobiaceae forming net-like colonies. Arch Microbiol 152:401–406. doi: 10.1007/BF00425181. [DOI] [Google Scholar]
- 29.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cardinale M, Brusetti L, Quatrini P, Borin S, Puglia AM, Rizzi A, Zanardini E, Sorlini C, Corselli C, Daffonchio D. 2004. Comparison of different primer sets for use in automated ribosomal intergenic spacer analysis of complex bacterial communities. Appl Environ Microbiol 70:6147–6156. doi: 10.1128/AEM.70.10.6147-6156.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 32.Luther GW III, Glazer BT, Ma S, Trouwborst RE, Moore TS, Metzger E, Kraiya C, Waite TJ, Druschel G, Sundby B, Taillefert M, Nuzzio DB, Shank TM, Lewis BL, Brendel PJ. 2008. Use of voltammetric solid-state (micro)electrodes for studying biogeochemical processes: laboratory measurements to real time measurements with an in situ electrochemical analyzer (ISEA). Mar Chem 108:221–235. doi: 10.1016/j.marchem.2007.03.002. [DOI] [Google Scholar]
- 33.Luther G, Ferdelman T, Tsamakis E. 1988. Evidence suggesting anaerobic oxidation of the bisulfide ion in Chesapeake Bay. Estuaries 11:281–285. doi: 10.2307/1352015. [DOI] [Google Scholar]
- 34.Luong JHT. 1986. Generalization of monod kinetics for analysis of growth data with substrate inhibition. Biotechnol Bioeng 29:242–248. [DOI] [PubMed] [Google Scholar]
- 35.De Levie R. 2004. Advanced Excel for scientific data analysis. Oxford University Press, New York, NY. [Google Scholar]
- 36.Van Gemerden H. 1984. The sulfide affinity of phototrophic bacteria in relation to the location of elemental sulfur. Arch Microbiol 139:289–294. doi: 10.1007/BF00408368. [DOI] [Google Scholar]
- 37.Gloe A, Pfennig N, Brockmann H Jr, Trowitzsch W. 1975. A new bacteriochlorophyll from brown-colored chlorobiaceae. Arch Microbiol 102:103–109. doi: 10.1007/BF00428353. [DOI] [PubMed] [Google Scholar]
- 38.Findlay AJ, Gartman A, MacDonald D, Hanson TE, Shaw T, Luther GW III. 2014. Distribution and size fractionation of elemental sulfur in aqueous environments: the Chesapeake Bay and Mid-Atlantic Ridge. Geochim Cosmochim Acta 142:334–348. doi: 10.1016/j.gca.2014.07.032. [DOI] [Google Scholar]
- 39.Rozan TF, Theberge SM, Luther GW III. 2000. Quantifying elemental sulfur (S0), bisulfide (HS−) and polysulfides (Sx2−) using a voltammetric method. Geochim Cosmochim Acta 415:175–184. [Google Scholar]
- 40.Luther GW III, Glazer BT, Hohmann L, Popp JI, Taillefert M, Rozan TF, Brendel PJ, Theberge SM, Nuzzio DB. 2001. Sulfur speciation monitored in situ with solid state gold amalgam voltammetric microelectrodes: polysulfides as a special case in sediments, microbial mats and hydrothermal vent waters. J Environ Monit 3:61–66. doi: 10.1039/b006499h. [DOI] [PubMed] [Google Scholar]
- 41.Prange A, Chauvistre R, Modrow H, Hormes J, Truper HG, Dahl C. 2002. Quantitative speciation of sulfur in bacterial sulfur globules: X-ray absorption spectroscopy reveals at least three different species of sulfur. Microbiology 148:267–276. doi: 10.1099/00221287-148-1-267. [DOI] [PubMed] [Google Scholar]
- 42.Griesbeck C, Hauska G, Schütz M. 2002. Biological sulfide oxidation: sulfide-quinone reductase (SQR), the primary reaction, p 179–203. In Pandalai SG. (ed), Recent research developments in microbiology, vol 4 Research Signpost, Trivandrum, India. [Google Scholar]
- 43.Riahi S, Rowley CN. 2014. Why can hydrogen sulfide permeate cell membranes? J Am Chem Soc 136:15111–15113. doi: 10.1021/ja508063s. [DOI] [PubMed] [Google Scholar]
- 44.VanGemerden H, Mas J. 1995. Ecology of phototrophic sulfur bacteria, p 49–85. In Blankenship RE, Madigan MT, Bauer CE (ed), Anoxygenic photosynthetic bacteria, 1st ed, vol 2 Springer, Dordrecht, The Netherlands. [Google Scholar]
- 45.Shahak Y, Hauska G. 2008. Sulfide oxidation from cyanobacteria to humans: sulfide-quinone oxidoreductase (SQR), p 319–335. In Hell R, Dahl C, Knaff DB, Leustek T (ed), Sulfur metabolism in phototrophic organisms, 1st ed, vol 27 Springer, Dordrecht, The Netherlands. [Google Scholar]
- 46.Manske AK, Glaeser J, Kuypers MM, Overmann J. 2005. Physiology and phylogeny of green sulfur bacteria forming a monospecific phototrophic assemblage at a depth of 100 meters in the Black Sea. Appl Environ Microbiol 71:8049–8060. doi: 10.1128/AEM.71.12.8049-8060.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marschall E, Jogler M, Hessge U, Overmann J. 2010. Large-scale distribution and activity patterns of an extremely low-light-adapted population of green sulfur bacteria in the Black Sea. Environ Microbiol 12:1348–1362. doi: 10.1111/j.1462-2920.2010.02178.x. [DOI] [PubMed] [Google Scholar]
- 48.Yao W, Millero FJ. 1993. The rate of sulfide oxidation by δMnO2 in seawater. Geochim Cosmochim Acta 57:3359–3365. doi: 10.1016/0016-7037(93)90544-7. [DOI] [Google Scholar]
- 49.Broch-Due M, Ormerod JG, Fjerdingen BS. 1978. Effect of light intensity on vesicle formation in Chlorobium. Arch Microbiol 116:269–274. doi: 10.1007/BF00417850. [DOI] [PubMed] [Google Scholar]
- 50.Zubkov MV, Fuchs BM, Eilers H, Burkill PH, Amann R. 1999. Determination of total protein content of bacterial cells by SYPRO staining and flow cytometry. Appl Environ Microbiol 65:3251–3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kan J, Crump BC, Wang K, Chen F. 2006. Bacterioplankton community in Chesapeake Bay: predictable or random assemblages. Limnol Oceanogr 51:2157–2169. doi: 10.4319/lo.2006.51.5.2157. [DOI] [Google Scholar]
- 52.Beatty JT. 2005. An obligately photosynthetic bacterial anaerobe from a deep-sea hydrothermal vent. Proc Natl Acad Sci U S A 102:9306–9310. doi: 10.1073/pnas.0503674102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Imhoff JF, Thiel V. 2010. Phylogeny and taxonomy of Chlorobiaceae. Photosynth Res 104:123–136. doi: 10.1007/s11120-009-9510-7. [DOI] [PubMed] [Google Scholar]