Abstract
Cell wall-active antibiotics cause induction of a locus that leads to elevated synthesis of two methionine sulfoxide reductases (MsrA1 and MsrB) in Staphylococcus aureus. To understand the regulation of this locus, reporter strains were constructed by integrating a DNA fragment consisting of the msrA1/msrB promoter in front of a promoterless lacZ gene in the chromosome of wild-type and MsrA1-, MsrB-, MsrA1/MsrB-, and SigB-deficient methicillin-sensitive S. aureus strain SH1000 and methicillin-resistant S. aureus strain COL. These reporter strains were cultured in TSB and the cellular levels of β-galactosidase activity in these cultures were assayed during different growth phases. β-galactosidase activity assays demonstrated that the lack of MsrA1, MsrB, and SigB upregulated the msrA1/msrB promoter in S. aureus strain SH1000. In S. aureus strain COL, the highest level of β-galactosidase activity was observed under the conditions when both MsrA1 and MsrB proteins were absent. The data suggest that the msrA1/msrB locus, in part, is negatively regulated by MsrA1, MsrB, and SigB in S. aureus.
1. Introduction
Staphylococcus aureus is part of the microbiome of roughly 30% of people who show no clinical symptoms [1]. It is an opportunistic human pathogen that can cause a wide variety of diseases and can involve any organ system in the human body. Diseases caused by S. aureus may include mild skin infections such as folliculitis and impetigo to fatal conditions such as pneumonia, osteomyelitis, and endocarditis [2]. Treatment of S. aureus infections has become problematic as it has developed numerous mechanisms to become resistant to almost all known antibiotics [3, 4].
It was previously reported that exposure of S. aureus to oxacillin and other cell wall-active antibiotics increases the expression of msrA1 and msrB both at the transcriptional and at the protein level [5, 6]. Pathogenic bacterial species are exposed to a variety of extremely potent reactive oxygen species (ROS) by the host phagocytic cells during the course of phagocytosis that are damaging to all cellular macromolecules. ROS can cause damage to proteins by the oxidation of sulfhydryl groups, reduction of disulfides, oxidative adduction of amino acid residues close to metal-binding sites, and peptide fragmentation [7]. In particular, ROS oxidize the sulfur atom of protein-bound methionine residues resulting in methionine sulfoxide (MetO) and loss of protein function. However, almost all biological species possess the ability to reduce oxidized methionines [8]. MsrA and MsrB proteins reduce S- and R-epimers of methionine sulfoxides (MetO), respectively [8].
In S. aureus, genes encoding MsrA1 and MsrB are the first and second genes of a four-gene cluster that are cotranscribed [6]. A mutation in the msrA1 gene increased the susceptibility of S. aureus to oxidative stress [6, 9]. More recently, it was shown that the MsrA1 protein was critical for S. aureus in establishing an infection in mice [10]. Interestingly, the MsrA1-deficient S. aureus was shown to possess an elevated level of MsrB [9] giving rise to the speculation of autoregulation of the msrA1/msrB locus. Additionally, sigma factor B (SigB) is an alternative sigma factor that is involved in regulating the expression of stress response genes in S. aureus [11]. Thus, it seems plausible that SigB may have a role in the regulation of the msrA1/msrB locus. Findings of this study provide evidence that the msrA1/msrB locus is negatively regulated by the products of this locus and SigB.
2. Materials and Methods
2.1. Bacterial Strains, Antibiotics, and Growth Conditions
The bacterial strains used in this study are shown in Table 1. S. aureus cultures were grown aerobically at 37°C in tryptic soy broth (TSB) in a shaking incubator (220 rpm) or on tryptic soy agar (TSA) by incubation for 24–48 h. Overnight cultures of S. aureus reporter strains were prepared in the presence of erythromycin at 10 μg mL−1. Oligonucleotide primers used in this study were obtained from Eurofins and are shown in Table 2.
Table 1.
Bacterial strains used in this study.
| Strains | Characteristics | Reference |
|---|---|---|
| SH1000 | S. aureus strain 8325-4 with functional RsbU | [11] |
| COL | Homogeneous in methicillin-resistance expression | [37] |
| SH1000ΔmsrA1 | msrA1 mutant of SH1000 | [10] |
| SH1000ΔmsrB | msrB mutant of SH1000 | [10] |
| SH1000ΔmsrA1-msrB | msrA1-msrB double mutant of SH1000 | [10] |
| SH1000ΔsigB | sigB mutant of SH1000 | [14] |
| COLΔmsrA1 | msrA1 mutant of SH1000 | [6] |
| COLΔmsrB | msrB mutant of SH1000 | This study |
| COLΔmsrA1-msrB | msrA1-msrB double mutant of SH1000 | This study |
| COLΔsigB | sigB mutant of SH1000 | [38] |
| SH1000-(A1/B)P-lacZ | msrA1/msrB promoter-lacZ fusion in SH1000 (ErmR) | [13] |
| SH1000ΔmsrA1-(A1/B)P-lacZ | msrA1/msrB promoter-lacZ fusion in msrA1 mutant of SH1000 (KanR, ErmR) | This study |
| SH1000ΔmsrB-(A1/B)P-lacZ | msrA1/msrB promoter-lacZ fusion in msrB mutant of SH1000 (KanR, ErmR) | This study |
| SH1000ΔmsrA1-msrB-(A1/B)P-lacZ | msrA1/msrB promoter-lacZ fusion in msr1-msrB mutant of SH1000 (KanR, ErmR) | This study |
| SH1000ΔsigB-(A1/B)P-lacZ | msrA1/msrB promoter-lacZ fusion in sigB mutant of SH1000 (KanR, ErmR) | This study |
| COL-(A1/B)P-lacZ | msrA1/msrB promoter-lacZ fusion in COL (ErmR) | This study |
| COLΔmsrA1-(A1/B)P-lacZ | msrA1/msrB promoter-lacZ fusion in msrA1 mutant of COL (KanR, ErmR) | This study |
| COLΔmsrB-(A1/B)P-lacZ | msrA1/msrB promoter-lacZ fusion in msrB mutant of COL (KanR, ErmR) | This study |
| COLΔmsrA1-msrB-(A1/B)P-lacZ | msrA1/msrB promoter-lacZ fusion in msrA1-msrB mutant of COL (KanR, ErmR) | This study |
| COLΔsigB-(A1/B)P-lacZ | msrA1/msrB promoter-lacZ fusion in sigB mutant of COL (KanR, ErmR) | This study |
KanR: kanamycin resistant; ErmR: erythromycin resistant.
Table 2.
Oligonucleotide primers used in this study.
| Oligo | Sequence (5′ → 3′) |
|---|---|
| P1 | GCTAACGTCATTGAATATG |
| P2 | GGAAGTAACCTCTGGATCA |
| P3 | GTTACACAAGAAAACGGCA |
| P4 | TCATCATCGTGTTTTGGG |
| P5 | AGGATGTTTCTGGTGCATGG |
| P6 | GACACAACTTCTCCTTCAGT |
| P7 | CCTTTGAACGGAAGTTTGA |
| P8 | TCTAATAGCAACCCACCT |
| P9 | GCTAACGTCATTGAATATG |
| P10 | GGATGGTTCGGATAATGC |
| P11 | GATTGGGATCATAGCGTCA |
| P12 | CTTCAGAGTTAATGGGACCA |
| P13 | AGGCATCAAGTCAGTCGTATC |
| P14 | GAAGTAACCTCTGGATCAAACG |
| P15 | GGTATGGTAAGAACTGAAGTGC |
| P16 | ATTGCAGCGGAATTGATACAG |
| P17 | TCTCCAATTGCAGGACGTGT |
| P18 | ACACTTCAAATCCTTCACCGTCT |
| P19 | TCCACAAGTCGCACGTACAG |
| P20 | GGAAGGCTTGCTACATCTAACG |
2.2. Transduction of msrA1/msrB Promoter-lacZ into S. aureus Strains
Construction of msrA1/msrB promoter-lacZ reporter strain has been previously described [6]. In this construct, a 1.3 kb DNA fragment starting 44 nucleotides downstream and going upstream of the msrA1 gene cloned in front of a promoterless lacZ gene in the vector pAZ106 [12] was integrated in the chromosome of S. aureus strain RN450 [5, 6]. The msrA1/msrB promoter-lacZ reporter was transduced into various strains of S. aureus using a phage 80α transduction procedure. Strains used in this study were verified by PCR.
2.3. Determination of the msrA1/msrB Promoter Strength in S. aureus
To determine if the msrA1/msrB locus is autoregulated, the expression of lacZ from the msrA1/msrB promoter-lacZ fusion was investigated in MsrA1-, MsrB-, and MsrA1-MsrB-deficient strains of S. aureus strains SH1000 and COL. In addition, SigB is a major regulator of stress response in S. aureus. Therefore, the strength of the msrA1/msrB promoter was also assessed in a sigB mutant. Overnight cultures of these strains were diluted (1 : 100) and grown at 37°C with shaking. These cultures were grown to OD600 = 0.5 that was considered time 0 and the levels of β-galactosidase activity in these cultures were measured at different time points (0, 90, 180, 270, and 360 min) as an indicator of the strength of the msrA1/msrB promoter.
2.4. Expression of msrA1/msrB Promoter in the Presence of a Cell Wall-Active Antibiotic, Oxacillin
Previous studies [5, 6, 9, 10, 13] have shown that, in the presence of oxacillin, there is an increased production of MsrA1 and MsrB in S. aureus. To further investigate the regulation of the msrA1/msrB locus and to see if it can be magnified in the presence of oxacillin, overnight cultures of wild-type and the derivative msrA1-msrB mutant of S. aureus strain COL were diluted (1 : 100) in fresh TSB and grown to OD600 of 0.5. 10.0 mL of the culture was split into two 15 mL tubes. To one of the cultures, oxacillin was added to the final concentration of 1.0 mg mL−1. Both cultures with and without oxacillin were allowed to grow for an additional 2 h at 37°C with shaking. Bacterial cells were harvested by centrifugation and β-galactosidase activities in these cells were measured.
2.5. Measurement of β-Galactosidase Activity
The OD600 of the culture was determined as a measure of cell density and cells were subsequently collected by centrifugation. For precise optical density readings, cultures were diluted appropriately to bring density into measurable range. The cell pellet was used to measure β-galactosidase activity as described previously using O-nitrophenyl-β-D-galactopyranoside (ONPG) as the substrate [5, 6, 13].
2.6. Quantitative Real-Time PCR (qRT-PCR) Assays
qRT-PCR assays were used to verify induced expression of the genes of the msrA1/msrB locus under oxacillin stress and to validate the lacZ reporter expression data in sigB mutants. Cultures of S. aureus strain COL were grown to OD600 = 0.3 and divided into two tubes. One tube was stressed with oxacillin at a concentration of 1.0 mg mL−1 for 2 h. Total RNA was extracted from these oxacillin stressed and control cultures as described previously [14]. For the validation of lacZ data, the wild-type and sigB mutant strains of S. aureus were allowed to grow for 90 min and 6 h after reaching the OD600 = 0.5 and total RNA from these cultures were extracted. cDNA from DNase treated 0.5 μg of total RNA was synthesized in a 20 μL reverse transcription reaction containing random hexamers and SuperScript III reverse transcriptase (Invitrogen). All real-time PCR reactions were carried out with Bio-Rad iCycler (iQ5 system). The transcript level of msrA1 was quantified using primers P13 and P14, that of msrB was quantified using P15 and P16, and that of the gene encoding the IIa(PTS) was quantified using primers P17 and P18. Transcript levels of genes were normalized to DNA gyrase mRNA using primers P19 and P20 based on a previous report [15, 16]. Changes in gene expression were calculated using the formula 2−ΔΔCq as described [17].
2.7. Statistical Analysis
All results are reported as the mean ± SE of at least three independent experiments. Data were analyzed with Student's t-test using R Studio for Windows (version 0.98.1103, 3.1.3). Statistical significance was set at p ≤ 0.05.
3. Results
3.1. Construction of msrA1/msrB Promoter-lacZ Reporter in Wild-Type and msrA1, msrB, msrA1-msrB, and sigB Mutants of S. aureus
Previously created msrA1, msrB, msrA1-msrB, and sigB knockout mutants of S. aureus strain SH1000 [6, 9, 10] were transduced in the methicillin-resistant S. aureus strain COL. These mutants and the presence of mecA gene in these strains were verified by PCR (see Supplemental Figures S1-S2 in Supplementary Material available online at http://dx.doi.org/10.1155/2015/617925). The msrA1/msrB promoter-lacZ fusion was subsequently integrated into the chromosome of these mutant strains using a bacteriophage transduction procedure. Overall, five msrA1/msrB promoter-lacZ reporter strains were created in methicillin-resistant as well as methicillin-sensitive S. aureus backgrounds. Proper integration of the msrA1/msrB promoter-lacZ fusion was also confirmed by PCR (Supplemental Figure S3).
3.2. Regulation of msrA1/msrB Locus in S. aureus
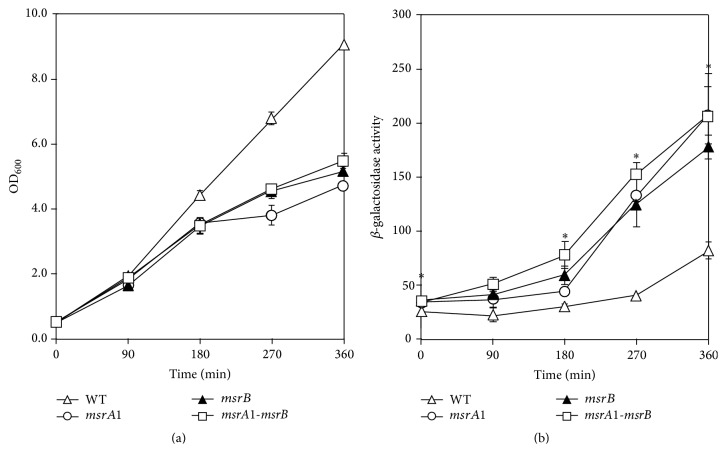
Previously, we reported higher MsrB levels in MsrA1-deficient S. aureus cells [9, 10]. This led to the speculation that the msrA1/msrB locus may in part be regulated by the products of this locus. To investigate this possibility, the level of β-galactosidase was measured in MsrA1-, MsrB-, and MsrA1-MsrB-deficient strains of S. aureus. β-galactosidase activity levels were higher in these strains compared to the activity level in the wild-type S. aureus strain SH1000 (Figure 1). The msrA1/msrB promoter-lacZ reporter was also studied in the methicillin-resistant strain COL. Overall, the expression of lacZ was lower in methicillin-resistant S. aureus compared to the methicillin-sensitive S. aureus (Figures 1(b) and 2(b)). In addition, β-galactosidase activity comparison revealed that only the msrA1-msrB double mutant strains had higher activity levels compared to the wild-type COL at the various time points (Figure 2(b)). In the individual msrA1 or msrB mutant strains, a significant increase in β-galactosidase activity was not observed compared to wild-type S. aureus COL (Figure 2(b)).
Figure 1.
Regulation of msrA1/msrB locus in a methicillin-sensitive S. aureus strain SH1000. The msrA1/msrB promoter-lacZ reporter strains were cultured in TSB and growth was measured as OD600 (a). β-galactosidase activity levels were measured in wild-type S. aureus strain SH1000 (open triangles) and its derivatives msrA1 (open circles), msrB (closed triangles), and msrA1-msrB (open square) mutants during different stages of growth (b). Values indicate averages of data from at least three independent experiments ± standard error (SE) (∗ significant at p ≤ 0.05).
Figure 2.
Regulation of msrA1/msrB locus in a methicillin-resistant S. aureus strain COL. The msrA1/msrB promoter-lacZ reporter strains were cultured in TSB and growth was measured as OD600 (a). β-galactosidase activity levels were measured in wild-type S. aureus strain COL (open triangles) and its derivatives msrA1 (open circles), msrB (closed triangles), and msrA1-msrB (open square) mutants during different stages of growth (b). Values indicate averages of data from at least three independent experiments ± standard error (∗ significant at p ≤ 0.05).
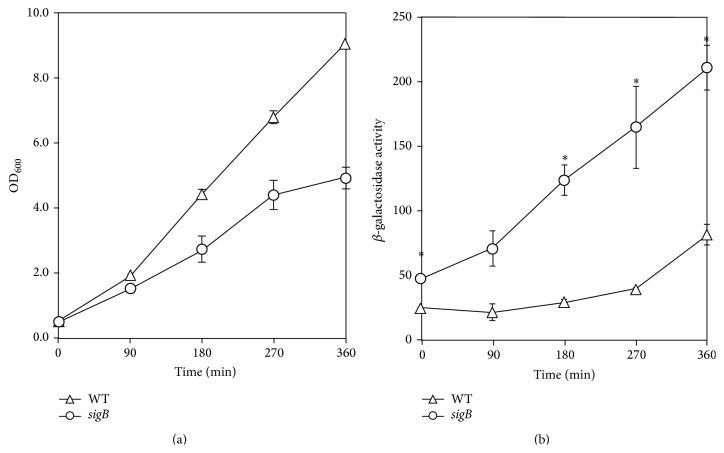
3.3. Role of SigB in the Regulation of msrA1/msrB Locus in S. aureus
Measurement of β-galactosidase activity demonstrated that there was increased expression of lacZ from the msrA1/msrB promoter when S. aureus was deficient of SigB in strain SH1000 (Figure 3(b)). However, in S. aureus COL, no such increase in the expression of lacZ was observed from the msrA1/msrB promoter under SigB-deficient conditions compared to the wild-type strain (Figure 4(b)). In qRT-PCR assays, a relatively higher level of msrA1 transcripts was observed in sigB mutant of S. aureus strain SH1000 compared to the wild-type strain (Table 3). However, this increase in msrA1 gene expression was not evident in the sigB mutant of S. aureus strain COL (Table 3) supporting the findings of the msrA1/msrB promoter-lacZ data in sigB mutant strains.
Figure 3.
Regulation of msrA1/msrB locus in a methicillin-sensitive S. aureus strain SH1000 by SigB. The msrA1/msrB promoter-lacZ reporter strains were cultured in TSB and growth was measured as OD600 (a). β-galactosidase activity levels were measured in wild-type S. aureus strain SH1000 (open triangles) and its derivative sigB mutant (closed squares) during different stages of growth (b). Values indicate averages of data from at least three independent experiments ± standard error (∗ significant at p ≤ 0.05).
Figure 4.
Regulation of msrA1/msrB locus in a methicillin-resistant S. aureus strain COL by SigB. The msrA1/msrB promoter-lacZ reporter strains were cultured in TSB and growth was measured as OD600 (a). β-galactosidase activity levels were measured in S. aureus strain COL (open triangles) and its derivative sigB mutant (closed squares) during different stages of growth (b). Values indicate averages of data from at least three independent experiments ± standard error (∗ significant at p ≤ 0.05).
Table 3.
Expression levels of msrA1 in sigB mutants relative to wild-type S. aureus strains SH1000 and COL.
| Strain | Fold increase in expression | |
|---|---|---|
| 90 min | 6 h | |
| SH1000ΔsigB1.30 | 3.16 | |
| COLΔsigB | 0.98 | 1.52 |
Values indicate averages of three independent experiments.
3.4. Induction of msrA1/msrB Locus in the Presence of Cell Wall-Active Antibiotic, Oxacillin
Previous studies have shown that the msrA1/msrB locus is induced by the cell wall-active antibiotics, oxacillin, vancomycin, and D-cycloserine, in a methicillin-sensitive S. aureus strain [5]. In a later study, while the msrA1/msrB locus remained inducible in the presence of D-cycloserine and vancomycin, no induction of this locus was noted in the presence of oxacillin, when similar experiments were carried out in a methicillin-resistant S. aureus strain COL [18]. However, in our experiments, a significantly increased β-galactosidase activity clearly indicates a significant induction of msrA1/msrB locus in the presence of oxacillin, even in a methicillin-resistant S. aureus (Figure 5). We also investigated the expression of the downstream genes of msrA1 locus in qRT-PCR assays. We determined that the oxacillin stress dramatically induced the expression of msrA1, msrB, and the gene encoding IIa(PTS) (Table 4). The expression level of the fourth gene of this locus was not investigated due to its very small size. This finding further supports our previous observation of cotranscription of the four genes of the msrA1/msrB locus [5, 6].
Figure 5.
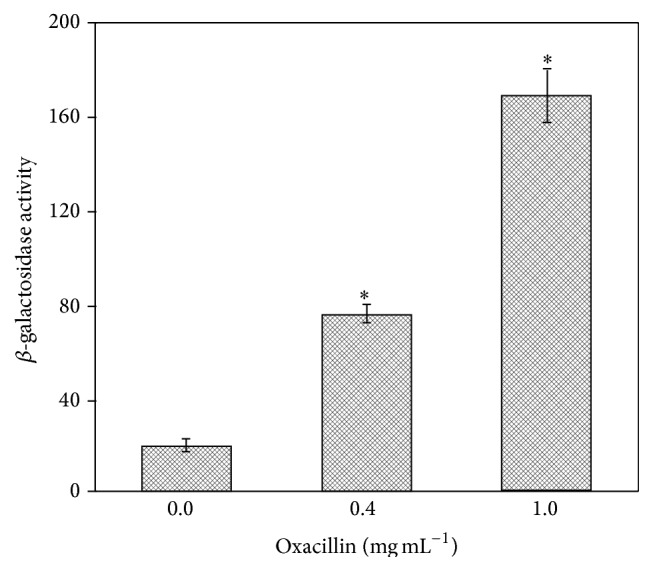
Analysis of msrA1/msrB promoter-lacZ fusion in S. aureus COL in response to oxacillin. Bacterial culture was grown in TSB to OD600 = 0.3 and then exposed to oxacillin (0.4 and 1.0 mg mL−1, resp.) for 2 h. Subsequently, cells were collected via centrifugation and the β-galactosidase activity was determined. Values indicate averages of data from at least three independent experiments ± standard error (∗ significant at p ≤ 0.05).
Table 4.
Induced expression of msrA1/msrB locus genes in S. aureus strain COL under oxacillin stress.
| Gene | Fold increase in expression under oxacillin stress |
|---|---|
| msrA1 | 22.9 |
| msrB | 18.97 |
| IIa(PTS) | 13.45 |
Values indicate averages of three independent experiments.
3.5. Expression of msrA1/msrB Locus in MsrA1-MsrB-Deficient S. aureus Strain COL in the Presence of Oxacillin
While studying the regulation of the msrA1/msrB locus in a methicillin-resistant S. aureus strain COL, oxacillin was added during the growth of the msrA1/msrB promoter-lacZ reporter to investigate any magnification of the regulation. In these studies, while an increased lacZ expression was observed in wild-type S. aureus strain COL after oxacillin treatment, a more dramatic increase in the lacZ expression in response to oxacillin was seen in MsrA1-MsrB-deficient COL (Figure 6).
Figure 6.
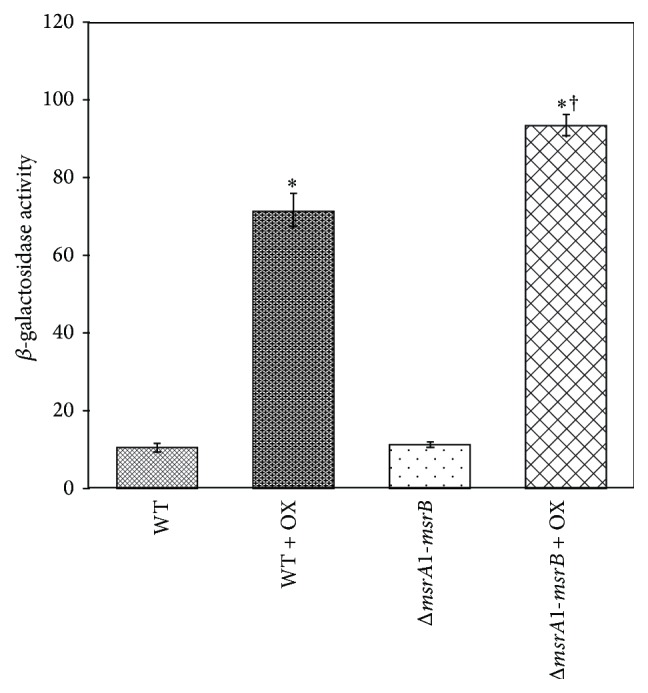
Analysis of msrA1/msrB promoter-lacZ fusion in wild-type and msrA1-msrB double mutant of S. aureus COL in response to oxacillin. At OD600 = 0.5, cells were treated with oxacillin for 2 h and the β-galactosidase activity levels were measured. Values indicate averages of data from at least three independent experiments ± standard error (∗ significant between samples with and without oxacillin at p ≤ 0.05; † significant between oxacillin treated wild-type and the oxacillin treated msrA1-msrB double mutant at p ≤ 0.05).
4. Discussion
Cell wall-active antibiotics have been used extensively for the treatment of infections caused by bacterial pathogens. S. aureus is a major human pathogen and is resistant to most commonly available antibiotics. Interestingly, cell wall-active antibiotics cause induction of a locus in S. aureus that leads to elevated synthesis of two methionine sulfoxide reductases (MsrA1 and MsrB) [5, 6]. These enzymes reduce methionine sulfoxide and play important roles in maintaining protein integrity and function particularly under oxidative stress. These two proteins have also been shown to have roles in the virulence of bacterial pathogens [19–23]. Msr-deficient bacterial mutants show a reduction in the ability to adhere to eukaryotic cells and are thus less likely to establish an inflection [21, 22, 24, 25]. It is speculated that the lack of the Msr enzymes compromises the integrity of the bacterial surface proteins responsible for adherence to eukaryotic cells. Reduced Msr activity decreases bacterial survival inside the phagocytic cells [20]. In addition to increased levels of MsrA1 and MsrB specifically in response to cell wall-active antibiotics, these proteins in S. aureus have been shown to play roles in the survival of bacterial cells under oxidative stress as well as in mice [6, 10].
We previously demonstrated that when the msrA1 gene is deleted in S. aureus, there is an increase in MsrB synthesis suggesting a possible role in the regulation of this locus [9]. Findings of this study suggest that, in a methicillin-sensitive S. aureus strain SH1000, MsrA1 and MsrB individually can downregulate the msrA1/msrB locus. However, in methicillin-resistant S. aureus strain COL, MsrA1 and MsrB both are needed to downregulate the expression of the msrA1/msrB locus. It is speculated that the msrA1/msrB locus, to some extent, is differentially regulated between methicillin-resistant and methicillin-sensitive S. aureus strains. It is not uncommon to observe a differential gene expression pattern between different S. aureus strains. It has been demonstrated that the growth of methicillin-resistant S. aureus is slower than that of methicillin-sensitive S. aureus in the lag phase but not during the exponential phase and that the alterations in virulence between these two strains may at least partially be due to the growth rate differences [26]. Deletion of a gene encoding nitric oxide synthase (NOS) in a methicillin-resistant S. aureus reduced virulence as seen by decreased bacterial survival and smaller abscess formation [27]. However, NOS was shown to have a limited role in a methicillin-sensitive S. aureus [28]. Expression of genes encoding staphylococcal superantigen-like (SSL) proteins also varies between S. aureus strains [29, 30]. Significant differences were also noted between the protein profiles of the methicillin-resistant and methicillin-sensitive S. aureus strains exposed to Triton X-100 [31].
It is well established that the msrA1/msrB locus is selectively induced in the presence of cell wall-active antibiotics [5]. These antibiotics interfere with the bacterial cell wall synthesis and, as a result, the cells become fragile and susceptible to lysis. Expression of msrA1/msrB locus is not induced by antibiotics that target other bacterial metabolic pathways [5]. In a previous report, it was shown that the msrA1/msrB locus was not induced by the presence of oxacillin but was induced by the presence of D-cycloserine and vancomycin in a methicillin-resistant S. aureus [18]. However, data from our study provide clear evidence that oxacillin does in fact induce the msrA1/msrB locus in a methicillin-resistant background of S. aureus. The previous report [18] did not observe any induction because the bacterialcells were not exposed to a high enough concentration to impose antibiotic stress in a methicillin-resistant S. aureus strain. Furthermore, we explored the induction of msrA1/msrB genes in msrA1-msrB double mutant in methicillin-resistant strain COL. An increase in induction of the msrA1/msrB locus was further magnified in msrA1-msrB double mutant exposed to oxacillin compared to the wild-type S. aureus COL in response to oxacillin. This further confirms the notion of downregulation of the msrA1/msrB locus by MsrA1 and MsrB and this is more likely an indirect effect. This speculation of an indirect regulation is based on the fact that, after conducting a protein domain search (http://prosite.expasy.org/), no specific DNA-binding domain was observed in MsrA1 and MsrB proteins. It is possible that the MsrA1 and MsrB enzymes are critical in maintaining the integrity of a cytoplasmic transcriptional regulator that is involved in the regulation of expression of this locus.
In recent years, regulation of msrA and msrB has been studied extensively across multiple species; however, none have shown that MsrA or MsrB directly or indirectly regulates its own expression. It has been demonstrated that RynB regulates the synthesis of Escherichia coli MsrB but not MsrA by binding to the 5′ untranslated region of msrB mRNA and interfering with its binding to the ribosome [32]. Nitric oxide, which is induced in Ulva fasciata upon exposure to light, upregulates the expression of msr genes in the intertidal macroalga [33]. In Saccharomyces cerevisiae, calcium phospholipid binding protein (CPBP) interacts with the msrA promoter and enhances its expression [34]. In Bacillus subtilis, a transcriptional regulator, Spx, is shown to significantly upregulate the expression of msrA and msrB [35]. Spx also upregulates msrA1 expression in S. aureus. Teicoplanin induces msrA1/msrB expression in S. aureus. However, in S. aureus spx mutant, teicoplanin exposure resulted in no significant induction of this locus, whereas, in the spx mutant strain complemented with the wild-type spx gene, msrA1/msrB induction in response to teicoplanin exposure was restored [36]. Additionally, in the spx mutant, basal msrA1 mRNA was significantly lower than spx complemented strain [36].
SigB is the alternative sigma factor in S. aureus that plays a role in the regulation of expression of stress responsive genes in S. aureus [11]. In addition, SigB is also associated with the regulation of expression of the virulence genes in S. aureus [11]. In a previous report, the level of expression of msrA1/msrB locus was investigated between RN450 (SigB−) and SH1000 (SigB+) [18]. It was shown that, in S. aureus strain SH1000, msrA1/msrB expression was 30% more induced than in S. aureus strain RN450 in the presence of oxacillin [18]. In contrast, our study shows that SigB in fact downregulates the expression of msrA1/msrB locus in S. aureus in the methicillin-sensitive S. aureus strain SH1000 and plays no role in the regulation of this locus in methicillin-resistant strain COL.
In summary, this study provides evidence that the expression of the msrA1/msrB locus is enhanced when S. aureus is deficient in MsrA1, MsrB, or both in a methicillin-sensitive S. aureus. However, in methicillin-resistant S. aureus, increased expression of the msrA1/msrB locus was apparent only when the bacterial cells were deficient in both MsrA1 and MsrB. In addition, SigB also in part downregulates the expression of this locus in methicillin-sensitive S. aureus but not in methicillin-resistant S. aureus.
Supplementary Material
“The supplemental material contains PCR verification of the bacterial strains used in this study”.
Acknowledgments
This work was supported in part by Warner/Fermaturo and ATSU Board of Trustees Research Funds and Grant 1R15AI090680-01 from the National Institutes of Health to Vineet K. Singh and a grant from KCOM Biomedical Sciences Graduate Program to Kyle R. Baum.
Conflict of Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
References
- 1.Kuehnert M. J., Kruszon-Moran D., Hill H. A., et al. Prevalence of Staphylococcus aureus nasal colonization in the United States, 2001–2002. The Journal of Infectious Diseases. 2006;193(2):172–179. doi: 10.1086/499632. [DOI] [PubMed] [Google Scholar]
- 2.Archer G. L. Staphylococcus aureus: a well-armed pathogen. Clinical Infectious Diseases. 1998;26(5):1179–1181. doi: 10.1086/520289. [DOI] [PubMed] [Google Scholar]
- 3.Lowy F. D. Staphylococcus aureus infections. The New England Journal of Medicine. 1998;339(8):520–532. doi: 10.1056/nejm199808203390806. [DOI] [PubMed] [Google Scholar]
- 4.Morell E. A., Balkin D. M. Methicillin-resistant Staphylococcus aureus: a pervasive pathogen highlights the need for new antimicrobial development. Yale Journal of Biology and Medicine. 2010;83(4):223–233. [PMC free article] [PubMed] [Google Scholar]
- 5.Singh V. K., Jayaswal R. K., Wilkinson B. J. Cell wall-active antibiotic induced proteins of Staphylococcus aureus identified using a proteomic approach. FEMS Microbiology Letters. 2001;199(1):79–84. doi: 10.1016/s0378-1097(01)00163-x. [DOI] [PubMed] [Google Scholar]
- 6.Singh V. K., Moskovitz J., Wilkinson B. J., Jayaswal R. K. Molecular characterization of a chromosomal locus in Staphylococcus aureus that contributes to oxidative defence and is highly induced by the cell-wall-active antibiotic oxacillin. Microbiology. 2001;147, part 11:3037–3045. doi: 10.1099/00221287-147-11-3037. [DOI] [PubMed] [Google Scholar]
- 7.Cabiscol E., Tamarit J., Ros J. Oxidative stress in bacteria and protein damage by reactive oxygen species. International Microbiology. 2000;3(1):3–8. [PubMed] [Google Scholar]
- 8.Moskovitz J., Singh V. K., Requena J., Wilkinson B. J., Jayaswal R. K., Stadtman E. R. Purification and characterization of methionine sulfoxide reductases from mouse and Staphylococcus aureus and their substrate stereospecificity. Biochemical and Biophysical Research Communications. 2002;290(1):62–65. doi: 10.1006/bbrc.2001.6171. [DOI] [PubMed] [Google Scholar]
- 9.Singh V. K., Moskovitz J. Multiple methionine sulfoxide reductase genes in Staphylococcus aureus: expression of activity and roles in tolerance of oxidative stress. Microbiology. 2003;149(10):2739–2747. doi: 10.1099/mic.0.26442-0. [DOI] [PubMed] [Google Scholar]
- 10.Singh V. K., Vaish M., Johansson T. R., et al. Significance of four methionine sulfoxide reductases in Staphylococcus aureus . PLoS ONE. 2015;10(2) doi: 10.1371/journal.pone.0117594.e0117594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Horsburgh M. J., Aish J. L., White I. J., Shaw L., Lithgow J. K., Foster S. J. δ B modulates virulence determinant expression and stress resistance: characterization of a functional rsbU strain derived from Staphylococcus aureus 8325-4. Journal of Bacteriology. 2002;184(19):5457–5467. doi: 10.1128/jb.184.19.5457-5467.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chan P. F., Foster S. J., Ingham E., Clements M. O. The Staphylococcus aureus alternative sigma factor sigmaB controls the environmental stress response but not starvation survival or pathogenicity in a mouse abscess model. Journal of Bacteriology. 1998;180(23):6082–6089. doi: 10.1128/jb.180.23.6082-6089.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Singh K., Singh V. K. Expression of four methionine sulfoxide reductases in Staphylococcus aureus . International Journal of Microbiology. 2012;2012:8. doi: 10.1155/2012/719594.719594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Singh V. K., Syring M., Singh A., Singhal K., Dalecki A., Johansson T. An insight into the significance of the DnaK heat shock system in Staphylococcus aureus . International Journal of Medical Microbiology. 2012;302(6):242–252. doi: 10.1016/j.ijmm.2012.05.001. [DOI] [PubMed] [Google Scholar]
- 15.Eleaume H., Jabbouri S. Comparison of two standardisation methods in real-time quantitative RT-PCR to follow Staphylococcus aureus genes expression during in vitro growth. Journal of Microbiological Methods. 2004;59(3):363–370. doi: 10.1016/j.mimet.2004.07.015. [DOI] [PubMed] [Google Scholar]
- 16.Goerke C., Campana S., Bayer M. G., Döring G., Botzenhart K., Wolz C. Direct quantitative transcript analysis of the agr regulon of Staphylococcus aureus during human infection in comparison to the expression profile in vitro. Infection and Immunity. 2000;68(3):1304–1311. doi: 10.1128/iai.68.3.1304-1311.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Livak K. J., Schmittgen T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 18.Pechous R., Ledala N., Wilkinson B. J., Jayaswal R. K. Regulation of the expression of cell wall stress stimulon member gene msrA1 in methicillin-susceptible or -resistant Staphylococcus aureus . Antimicrobial Agents and Chemotherapy. 2004;48(8):3057–3063. doi: 10.1128/aac.48.8.3057-3063.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Denkel L. A., Horst S. A., Rouf S. F., et al. Methionine sulfoxide reductases are essential for virulence of Salmonella typhimurium. PLoS ONE. 2011;6(11) doi: 10.1371/journal.pone.0026974.e26974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pang Y. Y., Schwartz J., Bloomberg S., Boyd J. M., Horswill A. R., Nauseef W. M. Methionine sulfoxide reductases protect against oxidative stress in Staphylococcus aureus encountering exogenous oxidants and human neutrophils. Journal of Innate Immunity. 2014;6(3):353–364. doi: 10.1159/000355915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saleh M., Bartual S. G., Abdullah M. R., et al. Molecular architecture of Streptococcus pneumoniae surface thioredoxin-fold lipoproteins crucial for extracellular oxidative stress resistance and maintenance of virulence. EMBO Molecular Medicine. 2013;5(12):1852–1870. doi: 10.1002/emmm.201202435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sasindran S. J., Saikolappan S., Dhandayuthapani S. Methionine sulfoxide reductases and virulence of bacterial pathogens. Future Microbiology. 2007;2(6):619–630. doi: 10.2217/17460913.2.6.619. [DOI] [PubMed] [Google Scholar]
- 23.Zhao C., Hartke A., La Sorda M., et al. Role of methionine sulfoxide reductases A and B of Enterococcus faecalis in oxidative stress and virulence. Infection and Immunity. 2010;78(9):3889–3897. doi: 10.1128/iai.00165-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dhandayuthapani S., Blaylock M. W., Bebear C. M., Rasmussen W. G., Baseman J. B. Peptide methionine sulfoxide reductase (MsrA) is a virulence determinant in Mycoplasma genitalium . Journal of Bacteriology. 2001;183(19):5645–5650. doi: 10.1128/jb.183.19.5645-5650.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wizemann T. M., Moskovitz J., Pearce B. J., et al. Peptide methionine sulfoxide reductase contributes to the maintenance of adhesins in three major pathogens. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(15):7985–7990. doi: 10.1073/pnas.93.15.7985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mizobuchi S., Minami J., Jin F., Matsushita O., Okabe A. Comparison of the virulence of methicillin-resistant and methicillin-sensitive Staphylococcus aureus . Microbiology and Immunology. 1994;38(8):599–605. doi: 10.1111/j.1348-0421.1994.tb01829.x. [DOI] [PubMed] [Google Scholar]
- 27.van Sorge N. M., Beasley F. C., Gusarov I., et al. Methicillin-resistant Staphylococcus aureus bacterial nitric-oxide synthase affects antibiotic sensitivity and skin abscess development. The Journal of Biological Chemistry. 2013;288(9):6417–6426. doi: 10.1074/jbc.m112.448738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vaish M., Singh V. K. Antioxidant functions of nitric oxide synthase in a methicillin sensitive Staphylococcus aureus . International Journal of Microbiology. 2013;2013:6. doi: 10.1155/2013/312146.312146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pantrangi M., Singh V. K., Shukla S. K. Regulation of Staphylococcal superantigen-like gene, ssl8, expression in Staphylococcus aureus strain, RN6390. Clinical Medicine & Research. 2015;13(1):7–11. doi: 10.3121/cmr.2014.1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pantrangi M., Singh V. K., Wolz C., Shukla S. K. Staphylococcal superantigen-like genes, ssl5 and ssl8, are positively regulated by Sae and negatively by Agr in the Newman strain. FEMS Microbiology Letters. 2010;308(2):175–184. doi: 10.1111/j.1574-6968.2010.02012.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cordwell S. J., Larsen M. R., Cole R. T., Walsh B. J. Comparative proteomics of Staphylococcus aureus and the response of methicillin-resistant and methicillin-sensitive strains to Triton X-100. Microbiology. 2002;148(9):2765–2781. doi: 10.1099/00221287-148-9-2765. [DOI] [PubMed] [Google Scholar]
- 32.Bos J., Duverger Y., Thouvenot B., et al. The sRNA RyhB regulates the synthesis of the Escherichia coli methionine sulfoxide reductase MsrB but not MsrA. PLoS ONE. 2013;8(5) doi: 10.1371/journal.pone.0063647.e63647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hsu Y.-T., Lee T.-M. Nitric oxide up-regulates the expression of methionine sulfoxide reductase genes in the intertidal macroalga Ulva fasciata for high light acclimation. Plant and Cell Physiology. 2012;53(2):445–456. doi: 10.1093/pcp/pcr190. [DOI] [PubMed] [Google Scholar]
- 34.Hanbauer I., Boja E. S., Moskovitz J. A homologue of elongation factor 1 gamma regulates methionine sulfoxide reductase a gene expression in Saccharomyces cerevisiae . Proceedings of the National Academy of Sciences of the United States of America. 2003;100(14):8199–8204. doi: 10.1073/pnas.1432898100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.You C., Sekowska A., Francetic O., Martin-Verstraete I., Wang Y., Danchin A. Spx mediates oxidative stress regulation of the methionine sulfoxide reductases operon in Bacillus subtilis . BMC Microbiology. 2008;8, article 128 doi: 10.1186/1471-2180-8-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Renzoni A., Andrey D. O., Jousselin A., et al. Whole genome sequencing and complete genetic analysis reveals novel pathways to glycopeptide resistance in Staphylococcus aureus . PLoS ONE. 2011;6(6) doi: 10.1371/journal.pone.0021577.e21577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pfeltz R. F., Singh V. K., Schmidt J. L., et al. Characterization of passage-selected vancomycin-resistant Staphylococcus aureus strains of diverse parental backgrounds. Antimicrobial Agents and Chemotherapy. 2000;44(2):294–303. doi: 10.1128/aac.44.2.294-303.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Singh V. K., Schmidt J. L., Jayaswal R. K., Wilkinson B. J. Impact of sigB mutation on Staphylococcus aureus oxacillin and vancomycin resistance varies with parental background and method of assessment. International Journal of Antimicrobial Agents. 2003;21(3):256–261. doi: 10.1016/s0924-8579(02)00359-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
“The supplemental material contains PCR verification of the bacterial strains used in this study”.