Abstract
Background
India has the world’s largest number of diabetics. Non-traumatic lower limb amputation is the most common devastating complication of diabetes, primarily due to diabetic foot ulcers (DFU) and diabetic foot infections (DFI). In India, the incidence of foot ulcers ranges from 8–17 per cent. DFIs are predominantly polymicrobial and multidrug-resistant (MDR) with the ability to form biofilm, which is an important virulence factor and results in treatment failure.
Aims
The main objectives of the study are to identify the spectrum of multidrug-resistant bacteria associated with these infections, their antibiotic sensitivity pattern, and to detect the biofilm formation.
Methods
This was a prospective study at a tertiary care hospital. One hundred patients over the age of 18, having chronic diabetic foot ulcer, and attending the surgery outpatient department were included. Samples of pus were collected from deep wounds and processed using standard techniques for culture and sensitivity. Biofilm detection was done. Results were compiled and statistically analysed.
Results
One hundred samples were processed and 82 yielded positive cultures. Staphylococcus aureus was the predominant organism, followed by Pseudomonas aeruginosa. Biofilm formation was seen in 38 (46.34 per cent) of the organisms. Biofilms were formed predominantly by Staphylococcus aureus (20 per cent).
Conclusion
The organisms causing chronic diabetic foot ulcers were commonly multidrug-resistant; this was also observed among biofilm formers. Therefore, screening for biofilm formation, along with the usual antibiogram, needs to be performed as a routine procedure in chronic diabetic ulcers to formulate effective treatment strategies for these patients.
Keywords: Biofilm, diabetic foot ulcers, multidrug-resistant bacteria
What this study adds:
-
What is known about this subject?
Biofilm formation is widespread in chronic wounds such as diabetic foot ulcers (DFUs). The biofilm phenotypes give rise to multidrug-resistant strains resulting in treatment failure. Most of these infections are polymicrobial.
-
What new information is offered in this study?
The organisms causing chronic diabetic foot ulcers were commonly multidrug-resistant, which was also observed among biofilm formers. Most of the infections were monomicrobial.
-
What are the implications for research, policy, or practice?
To develop effective treatment strategies for diabetics with chronic diabetic foot ulcers, screening for biofilm formation, along with the usual antibiogram, needs to be performed as a routine procedure. Regular wound debridement and stringent antibiotic usage policy also needs to be implemented.
Background
Bacteria within biofilms are sheltered from various stresses, including immune responses and antimicrobial agents. The biofilm-forming ability of bacteria has been associated with increased antibiotic resistance and chronic recurrent infections especially in diabetics.1 Diabetes is a common disease in India with a prevalence rate of 12–17 per cent in the urban population and 2.5 per cent in the rural population.1 A common devastating complication is non-traumatic lower limb amputation mostly due to diabetic foot ulcers (DFUs) and diabetic foot infections (DFIs).1 The incidence of foot ulcers ranges from 8–17 per cent in India.2 The presence of peripheral neuropathy seems to contribute to the development of ulceration and those with pre-ulceration, callosities, and deformity seem to be at increased risk.2
Diabetics with ulcers commonly experience infection with gram-positive organisms such as Staphylococcus aureus, Enterococcus, and gram-negative organisms like Pseudomonas aeruginosa, Escherichia coli, Klebsiella species, Proteus species, etc., and anaerobes.3 These organisms also show multi-drug resistance.4 The microorganism that colonises the surface wound also provides an ideal niche for further invasion resulting in these infections.
These different microorganisms can exist independently or combine together to form micro-communities within a matrix of extracellular polymeric substances called biofilms. The ability of a microorganism to form biofilm is an important virulence factor as it establishes a protective environment for the organisms to survive and evade antibiotics. These biofilms are the main cause of many chronic infections such as diabetic foot ulcers, and they pave the way for the re-emergence of multidrug-resistant strains and result in treatment failure.1 Biolfilms are difficult to eradicate using conventional antibiotics, hence the identification of biofilm producers among clinical isolates may lead to better management of wound infections in diabetics who, in spite of repeated antibiotic treatment, fail to respond to treatment because biofilms are not being tested for routinely.
Method
This prospective study was conducted at the Department of Microbiology, in a tertiary care research and referral hospital attached to a medical college and research institute. One hundred patients attending the surgery outpatient department of the hospitals were included in the study. Institutional ethical clearance was taken and informed consent was obtained from the subjects in their own language.
All patients over 18 years of age having chronic diabetic foot ulcers where ulcer duration is greater than three months were included in the study.5 These patients had received antibiotics earlier. Children (<18 years), pregnant women, and patients with other comorbid conditions like HIV infection, chronic venous insufficiency, and osteomyelitis were excluded.
The patients were assessed through detailed history and clinical examination. Surgeons assessed the ulcers, and after debridement material for culture was collected with a cotton-tipped sterile swab from the deeper parts of the foot ulcer. The ulcers were not demarcated as per the Wegner classification of ulcers. The swabs were transported immediately to the Department of Microbiology for culture and sensitivity and biofilm formation. Swabs received were cultured on blood agar and McConkey agar and the plates were incubated overnight at 37°C. Colonies obtained were identified by using standard techniques.6 Antibiotic sensitivity was done using Kirby Bauer’s disc diffusion technique method as described in the Clinical Laboratory Standard Institute (CLSI) guidelines 2012. Multidrug-resistant organisms for gram-positive and gram-negative bacteria are resistant to three or more antimicrobial classes as per the guidelines.7 No concomitant blood cultures were collected.
The biofilm formation was detected by Congo Red method as described by Freeman et al.8 A specially prepared medium composed of Brain Heart Infusion (BHI) broth (37gm/L), sucrose (50gm/L), agar no.1 (10gm/L) and Congo Red stain (0.8gm/L) was used. Congo Red was prepared as concentrated aqueous solution and autoclaved at 121°C for 15 minutes, separately from other medium constituents and was added when the agar had cooled to 55°C. Plates were inoculated and incubated aerobically for 24–48 hours at 37°C. Biofilm formers produced black colonies with a dry crystalline consistency, while weak slime producers usually remained pink, though occasional darkening at the centres of colonies was observed. Indeterminate results were characterised by darkening of the colonies with the absence of a dry crystalline colonial morphology. The tests were carried out in triplicate and repeated three times.9
Stepanovic et al. described the tissue culture plate method in plastic microtitre plates.10 On a sterile 96 well flat-bottomed polystyrene microtitre plate, 230μl of Trypticase Soya Broth (TSB) was added. Also, 20μl of overnight bacterial culture was added to the corresponding well (each strain in three successive wells). The negative control wells contained broth only. The plates were incubated aerobically for 24 hours at 35°C. The content of the wells was poured off and the wells were washed three times with 300μl of sterile distilled water. The bacteria adhering to the wells were fixed with 250μl of methanol for 15 minutes. Then the wells were stained with 250μl of one per cent solution of crystal violet for five minutes. Excess stain was removed by washing and the wells were air-dried. The dye bound to the wells was solubilised with 250μl of 33 per cent (v/v) glacial acetic acid. Theoptical density (O.D.) of each well was measured at 490nm using an ELISA auto reader.
The tests were carried out in triplicate and the results were averaged. The cut-off O.D (O.D.c) was determined as three standard deviations above the mean O.D. of the negative control. Strains were classified as biofilm producer and no biofilm producer. Data was compiled and descriptive statistics were applied using Microsoft Excel 2010 Edition (Microsoft, Seattle, WA).
Results
One hundred samples were collected from patients with chronic diabetic foot ulcers. The study group comprised 84 male patients and 16 female patients, whose ages ranged from 35–80 years. From these samples, 82 isolates were obtained. No polymicrobial infections were noted. Overall, 20 organisms (24.4 per cent) were gram-positive and 62 organisms (75.6 per cent) were gram-negative. Staphylococcus aureus and Escherichia coli were the most commonly isolated organisms (24.4 per cent each) followed by Pseudomonas aeruginosa (17.1 per cent), Citrobacter sp. (12.1 per cent), Klebsiella oxytoca (12.1 per cent), and Proteus sp. (9.8 per cent). This is depicted in Figure 1. With reference to the gram-negative organisms, 53.6 per cent of the organisms were extended spectrum beta lactamase (ESBL) producers, with the highest production by E. coli.
Figure 1: Comparison of biofilm-forming organisms.
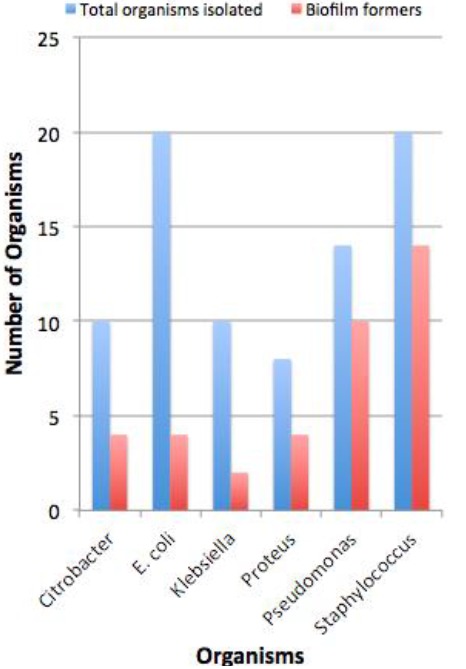
Thirty-eight (46.34 per cent) of the isolates showed biofilm formation. Staphylococcus aureus was the predominant biofilm former, with 14 (38.8 per cent) of the isolates testing positive for biofilm formation. All 10 (100 per cent) of the MRSA isolates were biofilm formers, while only four (40 per cent) of the MSSA isolates formed biofilm. The second highest biofilm formation was by Pseudomonas aeruginosa (26.5 per cent) followed by Citrobacter sp. (10.5 per cent), E. coli (10.5 per cent), Proteus sp. (10.5 per cent), and Klebsiella oxytoca (5.26 per cent). Eighty isolates (97.56 per cent) were MDR with 37 (46.3 per cent) of the MDR isolates also showing biofilm formation (Figure 2).
Figure 2: Congo-red agar – detection of biofilm.
The limitations of this study were that anaerobic culture and MIC were not performed due to logistical issues.
Discussion
The annual population-based incidence of diabetic foot ulcers is estimated to be 1.0–4.1 per cent, while the lifetime rate extends to around 25 per cent.11 A common complication of these ulcers is infection, which if left untreated, results in the need for distal limb amputation.12 According to the United States National Institutes of Health (NIH), more than 80 per cent of chronic bacterial infections are associated with biofilms.13
In the present study, all the samples yielded monomicrobial isolates. This is significantly different from most study results in which DFUs are polymicrobial in nature.14,15 However, some studies have shown lower than expected rates of polymicrobial infection.16 The monomicrobial nature of infection is associated with the duration of the ulcer and antimicrobial treatment. Earlier on in the infection, the monomicrobial state prevails and as the infection progresses with time, a polymicrobial state arises. Also, ulcers that are shallower and that have a lesser degree of necrosis tend to be monomicrobial.4 It is necessary to note that studies have shown that in polymicrobial infections not all isolates have to be eradicated to ensure an improvement in the ulcer’s healing process. In our study, it could also be attributed to the fact that all the patients were on antimicrobial treatment during sampling and only the multidrug-resistant organisms not responding to the treatment would have been cultured.
Of the isolates, 24.4 per cent were found to be gram-positive while 75.6 per cent were gram-negative. This corresponds with the findings of Bhansal et al.,14 in which 76 per cent of the microbes were gram-negative and 24 per cent were gram-positive. The predominance of gram-negative organisms has been noted in several studies.4,17 However, certain studies1,18,19 have established a higher proportion of gram-positive organisms. In this study, Staphylococcus aureus and Escherichia coli were the most commonly isolated organisms (24.40 per cent each) followed by Pseudomonas aeruginosa (17.07 per cent). These results were similar to those obtained by Bhansal et al.14 However, a study in Malaysia reported Proteus sp. to be the predominant gram-negative organism.16
Amongst the Staphylococcus aureus, MRSA (12.2 per cent) and MSSA (12.2 per cent) were obtained. MSSA exhibited resistance (100 per cent) to penicillin G, cotrimoxazole (60 per cent), and ciprofloxacin (60 per cent). The prevalence of MRSA was similar to a Malaysian study,16 but was lower when compared to prior studies in which it was from 40–69.8 per cent.14,15,20,21 The MRSA displayed a high level of resistance to clindamycin (80 per cent), erythromycin (80 per cent), and penicillin G (80 per cent). All of the organisms were sensitive to linezolid, vancomycin, and imipenem (100 per cent). This is similar to the study by Rani et al., where the gram-positive organisms showed complete sensitivity to vancomycin, linezolid, and teicoplanin.18
The gram-negative organisms in our study showed a high level of resistance to amoxicillin+clavulanic acid (56.1 per cent), ceftazidime (53.66 per cent), ciprofloxacin (46.34 per cent), cefoxitin, cephalothin, and cefuroxime (43.9 per cent each). The organisms were most sensitive to piperacillin+tazobactam (90.2 per cent) and imepenam (100 per cent). This corresponds with the findings of Rani et al. in which imepenam, cefaperazone+sulbactam, cefepime+tazobactam, and piperacilllin+tazobactam are reported as the most effective drugs against ESBL-producing gram-negative bacilli18 and those of Aasha et al.22 Of the organisms, 53.7 per cent of the organisms were ESBL producers, with the highest production by E. coli. This is similar to other studies in which 44.7–57.4 per cent are ESBL producers,16,21 but significantly lower than the 80 per cent ESBL formers found by Mamdouh et al.23
Studies have shown that biofilm-associated microorganisms can be up to 1,000 times more resistant to antibiotics than free-floating planktonic bacteria.13 In the present study, 80 isolates (97.6 per cent) were multidrug resistant with 37 (46.3 per cent) of the MDR isolates also showing biofilm formation. Swarna et al. reported that 80.4 per cent of the MDR organisms were biofilm formers,1 and this is a significantly larger percentage in comparison to the present result.
The biofilm structure has been analysed microscopically and biochemically to show multiple layers of bacteria encased in a biofilm matrix containing proteins, DNA, and polysaccharides. The mechanism of multidrug resistance in biofilm-forming organisms is believed to be a direct result of close cell-cell contact in the biofilm, which allows for easy transfer of plasmids containing MDR genes amongst one another.24 Organisms, which form biofilms, are also characterised by tolerance, which is a temporary, non-heritable characteristic. The mechanisms for tolerance are: (1) Antibiotics whose mechanism of action depends on the division of cells are inactive against microbes in a biofilm, which are in a slow-growing, dormant state.25 (2) Drug permeation is hindered the polysaccharide matrix of the biofilm. 23(3) Drug efficacy is altered in the microenvironment of the biofilm (pH and osmotic variations).26 In addition to their effect on antimicrobial agents, biofilms also block host defences. They have an anti-phagocytic property, which inactivates leukocytes in the polysaccharide matrix. There is also an element within the matrix that disables both complement and host antibodies.27
In our study 46.3 per cent of the isolates showed biofilm formation. This was lower compared to prior studies in which it ranged from 73–77.1 per cent.1,28 A study by James et al. recorded a rate of 60 per cent in chronic wounds, and 6 per cent in acute wounds.10 Such a deviation from the norm could be due to effective debridement procedures29 or shorter duration of ulcer in the patients. Staphylococcus aureus was the predominant biofilm former, with 38.8 per cent of the isolates testing positive for biofilm formation. This is an expected result, with existing literature supporting the biofilm forming nature of Staphylococci.30 Staphylococcus aureus is followed by Pseudomonas aeruginosa with 26 per cent. Studies have reported Pseudomonas aeruginosa to form biofilms more readily in the diabetic wound environment.13
Conclusion
Difficulty in eradicating a chronic diabetic foot infection associated with biofilm formation has been reported, and biofilm-producing bacteria have been shown to resist higher antibiotic and disinfectant concentrations than non-biofilm producing bacteria. Therefore, additional screening of multidrug-resistant organisms as well as non-resistant organisms like MSSA often associated with biofilms should be considered. Detection of biofilm formation is an easy and cost-effective test that can be performed routinely in the laboratory. Detection of biofilm will help surgeons to effectively manage these infections by providing more aggressive source control and appropriate antibiotics resulting in decrease mortality and the morbidity in patients.
ACKNOWLEDGEMENTS
None
Footnotes
PEER REVIEW
Not commissioned. Externally peer reviewed.
CONFLICTS OF INTEREST
The authors declare that they have no competing interests
FUNDING
None
ETHICS COMMITTEE APPROVAL
Bangalore Medical College and Research Institute Ethics Committee Reference Number: BMCRI/PS/224/2014-15
Please cite this paper as: Banu A, Noorul Hassan MM, Rajkumar J, Srinivasa S. Spectrum of bacteria associated with diabetic foot ulcer and biofilm formation: A prospective study. AMJ 2015;8(9): 280–285. http//dx.doi.org/10.4066/AMJ.2015.2422
References
- 1.Swarna SR, Radha M, Gomathi S. et al. A study of Biofilm on Diabetic Foot Ulcer. International Journal of Research in Pharmaceutical and Biomedical Sciences. 2012;3(4):1809–14. [Google Scholar]
- 2.Alex R, Ratnaraj B, Winston B. et al. Risk Factors for Foot Ulcers in Patients with Diabetes Mellitus – A Short Report from Vellore, South India. Indian Journal of Community Medicine : Official Publication of Indian Association of Preventive & Social Medicine. 2010;35(1):183–5. doi: 10.4103/0970-0218.62582. doi:10.4103/0970-0218.62582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shankar EM, Mohan V, Premalatha G. et al. Bacterial etiology of diabetic foot infections in South India. Eur J InternMed. 2005;16:56770. doi: 10.1016/j.ejim.2005.06.016. doi:http://dx.doi.org/10.1016/j.ejim.2005.06.016. [DOI] [PubMed] [Google Scholar]
- 4.Banu A, Noorul Hassan MM, Rajkumar J. et al. Prospective study of Multidrug Resistant Bacteria causing Diabetic Foot Ulcers in South India. Journal of Science. 2015;5(8):626–9. [Google Scholar]
- 5.Lauren C, Samina S. Diagnosis and Treatment of Venous Ulcers. Am Fam Physician. 2010;81(8):989–96. [PubMed] [Google Scholar]
- 6.Collee JG, Fraser AG, Marmion BP. et al. Practical Medical Microbiology. 14thed. New York: Churchill Livingstone; 2006. [Google Scholar]
- 7.Clinical and Laboratory Standards Institute. Performance standards for Antimicrobial Susceptibility testing; Twenty First Informational Supplement. CLSI document M100-S21. Wayne, PA: 2012. [Google Scholar]
- 8.Freeman DJ, Falkiner FR, Keane CT. New method for detecting slime production by coagulase negative staphylococci. J Clin Pathol. 1989;42:872–4. doi: 10.1136/jcp.42.8.872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mathur T, Singhal S, Khan S. et al. Detection of biofilm formation among the clinical isolates of Staphylococci: An evaluation of three different screening methods. Indian J Med Microbiol. 2006;24(1):25–9. doi: 10.4103/0255-0857.19890. doi: 10.4103/0255-0857.19890. [DOI] [PubMed] [Google Scholar]
- 10.James G, Swogger E, Wolcott R. et al. Biofilms in Chronic wounds. Wound Repair Regen. 2008 Jan-Feb;16(1):37–44. doi: 10.1111/j.1524-475X.2007.00321.x. Epub 2007 Dec 13. [DOI] [PubMed] [Google Scholar]
- 11.Wu SC, Driver VR, Wrobel JS. et al. Foot ulcers in the diabetic patient, prevention and treatment. Vasc Health Risk Manag. 2007;3(1):65–76. [PMC free article] [PubMed] [Google Scholar]
- 12.Singh N, Armstrong DG, Lipsky BA. Preventing Foot Ulcers in patients with Diabetes. JAMA. 2005;293:217–28. doi: 10.1001/jama.293.2.217. [DOI] [PubMed] [Google Scholar]
- 13.Ashok D. Why Diabetic Foot Ulcers do not heal? JIMSA. 2011 Oct-Dec;24(4):205. [Google Scholar]
- 14.Bansal E, Garg A, Bhatia S. Spectrum of microbial flora in diabetic foot ulcers. Indian J Pathol Microbiol. 2008;51:204–8. doi: 10.4103/0377-4929.41685. [DOI] [PubMed] [Google Scholar]
- 15.Pathare NA, Bal A, Talwalkar GV. et al. Diabetic foot infections: A study of microorganisms associated with the different Wagner Grades. Indian J Pathol Microbiol. 1998;41:437–41. [PubMed] [Google Scholar]
- 16.Raja NS. Microbiology of diabetic foot infections in a teaching hospital in Malaysia: a retrospective study of 194 cases. J Microbiol Immunol. Infect. 2007;14(1):45–9. [PubMed] [Google Scholar]
- 17.Gadepalli R, Dhawan B, Sreenivas V. et al. Clinico-Microbiological study of Diabetic foot ulcers in an Indian tertiary care hospital. Diabetes Care. 2006;29:1727–32. doi: 10.2337/dc06-0116. doi: 10.2337/dc06-0116. [DOI] [PubMed] [Google Scholar]
- 18.Rani V, Nithyalakshmi J. A comparative study of Diabetic and Non-diabetic wound infections with special reference to MRSA and ESBL. Int J Curr Microbiol App Sci. 2014;3(12):546–54. [Google Scholar]
- 19.Lipsky BA, Pecoraro RE, Chen MS. et al. Factors affecting Staphylococcal colonization among NIDDM outpatients. Diabetes Care. 1987;10:483–6. doi: 10.2337/diacare.10.4.483. [DOI] [PubMed] [Google Scholar]
- 20.Adel A, Ibrahim BZ, Al-Shamali AA. et al. Bacteriological study of diabetic foot infections. J. Diabet Complications. 2005;19(3):138–41. doi: 10.1016/j.jdiacomp.2004.06.001. doi: http://dx.doi.org/10.1016/j.jdiacomp.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 21.Viswanathan V, Jasmine JJ, Snehalatha C. et al. Prevalence of pathogens in diabetic foot infection in South India type 2 diabetic patients. J Assoc Physicians India. 2002;50:1013–6. [PubMed] [Google Scholar]
- 22.Aasha H, Kairavi JD, Ravindra J. et al. A study of aerobic and anaerobic bacteria in diabetic foot ulcer and in vitro sensitivity of antimicrobial agent. Int J Med Sci Public Health. 2014;3(7):818–21. doi: 10.5455/ijmsph.2014.220420145. [Google Scholar]
- 23.Mamdouh ME, Saif-AI A. Diabetic foot infection: Bacteriological causes and antimicrobial therapy. Journal of American Science. 2012;8(10):389–93. [Google Scholar]
- 24.Mah T, O’Toole G. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 2001;9:34–9. doi: 10.1016/s0966-842x(00)01913-2. doi:10.1016/S0966-842X(00)01913-2. [DOI] [PubMed] [Google Scholar]
- 25.Mertz P. Cutaneous biofilms: friend or foe? Wounds. 2003;15:129–32. [Google Scholar]
- 26.Trivedi U, Parameswaran S, Armstrong A. et al. Prevalence of Multiple Antibiotic Resistant Infections in Diabetic versus Nondiabetic Wounds. J Pathog. 2014;2014:173053. doi: 10.1155/2014/173053. doi: 10.1155/2014/173053. Epub 2014 Jun 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leid J. Human leukocytes adhere to, penetrate, and respond to Staphylococcus aureus biofilms. Infect Immun. 2002 Nov;70(11):6339–45. doi: 10.1128/IAI.70.11.6339-6345.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Percival L, Bowler P. Biofilms and their potential role in wound healing. Wounds. 2004;16:234–40. [Google Scholar]
- 29.Zubair M, Malik A, Ahmad J. et al. A study of biofilm production by gram-negative organisms isolated from diabetic foot ulcer patients. Biol Med. 2011;3(2):147–57. [Google Scholar]
- 30.Gordon RJ, Lowy FD. Pathogenesis of methicillin-resistant Staphylococcus aureus infection. Clin Infect Dis. doi: 10.1086/533591. 2008 Jun 1;46 Suppl 5:S350-9. doi: 10.1086/533591. Review. [DOI] [PMC free article] [PubMed] [Google Scholar]


