Abstract
Genome defense likely evolved to curtail the spread of transposable elements and invading viruses. A combination of effective defense mechanisms has been shown to limit colonization of the Neurospora crassa genome by transposable elements. A novel DNA transposon named Sly1-1 was discovered in the genome of the most widely used laboratory “wild-type” strain FGSC 2489 (OR74A). Meiotic silencing by unpaired DNA, also simply called meiotic silencing, prevents the expression of regions of the genome that are unpaired during karyogamy. This mechanism is posttranscriptional and is proposed to involve the production of small RNA, so-called masiRNAs, by proteins homologous to those involved in RNA interference−silencing pathways in animals, fungi, and plants. Here, we demonstrate production of small RNAs when Sly1-1 was unpaired in a cross between two wild-type strains. These small RNAs are dependent on SAD-1, an RNA-dependent RNA polymerase necessary for meiotic silencing. We present the first case of endogenously produced masiRNA from a novel N. crassa DNA transposable element.
Keywords: meiotic silencing, small RNA, transposon, genome defense, Neurospora
Genome protection is of paramount importance during sexual reproduction when DNA is replicated for packaging into gametes because, at this time, it is particularly susceptible to invasion by viruses and transposable elements (TEs) (Calvi and Gelbart 1994; Collins et al. 1987; Pelisson et al. 2002; Prudhomme et al. 2005; Shapiro 2005). To protect host genomes from attack by foreign elements, “genome defense” mechanisms have evolved that use gene silencing or targeted mutations to combat nonself elements (Aravin et al. 2003; Goodier and Kazazian 2008; Lau 2010). The fungus Neurospora crassa has at least three genome defense mechanisms that have limited the colonization of its genome by selfish elements (Galagan et al. 2003). The genome defense systems include the irreversible repeat-induced point mutation (RIP) (Selker and Garrett 1988; Cambareri et al. 1989) and two reversible posttranscriptional mechanisms, the RNA interference (RNAi)-like “quelling” (Romano and Macino 1992; Cogoni et al. 1994) and meiotic silencing (Aramayo and Metzenberg 1996; Shiu et al. 2001; Shiu and Metzenberg 2002). RIP is a premeiotic hypermutation process that targets duplicated segments of DNA (Selker and Garrett 1988; Cambareri et al. 1989) by converting C:G to T:A in both copies of the duplicated regions. Quelling is a posttranscriptional, small RNA-based gene-silencing pathway that has so far been only studied in detail in the asexual stages of the life cycle (Fulci and Macino 2007). The third genome defense system, first considered a form of “transvection” (Aramayo and Metzenberg 1996) and later called meiotic silencing by unpaired DNA (MSUD) (Shiu and Metzenberg 2002; Shiu et al. 2001) or simply meiotic silencing (Kelly and Aramayo 2007), occurs after karyogamy and targets transcripts that originate from regions with dissimilar DNA sequence and are therefore are unpaired. The system also affects RNA that is produced from additional paired alleles (Aramayo and Metzenberg 1996; Shiu et al. 2001).
The mechanism for detection of unpaired regions remains elusive, although DNA repair components have been linked to its efficiency (Samarajeewa et al. 2014). Genetic crosses of strains with unpaired regions show transient silencing of transcripts from genes in these region (Shiu et al. 2001; Lee et al. 2004; Shiu et al. 2006; Alexander et al. 2008), and this silencing is limited to stages from early karyogamy until ascospore, as tracked by expression of histone H1-green fluorescence protein fusion genes (Jacobson et al. 2008). It is hypothesized that RNAs produced from unpaired regions are detected as “aberrant” and subject to RNAi-mediated silencing (Lee et al. 2004). Many mutated genes affecting meiotic silencing are homologous to genes in RNAi pathways in plants, fungi, and animals. These genes include sad-1, a putative RNA-dependent RNA polymerase (Shiu and Metzenberg 2002; Shiu et al. 2001), dcl-1/sms-3, a Dicer-type exonuclease (Alexander et al. 2008), sms-2, an Argonaute homolog (Lee et al. 2003), QIP, which converts duplex RNA into siRNAS (Xiao et al. 2010; Lee et al. 2010a), and additional scaffold proteins and components SAD-2, SAD-3, SAD-4, SAD-5, and SAD-6 (Xiao et al. 2010; Hammond et al. 2011, 2013b; Samarajeewa et al. 2014; Decker et al. 2015). Suppression of meiotic silencing in some cases has enabled meiotic drive elements such as Spore killer (Raju et al. 2007; Hammond et al. 2012; Harvey et al. 2014). Recent work in support of the hypothesis that RNAi is involved in meiotic silencing used an engineered deletion at the Rsp locus to show that small RNAs are produced from this unpaired region during meiosis (Hammond et al. 2013a). However, small RNAs have not yet been reported from matings between wild-type strains with unpaired regions segregating in natural populations.
TEs present in only one parent will be unpaired during sexual crosses and thus become natural substrates for meiotic silencing. One proposed role for meiotic silencing and other genome defense mechanisms has been to control the spread of TEs (Nolan et al. 2005; Catalanotto et al. 2006; Girard and Hannon 2008). So far, however, there has been no direct demonstration for a role of this genome defense system in the control of TEs. This lack can be explained, in part, by the few active TEs in N. crassa. Currently only Tad (Transposon in Adiopodumé), a long interspersed element−like retroelement found intact and active in the Adiopodumé strain, has been demonstrated to transpose (Kinsey 1989; Kinsey and Helber 1989; Kinsey et al. 1994; Zhou et al. 2001). In addition, relics of TEs that have accumulated as a consequence of RIP have been described in the N. crassa reference genome derived exclusively from FGSC 2489 (OR74A) (Selker et al. 2003).
By comparing genomes of several laboratory strains, multiple loci in the reference 2489 from the Fungal Genetics Stock Center (FGSC, University of Missouri, Kansas City, MO) were identified to be missing among individuals in this pedigree. One of the largest of these detected insertion/deletions is a TE we named Sly1-1. When we sequenced small RNAs from three stages during premeiotic and meiotic development, we detected “meiotic-silencing-associated small interfering RNAs” (masiRNAs) that originated from Sly1-1. Here, we present evidence that Sly1-1 is an active DNA-type transposon that is recognized by meiotic silencing when unpaired during meiosis. Furthermore, we confirm that the meiotic silencing machinery is required for the production of masiRNAs emanating from Sly1-1. Thus we provide support for the role of meiotic silencing in genome defense through detection of meiotic small RNAs targeting a TE in an unpaired state.
Materials and Methods
Strains and growth conditions
Neurospora strains used in this study are listed in Supporting Information, Table S1. All strains were originally obtained from the FGSC and are maintained in the senior author’s laboratories. Mycelia of strains used for DNA extraction were collected after growth in liquid Vogel’s medium N (Vogel 1956) at 25° for 3 d. Vegetative tissues of FGSC 2489 for RNA extraction was collected after growth on solid Vogel’s medium N in the dark at 30° for 3 d, followed by growth in the light at room temperature for 2 d. For tissue collection during sexual development, FGSC 2489 was first grown on synthetic crossing medium (Westergaard and Mitchell 1947) covered with cellophane (Midsci, St. Louis, MO) in 245 × 245 × 25-mm bioassay dishes (Thermo Scientific, Hvidovre, Denmark). After 10 d of growth, at room temperature in light/dark conditions, we looked for protoperithecia (PP) under a dissecting microscope, and then harvested tissues enriched with PP by scraping from the cellophane, followed by flash-freezing in liquid nitrogen. Additional plates of PP were crossed with either the wild-type FGSC 8820 or Sad-1Δ strains. Regions enriched with perithecia were cut by sterile razor blades after 2, 4, and 6 d postfertilization (PF). Tissues from these regions were scraped from cellophane and flash-frozen in liquid nitrogen. All collected tissues were stored at −80° until further use.
RNA extraction and small RNA Northern blots
Harvested sexual tissue was processed for RNA extracted for northern blots or small RNA sequencing at least four separate times during the project each from new crosses of the FGSC 2489 and FGSC 8820 strains. Tissues were ground by mortar and pestle in a liquid nitrogen bath and transferred to a Falcon tube. Ground tissues were homogenized with at least 1 mL of Trizol reagent (Ambion, Carlsbad, CA) per 50−100 mg of tissue and vortexed thoroughly. To each 1 mL of Trizol, 200 μL of chloroform was added, vortexed for 15–30 sec, and incubated at room temperature for 5 min. Samples were centrifuged at 13,000g for 15 min at 4°, the aqueous phase was transferred to a clean tube, and 500 μL of isopropanol for each 1 mL of Trizol was added. Precipitated RNA formed a compacted pellet after centrifuging at 13,000g for 20 min at 4° and the supernatant was removed. The pellet was washed with 80% ethanol, vortexed, centrifuged at 7500g for 5 min, and allowed to air dry for 10 min. Total RNA solution was obtained after dissolving the pellet in diethylpyrocarbonate-treated water. Polyethylene glycol8000 and NaCl were added into total RNA with final concentration 5% polyethylene glycol and 0.5 M NaCl to differentially precipitated high-molecular-weight RNAs followed by sitting on ice for 2 hr. Low-molecular-weight RNAs were recovered from the supernatant by ethanol precipitation, resolved by diethylpyrocarbonate-treated water and quantified in a NanoDrop 2000c spectrophotometer (Thermo Scientific, Waltham, WA). Approximately 10 μg of isolated low-molecular- RNAs were separated on a 15% denaturing polyacrylamide-urea gel in with a miRNA marker was used as molecular mass standard (New England Biolabs, Ipswich, MA). RNA was transferred to a Hybond-NX membrane (Amersham Biosciences, Freiburg, Germany) in 0.5× TBE using Trans-Blot Electrophoretic Transfer Cell apparatus (Bio-Rad, Hercules, CA) at 14V overnight. Constitutively expressed 18S ribosomal RNA was used as a control to test for equal loading of RNA by staining membranes with ethidium bromide for visualization. Carbodiimide-mediated crosslinking for 2 hr at 60° was used to crosslink RNA to Hybond-NX membranes followed by baking at 80° for 1 hr (Pall et al. 2007).
Twelve primer pairs were used to amplify the NCU09969 locus (Table S4). The polymerase chain reaction (PCR) products were ∼500 bp and arranged end to end. Each of the amplicons were verified by sequencing and mixed together as templates to make 32P-labeled DNA probes. Prehybridization and hybridization was performed in PerfectHyb Plus hybridization buffer (Sigma-Aldrich, St. Louis, MO), at 42° overnight. To remove unspecific background, the membrane was washed twice in 2× saline sodium citrate (SSC; 0.3M NaCl, 30mM sodium citrate, pH 7.0) and 0.1% sodium dodecyl sulfate (SDS) at 40° for 15 min, and once in 0.5× SSC (75 mM NaCl, 7.5 mM sodium citrate, pH 7.0) and 0.1% SDS at 40° for 15 min. Finally, the membranes were exposed to a phosphoimager screen and scanned after 24 hr on a Typhoon 941 phosphoimager. Results were analyzed by Image Quant TL (version 7.0) software.
DNA extraction and Southern blots
Mycelia from strains were grown in liquid culture, filtered on Whatman paper, and air dried in a Buchner funnel by vacuum suction and weighed. Approximately 1 mg of mycelium of each strain was ground by mortar and pestle in a liquid nitrogen bath and homogenized with 600 μL of cell lysis buffer (QIAGEN, Valencia, CA) and 3 μL of Proteinase K (QIAGEN) at 60° for 1−2 hr. After cooling to room temperature, 200 μL protein precipitation solution (QIAGEN) was added to each sample and the solution kept on ice for 10 min. Samples were centrifuged at 13,000g for 15 min and 500 μL of clear supernatant was transferred to a new tube followed by adding 500 μL of isopropanol for DNA precipitation. Samples were centrifuged at 13,000g for 10 min and a compacted DNA pellet formed, which was washed with 1 mL of cold 70% ethanol, followed by air-drying and resuspension in sterile water.
The restriction enzymes XhoI, AflIII, Ncol, DraIII, and Styl were used separately to digest ∼20 μg of genomic DNA from various strains. Digested products were separated in 0.8% agarose gels, followed by blotting onto nylon membranes (GE Healthcare Bio-Sciences, Pittsburgh, PA). Four PCR products were used as templates to synthesize four 32P-labeled DNA probes (probes A, B, C, and D, respectively; Table S5). Probes A, B, and C were hybridized to blots with XhoI-digested DNA. Probe D was hybridized to blots with AflIII-, Ncol-, and DraIII-digested DNA. Prehybridization and hybridization was performed in PerfectHyb Plus hybridization buffer (Sigma, Deisenhofen, Germany) at 65° overnight. Membranes were washed one time at 65° in four solutions [2× SSC (0.3 M NaCl, 30 mM sodium citrate, pH 7.0) and 0.1% SDS, 1× SSC (0.15 M NaCl, 15 mM sodium citrate, pH 7.0) and 0.1% SDS, 0.5× SSC (75 mM NaCl, 7.5 mM sodium citrate, pH 7.0), and 10% SDS, 0.1× SSC (7.5 mM NaCl, 1.5 mM sodium citrate, pH 7.0)] and 0.1% SDS for 15 min, respectively, to minimize unspecific background. The membranes were exposed to a phosphoimager screen and scanned after 24 hr on a Typhoon 941 phosphorimager. Results were analyzed by Image Quant TL (version 7.0) software.
Real-time quantitative (qRT)-PCR
Equal amounts (∼2 μg) of DNase I (Invitrogen, Carlsbad, CA) treated total RNAs were reverse transcribed with SuperScript II reverse transcriptase (Invitrogen) using random hexamers. The 10-μL qRT-PCR system was used, including ∼50 ng of cDNA, 10 μL of iQ SYBR Green Supermix (Bio-Rad, Hercules, CA), and 150 nM primers. The N. crassa β-tubulin gene (NCU04054) was used as an internal control for qRT-PCR. Each reaction was in triplicate and performed in a Bio-Rad CFX 96 Real-Time PCR machine. Primer sequences are listed in Table S5. Data analysis was performed using CFX Manager Software v3.1 to calculate the fold change using delta-delta Ct values. A sample from vegetative growth was used as control sample to calculate the relative RNA levels.
Small RNA sequencing and FGSC 8820 genome sequencing
Total RNAs from samples of three time points (PP, 2d PF, 4d PF) were extracted with an miRNeasy Mini kit (QIAGEN). Small RNA sequencing library are constructed by following the standard protocols of Illumina TruSeq Small RNA Sample Prep kit in the University of Utah Sequencing Core. Size from 145 to 160 bp small RNA with adaptors (118 bp) were isolated and sequenced on an Illumina Genome Analyzer IIx to generate 50-nt single end reads. Sequence reads from three time points (PP, 2d PF, 4d PF) are deposited in the SRA database under project accession number SRP021051. Additional pilot sequencing of the 4d PF time point, which had been previously prepared in a similar fashion and sequenced on an Illumina Genome Analyzer at University of British Columbia Sequencing center serves as replicate for comparison, though with considerably lower sequencing coverage.
The genomic sequencing library of strain FGSC 8820 was constructed by using Nextera Illumina DNA preparation kit with dual indexing primers and sequenced in the Genomic Core at the Institute of Integrative Genome Biology, University of California, Riverside, on an Illumina HiSeq2000 genome analyzer. The sequence coverage was approximately 80X, and the reads are deposited in the SRA database under project accession SRP021049.
Small RNA sequence analysis
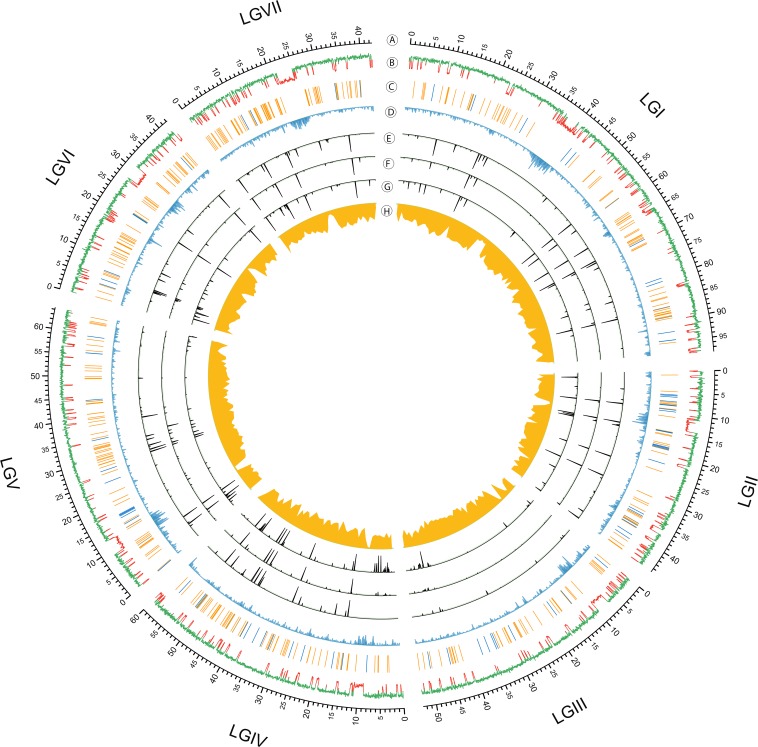
Illumina sequence reads in FASTQ format were processed to remove low quality and artifactual reads, trim Illumina adapter sequences with the fastx_toolkit. (http://hannonlab.cshl.edu/fastx_toolkit/). Reads longer than 17 nt and smaller than 30 nt were mapped to the reference genome assembly (N. crassa version 12 – accession AABX03000000.3) by Bowtie v2.1.0 (Langmead and Salzberg 2012), allowing for no mismatches. SAM and BAM files were manipulated with SAMtools v1.1 (Li et al. 2009) and Picard v1.81 (http://broadinstitute.github.io/picard). Identification of reads aligning to unpaired regions was performed using BEDtools v2.17.0 (Quinlan and Hall 2010) and custom scripts written in Perl (v5.10.1) (http://github.com/stajichlab/neurospora_MSUD). An annotation file of noncoding RNAs was created by aligning the known sequences for ribosomal RNA (rRNA; accession FJ360521.1), small nucleolar RNAs (Liu et al. 2009) microRNA-like (milRNA) and Dicer-independent small interfering RNA loci [Table S2 and Table S3 from Lee et al. (2010b)] to Nc12 assembly with BLAT (Kent 2002). The transfer RNAs (tRNAs) were predicted with tRNAScan-SE (Lowe and Eddy 1997). Small RNAs that aligned to the mitochondrial genome were classified as mitochondrial RNA; other small RNAs that aligned to genome regions without any gene or repetitive element annotation were grouped into the “other” category. The genome representation as a Circos plot (Figure 1) was generated with Circos version 0.66 (Krzywinski et al. 2009); the configuration scripts are available at http://github.com/stajichlab/neurospora_MSUD. Figures displaying the distribution of smallRNA classes, read length, 5′ base preference, and strand specificity were made in R (http://r-project.org) and postprocessed with Adobe Illustrator.
Figure 1.
Circular genome visualization and data visualization with Circos. From outside in (A) seven linkage groups of N. crassa; (B) global profile of RIP shown by the composite repeat-induced point mutation (RIP) index [CRI; (Lewis et al. 2009)]; positive and negative CRI values imply that DNA has been subjected to RIP or not, presented by red and green on the plot, respectively; (C) rRNA (blue bar) and tRNA locus (orange bar); (D) repeat density based on a library of repeats from RepBase and a curated collection derived from multiple Neurospora species (Gioti et al., 2013); (E−G) small RNA sequence profiles from three time points in sexual development: protoperithecia of FGSC 2489, 2 d and 4 d postfertilization from a cross between FGSC 2489 and FGSC 8820; (H). Read depth of FGSC 8820 genome sequencing aligned to the FGSC 2489 genome.
Analysis of resequencing data
Analysis of Illumina genome sequencing of strains FGSC 2489, FGSC 8820, and 19 classic mutants (McCluskey et al. 2011) was performed to test if Sly1-1 could be detected in any other strains. FGSC 2489 sequencing data were taken from public SRA accessions (SRR018138-SRR018144). Chromatin immunoprecipitation sequencing of the NMF229 strain also was analyzed in the same fashion. Sequences were trimmed for quality with sickle (http://github.com/najoshi/sickle) requiring at least phred quality of 30 and length of 25 bp. Paired end data processing required both pairs were required to pass filtering and quality control or else the entire pair was discarded. Trimmed and filtered short read DNA sequences were aligned to the genome with BWA v0.7.9-r783 using ‘bwa aln –q 20,’ which was further processed to produce SAM and sorted BAM files with SAMtools and Picard. Multiple matches were allowed to support the presence of multiple Sly-1 loci in the genome. Visualization of the read depth, and presence or absence of reads in the Sly1-1 locus was performed in the Genome Browser and computed using BEDtools v2.17.0 (Quinlan and Hall 2010).
Transposon bioinformatics analysis
The superfamily classification of Sly1-1 was performed by searching the sequence against a curated library of DNA transposase domains (Yuan and Wessler 2011) to identify it as likely member of the CMC superfamily (CACTA – Mirage – Chapev and Transib). A multiple alignment was constructed of these homologs and the Ch. globosum and Co. immitis copies and a phylogenetic tree constructed with RAxML (v7.3.2) (Stamatakis 2006) using a BLOSUM62 model with empirical frequencies. Internal node support in the tree was estimated from 100 bootstrap replicates. Identification of the specific D-D-E residues was performed by predicting secondary structure of the protein using PSIPRED of the DDE domain (Buchan et al. 2013; Jones 1999) and identifying the residues falling within the correct range of expected folds (Yuan and Wessler 2011). The distance between the DDE triad and the range of signature motifs C(2)C and H(3)H are consistent with the distance observed in members of the CMC family.
Terminal inverted repeat (TIR) and target-site duplication (TSD) analysis methods
Terminal inverted repeat 1 and 2 (TIR1 and TIR2) were identified by self-aligning putative regions of Sly1-1 including its 3-kb flanking DNA sequences upstream and downstream, using NCBI-BLASTN (Altschul et al. 1997). We then detected clear breaks in reads adjacent to the upstream TIR1 and downstream TIR2 in strains from with Sly1-1 is absent. These were considered breakpoints between TIRs and TSDs. In the strains with Sly1-1 the reads located at breakpoints are continuous. Sequences of TSDs were determined according to these continuous reads by detecting their common sequences adjacent to the TIRs.
Annotation of regions that have undergone RIP
Testing for copies of Sly1-1 that have undergone RIP was done by RIPCAL (Hane and Oliver 2008) based on a multiple alignment of all homologs of the TE sequence. In addition, whole genome RIP analysis was performed with the script RIP_index_calculation.pl (http://github.com/hyphaltip/fungaltools) by computing RIP indices in sliding windows across all chromosomes. The RIP index for each locus was evaluated visually as a track in the Stajich lab Genome Browser (http://gb2.fungalgenomes.org).
Data visualization
Genome sequence, annotation, and BAM files for aligned reads were loaded into the Generic Genome Browser (Stein et al. 2002), hosted by the Stajich lab at http://gb2.fungalgenomes.org for visualization.
Data availability
Whole-genome and small RNA sequencing reads are deposited in the Short Read Archive under accession numbers: SRP021049 and SRP021051. Perl scripts and processed data files used in this project are available from http://github.com/stajichlab/neurospora_MSUD. Trimmed short reads and alignments are available at http://fungalgenomes.org/public/neurospora/data/support_files/Wang_MSUD.
Results and Discussion
Detection of small RNAs during the sexual cycle
To identify small RNAs produced from unpaired regions during meiotic silencing (masiRNAs; Hammond et al. 2013a), we crossed the laboratory wild-type strain, FGSC 2489, with FGSC 8820, a progeny from crosses of wild collected strains. We isolated pools of small RNA from tissues at three different times: before fertilization ( PP), 2d PF, and 4d PF. After trimming adaptor sequences and eliminating spurious RNAs or degradation products, we obtained abundant short read sequences from the samples (PP 22,538,276; 2d PF 21,562,565; 4d PF 22,731,295). These reads were mapped to the N. crassa reference genome assembly 12 (AABX00000000.3) to identify genomic origins of small RNAs. Analysis of read coverage indicated that some genomic regions were highly enriched in small RNA production, including subtelomeric, centromeric, and many rRNA and tRNA regions (Figure 1). Analysis of small RNA features showed that these identified small RNAs had a strong preference for 5′ uridine and their size peaked at 20 nt at all three time points (Figure S1A).
We next classified reads based on their match to genomic features into eight pools of small RNAs, namely rRNA, tRNA, small nucleolar RNAs, milRNA (Lee et al. 2010b), Dicer-independent small interfering RNA (Lee et al. 2010b), masiRNA, mitochondrial RNA, and other unspecified RNAs (Figure S1B). Most 20-nt long RNAs were identified to originate from ribosomal DNA loci (Figure S1C). We noted that the abundance of reads mapping to tRNAs decreased dramatically from 41.5% in PP to 16.9% in 2d PF and then increased slightly to 23.5% in 4d PF. It has been shown that the microRNA-like milRNA-4 is derived from the precursor of tRNA in vegetative tissue (Yang et al. 2013), suggesting that other unknown milRNA genes may account for the high percentage of tRNA in the PP sample. The masiRNA class had a relative large variance of abundance in three time points, ranging from low frequencies of 0.1% at PP and 0.2% at 2d PF to 10-fold greater frequency of 1.9% at 4d PF. This class of small RNAs, with sizes peaking at 25 nt, was primarily derived from 31 loci unique to FGSC 2489 (Table S2). This class also showed preference for a 5′ uridine (Figure S2). This is expected for small RNAs processed by Argonaute-like proteins (Mi et al. 2008). In addition, the observed size of 25 nt for masiRNAs is identical to the size of small RNAs generated in vitro by N. crassa DCL-1, which has been shown to be required for meiotic silencing (Catalanotto et al. 2004; Macrae et al. 2006; Alexander et al. 2008), suggesting that masiRNAs are DCL-1 products. No strand preference for the production of these small RNAs was observed (Table S2), suggesting that the formation of double-stranded RNA (dsRNA) is required.
Unpaired regions that may undergo meiotic silencing
Analysis of the genomes of FGSC 2489 and FGSC 8820 was performed to identify regions unique to FGSC 2489 that could potentially form unpaired loops and trigger meiotic silencing. Previous work has shown that unpaired regions larger than 700 bp are needed to efficiently trigger meiotic silencing (Lee et al. 2004). To identify candidate loci targeted by meiotic silencing we applied the following filters: (1) the region is unique to strain FGSC 2489; (2) the size of unpaired region is larger than 700 bp; (3) small RNAs are only enriched at 4d PF; (4) the ratio of small RNA production to the size of the unpaired region is greater than 1. There were 31 regions that met all criteria. Twenty-four of these contain predicted genes, all of which encode “hypothetical proteins”. The regions ranged in size from 1 kb to 15 kb with 19 smaller than 5 kb (59%), nine between 5 kb and 10 kb, and four longer than 10 kb. The masiRNAs were primarily derived from these 31 unique regions and share similar properties: they are 24∼26 bp long with a peak frequency at 25 bp, they show no specific strand bias, and enrichment for RNAs with a 5′ uridine (Figure S2 and Table S2). Similarity analysis using a curated set of repetitive elements found eight of the regions contained the repeated sequences or remnants of TEs (Table S2).
Identification of a novel DNA transposon
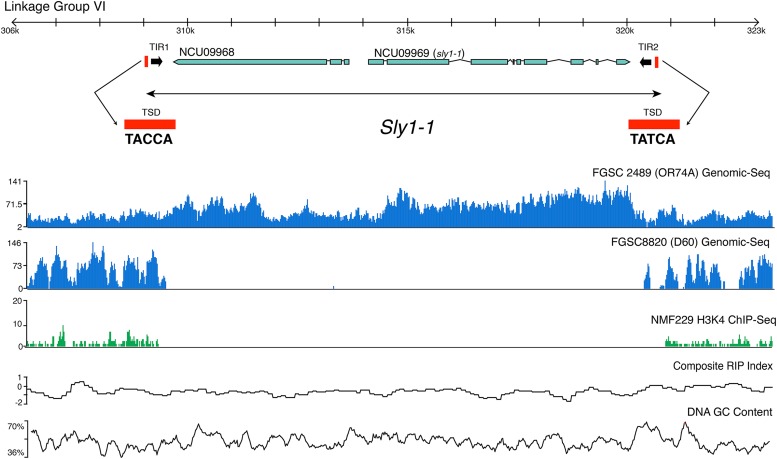
To analyze endogenous masiRNA in more detail, we focused our analyses on a single 10-kb region unique to FGSC 2489 (Table S2). This region, located on linkage group (LG) VI (309,012 – 320,545 nt), had a high abundance of small RNAs and contained two hypothetical genes, NCU09968 and NCU09969. We noticed that the read coverage of genomic sequencing of this locus in FGSC 2489 was greater than the flanking regions, which indicated that multiple copies of this region were present in the reference genome (Figure 2). Inspection, first by PCR and later by analysis of available genomic sequencing and Southern blots of N. crassa strains (Figure 3), indicated that this region was present in the genome of only a few laboratory wild-type strains (FGSC 352), and none of the resequenced laboratory strains (McCluskey et al. 2011). This difference was initially noticed in comparison of chromatin immunoprecipitation sequencing samples of strain NMF229 (Smith et al. 2011) to the reference genome of the lab wild-type strain, FGSC 2489 (Galagan et al. 2003). We named this putative transposon Sly1-1 and this new DNA transposon family Sly, for Silently.
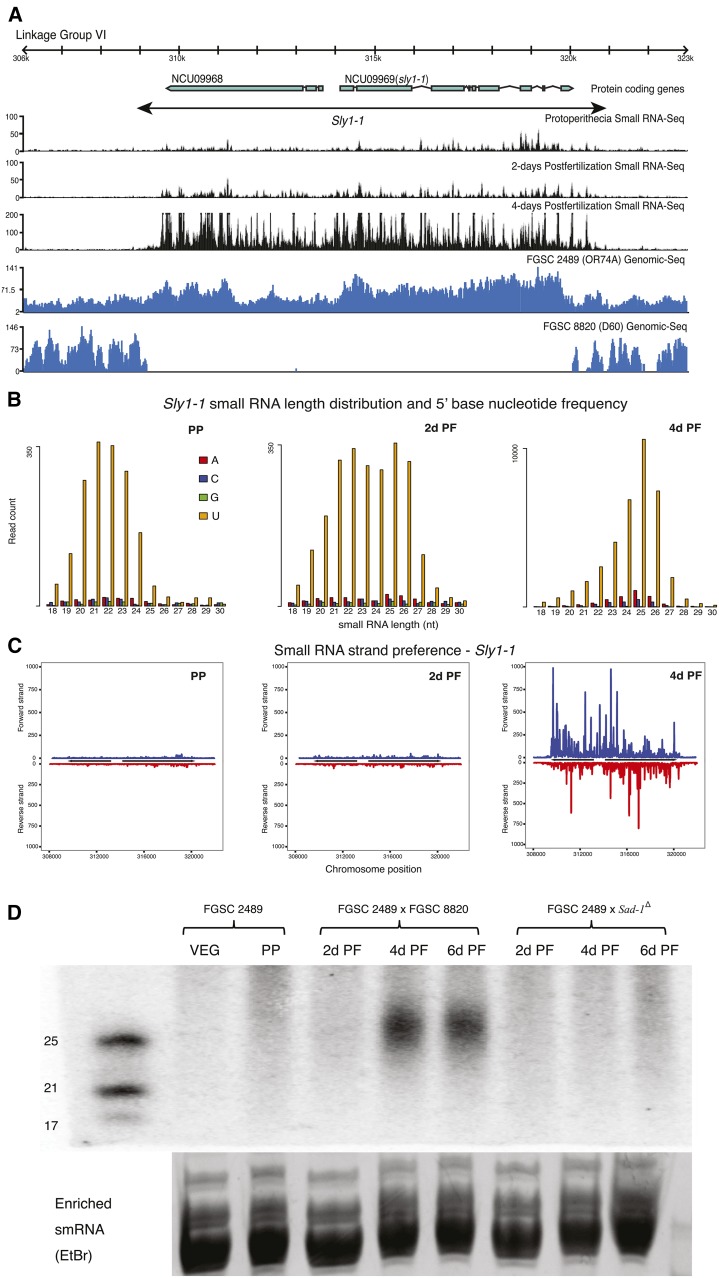
Figure 2.
Organization of the Sly1-1 locus. Schematic showing position and sequences of terminal inverted repeats (TIRs) and target-site duplications (TSD) of Sly1-1, and resequencing data from the reference strain FGSC 2489 (mat A) and strain FGSC8820 (mat a), the partner in the wild-type sexual cross. Chromatin immunoprecipitation sequencing (ChIP-seq) of NMF229 shows absence polymorphism of the Sly1-1 locus. Repeat-induced point mutation (RIP) and DNA GC% plots indicate no A+T nucleotide skew that would be observed in a region mutated by RIP. The FGSC 2489 resequencing data indicate increased coverage in Sly1-1 relative to the flanking genomic region, suggesting multiple copies of the element. Clear and almost identical boundaries can be observed where Sly1-1 is missing in the H3K4me2 ChIP-seq of strain NMF229 and genome sequence of FGSC 8820. We identified genomic sequence reads from FGSC 8820 that perfectly spanned the Sly1-1 insertion present in FGSC 2489.
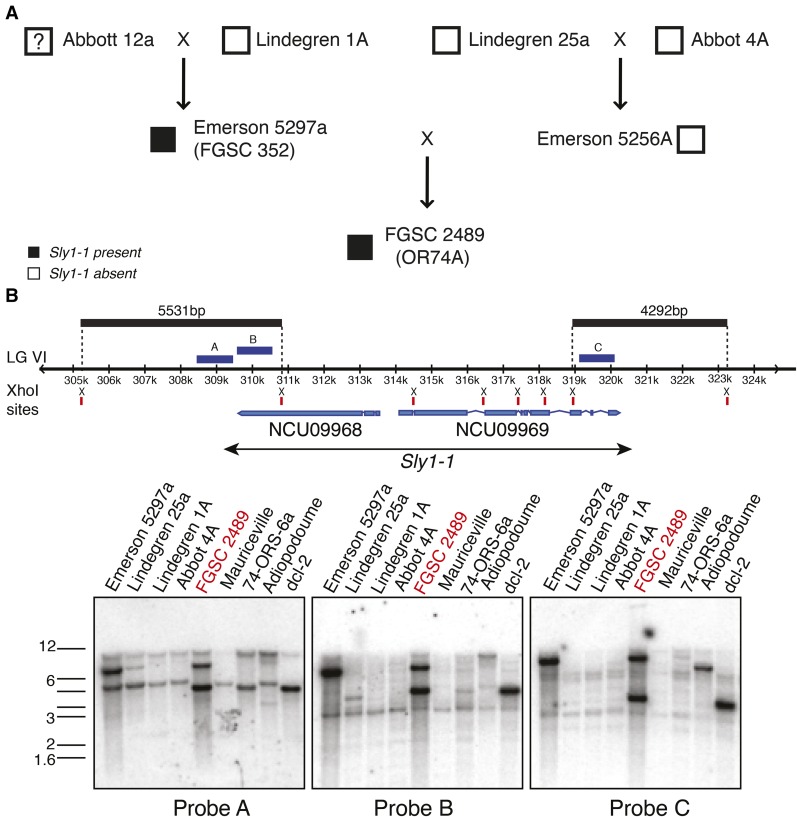
Figure 3.
Mapping the presence of Sly1-1 in the pedigree of FGSC 2489 and related strains. A cross of Lindegren 1A and the presumed Abbott 12a strain (see Newmeyer et al. 1987) yielded Emerson 5297a. One copy of Sly1-1 is present in Emerson 5297a as shown by Southern blot analysis; it remains unknown if Sly1-1 was introduced from Abbott 12a as the strain may be lost. Lindegren 25a and Abbott 4A produced Emerson 5256A and neither of them carries Sly1-1. FGSC 2489 was derived via several backcrosses from a cross between Emerson 5297a and Emerson 5256a. Two copies of Sly1-1, instead of just one in Emerson 5297a, are present in the genome of FGSC 2489, as shown in the Southern blots, indicating that Sly1-1 is potentially active and may have transposed at least once. The FGSC 2489 lineage resulted from multiple back-crosses to Emerson parents, indicated by the dotted line. Genomic DNAs were digested by the restriction endonuclease XhoI for Southern blotting analysis. Digested DNA fragments were hybridized with three probes whose positions are illustrated: probe A and probe B hybridized with the 5531-bp fragment of Sly1-1 at the 5′ end; probe C hybridized with the 4292-bp fragment of Sly1-1 at the 3′ end.
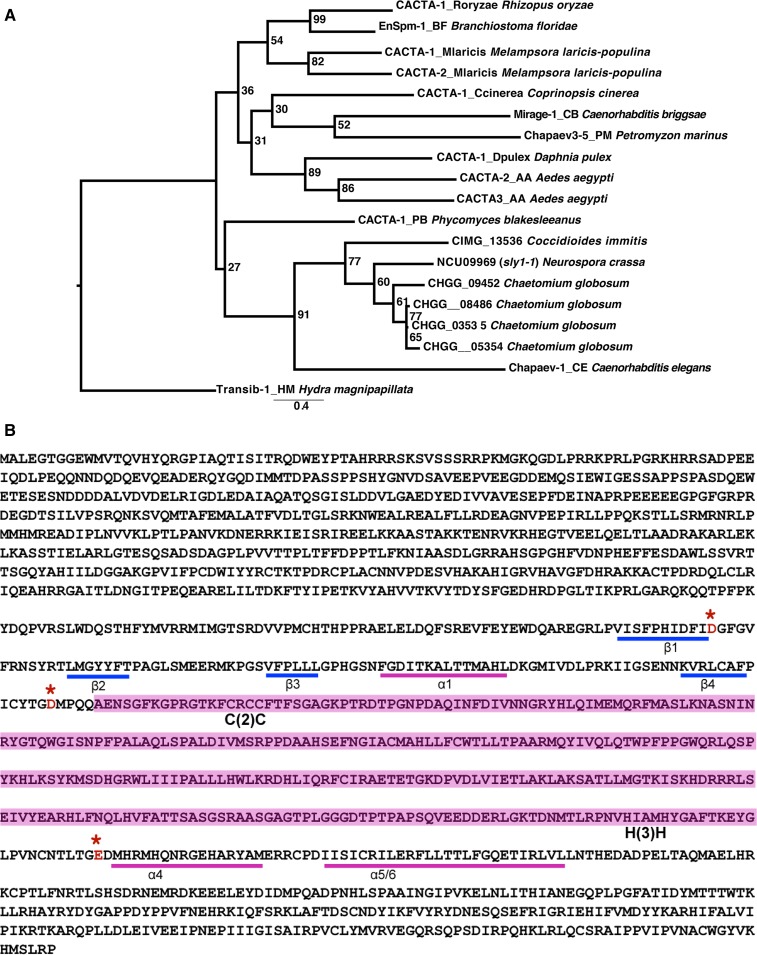
The original locus on LG VI contains a complete copy of Sly1-1. The region is 11,534 bp long and includes an intact transposase domain-containing gene, NCU09969, named sly1-1, and a gene of unknown function, NCU09968, which has similarity to Chaetomium globosum CHGG_09452 (Figure 2). Phylogenetic analysis of NCU09969 identified it as member of the CMC (CACTA-Mirage-Chapaev) superfamily of DNA transposons (Figure 4A), based on conservation of DDE motifs found in the transposase domain and other conserved residues (Figure 4B) (Yuan and Wessler 2011). The arrangement of genes within Sly1-1 is conserved in other species. Homologs of both ORFs were found as adjacent and divergently transcribed genes in Ch. globosum (CHGG_09451 and CHGG_09452) while in Coccidioides immitis only the Sly transposase could be identified as gene (CIMG_13536).
Figure 4.
Phylogenetic tree of CMC family and DDE motif present in sly1-1. (A) A maximum likelihood phylogenetic tree of CMC family transposases homologous to sly1-1. Copies originate from animals and fungi with the species names indicated in the name. (B) Identification of the DDE motif based on expected motif patterns following Yuan and Wessler (2011). The figure shows locations of the beta sheets (β) or alpha helices (α) protein folds, the presence of Cysteine-XX-Cysteine [C(2)C] and Histidine-XXX-Histidine [H(3)H] motifs for the CMC family, and the location of the acidic residues adjacent to the predicted protein folds. The area highlighted in pink between the D and E contains multiple alpha helices.
TIRs and TSDs are hallmarks of DNA transposons (Potter et al. 1980). We identified two TIRs (TIR1 and TIR2) and TSDs (Figure 2) at the boundary of Sly1-1 by comparing Sly1-1 with non-Sly1-1−containing strains (FGSC8820; FGSC7022; FGSC1363; FGSC106; D48, FGSC8088; D106, FGSC8866; NMF229). The comparison between strains with no Sly1-1 and FGSC 2489 helped define the precise boundary of the transposon and the locations of the TIRs and TSDs and we observe that small RNA read abundance drops off sharply outside of the delineated locus (Figure 5A). The sequences of two TSD located at the boundaries are not exactly the same, likely due to RIP as the third nucleotide of TSD1, cytosine, is replaced by thymine.
Figure 5.
Illustration and analysis of small RNAs from Sly1-1. (A) Small RNA sequence profiles from three sexual developmental time points are aligned to genomic sequencing reads from FGSC 2489 and FGSC 8820 and a diagram to show the structure of Sly1-1. Small RNA production precisely matched the boundaries of Sly1-1. (B) Length distribution and 5′ base nucleotide frequency of small RNAs produced from Sly1-1 at the same three time points as above [protoperithecial (PP); 2 d postfertilization (PF) 2d PF and 4d PF]. Most of the small RNAs at 4d PF are 25 nt long and mostly have uridine as 5′ base. (C) There is no obvious strand preference of small RNA from Sly1-1 at the same three time points. Few small RNAs are produced at PP and 2d PF; abundant small RNAs are produced at 4d PF. (D) Small RNA northern blot of small RNAs from a cross (FGSC 2489 × FGSC 8820) in which meiotic silencing should occur and a cross (FGSC 2489 × Sad-1Δ) in which meiotic silencing should be absent. Strong signals of small RNA (25 nt long) were detected at 4d PF and 6d PF in the wild-type cross but not in the mutant cross that lacks SAD-1.
To survey the presence of Sly1-1 in N. crassa laboratory strains we carried out Southern blotting, which revealed that Sly1-1 is not common and is present at low copy number (Figure 4B and Figure S3). Only strains in the FGSC 2489 lineage (Gavric and Griffiths 2004; Newmeyer et al. 1987), including FGSC 352 (Emerson 5279a) and strains derived from crosses with FGSC 2489, such as the collection of single-gene deletion strains (Colot et al. 2006), contain at least two copies of Sly1-1 (Figure 4B and Figure S3). Our survey of all available re-sequencing data of N. crassa strains (McCluskey et al. 2011), including the Mauriceville strain (Pomraning 2012), N. tetrasperma (Ellison et al. 2011), N. discreta (a homothallic Neurospora species) (Gioti et al. 2013), and Sordaria macrospora (Nowrousian et al. 2010), suggests that Sly1-1 is largely absent from the tested strains (data not shown); inspection of unreleased genome data from the pedigree of FGSC 2489 showed presence of Sly1-1 only in FGSC4200, FGSC9718, and FGSC987 (S. Baker, K. McCluskey, I. Grigoriev, and J. Stajich, unpublished results). The absence of Sly1-1 in many of these strains suggests that it arrived or was activated only recently in N. crassa.
We used BLAST (Altschul et al. 1997) to search the FGSC 2489 genome with the Sly1-1 sequence to identify the additional copy inferred by the second band in the Southern blots. A partial copy, which we named Sly1-2, is present in assembly 12 within centromeric DNA on LG II (1,151,460–1,155,073 nt). In FGSC 2489 we found no evidence for RIP in the two Sly copies. The second copy may represent evidence of recent transposition. However, confidence in the assembly of the genome in this region is not high as repetitive regions are a hallmark of centromeric DNA and difficult to assemble. Because probes for both ends of Sly1-1 in the Southern blot showed evidence for two copies it is likely Sly1-2 is intact, just not assembled correctly (Figure 3B). The original and transposed positions of Sly1-1 are similar in that both show cytosine DNA methylation in the Sly1-1 flanks, detected by high-throughput sequencing of immunopurified methylated DNA (Smith et al. 2011). Both regions also show absence of the centromere-specific H3, CenH3 and histone H3 lysine 9 trimethylation (H3K9me3) but presence of H3K4me2 (Smith et al. 2011). The identification of two, nearly identical stretches of DNA in the N. crassa genome that have not been RIPed is unexpected as previous work had identified only few regions with predicted genes that showed high similarity (Galagan et al. 2003; Smith et al. 2011).
Searching the FGSC 2489 reference genome with TIR1 and TIR2 of Sly1-1 showed that Sly1-2 contained the Sly1-1 TIR1 but lacked TIR2 (Figure S4A). We also detected two additional loci that contained nearly identical TIR pairs, located on LG V and LG I; these were named Sly1-3 and Sly1-4, respectively (Figure S4A and Table S3). Sly1-3 and Sly1-4 are both absent in the genome of FGSC 8820. Sly1-2 is only about 3614 bp long, 100% identical to Sly1-1, has the same TSD as Sly1-1 adjacent to the sequence of TIR1 and contains a putative gene, NCU16528. The 3′ end of Sly1-2 is not completely assembled and stretches across two contigs. It is possible that Sly1-2 is larger in size and that its TIR2 is located in an unsequenced region. The unequal read coverage observed for the two loci in Sly1-1 (Figure 2) may arise because the total reads aligned are titrated between two chromosome locations for NCU09968 (e.g., Sly1-1 and Sly1-2) while only one NCU09969 sequence is in the reference assembly at Sly1-1 which results in higher computed read coverage. Sly1-3 and Sly1-4 are very similar to each other: BLASTN showed query coverage ∼95% with identity ∼85% (E value < 0.001), both are ∼12 kb long, contain no identified open reading frames, and have nearly the same TSDs adjacent to TIRs. RIPCAL (Hane and Oliver 2008) indicates that Sly1-3 and Sly1-4 have undergone RIP. This situation is consistent with the observation that alignments of Sly1-3 and Sly1-4 to Sly1-1 cover 94% and 91% of the sequences, but only have 67% and 72% identity, respectively.
BLASTX searches with Sly1-2 identify significant similarity to NCU09968 (query coverage = 61%; E value = 0 and identity = 99%). The translated search of Sly1-3 and Sly1-4 also reveal similarity to both NCU09968 (Sly1-3: query coverage = 29%; E value < 0.001; identity = 54%; Sly1-4: query coverage = 30%; E value < 0.001; identity = 60%) and NCU09969 (Sly1-3: query coverage = 27%; E value < 0.001; identity = 53%; Sly1-4: query coverage = 28%; E value < 0.001; identity = 54%), respectively. Summarizing all of these results, we conclude that Sly1-1 is a recently active TE, with at least two inactivated copies that underwent RIP (Sly1-3 and Sly1-4), but a partial copy which is identical in sequence and thus not mutated by RIP, Sly1-2, that may be active. Detection of similar but mutated copies of both genes, NCU09968 and sly1-1, in Sly1-3 and Sly1-4 suggests that the entire Sly1-1 locus transposed and not just the transposase, sly1-1.
Meiotic silencing machinery is required for the production of Sly1-1 small RNAs
To characterize transcription and small RNAs produced from Sly1-1 as a consequence of meiotic silencing, we first determined when the sly1-1 transposase gene (NCU09969) was expressed. Analysis of published transcriptome data (Wang et al. 2012) detected no sly1-1 transcript during vegetative growth. However, during the sexual cycle, gene expression was detected at 0, 2, 24, 48, 72, 96, 120, and 144 hr after crossing FGSC 2489 with FGSC 4200. The RPKM (reads per kilobase per million mapped) values for sly1-1 from RNA-seq ranged from 6 to 10 (Wang et al. 2014), indicating that it is expressed during sexual development, if not at a high level.
We also examined the small RNAs produced from Sly1-1 during later stages of sexual development. Karyogamy occurs in perithecia 3−4 d after fertilization (Raju 1980) and meiotic silencing is first detected at karyogamy (Shiu et al. 2001). Our small RNA profiles support these previous results but show small RNA production occurring at the same time as karyogamy, not after, which is when MSUD is thought to occur (Shiu et al. 2001). This difference may reflect a delay between when small RNAs are produced, the process of aberrant RNA recognition, and when silencing machinery acts on transcripts. We observed an abundance of small RNAs produced from Sly1-1 at 4d PF, compared with samples collected at PP stages or 2d PF (Figure 5, C and D). Similar to the small RNAs described previously and to those identified to be from the Roundspore (Rsp) locus in an unpaired and MSUD inducing cross (Hammond et al. 2013a), the small RNAs from Sly1-1 have features typical of those produced during quelling (Figure 5, B and C and Table S2). The size and expression pattern of Sly1-1 derived small RNAs were also confirmed by small RNA northern blots (Figure 5D). Samples from PP, 2d PF, 4d PF, and 6d PF were examined and small RNAs were detected at 4d and 6d PF, indicating that the production of small RNAs was sustained.
Previous work showed that sad-1 is essential for meiotic silencing and is specifically expressed during sexual development (Shiu et al. 2001, 2006). SAD-1 is homologous to the RNA-dependent RNA polymerase, QDE-1. As noted previously, QDE-1 is involved in quelling (Romano and Macino 1992; Cogoni and Macino 1997; Fulci and Macino 2007), whereas SAD-1 operates during meiosis, where it is thought to convert single-stranded aberrant RNA transcribed from unpaired regions into dsRNA, producing the substrate that is cleaved by DCL-1 to generate small interference RNA (Kelly and Aramayo 2007; Aramayo and Pratt 2010; Aramayo and Selker 2013). To test whether the small RNA production observed from this unpaired region was dependent on SAD-1, we crossed the dominant mutant allele Sad-1Δ (present in a genetic background lacking Sly1-1; FGSC 8740), to FGSC 2489. RNA collected at 2d, 4d, and 6d PF was tested by northern analyses. No small RNA from the Sly1-1 locus was detected under these conditions (Figure 5D). This result suggested a relationship between synthesis of dsRNA and small RNA production, both dependent on SAD-1.
We also tested whether the production of unprocessed sly1-1 RNAs was dependent on SAD-1. We used a method developed to detect qiRNA (Lee et al. 2009) and examined transcript levels from the intergenic regions outside of the sly1-1 gene (NCU09969) to test for aberrant RNA production. Quantitative PCR showed that transcripts originating from downstream regions of the sly1-1 locus were highly induced from 2d PF to 6d PF (Figure S5). This observation suggested that RNA is required to initiate and maintain the production of aberrant and, presumably, small RNAs. In the cross between Sad-1Δ and FGSC 2489, transcripts accumulated at a high level at 2d PF, but decreased dramatically at 4d PF and 6d PF (Figure S5). This decrease in transcript levels indicated that the lack of SAD-1 did not block the production of aberrant, or any, RNA initially, but may have stalled dsRNA production, which resulted in the suppression of the production of small RNAs. The exact mechanisms underlying this surprising finding will need to be addressed by future experiments.
In summary, our study provides evidence for a novel and apparently intact TE in the widely used laboratory wild-type strain of N. crassa, a genome thought to be lacking active transposons (Selker et al. 2003). Based on Southern blots examining the pedigree and genome of FGSC 2489, and further validated by examination of whole genome sequences of most of the strains in this pedigree, it appears that Sly1-1 was acquired recently and has actively transposed, with evidence for at least two intact copies. Examination of additional wild strains from ongoing resequencing efforts (S. Baker, K. McCluskey, I. Grigoriev, J. Stajich, and unpublished data) provided a better understanding of the origins and timing of acquisition of this newly described selfish genetic element. Our study also provides evidence for the original hypothesis that meiotic silencing targets unpaired DNA created by a transposon insertion and is an effective genome defense mechanism. The unpaired region triggers the production of masiRNA during sexual development, approximately 4−6 d after fertilization, when karyogamy is expected to occur.
Acknowledgments
We are pleased to acknowledge use of materials from the Fungal Genetics Stock Center and P01GM068087 “Functional analysis of a Model Filamentous Fungus.” We thank Christopher Ellison for assistance in pilot experiments; Damon Lisch, Yaowu Yuan, and Susan Wessler for transposase classification and helpful discussions; and Glen Hicks and the IIGB Genomics core for donating Nextera kits and sequencing of the FGSC 8820 genome. We thank Katherine Borkovich, Hailing Jin, Xuemei Chen, Shou-wei Ding, Shengben Li, and Peng Du for suggestions, assistance, support in experimental procedures, and help with small RNA northern blots. J.E.S. and Y.W. were supported by initial complement funds from the College of Natural and Agricultural Sciences, University of California, Riverside, and the National Science Foundation grant #IOS-1027542 to J.E.S. and S.R. Wessler. K.M.S. was partially supported by a grant from the National Institutes of Health (P01GM068087). This work was also partially supported by grants from the ACS (RSG-08-030-01-CCG) and National Institutes of Health (R01GM097637) to M.F.
Y.W., K.M.S., M.F., J.W.T., and J.E.S conceived of the experiments; Y.W. and K.M.S. carried out experiments; Y.W., K.M.S., M.F., and J.E.S. performed data analyses; and J.E.S., Y.W., K.M.S., J.W.T., and M.F. wrote the paper.
Footnotes
Supporting information is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.115.017921/-/DC1
Communicating editor: M. S. Sachs
Literature Cited
- Alexander W. G., Raju N. B., Xiao H., Hammond T. M., Perdue T. D., et al. , 2008. DCL-1 colocalizes with other components of the MSUD machinery and is required for silencing. Fungal Genet. Biol. 45: 719–727. [DOI] [PubMed] [Google Scholar]
- Altschul S. F., Madden T. L., Schaffer A. A., Zhang J., Zhang Z., et al. , 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aramayo R., Metzenberg R. L., 1996. Meiotic transvection in fungi. Cell 86: 103–113. [DOI] [PubMed] [Google Scholar]
- Aramayo R., Pratt R. J., 2010. Meiotic trans-sensing and silencing in Neurospora, pp. 132–144 in Cellular and Molecular Biology of Filamentous Fungi, edited by Borkovich K. A., Ebbole D. J. ASM Press, Washington, DC. [Google Scholar]
- Aramayo R., Selker E. U., 2013. Neurospora crassa, a model system for epigenetics research. Cold Spring Harb. Perspect. Biol. 5: a017921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin A. A., Lagos-Quintana M., Yalcin A., Zavolan M., Marks D., et al. , 2003. The small RNA profile during Drosophila melanogaster development. Dev. Cell 5: 337–350. [DOI] [PubMed] [Google Scholar]
- Buchan, D. W., F. Minneci, T. C. Nugent, K. Bryson, and D. T. Jones, 2013 Scalable web services for the PSIPRED Protein Analysis Workbench. Nucleic Acids Res. 41 (Web Server issue): W349–357. [DOI] [PMC free article] [PubMed]
- Calvi B. R., Gelbart W. M., 1994. The basis for germline specificity of the hobo transposable element in Drosophila melanogaster. EMBO J. 13: 1636–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cambareri E. B., Jensen B. C., Schabtach E., Selker E. U., 1989. Repeat-induced G-C to A-T mutations in Neurospora. Science 244: 1571–1575. [DOI] [PubMed] [Google Scholar]
- Catalanotto C., Pallotta M., ReFalo P., Sachs M. S., Vayssie L., et al. , 2004. Redundancy of the two dicer genes in transgene-induced posttranscriptional gene silencing in Neurospora crassa. Mol. Cell. Biol. 24: 2536–2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalanotto C., Nolan T., Cogoni C., 2006. Homology effects in Neurospora crassa. FEMS Microbiol. Lett. 254: 182–189. [DOI] [PubMed] [Google Scholar]
- Cogoni C., Macino G., 1997. Isolation of quelling-defective (qde) mutants impaired in posttranscriptional transgene-induced gene silencing in Neurospora crassa. Proc. Natl. Acad. Sci. USA 94: 10233–10238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogoni C., Romano N., Macino G., 1994. Suppression of gene expression by homologous transgenes. Antonie van Leeuwenhoek 65: 205–209. [DOI] [PubMed] [Google Scholar]
- Collins J., Saari B., Anderson P., 1987. Activation of a transposable element in the germ line but not the soma of Caenorhabditis elegans. Nature 328: 726–728. [DOI] [PubMed] [Google Scholar]
- Colot H. V., Park G., Turner G. E., Ringelberg C., Crew C. M., et al. , 2006. A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc. Natl. Acad. Sci. USA 103: 10352–10357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker L. M., Boone E. C., Xiao H., Shanker B. S., Boone S. F., et al. , 2015. Complex formation of RNA silencing proteins in the perinuclear region of Neurospora crassa. Genetics 199: 1017–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellison C. E., Stajich J. E., Jacobson D. J., Natvig D. O., Lapidus A., et al. , 2011. Massive changes in genome architecture accompany the transition to self-fertility in the filamentous fungus Neurospora tetrasperma. Genetics 189: 55–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulci V., Macino G., 2007. Quelling: post-transcriptional gene silencing guided by small RNAs in Neurospora crassa. Curr. Opin. Microbiol. 10: 199–203. [DOI] [PubMed] [Google Scholar]
- Galagan J. E., Calvo S. E., Borkovich K. A., Selker E. U., Read N. D., et al. , 2003. The genome sequence of the filamentous fungus Neurospora crassa. Nature 422: 859–868. [DOI] [PubMed] [Google Scholar]
- Gavric O., Griffiths A., 2004. Another inconsistency in the pedigree of the Oak Ridge wild types of Neurospora crassa. Fungal Genet. Newsl. 51: 9–12. [Google Scholar]
- Gioti A., Stajich J. E., Johannesson H., 2013. Neurospora and the dead-end hypothesis: genomic consequences of selfing in the model genus. Evolution 67: 3600–3616. [DOI] [PubMed] [Google Scholar]
- Girard A., Hannon G. J., 2008. Conserved themes in small-RNA−mediated transposon control. Trends Cell Biol. 18: 136–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodier J. L., Kazazian H. H., Jr, 2008. Retrotransposons revisited: the restraint and rehabilitation of parasites. Cell 135: 23–35. [DOI] [PubMed] [Google Scholar]
- Hammond T. M., Xiao H., Boone E. C., Perdue T. D., Pukkila P. J., et al. , 2011. SAD-3, a putative helicase required for meiotic silencing by unpaired DNA, interacts with other components of the silencing machinery. G3 (Bethesda) 1: 369–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond T. M., Rehard D. G., Xiao H., Shiu P. K., 2012. Molecular dissection of Neurospora Spore killer meiotic drive elements. Proc. Natl. Acad. Sci. USA 109: 12093–12098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond T. M., Spollen W. G., Decker L. M., Blake S. M., Springer G. K., et al. , 2013a Identification of small RNAs associated with meiotic silencing by unpaired DNA. Genetics 194: 279–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond T. M., Xiao H., Boone E. C., Decker L. M., Lee S. A., et al. , 2013b Novel proteins required for meiotic silencing by unpaired DNA (MSUD) and siRNA generation in Neurospora crassa. Genetics 194: 91–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hane J. K., Oliver R. P., 2008. RIPCAL: a tool for alignment-based analysis of repeat-induced point mutations in fungal genomic sequences. BMC Bioinformatics 9: 478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey A. M., Rehard D. G., Groskreutz K. M., Kuntz D. R., Sharp K. J., et al. , 2014. A critical component of meiotic drive in Neurospora is located near a chromosome rearrangement. Genetics 197: 1165–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson D. J., Raju N. B., Freitag M., 2008. Evidence for the absence of meiotic silencing by unpaired DNA in Neurospora tetrasperma. Fungal Genet. Biol. 45: 351–362. [DOI] [PubMed] [Google Scholar]
- Jones D. T., 1999. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292: 195–202. [DOI] [PubMed] [Google Scholar]
- Kelly W. G., Aramayo R., 2007. Meiotic silencing and the epigenetics of sex. Chromosome Res. 15: 633–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent W. J., 2002. BLAT—the BLAST-like alignment tool. Genome Res. 12: 656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinsey J. A., 1989. Restricted distribution of the Tad transposon in strains of Neurospora. Curr. Genet. 15: 271–275. [DOI] [PubMed] [Google Scholar]
- Kinsey J. A., Helber J., 1989. Isolation of a transposable element from Neurospora crassa. Proc. Natl. Acad. Sci. USA 86: 1929–1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinsey J. A., Garrett-Engele P. W., Cambareri E. B., Selker E. U., 1994. The Neurospora transposon Tad is sensitive to repeat-induced point mutation (RIP). Genetics 138: 657–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M., Schein J., Birol I., Connors J., Gascoyne R., et al. , 2009. Circos: an information aesthetic for comparative genomics. Genome Res. 19: 1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S. L., 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau N. C., 2010. Small RNAs in the animal gonad: guarding genomes and guiding development. Int. J. Biochem. Cell Biol. 42: 1334–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D. W., Pratt R. J., McLaughlin M., Aramayo R., 2003. An Argonaute-like protein is required for meiotic silencing. Genetics 164: 821–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D. W., Seong K. Y., Pratt R. J., Baker K., Aramayo R., 2004. Properties of unpaired DNA required for efficient silencing in Neurospora crassa. Genetics 167: 131–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H. C., Chang S. S., Choudhary S., Aalto A. P., Maiti M., et al. , 2009. qiRNA is a new type of small interfering RNA induced by DNA damage. Nature 459: 274–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H. C., Li L., Gu W., Xue Z., Crosthwaite S. K., et al. , 2010b Diverse pathways generate microRNA-like RNAs and Dicer-independent small interfering RNAs in fungi. Mol. Cell 38: 803–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D. W., Millimaki R., Aramayo R., 2010a QIP, a component of the vegetative RNA silencing pathway, is essential for meiosis and suppresses meiotic silencing in Neurospora crassa. Genetics 186: 127–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis Z. A., Honda S., Khlafallah T. K., Jeffress J. K., Freitag M., et al. , 2009. Relics of repeat-induced point mutation direct heterochromatin formation in Neurospora crassa. Genome Res. 19: 427–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., et al. , 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N., Xiao Z. D., Yu C. H., Shao P., Liang Y. T., et al. , 2009. SnoRNAs from the filamentous fungus Neurospora crassa: structural, functional and evolutionary insights. BMC Genomics 10: 515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe T. M., Eddy S. R., 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25: 955–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macrae I. J., Zhou K., Li F., Repic A., Brooks A. N., et al. , 2006. Structural basis for double-stranded RNA processing by Dicer. Science 311: 195–198. [DOI] [PubMed] [Google Scholar]
- McCluskey K., Wiest A. E., Grigoriev I. V., Lipzen A., Martin J., et al. , 2011. Rediscovery by whole genome sequencing: classical mutations and genome polymorphisms in Neurospora crassa. G3 (Bethesda) 1: 303–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi S., Cai T., Hu Y., Chen Y., Hodges E., et al. , 2008. Sorting of small RNAs into Arabidopsis Argonaute complexes is directed by the 5′ terminal nucleotide. Cell 133: 116–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newmeyer D. D., Perkins D. D., Barry E. G., 1987. An annotated pedigree of Neurospora crassa laboratory wild types, showing the probable origin of the nucleolus satellite and showing that certain stocks are not authentic. Fungal Genet. Newsl. 34: 46–51. [Google Scholar]
- Nolan T., Braccini L., Azzalin G., De Toni A., Macino G., et al. , 2005. The post-transcriptional gene silencing machinery functions independently of DNA methylation to repress a LINE1-like retrotransposon in Neurospora crassa. Nucleic Acids Res. 33: 1564–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowrousian M., Stajich J. E., Chu M., Engh I., Espagne E., et al. , 2010. De novo assembly of a 40 Mb eukaryotic genome from short sequence reads: Sordaria macrospora, a model organism for fungal morphogenesis. PLoS Genet. 6: e1000891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pall G. S., Codony-Servat C., Byrne J., Ritchie L., Hamilton A., 2007. Carbodiimide-mediated cross-linking of RNA to nylon membranes improves the detection of siRNA, miRNA and piRNA by northern blot. Nucleic Acids Res. 35: e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelisson A., Mejlumian L., Robert V., Terzian C., Bucheton A., 2002. Drosophila germline invasion by the endogenous retrovirus gypsy: involvement of the viral env gene. Insect Biochem. Mol. Biol. 32: 1249–1256. [DOI] [PubMed] [Google Scholar]
- Pomraning, K. R., 2012 Characterization of Neurospora crassa and Fusarium graminearum mutants defective in repeat-induced point mutation (RIP). Ph.D. Thesis, Oregon State University. [Google Scholar]
- Potter S., Truett M., Phillips M., Maher A., 1980. Eucaryotic transposable genetic elements with inverted terminal repeats. Cell 20: 639–647. [DOI] [PubMed] [Google Scholar]
- Prudhomme S., Bonnaud B., Mallet F., 2005. Endogenous retroviruses and animal reproduction. Cytogenet. Genome Res. 110: 353–364. [DOI] [PubMed] [Google Scholar]
- Quinlan A. R., Hall I. M., 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raju N. B., 1980. Meiosis and ascospore genesis in Neurospora. Eur. J. Cell Biol. 23: 208–223. [PubMed] [Google Scholar]
- Raju N. B., Metzenberg R. L., Shiu P. K., 2007. Neurospora spore killers Sk-2 and Sk-3 suppress meiotic silencing by unpaired DNA. Genetics 176: 43–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano N., Macino G., 1992. Quelling: transient inactivation of gene expression in Neurospora crassa by transformation with homologous sequences. Mol. Microbiol. 6: 3343–3353. [DOI] [PubMed] [Google Scholar]
- Samarajeewa D. A., Sauls P. A., Sharp K. J., Smith Z. J., Xiao H., et al. , 2014. Efficient detection of unpaired DNA requires a member of the Rad54-like family of homologous recombination proteins. Genetics 198: 895–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selker E. U., Garrett P. W., 1988. DNA sequence duplications trigger gene inactivation in Neurospora crassa. Proc. Natl. Acad. Sci. USA 85: 6870–6874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selker E. U., Tountas N. A., Cross S. H., Margolin B. S., Murphy J. G., et al. , 2003. The methylated component of the Neurospora crassa genome. Nature 422: 893–897. [DOI] [PubMed] [Google Scholar]
- Shapiro J. A., 2005. Retrotransposons and regulatory suites. BioEssays 27: 122–125. [DOI] [PubMed] [Google Scholar]
- Shiu P. K., Metzenberg R. L., 2002. Meiotic silencing by unpaired DNA: properties, regulation and suppression. Genetics 161: 1483–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu P. K., Raju N. B., Zickler D., Metzenberg R. L., 2001. Meiotic silencing by unpaired DNA. Cell 107: 905–916. [DOI] [PubMed] [Google Scholar]
- Shiu P. K., Zickler D., Raju N. B., Ruprich-Robert G., Metzenberg R. L., 2006. SAD-2 is required for meiotic silencing by unpaired DNA and perinuclear localization of SAD-1 RNA-directed RNA polymerase. Proc. Natl. Acad. Sci. USA 103: 2243–2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith K. M., Phatale P. A., Sullivan C. M., Pomraning K. R., Freitag M., 2011. Heterochromatin is required for normal distribution of Neurospora crassa CenH3. Mol. Cell. Biol. 31: 2528–2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A., 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688–2690. [DOI] [PubMed] [Google Scholar]
- Stein L. D., Mungall C., Shu S., Caudy M., Mangone M., et al. , 2002. The generic genome browser: a building block for a model organism system database. Genome Res. 12: 1599–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel H. J., 1956. A convenient growth medium for Neurospora. Microbial Genetics Bulletin 13: 42–43. [Google Scholar]
- Wang Z., Kin K., Lopez-Giraldez F., Johannesson H., Townsend J. P., 2012. Sex-specific gene expression during asexual development of Neurospora crassa. Fungal Genet. Biol. 49: 533–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Lopez-Giraldez F., Lehr N., Farre M., Common R., et al. , 2014. Global gene expression and focused knockout analysis reveals genes associated with fungal fruiting body development in Neurospora crassa. Eukaryot. Cell 13: 154–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westergaard M., Mitchell H. K., 1947. Neurospora V. A synthetic medium favoring sexual reproduction. Am. J. Bot. 34: 573–577. [Google Scholar]
- Xiao H., Alexander W. G., Hammond T. M., Boone E. C., Perdue T. D., et al. , 2010. QIP, a protein that converts duplex siRNA into single strands, is required for meiotic silencing by unpaired DNA. Genetics 186: 119–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q., Li L., Xue Z., Ye Q., Zhang L., et al. , 2013. Transcription of the major Neurospora crassa microRNA-like small RNAs relies on RNA polymerase III. PLoS Genet. 9: e1003227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y. W., Wessler S. R., 2011. The catalytic domain of all eukaryotic cut-and-paste transposase superfamilies. Proc. Natl. Acad. Sci. USA 108: 7884–7889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Cambareri E. B., Kinsey J. A., 2001. DNA methylation inhibits expression and transposition of the Neurospora Tad retrotransposon. Mol. Genet. Genomics 265: 748–754. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Whole-genome and small RNA sequencing reads are deposited in the Short Read Archive under accession numbers: SRP021049 and SRP021051. Perl scripts and processed data files used in this project are available from http://github.com/stajichlab/neurospora_MSUD. Trimmed short reads and alignments are available at http://fungalgenomes.org/public/neurospora/data/support_files/Wang_MSUD.