Abstract
Amino acids typically are encoded by multiple synonymous codons that are not used with the same frequency. Codon usage bias has drawn considerable attention, and several explanations have been offered, including variation in GC-content between species. Focusing on a simple parameter—combined GC proportion of all the synonymous codons for a particular amino acid, termed GCsyn—we try to deepen our understanding of the relationship between GC-content and amino acid/codon usage in more details. We analyzed 65 widely distributed representative species and found a close association between GCsyn, GC-content, and amino acids usage. The overall usages of the four amino acids with the greatest GCsyn and the five amino acids with the lowest GCsyn both vary with the regional GC-content, whereas the usage of the remaining 11 amino acids with intermediate GCsyn is less variable. More interesting, we discovered that codon usage frequencies are nearly constant in regions with similar GC-content. We further quantified the effects of regional GC-content variation (low to high) on amino acid usage and found that GC-content determines the usage variation of amino acids, especially those with extremely high GCsyn, which accounts for 76.7% of the changed GC-content for those regions. Our results suggest that GCsyn correlates with GC-content and has impact on codon/amino acid usage. These findings suggest a novel approach to understanding the role of codon and amino acid usage in shaping genomic architecture and evolutionary patterns of organisms.
Keywords: GC-content, synonymous codon, amino acid usage, codon usage
The genetic code describes how the 64-nucleotide triplets specify 20 amino acids. Most amino acids have at least two synonymous codons that are, however, not used at the same frequencies in different genomes. Grantham et al. (1980) proposed the “genome hypothesis” in 1980 that assumed a species-specific pattern of codon usage. Interestingly, even in the same genome, the codon usage varies significantly among genes with different expression levels (Dos Reis et al. 2003), functions (Chiapello et al. 1998; Karlin et al. 1998; Liu et al. 2005), and tissue-specific patterns (Plotkin et al. 2004). Various factors have been suggested to affect codon usage bias, such as relative abundance of iso-accepting transfer RNAs, gene expression level, gene length, gene conversion, messenger RNA structure, and DNA base composition (Miyata et al. 1979; Ikemura 1981; Gouy and Gautier 1982; Sharp et al. 1986; Eyre-Walker 1996; Duret and Mouchiroud 1999; Sueoka and Kawanishi 2000; Maside et al. 2004). The most significant factor linked to the phenomenon of codon bias between different organisms is perhaps GC-content.
The great influence of GC-content on codon bias was first predicted by Sueoka in the 1960s (Sueoka 1961, 1962). With limited available nucleotide sequences during the 1980s and 1990s, intragenomic comparisons of heterologous DNA and protein sequences (Bernardi and Bernardi 1986; D’Onofrio et al. 1991; Collins and Jukes 1993; Berkhout and Van Hemert 1994; Porter 1995) and intergenomic comparisons of homologous gene sequences (Lobry 1997; Gu et al. 1998; Wilquet and Van De Casteele 1999; Lafay et al. 1999; D’Onofrio et al. 1999) were performed to confirm the correlations between the nucleotide composition of DNA and the amino acid content of the encoded proteins. Later, when more sequenced genomes became available, Knight et al. (2001) found similar results and suggested that GC-content is the drive for codon usage and that the correlation between GC-content and amino acid or codon usage is modulated by both mutation and selection. Another study showed that the genome-wide codon bias in eubacteria and archaea could be predicted from intergenic sequences that are not translated, suggesting that genome-wide codon bias is determined primarily by mutational processes throughout the genome (Chen et al. 2004). On the basis of the complete genome sequences, Singer and Hickey (2000) partitioned the universal codon table into GC-rich, AT-rich, and neutral codons. They further confirmed a prediction that GC-rich coding sequences (CDS) would encode amino acids with GC-rich codons, showing that biased DNA encodes biased proteins on a genome-wide scale. The finding that a positive correlation between the degree of amino acid bias and the magnitude of protein sequence divergence further support that mutational bias can have a major effect on the molecular evolution of proteins (Singer and Hickey 2000). The influence of GC-content to codon bias also was demonstrated by other studies (Karlin and Mrázek 1996; Kudla et al. 2006; Hildebrand et al. 2010; Nabiyouni et al. 2013; Bohlin et al. 2013).
Mutation and natural selection are suggested to be the two main forces shaping the genomic codon and amino acid usage patterns within and between species (Duret 2002; Chamary et al. 2006; Hershberg and Petrov 2008). The mutational explanation posits that codon bias arises from biases in nucleotides composition that are produced by point mutations, contextual biases in the point mutation rates or biases in repair. It is neutral without any fitness advantages. In contrast, the natural selection explanation suggests that synonymous mutations would influence the fitness of organisms and therefore be promoted or repressed during evolution. Despite the fact that both remain elusive, these two mechanisms are not mutually exclusive but can both play important roles in patterning the codon and amino acid usage in genomes (Bulmer 1991; Duret 2002; Hershberg and Petrov 2008).
Even though codon usage bias has been documented extensively (Karlin et al. 1998; Chen et al. 2004; Gustafsson et al. 2004; Liu et al. 2005; Hershberg and Petrov 2008), any further understanding of codon usage would still have important implications for molecular and genomic evolution. Here, we intend to investigate the impacts of GC-content on codon/amino acid usage quantitatively in more detail, starting from and focusing on a simple parameter, GC proportion of all the synonymous codons for a particular amino acid. We term this proportion the GCsyn. In this way, the GC-content of codons is defined quantitatively rather than qualitatively regarded as GC-rich or AT-rich. On the basis of GCsyn, the 20 amino acids are distinctly classified into three groups (four high-, 11 intermediate-, and five low-GCsyn amino acids), and their usage characteristics could be analyzed independently. Using 65 representative genomes from diverged species including bacteria, plants, and animals, we identified different associations between usage pattern and synonymous codon numbers for an amino acid from different GCsyn groups, indicative of adaptive evolution. More important, according to distinct usage patterns of the three groups of amino acids, we audaciously predict that identical codon/amino acid usage patterns exist when GC-content is similar, regardless of species or lineage. Comprehensive investigation of codon usage using three different units—50 consecutive codons, CDS, and genomes—in diverse prokaryotic and eukaryotic organisms indicates that our predictions are true, indicating that GC-content is a pronounced determinant of amino acid usage. In addition, we created an equation to estimate the degree of GC-content determinant to amino acid composition and GCsyn variation in different regions of the genomes. Overall, our results provide a novel view to understand codon bias and its role in the formation of genome architecture.
Materials and Methods
Selection of genome sequences
Fifteen eukaryotic and 50 prokaryotic genomes were selected on the basis of the following criteria: widely represented eukaryotic genomes according to phylogeny, and all the prokaryotic genomes greater than 4 Mb in the National Center for Biotechnology Information (ftp://ftp.ncbi.nlm.nih.gov). The genomes and annotations of animals were retrieved from the Ensembl database (www.ensembl.org), and those of Arabidopsis thaliana and Oryza sativa were downloaded from TIGR (http://www.tigr.org) and GRAMENE (http://www.gramene.org), respectively. The genomic information of other plants was obtained from JGI (ftp://ftp.jgi-psf.org/pub/JGI_data/). The CDS of each species were extracted with the annotations from corresponding, aforementioned Web sites by the use of Perl programs. Protein coding genes with ambiguous bases were eliminated. For the genes with multitranscripts, we chose the longest CDS.
Classification of the analysis units
To investigate the correlation between the frequency of amino acid (or codon) usage and GC-content, we used the whole-genome sequences of different species, CDS, or 50 consecutive codons as separate units. Of note, we also tested 30 consecutive codons and 100 consecutive codons as analysis units in comparison with 50 consecutive codons analyses. The results were highly similar so that we focused on the 50 consecutive codons analysis in the following work. For the “whole-genome sequence” analyze unit, the organisms were ranked according to their genomic GC-content from low to high. For the “CDS” analyze unit, we combined CDSs from 65 organisms and sorted them based on their GC-content, which ranged from 0.30 to 0.80, into 50 groups with an equal interval of 0.01 (second column of Figure 1). Therefore, every group of CDS contains sequences from multiple species instead of from only one genome, which eliminates potential genome biases since any genome may have a particular coding strategy as suggested in Grantham’s genome hypothesis (Grantham et al. 1980). For the “50 consecutive codon” units, each CDS in any genome was dissected sequentially into sets of 50 consecutive codons, whereas the remaining regions shorter than 50 codons were discarded. This largely diminishes the influence of special or functional motifs in a gene. These sequences also were sorted into 50 groups in the same way as the CDS with a GC interval of 0.01. Since the fractions of sequences whose GC-content is <0.3 or >0.8 were small (total < 1.30%), they were excluded from further analyses.
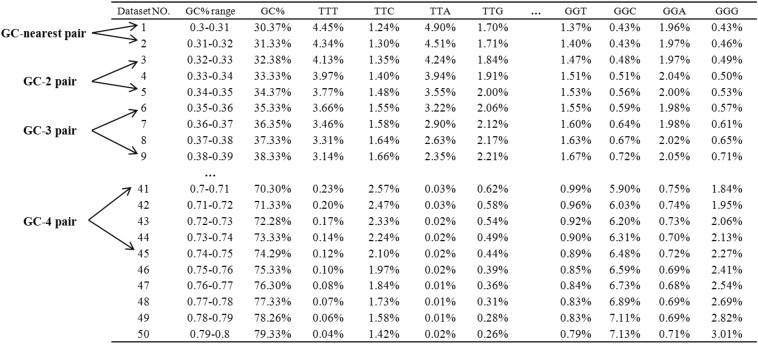
Figure 1.
The schematic diagram of codon usage analysis. As is shown in the figure, the table represents some of the codon usage results of 50-consecutive codon group and the table was sorted by the GC-content range of each group. The first column is the number of each dataset line; the second column lists the GC-content range of each 50 consecutive codon group; the third column is the average GC-content of each group; and the remaining columns show the usage frequencies of each synonymous codon. GC-nearest pair consists of two datasets with the nearest GC-content (i.e., the nearest dataset line number, such as datasets 1 and 2, 3 and 4, …, 49 and 50 in the table); GC-N pair consists of two datasets with larger GC-content divergence and N is the discrepancy between the number of any two datasets (e.g., GC-2 pairs contain the pairs of datasets 1 and 3, 2 and 4, 3 and 5…48 and 50; GC-3 pairs contain the pairs of datasets 1 and 4, 2 and 5, 3 and 6 … 47 and 50).
Linear regression analysis of codon usage
We calculated the usage of all the 61 codons (excluding the start and stop codons; of note, we excluded the first Met encoded by start codon in CDS, which would lead to biased analysis of amino acid usage) in all the 65 genomes that were first orderly ranked based on genomic GC-content. Similarly, for “CDS” and “50 consecutive codon” unit, we acquired codon usage data for each of the 50 groups (Figure 1 depicts the organization of the data). Then, we compared the codon usage in any possible pairs, such as two organisms, two groups of CDSs, and two groups of 50 consecutive codons segments. The codon usage data of two groups (i.e., to compare the linear regression correlation between two sets of the individual codon usage data as demonstrated in the last column of Figure 1) were plotted on x- and y-axes, respectively. More specifically, to analyze the group pairs with the nearest GC-content, we used the sorted codon usage tables and performed the linear regression analysis on the adjacent group pairs to get the slope and correlation coefficient (Figure 1 “GC-nearest pair”). GC-nearest pair consists of two datasets with the nearest GC-content (i.e., the nearest dataset line number, such as datasets 1 and 2, 3 and 4, …, 49 and 50 in the table). In addition, we also conducted regression analysis between groups with greater GC-content divergence and calculated slope and R2, which were defined as GC-2 pair, GC-3 pair... GC-N pair (Figure 1). GC-N pair consists of two datasets with larger GC-content divergence and N is the discrepancy between the number of any two datasets (e.g., GC-2 pairs contain the pairs of datasets 1 and 3, 2 and 4, 3 and 5…48 and 50; GC-3 pairs contain the pairs of datasets 1 and 4, 2 and 5, 3 and 6 … 47 and 50). The average values of the slope and R2 between GC-nearest pairs, GC-2 pairs, GC-3 pairs…GC-N pairs were calculated and plotted against the difference of GC-content.
The linear regression analysis was conducted in the software package R (version 2.14.1) to derive the correlation coefficient (R2) and the slope of the linear correlation between pairs. For every paired comparison, regression analyses were conducted for the usages of both 20 amino acids and 61 sense codons.
Estimation of the effects of regional GC-content
To estimate the effects of increased regional GC-content (i.e., ΔGC, the difference of GC-content between the regions with 0.79−0.80 and 0.30−0.31 GC-content), we split it into two components, which are (1) the GC fraction change due to the use of different amino acids and (2) the GC change due to the use of different synonymous codons for the same amino acid. The following formula was used:
where ΔGC indicates the increased GC-content of an amino acid from low (0.30−0.31) to high-GC (0.79−0.80) regions; GCsyn0.8 and GCsyn0.3 are the average GC-content of synonymous codons of an amino acid appearing at the regions with 0.79−0.80 and 0.30−0.31 GC-content, respectively; A0.8 and A0.3 are the usages of this amino acid in these two regions, respectively. (A0.8 − A0.3) × GCsyn0.8 explains the proportion of GC-content changes influencing the changes of amino acid usage. (GCsyn0.8 − GCsyn0.3) × A0.3 represents the proportion of GC changes influencing the GC changes in the use of high GC-content synonymous codons for that amino acid. It is supposed that the regional GC-content increase would affect the increased usage of an amino acid and its increased GCsyn. To give a conserved estimate of the contribution, only four high-GCsyn amino acids (Ala, Gly, Pro, and Arg) are included in our analysis. We can likewise infer that of an amino acid to the change of AT-content from high (0.79−0.80) to low GC (0.30−0.31) regions.
Simulation with random sequences
In addition using the real genome sequences, we repeated our analysis on simulated genomes with different level of GC-content as a control. These simulated genomes are composed of random DNA sequences. We used online software to generate random DNA sequences with different GC-content (Villesen 2007), which varies from 0.30 to 0.80 with an equal interval of 0.01, just the same as we described above. In each of these GC-content groups, we generated 10,000 random DNA sequences with different lengths ranging from 1200 to 1500 bp, which is almost the same as the average CDS length of our real sequence data.
Data availability
All the sequence data are available in public databases as described in Materials and Methods.
Results
Three types of amino acids based on GC-content of synonymous codons
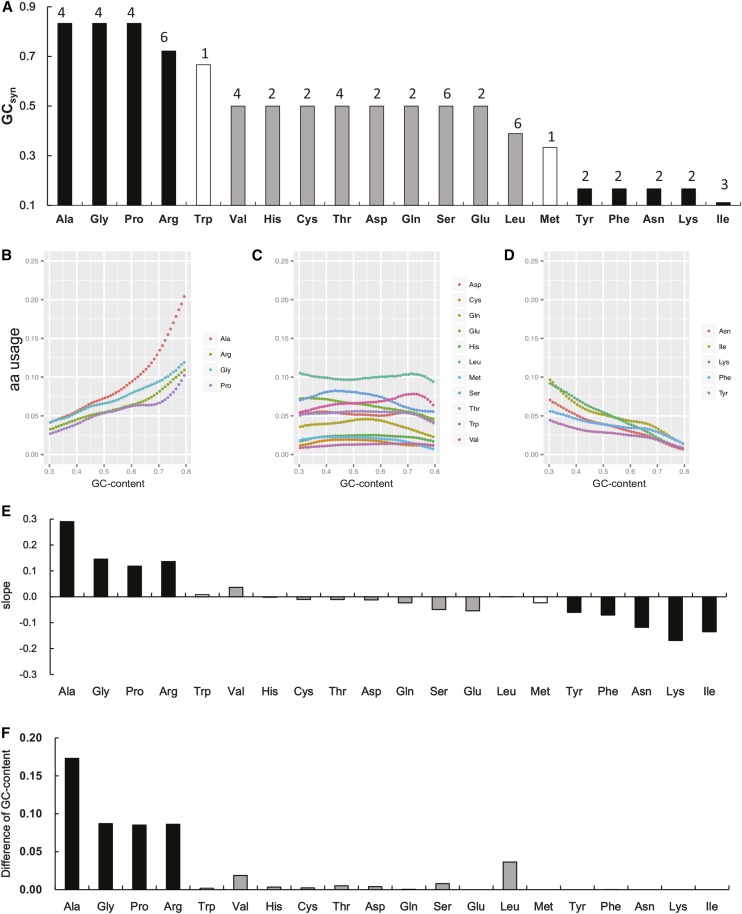
We calculated the average GC-content among the synonymous codons for each amino acid (GCsyn) by using the standard DNA codon table for nuclear genes. GCsyn was determined to range from 0.11 to 0.83 (Figure 2A). With respect to the GCsyn, there are three largely distinct groups of amino acids: four high-, 11 intermediate-, and five low-GCsyn amino acids. Within each group, GCsyn is the same for most amino acids but lower for one amino acid. Trp and Met are unusual in that they are midway between the high/intermediate and intermediate/low groups, respectively (Figure 2A).
Figure 2.
Patterns of amino acid usage and its GC-content of synonymous codons in 65 nuclear genomes in this study. (A) Average GC-content of codons for an amino acid based on the standard DNA genetic code table; the numbers on the bars are the number of synonymous codons for each amino acid; (B−D) Usage frequency variation of amino acids in three groups along with the regional GC-content (in 50 consecutive codons analyze unit), (E) is the slope value in a linear regression analysis for the lines in B−D, and (F) is for the absolute increased GC-content for each amino acid between regions with 0.3−0.31 and 0.79−0.80 GC-content. The black and nonblack bars represent the variably and the less-variably used amino acids, respectively.
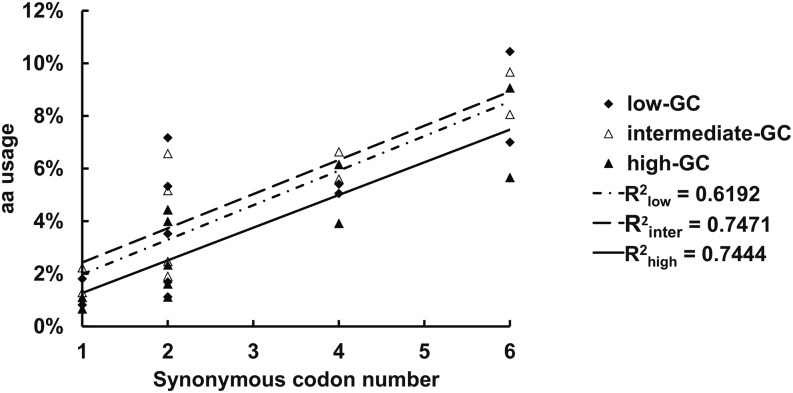
We used 65 representative genomes from bacteria, plants, and animals to investigate the amino acid usage and codon usage based on GCsyn (Supporting Information, Table S1). Our analyses show evidences for a connection between the usage and the number of synonymous codons for each amino acid. First, within each group of amino acid described previously, there is one amino acid (i.e., Arg, Leu, or Ile) for which the GCsyn is lower than other members (Figure 2A). Each of these three amino acids has the greatest number of synonymous codons in its group: 6, 6, and 3, respectively. Among these 15 codons are six of the eight least used sense codons (4 for Arg, 1 for Leu, and 1 for Ile) (Table S2). When these less frequently used codons are ignored, each of the three amino acids’ GCsyn becomes greater. Second, the single codon for both Trp and Met does not have a GCsyn that can fall into one of the three defined groups (Figure 2A), and they represent the least (0.0128, Trp) and the third least-used amino acids (0.0190, Met) in all 65 species (Table S2). Third, there is a positive correlation between amino acid usage and the number of codons for the 11 amino acids in the intermediate-GCsyn group, irrespective of the regional GC-content (Figure 3; r = 0.787−0.864, P < 0.01). Interestingly, Leu, which is encoded by six synonymous codons, always has the greatest usage (0.100) although the usage frequencies for each of the six individual codons are different. For all 20 amino acids, a significantly positive correlation (P < 0.05) is still present in the regions in which GC-content is greater than 0.35 (Table S3). These close connections between GCsyn, codon usage and the number of synonymous codons suggest possible mechanisms of adaptive evolution.
Figure 3.
The correlation between amino acid usage and the number of synonymous codons for the 11 less variably used amino acids with intermediate-GCsyn at high- (0.79−0.80), intermediate- (0.50−0.51), and low-GC (0.3−0.31) regions.
Three distinct usage patterns of amino acids
With three obvious types of amino acids based on GCsyn, one may ask whether this could be related to the different amino acid usage patterns in organisms. To address this question, we focused on the relationship among GCsyn, regional GC-content, and amino acid usage in different genomes, hypothesizing that the amino acids with high GCsyn could be used more frequently in high-GC regions and contrarily, amino acids with low GCsyn could be used more frequently in low-GC regions. In light of this hypothesis, amino acids with intermediate GCsyn are expected to be less sensitive to regional GC-content than those with extreme GCsyn. With the 65 genomes, genome sequences, CDS, and 50-consecutive codons were analyzed separately as a unit from each of these species. We calculated the GC-content and amino acid usage for each unit and classified the results from CDS and 50 consecutive codon analysis units with an interval of 0.01 GC-content from “0.30−0.31” to “0.79−0.80” (see Materials and Methods for details; whole-genome comparisons do not result in continuous GC range fractions from one another). Three distinct amino acid usage patterns, consistent with the three types grouped by GCsyn, were observed regardless of which unit was used (Figure 2, B−D for the 50 consecutive codons unit and Figure S1 for the other two units). The overall usages of the four amino acids with the highest GCsyn and the five amino acids with the lowest GCsyn both vary with the regional GC-content, whereas the usage of the remaining 11 amino acids with intermediate GCsyn is less variable. Of note, this analysis suggests that both Trp and Met fall in with the intermediate group although their GCsyn appear midway between adjacent groups (Figure 2A).
To further characterize the three types of amino acids, the slopes of linear regression analyses for the lines in Figure 2, B−D were then calculated to reveal the degree of inclination in response to the increase of GC-content. Interestingly, a steeper incline was always observed in the amino acids with extreme GCsyn (Figure 2E). The largest positive slopes (from 0.116 to 0.286; 0.170 on average) were observed in the four amino acids with the greatest GCsyn (Ala, Gly, Pro, and Arg; P < 0.001 for each regression), whereas the most negative ones (from −0.17 to −0.06; −0.110 on average) appeared in five amino acids with the lowest GCsyn (Tyr, Phe, Asn, Ile, and Lys; P < 0.001 for each). The other 11 amino acids with intermediate GCsyn exhibited relatively flat patterns (Figure 2C) (slope = 0.012 on average; P < 0.01). The minor usage changes across regions with different GC-content indicate that these 11 amino acids are less sensitive to the regional GC-content, consistent with our expectation. These results therefore suggest that there is a strong association between GCsyn for a given amino acid and its usage variation among regions with different GC-content.
Besides, given that the 65 species we investigated here have very different biology, life history traits, effective populations size, etc., which will greatly affect the selective pressures acting on a genome in general, and on synonymous codon and amino acid usage in particular, we performed the same analysis by separating prokaryotes from eukaryotes. Within eukaryotes, we also separated mammals and plants from other species. Indeed, we still observed the three distinct amino acid usage patterns in these different datasets (Figure S2). Such findings suggest that the correlation is independent of selective pressures and any specific species.
In addition to calculate regional GC-content by considering all the positions in CDS, we also computed GC-content from the third codon positions (GC3-content) because inside a codon, positions 1, 2, and 3 are not under similar evolutionary forces. We computed GC3-content and sorted the sequences based on their GC3-content, which ranged from 0.06 to 0.32, into 26 groups with an equal interval of 0.01. Then we checked the correlation between amino acid usage and GC3-content. We still observed the same three distinct usage patterns of amino acids (Figure S4, A and B). The GC3-content analysis shows that the pattern of amino acid usage and GC3-content is consistent with the results derived from whole GC-content.
The usages of amino acids and codons are nearly identical for similar GC-content regions
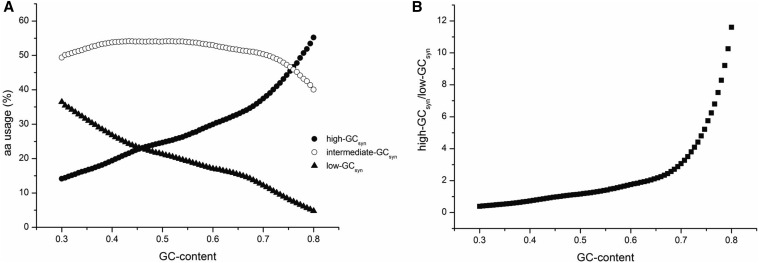
The regular usage patterns of high-, low- and intermediate- GCsyn amino acids (Figure 2, B−D) and the corresponding groups (Figure 4A) suggest their usage is near equilibrium for the given GC-content, regardless of their eukaryotic or prokaryotic origins. When regional GC-content is low, the usage ratio of high- to low-GCsyn amino acids is much less than 1, indicating the more frequent uses of low GCsyn amino acids. As the GC-content increases, there is a point where high- and low-GCsyn amino acids are used equally; after this point the usage ratio of high- to low-GCsyn continues to increase gradually (Figure 4B). The dramatic ratio variation reflects the dynamics of amino acid usage and may indicate that it could be associated with functional variations in these motifs. However, in contrast to the high- and low-GCsyn groups, the amino acid usage in the intermediate GCsyn group shows little variation when GC-content changes from 0.3 to 0.7 (Figure 4A). On the basis of these observations, the usage frequencies would be expected to be the same in different genes or motifs with similar GC-content. We then conducted another linear regression analysis in which we calculated 49 slopes and 49 R2s from the 20 pairs of amino acid usage frequency between every two adjacent GC-groups (0.010 GC-difference intervals means every GC nearest pair in Figure 1). Strikingly, both slopes and R2s are close to 1 (1.0123 ± 0.025 and 0.9984 ± 0.0006, respectively) when the regional GC-content is similar, strongly suggesting that the usage frequency of a certain amino acid is likely constant in regions with same GC-content.
Figure 4.
The total usage patterns of three amino acids types. (A) The usage frequencies of high-, low-, and intermediate-GCsyn amino acids change when GC-content increase from 0.30 to 0.80. (B) The amino acid usage ratios of the high-GCsyn to low-GCsyn types are shown.
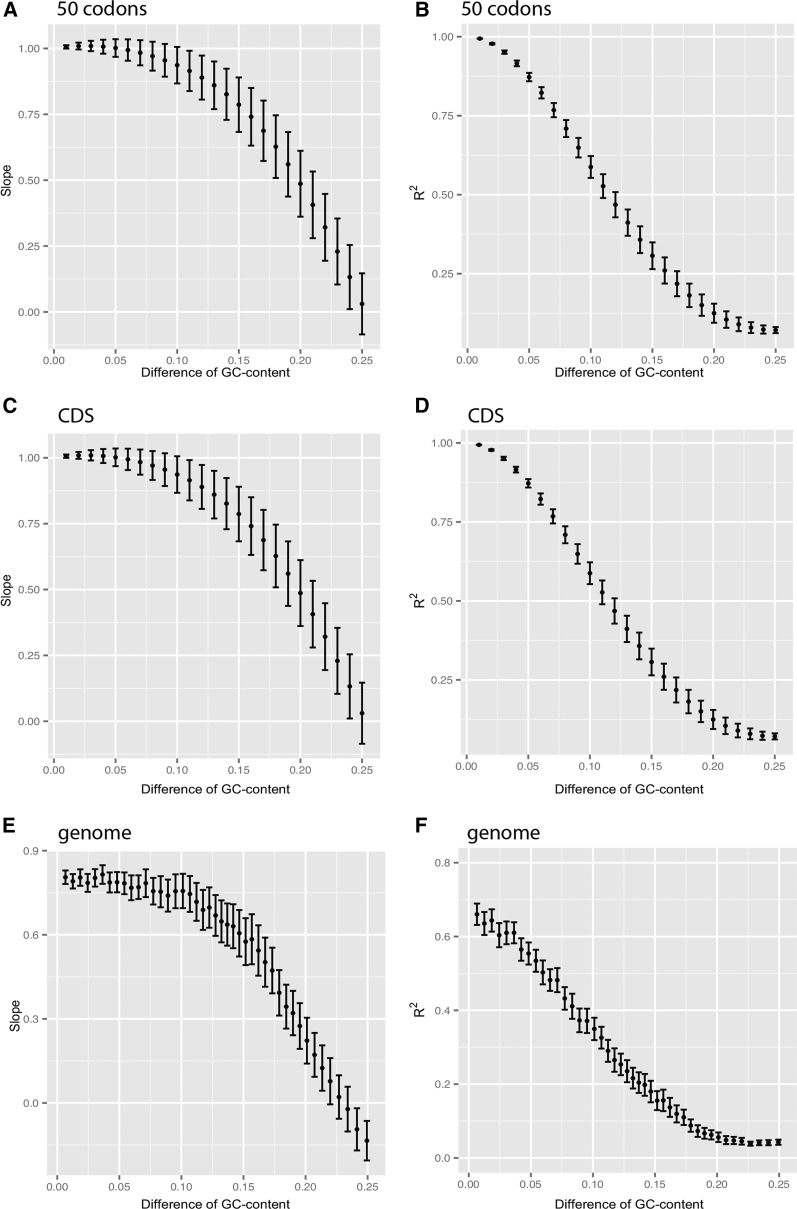
The comparable usage of amino acids at similar GC-content may essentially reflect a constant usage of codons. On the basis of the usage of each of the 61 sense codons in the same set of data as previously used for the amino acid usage analysis, regression analysis (Figure 5) reveals that both slopes and R2s are still close to 1 (1.0045 ± 0.0737 and 0.9946 ± 0.0038, respectively) when GC-content is similar. As the GC-content difference increases, the observed slope or R2 is expected to deviate from 1. To test this expectation, we calculated slope or R2 from all possible comparisons between two groups (see Materials and Methods for details; Figure 1). Shown in Figure 5, the average slope and R2 are reduced with the increase of GC difference between compared groups. For 50 consecutive codons and CDS analysis units, both the slope and R2 approach 1 when GC-difference is 0 (Figure 5). In comparison, we used the “genome” analysis unit as a control. The slope and R2 are 0.805 and 0.660 respectively in the “genome” unit when comparing two genomes with the smallest GC-content difference (0.0068). The lower values of slope and R2 for “genome” than for “50 consecutive codon” and “CDS” units are mainly contributed by the noncoding sequences. Therefore, these results further suggest that compared with the noncoding regions, the usage frequency of each codon (Figure 5) or amino acid (Figure S5) in different genes is more likely to be the same in regions of similar GC-content in the coding regions. In other words, there is a constant codon and amino acid usage when GC-content is similar. We also performed this regression analysis by separating prokaryotes, mammals, plants and other eukaryotes (Figure S3). The results also are consistent, indicating that our results are more generally distributed in living organisms rather than specific for one group. It is also true that when the GC3-content is the same, the R square and slope are both approaching 1 (Figure S4C).
Figure 5.
The patterns of the correlation slope (A, C, and E) and the coefficient of determination R2 values (B, D, and F) for codon usages between different GC groups. Fifty-codon (A and B), CDS (C and D), and whole genome (E and F) were used respectively as an analysis unit, with a GC-content difference less than 0.25 (the values are nearly 0 when ΔGC > 0.25). As shown in 50 consecutive codons (A and B) and CDS (C and D) analysis units, when the difference of GC-content from two compared units is 0 (x-axis), both the linear regression slope and the regression coefficient R2 are infinitely approaching 1 (P = 2.32e-54), which strongly suggests that codon usage frequencies are nearly constant in regions with similar GC-content.
To make our result more convincing, we performed an experiment as a control by repeating the analysis on simulated genomes with different level of GC-content and these simulated genomes are made up of random DNA sequences. We used online software to generate random DNA sequences with different GC-content (Villesen 2007). GC-content varies from 0.30 to 0.80 with an equal interval of 0.01. In each GC-content group, we generated 10,000 random DNA sequences with different lengths ranging from 1200 to 1500 bp. Using these random sequences, we repeated our analysis. Different from what we identified using real sequences, the regression result of random sequences (Figure S6) shows that when GC-content difference is 0.01, the R square is not equal to 1.0 (mean = 0.980, P = 0.1316, t-test). In comparison, with the real CDS, when GC-content difference is 0.01, the R square is almost 1.0 (mean = 0.994, P = 2.008e-12, t-test). We also performed a paired t-test between the real data and the simulated data, which showed significant differences between each other (P < 2.2e-16). This result indicates that when the GC-content is the same, the amino acid usage or codon usage is not constant in random sequences. In addition, we observed a correlation between amino acid usage and GC-content based on different GCsyn groups in random sequences as well (Figure S6C). Although the pattern of amino acid usage and GC-content appeared similar between the genomic data and simulated data, we found significant differences between them for the majority of the amino acids (Table S7). This simulation serves as a good control to our analysis of the coding regions and certifies our results: codon usage frequencies are nearly constant in regions with similar GC-content; and that intrinsically harbors some biological significance.
The influence of GC-content on amino acid composition
Although GC-content is considered widely to determine amino acid usage, it is interesting to quantitatively dissect how that changes the amino acid composition from one region to another. Besides, GC-content also would influence which synonymous codon for a certain amino acid would be used. Either GC-rich or AT-rich synonymous codons would be chosen on the basis of the variation of GC-content. To examine the effects of GC-content, we divided the contributions of GC-content’s effects into two parts: on the use of different amino acids or their synonymous codons that bears different GC fractions. Here we created an equation to quantify these two parameters due to the change of regional GC-content.
ΔGC indicates the increased GC-content of an amino acid from low to high GC-content regions (e.g., GC-content from 0.30−0.31 to 0.79−0.80), which could be split into two components: the amino acid’s increased usage (ΔA) and its increased GCsyn (ΔGCsyn) due to the usage of different synonymous codons. The first component reflects the influence of GC-content variation on amino acid usage. The second component indicates the influence on GCsyn change (see details in Materials and Methods). Here only the four high-GCsyn amino acids are taken into account because these four amino acids account for most of the increased GC-bases from low- to high-GC regions (Figure 2E), and the usage of each increases constantly as the regional GC-content increases (Figure 4A). The total increased GC-content is 0.490 for all the 20 amino acids between the regions with 0.30−0.31 and 0.79−0.80 GC-content. We show that the influence on the use of four high-GCsyn amino acids is 0.376, which accounts for 76.7% of the total increased GC-content whereas the influence on synonymous codons accounts for only 8.2% (Figure 2F; Table 1). Similarly, it is also possible to calculate the influence of low-GCsyn amino acids to the increase of AT-bases between the regions with 0.79−0.80 and 0.30−0.31 GC-content (Table S4). All these results suggest that high GC-content CDS produces genes with high GCsyn amino acids whereas low GC-content sequence encodes genes with low GCsyn amino acids. Therefore, GC-content not only determines the codon/amino acid usage but also has great influence on amino acid composition, especially the extreme-GCsyn amino acids in a region.
Table 1. The effects of GC-content by amino acids from low to high GC regions.
| Item | GC-Content | aa Usage | GCsyn | Effects on | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| GC0.3 | GC0.8 | ΔGC | A0.3 | A0.8 | ΔA | GCsyn0.3 | GCsyn0.8 | ΔGCsyn | aa | GCsyn | |
| Ala | 0.030 | 0.199 | 0.169 | 0.042 | 0.204 | 0.162 | 0.727 | 0.973 | 0.246 | 0.158 | 0.010 |
| Gly | 0.031 | 0.114 | 0.083 | 0.042 | 0.119 | 0.077 | 0.735 | 0.954 | 0.220 | 0.074 | 0.009 |
| Pro | 0.020 | 0.098 | 0.078 | 0.027 | 0.102 | 0.075 | 0.727 | 0.963 | 0.236 | 0.073 | 0.006 |
| Arg | 0.017 | 0.103 | 0.086 | 0.033 | 0.109 | 0.076 | 0.510 | 0.943 | 0.433 | 0.072 | 0.014 |
| Total | 0.097 | 0.514 | 0.416 | 0.143 | 0.534 | 0.391 | / | / | / | 0.376 | 0.040 |
| Percentage relative to 0.490 | / | / | / | / | / | 76.7% | 8.2% | ||||
Discussion
All of our findings initiated from a simple parameter, the GC-content of all the synonymous codons for an amino acid (GCsyn). On the basis of GCsyn, our results show that the 20 amino acids fall into three distinct groups. The usage of amino acids from different groups is distinct from each other. This result further led us to find constant amino acid usage when GC-content is determined. Therefore, GC-content reveals the profound influence on genomic architecture and divergence. In this point of view, it will shed light on our understanding of fundamental biological questions.
Genetic codons have long been considered to bear no inherent advantages (Francino and Ochman 1999; Yang and Nielsen 2008) and resulted from a “frozen accident” (Crick 1968). Codon reassignment could change the amino acid sequences of most proteins, resulting in a destructive impact on the organism. However, these ideas are inconsistent with the patterns observed in this report. The association between usage and synonymous codon numbers of amino acids with different GCsyn suggests that a specific codon reassignment could be associated with the usage variation of both the codon and amino acid. In this light, the least used codons presumably have a greater potential to be reassigned because this type of coding changes would have the least impact on the genome. Indeed, several lines of evidence support this point. First, the majority of nonuniversal codons in mitochondria genomes, ATA, AGA, AGG, and CTA (Li and Tzagoloff 1979; Barrell et al. 1979; Andersson and Kurland 1991; Knight et al. 2001), include four of the eight least used codons (Table S2) in our analyzed dataset. These four codons encode Ile, Arg, and Leu, which are exactly the same amino acids with the lowest GCsyn in each distinct GCsyn group. Second, Trp and Met that are each encoded by a single codon have an unusual GCsyn, and are used two times more frequently in mitochondrial genomes than in nucleic genomes (Table S5). Third, the bacterial Mycoplasma species translate UGA as Trp, which is a property also observed in animal mitochondria. Trp is among the least used amino acids and possesses a GCsyn different from other amino acids. These evidences confirm that GCsyn-based association has a great influence on the reassignment of specific codons and, subsequently, the encoded amino acids.
Besides the influence on reassignment, the association between usage pattern and synonymous codon numbers of amino acids with different GCsyn also may suggest an adaptive pattern in the course of evolution. For example, amino acids (Arg, Leu, or Ile) with a relatively low GCsyn in each group also have the most synonymous codons within that group, some of which are the least used codons. If these atypically used codon(s) are ignored, each of the three GCsyn becomes greater and is more similar to others in the corresponding group. Trp and Met both have unique GCsyn and also are used infrequently, suggesting that the single codon may only fit to the least-used amino acids because they do not have synonymous codons and consequently their GCsyn are fixed across genomic regions. In addition to these five amino acids, the usage of the eight less-variable amino acids with GCsyn of 0.5 is positively correlated with their codon numbers, which also provides evidence for adaptive evolution.
The most striking finding in our study is constant codon and amino acid usage in the regions with similar GC-content even though the organisms selected for genome comparisons in our study are widely represented. In other words, this conclusion is beyond the phylogenetic constraints and demonstrates a unique spectrum of amino acid usage that applies to all species, which is extremely critical to understanding amino acid usage and nucleotide sequences in poorly characterized taxa. In addition to this implication, this observation also can lead to an assumption that the fitness of the organism would be greater if the usage of codons and amino acids is in concordance with underlying regulations at the level of GC-content. A recent study in Escherichia coli using GFP genes varying in GC-content directly supports our assumption. In that study, E coli strains harboring GC-rich versions of GFP genes display a greater growth rate (Raghavan et al. 2012). We further analyzed their experimental data and were interested to discover that the strains showing a greater growth rate are the ones harboring GFP genes with a similar GC-content to that in the E. coli’s overall CDS (Table S6). The more GC-content in GFP deviated from that in intrinsic CDSs (0.519), the lower the growth rate displayed. The gene harboring the same GC-content as the genomic CDS might give rise to more efficient/accurate transcription and translation, resulting in greater fitness. The authors explain their result as a selective force driving genes toward greater GC-content, which is contrary to the pervasive mutational bias toward AT (Raghavan et al. 2012). We suggest an alternative hypothesis in which selection is strong when codon bias levels deviate from the equilibrium level but weak when it reaches equilibrium. Additional experiments are required to test our equilibrium model.
Another critical question we want to address is whether the GC-content variation across the genome is due to intrinsic mechanisms or external factors, and whether it is a result of a neutral process or selection (Eyre-Walker and Hurst 2001). As is well known, the genomic organization of GC-rich regions differs significantly from that of GC-poor regions, including gene densities (Lander et al. 2001), CpG island densities (Cross et al. 2000), and repetitive DNA elements densities (Bernardi 2000). Genes in GC-rich regions also have been argued to yield different proteins from those in GC-poor regions (D’Onofrio et al. 1991), and genes in GC-rich regions generally were found to have increased expression levels especially in mammalian cells. We provide a different approach to explain those findings. Using a formula to measure the influence of regional GC-content change on amino acid usage and GCsyn separately and quantitatively, we show in our results that GC-content produces the organization of high-, low-, and intermediate- GCsyn amino acids, particularly those with extreme GCsyn in any region. This may be related with gene functions. Tatarinova et al. (2010) demonstrated that GC-content is greater for genes from electron transport or energy pathways, response to abiotic or biotic stimuli, response to stress, transcription and signal transduction. Accompanying the requirement for GC-content variation, the synonymous codons have to be changed toward a higher (or lower) GC state. The usage changes in the amino acids by the gene’s GC-content also can facilitate and perhaps modulate the formation of genomic structures. Our study further suggests why a particular synonymous codon may be preferentially used. Due to functional requirements, the usage of amino acids in genes could be predetermined. Given that, the different GC-content of synonymous codons makes it possible to adjust more efficiently to the regional GC changes. A close correlation was previously revealed between exon GC-content and its surrounding intron GC-composition (Zhu et al. 2009). Our observation of functionally associated exon GC-composition provides good evidence for the influence of GC-content from exons to introns, and it will greatly further our understanding of the evolution of gene architecture.
Acknowledgments
We thank Hitoshi Araki for stimulating discussions during the study. We also thank Daniel Hartl, Danna Eickbush, Hitoshi Araki, Marjorie Lienard, the editor, and two un-known reviewers for helpful comments on the manuscript. This study was supported by National Natural Science Foundation of China (91331205 and 31571267) and PCSIRT (IRT1020) to D.T.
D.T designed the analysis. J.L. and J.Z. performed the research and analysis and wrote the manuscript. Y.W. helped to analyze the data. D.T. and S.Y. helped with the discussion and worked on the manuscript.
Footnotes
Supporting information is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.115.019877/-/DC1
Communicating editor: E. Akhunov
Literature Cited
- Andersson G. E., Kurland C. G., 1991. An extreme codon preference strategy: codon reassignment. Mol. Biol. Evol. 8: 530–544. [DOI] [PubMed] [Google Scholar]
- Barrell B. G., Bankier A. T., Drouin J., 1979. A different genetic code in human mitochondria. Nature 282: 189–194. [DOI] [PubMed] [Google Scholar]
- Berkhout B., van Hemert F. J., 1994. The unusual nucleotide content of the HIV RNA genome results in a biased amino acid composition of HIV proteins. Nucleic Acids Res. 22: 1705–1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardi G., 2000. The compositional evolution of vertebrate genomes. Gene 259: 31–43. [DOI] [PubMed] [Google Scholar]
- Bernardi G., Bernardi G., 1986. Compositional constraints and genome evolution. J. Mol. Evol. 24: 1–11. [DOI] [PubMed] [Google Scholar]
- Bohlin J., Brynildsrud O., Vesth T., Skjerve E., Ussery D. W., 2013. Amino acid usage is asymmetrically biased in AT- and GC-rich microbial genomes. PLoS One 8: e69878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulmer M., 1991. The selection-mutation-drift theory of synonymous codon usage. Genetics 129: 897–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamary J. V., Parmley J. L., Hurst L. D., 2006. Hearing silence: non-neutral evolution at synonymous sites in mammals. Nat. Rev. Genet. 7: 98–108. [DOI] [PubMed] [Google Scholar]
- Chen S. L., Lee W., Hottes A. K., Shapiro L., McAdams H. H., 2004. Codon usage between genomes is constrained by genome-wide mutational processes. Proc. Natl. Acad. Sci. USA 101: 3480–3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiapello H., Lisacek F., Caboche M., Hénaut A., 1998. Codon usage and gene function are related in sequences of Arabidopsis thaliana. Gene 209: GC1–GC38. [DOI] [PubMed] [Google Scholar]
- Collins D. W., Jukes T. H., 1993. Relationship between G + C in silent sites of codons and amino acid composition of human proteins. J. Mol. Evol. 36: 201–213. [DOI] [PubMed] [Google Scholar]
- Crick F. H., 1968. The origin of the genetic code. J. Mol. Biol. 38: 367–379. [DOI] [PubMed] [Google Scholar]
- Cross S. H., Clark V. H., Simmen M. W., Bickmore W. A., Maroon H., et al. , 2000. CpG island libraries from human chromosomes 18 and 22: landmarks for novel genes. Mamm. Genome Off. J. Int. Mamm. Genome Soc. 11: 373–383. [DOI] [PubMed] [Google Scholar]
- D’Onofrio G., Mouchiroud D., Aïssani B., Gautier C., Bernardi G., 1991. Correlations between the compositional properties of human genes, codon usage, and amino acid composition of proteins. J. Mol. Evol. 32: 504–510. [DOI] [PubMed] [Google Scholar]
- D’Onofrio G., Jabbari K., Musto H., Bernardi G., 1999. The correlation of protein hydropathy with the base composition of coding sequences. Gene 238: 3–14. [DOI] [PubMed] [Google Scholar]
- Dos Reis M., Wernisch L., Savva R., 2003. Unexpected correlations between gene expression and codon usage bias from microarray data for the whole Escherichia coli K-12 genome. Nucleic Acids Res. 31: 6976–6985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret L., 2002. Evolution of synonymous codon usage in metazoans. Curr. Opin. Genet. Dev. 12: 640–649. [DOI] [PubMed] [Google Scholar]
- Duret L., Mouchiroud D., 1999. Expression pattern and, surprisingly, gene length shape codon usage in Caenorhabditis, Drosophila, and Arabidopsis. Proc. Natl. Acad. Sci. USA 96: 4482–4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyre-Walker A., 1996. Synonymous codon bias is related to gene length in Escherichia coli: selection for translational accuracy? Mol. Biol. Evol. 13: 864–872. [DOI] [PubMed] [Google Scholar]
- Eyre-Walker A., Hurst L. D., 2001. The evolution of isochores. Nat. Rev. Genet. 2: 549–555. [DOI] [PubMed] [Google Scholar]
- Francino M. P., Ochman H., 1999. Isochores result from mutation not selection. Nature 400: 30–31. [DOI] [PubMed] [Google Scholar]
- Gouy M., Gautier C., 1982. Codon usage in bacteria: correlation with gene expressivity. Nucleic Acids Res. 10: 7055–7074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grantham R., Gautier C., Gouy M., Mercier R., Pavé A., 1980. Codon catalog usage and the genome hypothesis. Nucleic Acids Res. 8: r49–r62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X., Hewett-Emmett D., Li W. H., 1998. Directional mutational pressure affects the amino acid composition and hydrophobicity of proteins in bacteria. Genetica 102–103: 383–391. [PubMed] [Google Scholar]
- Gustafsson C., Govindarajan S., Minshull J., 2004. Codon bias and heterologous protein expression. Trends Biotechnol. 22: 346–353. [DOI] [PubMed] [Google Scholar]
- Hershberg R., Petrov D. A., 2008. Selection on codon bias. Annu. Rev. Genet. 42: 287–299. [DOI] [PubMed] [Google Scholar]
- Hildebrand F., Meyer A., Eyre-Walker A., 2010. Evidence of selection upon genomic GC-content in bacteria. PLoS Genet. 6: e1001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikemura T., 1981. Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes: a proposal for a synonymous codon choice that is optimal for the E. coli translational system. J. Mol. Biol. 151: 389–409. [DOI] [PubMed] [Google Scholar]
- Karlin S., Mrázek J., 1996. What drives codon choices in human genes? J. Mol. Biol. 262: 459–472. [DOI] [PubMed] [Google Scholar]
- Karlin S., Mrázek J., Campbell A. M., 1998. Codon usages in different gene classes of the Escherichia coli genome. Mol. Microbiol. 29: 1341–1355. [DOI] [PubMed] [Google Scholar]
- Knight R. D., Freeland S. J., Landweber L. F., 2001. A simple model based on mutation and selection explains trends in codon and amino-acid usage and GC composition within and across genomes. Genome Biol. 2: RESEARCH0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudla G., Lipinski L., Caffin F., Helwak A., Zylicz M., 2006. High guanine and cytosine content increases mRNA levels in mammalian cells. PLoS Biol. 4: e180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafay B., Lloyd A. T., McLean M. J., Devine K. M., Sharp P. M., et al. , 1999. Proteome composition and codon usage in spirochaetes: species-specific and DNA strand-specific mutational biases. Nucleic Acids Res. 27: 1642–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander E. S., Linton L. M., Birren B., Nusbaum C., Zody M. C., et al. , 2001. Initial sequencing and analysis of the human genome. Nature 409: 860–921. [DOI] [PubMed] [Google Scholar]
- Li M., Tzagoloff A., 1979. Assembly of the mitochondrial membrane system: sequences of yeast mitochondrial valine and an unusual threonine tRNA gene. Cell 18: 47–53. [DOI] [PubMed] [Google Scholar]
- Liu Q., Dou S., Ji Z., Xue Q., 2005. Synonymous codon usage and gene function are strongly related in Oryza sativa. Biosystems 80: 123–131. [DOI] [PubMed] [Google Scholar]
- Lobry J. R., 1997. Influence of genomic G+C content on average amino-acid composition of proteins from 59 bacterial species. Gene 205: 309–316. [DOI] [PubMed] [Google Scholar]
- Maside X., Lee A. W., Charlesworth B., 2004. Selection on codon usage in Drosophila americana. Curr. Biol. 14: 150–154. [DOI] [PubMed] [Google Scholar]
- Miyata T., Hayashida H., Yasunaga T., Hasegawa M., 1979. The preferential codon usages in variable and constant regions of immunoglobulin genes are quite distinct from each other. Nucleic Acids Res. 7: 2431–2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabiyouni M., Prakash A., Fedorov A., 2013. Vertebrate codon bias indicates a highly GC-rich ancestral genome. Gene 519: 113–119. [DOI] [PubMed] [Google Scholar]
- Plotkin J. B., Robins H., Levine A. J., 2004. Tissue-specific codon usage and the expression of human genes. Proc. Natl. Acad. Sci. USA 101: 12588–12591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter T. D., 1995. Correlation between codon usage, regional genomic nucleotide composition, and amino acid composition in the cytochrome P-450 gene superfamily. Biochim. Biophys. Acta 1261: 394–400. [DOI] [PubMed] [Google Scholar]
- Raghavan R., Kelkar Y. D., Ochman H., 2012. A selective force favoring increased G+C content in bacterial genes. Proc. Natl. Acad. Sci. USA 109: 14504–14507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. M., Tuohy T. M., Mosurski K. R., 1986. Codon usage in yeast: cluster analysis clearly differentiates highly and lowly expressed genes. Nucleic Acids Res. 14: 5125–5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer G. A. C., Hickey D. A., 2000. Nucleotide bias causes a genomewide bias in the amino acid composition of proteins. Mol. Biol. Evol. 17: 1581–1588. [DOI] [PubMed] [Google Scholar]
- Sueoka N., 1961. Correlation between base composition of deoxyribonucleic acid and amino acid composition of protein. Proc. Natl. Acad. Sci. USA 47: 1141–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sueoka N., 1962. On the genetic basis of variation and heterogeneity of DNA base composition. Proc. Natl. Acad. Sci. USA 48: 582–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sueoka N., Kawanishi Y., 2000. DNA G+C content of the third codon position and codon usage biases of human genes. Gene 261: 53–62. [DOI] [PubMed] [Google Scholar]
- Tatarinova T. V., Alexandrov N. N., Bouck J. B., Feldmann K. A., 2010. GC3 biology in corn, rice, sorghum and other grasses. BMC Genomics 11: 308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villesen P., 2007. FaBox: an online toolbox for fasta sequences. Mol. Ecol. Notes 7: 965–968. [Google Scholar]
- Wilquet V., Van de Casteele M., 1999. The role of the codon first letter in the relationship between genomic GC content and protein amino acid composition. Res. Microbiol. 150: 21–32. [DOI] [PubMed] [Google Scholar]
- Yang Z., Nielsen R., 2008. Mutation-selection models of codon substitution and their use to estimate selective strengths on codon usage. Mol. Biol. Evol. 25: 568–579. [DOI] [PubMed] [Google Scholar]
- Zhu L., Zhang Y., Zhang W., Yang S., Chen J.-Q., et al. , 2009. Patterns of exon-intron architecture variation of genes in eukaryotic genomes. BMC Genomics 10: 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All the sequence data are available in public databases as described in Materials and Methods.