Abstract
Two endoplasmic reticulum (ER) molecular chaperones [glucose-regulated protein 78 (grp78) and calreticulin (crt)] and three ER stress sensors [PKR-like ER kinase (perk), inositol requiring enzyme (ire)-1α, and activating transcription factor (atf)-6α] cDNAs were first characterized from yellow catfish, Pelteobagrus fulvidraco. The predicted amino acid sequences for the yellow catfish grp78, crt, perk, ire-1α, and atf-6α revealed that the proteins contained all of the structural features that were characteristic of the five genes in other species, including the KDEL motif, signal peptide, sensor domain, and effector domain. mRNAs of the five genes mentioned above were expressed in various tissues, but their mRNA levels varied among tissues. Dietary Cu excess, but not Cu deficiency, activated the chaperones (grp78 and crt) and folding sensors in ER, and the UPR signaling pathways (i.e., perk–eif2α and the ire1–xbp1) in a tissue-specific manner. For the first time, our study cloned grp78, crt, perk, ire-1α, and atf-6α genes in yellow catfish and demonstrated their differential expression among tissues. Moreover, the present study also indicated differential regulation of these ER stress–related genes by dietary Cu deficiency and excess, which will be beneficial for us to evaluate effects of dietary Cu levels in fish at the molecular level, based on the upstream pathway of lipid metabolism (the ER) and thus provide novel insights regarding the nutrition of Cu in fish.
Keywords: Pelteobagrus fulvidraco, molecular characterization, dietary Cu, ER stress, unfolded protein response
The endoplasmic reticulum (ER) is an important intracellular organelle responsible for protein and lipid synthesis, xenobiotic detoxification, and cellular calcium storage (Borgese et al. 2006; Gorlach et al. 2006). Conditions disrupting the ER homeostasis cause accumulation of unfolded and misfolded proteins in the ER lumen, create a state defined as ER stress, and activate a complex signaling network termed unfolded protein response (UPR) (Schröder and Kaufman 2005). UPR is known to trigger a unique signaling cascade from the ER to the nucleus, being characterized by enhanced expression of ER-resident chaperones, such as glucose-regulated protein 78 (Grp78) (Kozutsumi et al. 1988), and calreticulin (Crt) (Caspersen et al. 2000). In addition, UPR is characterized by downregulation of protein translation through three ER-transmembrane transducers: PKR-like ER kinase (Perk), inositol requiring enzyme (ire)-1α, and activating transcription factor (Atf)-6α (Harding et al. 1999). In the unstressed state, the ER lumenal domains of all three stress sensors are bound by the ER chaperone grp78. Upon ER stress, Grp78 binding to unfolded proteins causes dissociation from three ER-transmembrane transducers, leading to their activation (Bertolotti et al. 2000; Oikawa et al. 2009). Following activation, UPR signaling pathways act to induce expression of genes that encode functions to ameliorate the stressed state of the ER.
During the past few years, ER stress and UPR-related proteins have been extensively analyzed at the molecular level in mammals. Completely characterized cDNA and genomic sequences of Grp78, crt, Perk, Ire-1α, and Atf-6α have been previously reported in human, mouse, and rat species (Schröder and Kaufman 2005). However, in fish, to our knowledge, full-length cDNA sequences of the ER stress–related genes (grp78, crt, perk, ire-1α, and atf-6α) were only determined in very limited fish species, such as grass carp Ctenopharyngodon idella (grp78) (Zhu et al. 2013), rainbow trout Oncorhynchus mykiss (crt) (Kales et al. 2004), Asian seabass Lates calcarifer (crt) (Bai et al. 2012), and medaka Oryzias latipes (atf6) (Ishikawa et al. 2011), although some partial sequences have been reported for other fish species (Ojima et al. 2005; Komoike and Matsuoka 2013; Chen et al. 2014; Das et al. 2015; Li and Li 2015). Although their expression pattern was often covered in these works, study involved in their physiological function was often absent. Thus, identification of ER stress–related genes in other teleosts was a key step for characterizing the role and mechanism of ER stress.
Copper (Cu) is an essential micronutrient for vertebrate animals including fish (Watanabe et al. 1997). It has numerous functions in cellular biochemistry including vital roles in cellular respiration, and as a cofactor for more than 30 different enzymes (Watanabe et al. 1997). At present, optimal dietary Cu requirement has been determined in many fish species, ranging from 3 to 10 mg Cu kg−1 feed, which depends on the species, feeding regime, and life stage (National Research Council 2011). Studies have also shown that overloading of dietary Cu in fish caused toxic syndrome (Berntssen et al. 1999; Lundebye et al. 1999; Shiau and Ning 2003). In contrast, dietary Cu deficiency has been shown to reduce appetite and growth and cause anemia in several fish (Gatlin and Wilson 1986; Shiau and Ning 2003; Lin et al. 2008; Tan et al. 2011). Recently, our own studies have pointed out that dietary Cu deficiency and excess could result in the changes of lipid deposition and metabolism in fish (Chen et al. 2015). However, the molecular mechanisms of the upstream pathway of lipid metabolism underlying dietary Cu-induced alteration in lipid metabolism have not been elucidated. Studies have pointed out that ER stress is one of the cellular stresses reported to induce lipid accumulation (Lee et al. 2008), and ER stress played crucial roles in hepatic lipid metabolism in mammals (Sriburi et al. 2004; Rutkowski et al. 2008; Kammoun et al. 2009). Thus, considering the important role ER plays in lipid metabolism, we hypothesize that ER stress–dependent alteration in lipid homeostasis was the mechanism that underlies the change of lipid deposition of yellow catfish in responses to dietary copper levels.
Yellow catfish Pelteobagrus fulvidraco, an omnivorous freshwater fish, is widely distributed in the inland freshwater rivers in China. The fish is regarded as a potential model for studying lipid metabolism in fish because it has relatively high lipid accumulation under natural conditions (Zheng et al. 2013a). The cDNA sequences and molecular characterization of many genes have been elucidated in the fish species in our laboratory (Gong et al. 2013; Zheng et al. 2013a,b; Chen et al. 2014; Song et al. 2014). Recent studies in our laboratory indicated that dietary Cu deficiency and excess could influence lipid deposition and metabolism in P. fulvidraco (Chen et al. 2015). However, these studies only determined the change of lipid metabolism–related genes expression and enzyme activities after dietary Cu treatment. The molecular and regulatory mechanisms in the upstream pathway of lipid metabolism under dietary Cu treatment have not been explored. Clearly, an understanding of the molecular basis of ER stress could underpin efforts to address this problem. Therefore, in this study, the full-length cDNA sequences of two ER molecular chaperones (grp78 and crt) and three ER stress sensors (perk, ire-1α, and atf-6α) genes were first cloned and their tissue-specific expressions were determined. Meanwhile, the effect of dietary Cu deficiency and excess on their mRNA levels was explored. The results will be beneficial for us to elucidate the molecular basis of ER stress, and to further evaluate effects of dietary Cu levels in fish at a molecular level, based on the upstream pathway of lipid metabolism, which will greatly extend our understanding of the nutrition of Cu in fish.
Materials and Methods
Here, two experiments were conducted. The first experiment was involved in the cloning and tissue expression profile analysis of grp78, crt, perk, ire-1α, and atf-6α transcripts. The second experiment was designed to evaluate a possible transcriptional regulation of grp78, crt, perk, eif2α, ire-1α, xbp-1, and atf-6α by dietary Cu deficiency and excess. We ensured that the experiments performed on animals followed the ethical guidelines of Huazhong Agricultural University and Animal Care and Use Committee (IACUC) in Huazhong Agricultural University specifically approved this study.
Experiment 1: grp78, crt, perk, ire-1α, and atf-6α, cDNAs cloning and mRNA expression patterns of various tissues
Experimental animals:
Yellow catfish (1.83 ± 0.05 g, mean ± SEM) for cloning the grp78, crt, perk, ire-1α, and atf-6α genes were obtained from a local fish pond (Wuhan, China) and transferred to indoor fiberglass tanks (400 liters in water volume) for 2-wk acclimation. During the acclimation period, the fish were fed to apparent satiation twice daily with a commercial pellet diet. The experiment was conducted at ambient temperature and subjected to natural photoperiod (approximately 14 hr light/10 hr darkness). Water quality parameters were monitored twice per week in the morning. The following parameters were used: water temperature, 27.9 ± 1.2°; pH, 7.66 ± 0.11; dissolved oxygen, 6.34 ± 0.15 mg liter−1; and total hardness, 151.8 ± 1.1 mg liter−1 as CaCO3. Fish were anesthetized with 3-aminobenzoic acid ethyl ester methanesulfonate (MS-222 at 10 mg liter−1) and killed by decapitation. Skeletal muscle, gill, head kidney, mid-intestine, brain, spleen, liver, heart, testis, and ovary from six fish were dissected and immediately prepared for RNA isolation (n = 6).
RNA isolation and first-strand cDNA synthesis:
Each sample was pulverized with its own mortar and pestle, and the mortars and pestles were heated at 230° for 8 hr to inactivate RNases. Total RNA was extracted using TRIzol RNA reagent (Invitrogen, USA) based on the acid guanidinium thiocyanate-phenol-chloroform extraction method, according to the manufacturer’s instructions. The quality of total RNA was checked by agarose gel electrophoresis. The concentration and the ratio (between 1.8 and 2.0) of OD260/OD280 of total RNA were determined using a Nanodrop ND-2000 spectrophotometer (Thermo Fisher Scientific, USA). Total RNA (1 μg) was reverse-transcribed to cDNA with oligo-dT primers and a cDNA Synthesis Kit (TaKaRa, Japan) according to manufacturer’s instructions.
Cloning and sequencing of grp78, crt, perk, ire-1α, and atf-6α cDNAs:
In the present study, we used the liver of yellow catfish to clone the cDNA sequences of the five genes. Degenerate primers (Table 1) designed based on the most conserved regions of the fish grp78, crt, perk, ire-1α, and atf-6α sequences available in the GenBank and Ensembl databases were used to amplify partial cDNA fragments of grp78, crt, perk, ire-1α, and atf-6α. The PCR program consisted of initial denaturing for 4 min at 94° followed by 30 cycles of 94° for 30 sec, 55° for 30 sec, 72° for 1 min, and a final elongation step at 72° for 5 min, with 500 ng cDNA template, 0.2 μM primer, 5 μl 10 × Ex Taq Buffer, 2.5 mM dNTP, and 1.25 units Ex Taq polymerase (TaKaRa, Japan). The target fragments were purified using the EZNA gel extraction kit (Omega, USA) according to manufacturer’s instructions, and then subcloned into the pMD 19-T cloning vector (TaKaRa, Japan). Positive clones containing inserts of an expected size were sequenced with sanger dideoxy mediated chain termination method (Sangon, China).
Table 1. Nucleotide sequences of the primers used for the cDNAs cloning of grp78, crt, perk, ire-1α, and atf-6α genes.
| Primers | Sequences (5′-3′) |
|---|---|
| Primers for partial fragment | |
| grp78- F | AGAACACVGTYTTYGATGCC |
| grp78-R | TCCMACDGTCTCAATGCC |
| crt-F | RGATGCYCGYTTYTATGC |
| crt-R | GGYTTCCAYTCSCCCTKRTA |
| perk-F | AGTGYTTAGGTSGAGGCVG |
| perk-R | GAGSAGTGARAGAGGMTGG |
| ire-1α-F | ACTCTRACCTCCTYTTTTG |
| ire-1α-R | CCMCTRTCATTYACCTTCT |
| atf-6α-F | TGGTMCTTGYTTTCCTGGC |
| atf-6α-F | TCMTCASCTCAATCTCCTT |
| Primers for 3′ RACE PCR | |
| 3′ GS-grp78 01-F | TGAAGACTTTGACCAGCGTG |
| 3′ GS-grp78 11-F | CTGGACAGGAGGATACAGGAA |
| 3′ GS-crt 01-F | GGATGATGAATACACCCACC |
| 3′ GS-crt 11-F | GATGGATGGAGAATGGGAGC |
| 3′ GS-perk 01-F | CCAATCAGATGCGGTGTCC |
| 3′ GS-perk 11-F | GGACAAGTGGGTACTAAAC |
| 3′ GS-ire-1α 01-F | CAAACATCCCTTTTTCTGG |
| 3′ GS-ire-1α 11-F | ACATCCCGCTTTCCCCACCT |
| 3′ GS-atf-6α 01-F | CTTGAGGAACACCATCCCA |
| 3′ GS-atf-6α 11-F | CTGCTTATTCACTGCATCA |
| 3′ RACE Outer | TACCGTCGTTCCACTAGTGATTT |
| 3′ RACE Inner | CGCGGATCCTCCACTAGTGATTTCACTATAGG |
| Primers for 5′ RACE PCR | |
| 5′ GS-grp78 01-R | CAGACCATAGGCGATAGCAG |
| 5′ GS-grp78 11-R | TTTCCAAGGTAAGCCTCTGC |
| 5′GS-crt 01-R | TTACCTTTGTAGGCAGGATTG |
| 5′ GS-crt 11-R | CTCCCATTCTCCATCCATCT |
| 5′ GS-perk 01-R | CAGGGCTTTGGGTTTAGAT |
| 5′ GS-perk 11-R | TGTAGATTCCCAGACCATA |
| 5′ GS-ire-1α 01-R | CACCACGGGAGAGTCATAG |
| 5′ GS-ire-1α 11-R | TTTCGTCAGTCCCTCATTG |
| 5′ GS-atf-6α 01-R | TGAATGGGTCTCATAACAG |
| 5′ GS-atf-6α 11-R | AACTCTTCCGCATCTCCCA |
| 5′ RACE Outer | CATGGCTACATGCTGACAGCCTA |
| 5′ RACE Inner | CGCGGATCCACAGCCTACTGATGATCAGTCGATG |
Mixed bases: R-A/G; Y-C/T; M-A/C; K-G/T; S-G/C; W-A/T; H-A/T/C; B-G/T/C; V-G/A/C; D-G/A/T.
To obtain the 3′ and 5′ end sequences of grp78, crt, perk, ire-1α, and atf-6α, nested 3′ and 5′ RACE PCR were performed with a SMART RACE cDNA Amplification Kit (Clontech, USA) using the DNase treated total RNA, with 1 μl 10 × Reaction Buffer, 1 units DNase I for 1 μg RNA incubated at 37° for 15 min, then with 1 μl 25 mM EDTA incubated at 65° for 15 min to inactivate the DNase I. In the first PCR, the cDNA was amplified with two outer primer sets (Table 1) and Universal Primer Mix (provided in the kit) using Advantage 2 PCR Kit (Clontech, USA) with 200 ng cDNA template, 10 μM primer, 4 μl 10 × LA PCR Buffer, 25 mM MgCl2, and 1.25 units LA Taq (TaKaRa, Japan). In the second PCR, inner primer sets (Table 1) and Nested Universal Primer (provided in the kit) were used, with 1 μl 1st cDNA product, 2.5 mM dNTP, 10 μM primer, 5 μl 10 × LA PCR Buffer, 25 mM MgCl2, and 2.5 units LA Taq (TaKaRa, Japan). The PCR parameters were 30 cycles at 94° for 30 sec, 55° for 30 sec, and 72° for 1 min, with an additional initial 3-min denaturation at 94° and a 10-min final extension at 72°. The RACE products were sequenced as described above.
Sequence analysis:
The core fragment, 3′ end and 5′ end sequences were assembled using SeqMan II software in DNASTAR PACKAGE to obtain full-length grp78, crt, perk, ire-1α, and atf-6α cDNAs. The sequences were edited and analyzed using the program EDITSEQ of DNASTAR package to search for the open reading frame (ORF) and then translated into amino acid sequences using standard genetic codes. The nucleotide sequences were compared with DNA sequences present in the GenBank database using BLAST network service at the NCBI (http://blast.ncbi.nlm.nih.gov/). Sequence alignments and percentage of amino acid conservation were assessed with the Clustal-W multiple alignment algorithm. For phylogenetic analysis, multiple sequence alignments were made with MAFFT (Katoh et al. 2002) using an amino acid model at the GUIDANCE web server (http://guidance.tau.ac.il/) (Penn et al. 2010), which pruned unreliably aligned regions by rejecting columns with confidence scores below 0.93. The phylogenetic tree was constructed with MEGA 5.0 (Tamura et al. 2011) by the neighbor-joining (NJ) method based on the JTT+G model (Jones et al. 1992), the best-fit model of sequence evolution obtained by ML model selection. The confidence of each node was assessed by 1000 bootstrap replicates.
Experiment 2: responses of mRNA expression of grp78, crt, perk, eif2α, ire-1α, xbp-1, and atf-6α to dietary Cu deficiency and excess
Diet preparation:
Three experimental diets, in agreement with the study by Chen et al. (2015), were formulated with CuSO4⋅5H2O supplemented at levels of 0, 0.013, and 0.39 g kg−1 diet at the expense of cellulose (Table 2). Different Cu contents were added to the diets based on our recent study (Tan et al. 2011) to produce three different dietary Cu groups (Cu deficiency, adequate Cu, and Cu excess, respectively). The formulation of the experimental diets was detailed in the work by Chen et al. (2015). The formulated diets were dried at 80° in an oven until the moisture was reduced to less than 10%. The dry pellets were placed in plastic bags and stored at 20° until fed. Final Cu concentrations in the experimental diets were analyzed in triplicate using graphite furnace atomic absorption spectrometry (GFAAS) (Chen et al. 2015), and the contents were 0.76 (Cu deficiency), 4.18 (adequate Cu), and 92.45 (Cu excess) mg Cu kg−1 diet, respectively.
Table 2. Feed formulation and proximate analysis of experimental diets (Chen et al. 2015).
| Ingredients (g kg−1) | Adequate Cu | Cu Deficiency | Cu Excess |
|---|---|---|---|
| Casein | 320 | 320 | 320 |
| Gelatin | 80 | 80 | 80 |
| Fish oil | 25 | 25 | 25 |
| Corn oil | 50 | 50 | 50 |
| Wheat flour | 250 | 250 | 250 |
| Ascorbyl-2-polyphosphate | 10 | 10 | 10 |
| NaCl | 10 | 10 | 10 |
| NaH2PO4⋅2H2O | 10 | 10 | 10 |
| CuSO4⋅5H2O | 0.013 | 0 | 0.39 |
| Vitamin premix | 5 | 5 | 5 |
| Mineral premix (Cu-free) | 5 | 5 | 5 |
| Betaine | 10 | 10 | 10 |
| Cellulose | 224.987 | 225 | 224.61 |
| Proximate analysis (% dry matter basis) | |||
| Moisture | 8.63 | 8.58 | 8.92 |
| Crude protein | 34.38 | 34.21 | 34.29 |
| Lipid | 7.63 | 7.59 | 7.72 |
| Cu (mg kg−1) | 4.18 | 0.76 | 92.45 |
Vitamin premix according to Tan et al. (2010); Mineral premix according to Tan et al. (2010) without Cu addition. CuSO4·5H2O(≥99.0% in purity): Sinopharm Chemical Reagent Co. Ltd., Shanghai, China.
Experimental procedures:
Experimental procedures are in agreement with these in our parallel study (Chen et al. 2015). Briefly, yellow catfish were obtained from a local fish pond (Wuhan, China) and transferred to indoor fiberglass tanks (400 liters in water volume) for 2-wk acclimation. At the beginning of the experiment, 30 uniform-sized fish (1.83 ± 0.05 g; mean ± SEM) were stocked in each fiberglass tank (the loading of fish was approximately 0.183 g l−1) and subjected to natural photoperiod (approximately 14 hr light/10 hr darkness). Each diet was assigned to three tanks in a completely randomized design, with nine tanks for the experiment. Water in each tank was renewed 80% twice daily, before feeding. They were provided with continuous aeration to maintain the dissolved oxygen level near saturation. The fish were fed to apparent satiation twice daily at two equal meals (09:00 and 16:30 hr) during the week. Care was taken to ensure that no noneaten food remained in the tanks during feeding, thus leaching of CuSO4 into water was very low and negligible. Cu concentrations in water samples collected 10 min before and after feeding remained low throughout the experiment (1.7 ± 0.2 μg 1−1). The experiment continued for 8 wk. At the end of the 8-wk period, 24 hr after the last feeding, all fish were killed with MS-222 (10 mg l−1). Three fish per tank were randomly selected. After the skin was rapidly removed from muscle, the liver and skeletal muscle above the lateral line were removed immediately using sterile forceps, frozen in liquid nitrogen, and stored at −80° (not longer than 2 wk) for total RNA extraction (n = 3 replicate tanks, and three fish were sampled for each tank).
Quantitative real-time PCR:
Analyses on the distribution and mRNA expression levels of grp78, crt, perk, eif2α, ire-1α, xbp-1, and atf-6α were conducted by quantitative real-time PCR (qPCR) method. Primers used for qPCR are shown in Table 3. Total RNA extraction and first strand cDNA synthesis were performed according to previously described methods (Chen et al. 2014). The cDNA synthesis reactions were diluted to 200 μl in water. Real-time PCR was performed in a 20 μl reaction mixture including 10 μl SYBR Premix Ex TaqTM II (2×), 0.4 μl each primer (25 μM), 1 μl diluted first-strand cDNA (5 ng) product, and 8.2 μl ddH2O. Reactions were based on a three-step method as followed: at 95° for 30 sec, 40 cycles of 5 sec at 95°, 10 sec at 57°, and 30 sec at 72°. All reactions were performed in duplicate and each reaction was verified to contain a single product of the correct size by agarose gel electrophoresis. A nontemplate control and dissociation curve were performed to ensure that only one PCR product was amplified and that stock solutions were not contaminated. A 10-fold dilution series was created from a random pool of cDNA from our sample groups ranging from ×1 dilution to ×100,000 dilutions. The PCR efficiency and correlation coefficients of each primer pair were generated using the slopes of the standard curves. The amplification efficiencies of all genes were approximately equal and ranged from 98 to 103%. Because housekeeping gene sequences were not available in P. fulvidraco, a set of eight housekeeping genes (β-actin, gapdh, ef1A, 18s rRNA, hprt, b2m, tuba, and rpl17) were selected from the literature (Zhao et al. 2011) to test their transcription stability. The relative expression levels were calculated using the 2−ΔΔCt method (Livak and Schmittgen 2001) when normalizing to the geometric mean of the best combination of two genes (β-actin and gapdh, M = 0.27) as suggested by geNorm (Vandesompele et al. 2002). Prior to the analysis, experiments were performed to check the stability of housekeeping genes, from which β-actin and gapdh showed the most stable level of expression across the experimental conditions (Chen et al. 2014).
Table 3. Primers used for real-time PCR analysis.
| Genes | GenBank Accession No. | Forward Primer (5′-3′) | Reverse Primer (5′-3′) | Size (bp) |
|---|---|---|---|---|
| grp78 | KM114873 | GCTCCACTCGTAT | TCCGTAAGCCACA | 101 |
| CCCCAA | GCCTCA | |||
| crt | KM114875 | CGTGAAGAAGAG | AGAATAGGAGGCA | 185 |
| GACAAGA | TAGCAG | |||
| perk | KP687344 | GAAAAATAACATG | GCCGAGGCACCAT | 197 |
| GTGCCTCGG | GTTATTTTTC | |||
| eif2α | KR231690 | AGGATGTGGTGAT | CGATGCGGATAAG | 147 |
| GGTGAA | TTTGTT | |||
| ire-1α | KP687345 | CCTACTTCACATCC | AGTTCGCTTGACT | 168 |
| CGCTT | TTGCTC | |||
| xbp-1 | KR231691 | GTGCTTCTCATTTC | ACTCTGTTCTTCA | 143 |
| TTCATC | GTTTCC | |||
| atf-6 | KP687343 | CAGTAAGAAGGCG | TGGTGAGGGGCG | 102 |
| GAAGTG | TAGTAGAC | |||
| β-actin | EU161066 | GCACAGTAAAGGC | ACATCTGCTGGAA | 136 |
| GTTGTGA | GGTGGAC | |||
| gapdh | KP938521 | CACTGCCACCCAG | AGGGACACGGAA | 143 |
| AAGACA | AGCCAT |
Statistical analysis
Quantitative data were expressed as means ± SEM. Prior to statistical analysis, all data were tested for normality of distribution using the Kolmogorov–Smirnov test. The homogeneity of variances among the different tissues was tested using the Barlett’s test. Data were then subjected to one-way ANOVA and Tukey’s multiple range tests. Differences between control (adequate Cu) and individual Cu-treated groups (Cu deficiency and Cu excess) were analyzed by student’s t-test for independent samples. Difference was considered significant at P < 0.05. All statistics were performed using the SPSS 17.0 for Windows (SPSS, Chicago, IL, USA).
Data availability
File S1 contains detailed data about tissue expression mRNA of grp78, crt, perk, ire-1α, and atf-6α. File S2 contains detailed about effect of dietary Cu deficiency and excess on epression of grp78, crt, perk, eif2α, ire-1α, xbp-1α, and atf-6α.
Results
Molecular characterization of grp78, crt, perk, ire-1α, and atf-6α cDNAs
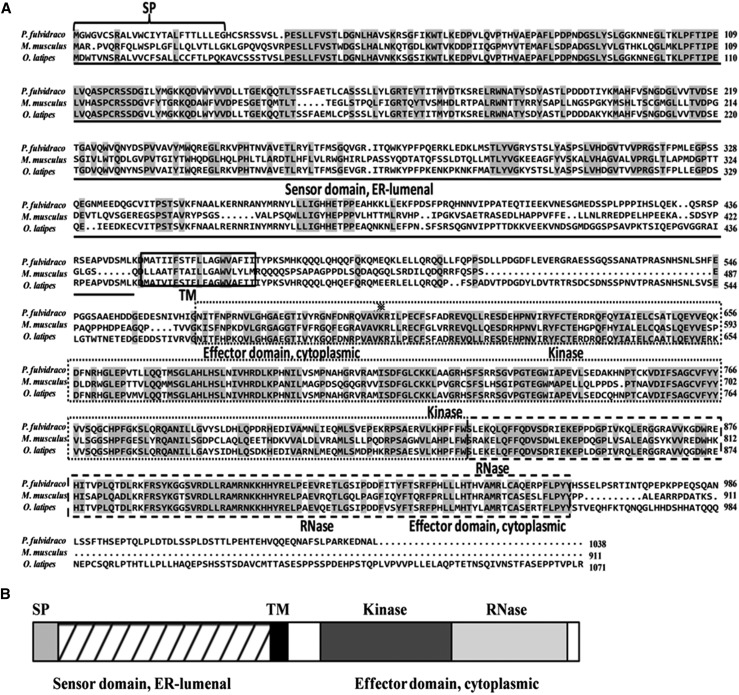
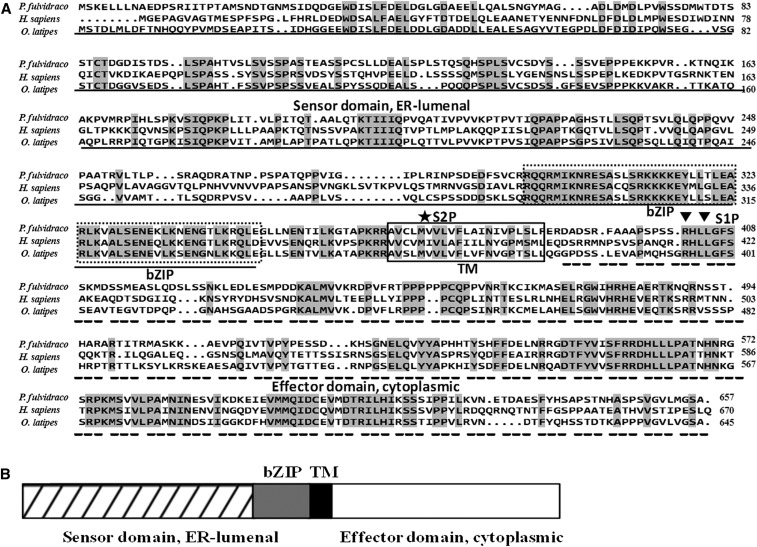
In the present study, by RT-PCR, 3′- and 5′-RACE PCR, we obtained the full-length cDNA sequences of grp78, crt, perk, ire-1α, and atf-6α genes, which were shown to be 2544 bp, 1678 bp, 3776 bp, 3440 bp, and 2785 bp, respectively (Table 4). Sequencing revealed that their cDNAs included an ORF of 1956 bp, 1254 bp, 3240 bp, 3113 bp, and 1971 bp, respectively, encoding a protein of 652 amino acids, 418 amino acids, 1080 amino acids, 1038 amino acids, and 657 amino acids, respectively. The protein sequence of P. fulvidraco grp78 possessed all the characteristic features of grp78, including signal peptide, C-terminal KDEL motif, ATPase domain, and substrate-binding domain (Figure 1). The protein sequence of yellow catfish crt possessed all the characteristic features of crt, including signal peptide, N-glycosylation site, and C-terminal KDEL motif (Figure 2). The predicted amino acid sequence for the yellow catfish perk, ire-1α, and atf-6α possessed all the characteristic features of ER stress–sensing genes, including signal peptide, transmembrane domain, protein kinase, ribonuclease, sensor domain, and effector domain (Figure 3, Figure 4, and Figure 5).
Table 4. Information for grp78, crt, perk, ire-1α, and atf-6α cDNAs cloned from yellow catfish.
| grp78 | crt | perk | ire-1α | atf-6α | |
|---|---|---|---|---|---|
| Length (bp) | 2544 | 1678 | 3776 | 3440 | 2785 |
| 5′- UTR (bp) | 143 | 126 | 181 | 157 | 106 |
| 3′- UTR (bp) | 445 | 298 | 355 | 169 | 708 |
| ORF (bp) | 1956 | 1254 | 3240 | 3113 | 1971 |
| No. of amino acids | 652 | 418 | 1080 | 1038 | 657 |
Figure 1.
(A) ClustalX alignment of the deduced amino acid sequence of grp78 from P. fulvidraco and other species. The identical residues are shaded dark gray. Characteristic features are denoted as follows: SP (signal peptide) brace; C-terminal KDEL motif, an ER-retrieval sequence, asterisk; ATPase domain, dotted line; substrate-binding domain, solid line. Deduced amino acid sequences were obtained from D. rerio (GenBank/EMBL accession no. ENSDART00000010079) and H. sapiens (M19645). (B) The predicted peptide features of grp78. SP, signal peptide.
Figure 2.
(A) ClustalX alignment of the deduced amino acid sequence of crt from P. fulvidraco and other species. The identical residues are shaded dark gray. Characteristic features are denoted as follows: SP (signal peptide) brace; N-glycosylation site, triangle; C-terminal KDEL motif, an ER-retrieval sequence, asterisk; N domain, dashed line; P domain, solid line; C domain, dotted line; triplicate repeats A, solid lines box; triplicate repeats B, dotted lines box. Deduced amino acid sequences were obtained from D. rerio (GenBank accession no. NM_131047) and H. sapiens (NM_004343). (B) The predicted peptide features of crt. SP, signal peptide.
Figure 3.
(A) ClustalX alignment of the deduced amino acid sequence of perk from P. fulvidraco and other species. The identical residues are shaded dark gray. Characteristic features are denoted as follows: SP (signal peptide) brace; N-linked glycosylation site, ¥; the invariant lysine, ※; TM (transmembrane domain), solid lines box; Kinase (protein kinase), dotted lines box; stress sensing domain, solid line; effector domain, dotted line. Transmembrane regions were predicted from a hydropathy plot taking peaks with scores above 1.6 using a scan window size of 18 (Kyte and Doolittle 1982). Deduced amino acid sequences were obtained from M. musculus (GenBank accession no. NM_010121) and D. rerio (XM_005156585). (B) The predicted peptide features of perk. SP, signal peptide; TM, transmembrane domain; Kinase, kinase protein.
Figure 4.
(A) ClustalX alignment of the deduced amino acid sequence of ire-1α from P. fulvidraco and other species. The identical residues are shaded dark gray. Characteristic features are denoted as follows: SP (signal peptide) brace; TM (transmembrane domain), solid lines box; Kinase (protein kinase), dotted lines box; RNase (ribonuclease), bold dotted lines box; the conserved lysine in kinase domain II, ※; stress sensing domain, solid line; effector domain, dotted line. Transmembrane regions were predicted from a hydropathy plot taking peaks with scores above 1.6 using a scan window size of 18 (Kyte and Doolittle 1982). Deduced amino acid sequences were obtained from M. musculus (GenBank accession no. AF071777) and O. latipes (AB667982). (B) The predicted peptide features of ire-1α. SP, signal peptide; TM, transmembrane domain; Kinase, protein kinase; RNase, ribonuclease.
Figure 5.
(A) ClustalX alignment of the deduced amino acid sequence of atf-6α from P. fulvidraco and other species. The identical residues are shaded dark gray. Characteristic features are denoted as follows: bZIP (basic leucine zipper), dotted lines box; TM (transmembrane domain), solid lines box; S1P, (Site-1 cleavage site) triangle; S2P, (Site-2 cleavage site), asterisk; stress sensing domain, solid line; effector domain, dotted line. Transmembrane regions were predicted from a hydropathy plot taking peaks with scores above 1.6 using a scan window size of 18 (Kyte and Doolittle 1982). Deduced amino acid sequences were obtained from H. sapiens (GenBank accession no. AB015856) and O. latipes (NM_001278901). (B) The predicted peptide features of atf-6α. bZIP, basic leucine zipper; TM, transmembrane domain.
Alignment of predicted polypeptide sequences showed that the amino acid sequences of yellow catfish grp78, crt, perk, ire-1α, and atf-6α were similar to those from other fish and mammals, exhibiting 91.9–94.9%, 69.9–84.8%, 56.8–74.4%, 65.1–81.7%, and 40.2–65.0% amino acid sequence identities, respectively (Table 5).
Table 5. Amino acid sequence identity of grp78, crt, perk, ire-1α, and atf-6α between yellow catfish and other species (%).
| Zebrafish | Medaka | Xenopus | Turkey | Horse | Mouse | Human | |
|---|---|---|---|---|---|---|---|
| grp78 | 93.2 | 92.6 | 91.1 | 94.9 | 91.2 | 91.1 | 91.3 |
| crt | 70.0 | 69.9 | 73.3 | 84.8 | 73.8 | 73.8 | 73.4 |
| perk | 74.4 | 68.6 | 56.8 | 61.9 | 61.5 | 57.9 | 58.6 |
| ire-1α | 81.1 | 81.7 | 65.1 | 72.3 | 74.4 | 72.0 | 73.6 |
| atf-6α | 65.0 | 57.5 | 40.4 | 44.2 | 43.8 | 44.1 | 43.8 |
EMBL databases accession numbers: grp78 (ENSDARG00000004665, ENSORLG00000006886, ENSXETG00000016838, ENSMGAG00000008510, ENSECAG00000024205, ENSMUSG00000026864, ENSG00000044574); crt (ENSDARG00000043276, ENSORLG00000002923, ENSXETG00000007937, ENSMGAG00000002470, ENSECAG00000008164, ENSMUSG00000003814, ENSG00000179218); perk (ENSDARG00000062139, ENSORLG00000017993, ENSXETG00000001926, ENSMGAG00000013189, ENSECAG00000020856, ENSMUSG00000031668, ENSG00000172071); ire-1α (ENSDARG00000013997, ENSORLG00000008940, ENSXETG00000030398, ENSMGAG00000007831, ENSECAG00000011262, ENSMUSG00000020715, ENSG00000178607); atf-6α (ENSDARG00000012656, ENSORLG00000016676, ENSXETG00000004747, ENSMGAG00000005185, ENSECAG00000022307, ENSMUSG00000026663, and ENSG00000118217). The order of accession numbers of each gene corresponds to zebrafish, medaka, xenopus, turkey, horse, mouse, and human, respectively.
The phylogenetic analysis showed that all teleost grp78 formed an independent cluster, whereas amphibian and mammalian grp78 formed another cluster. The P. fulvidraco grp78 was closer with Astyanax mexicanus and more distant from Oryzias latipes (Figure 6A). Yellow catfish crt clustered most closely with Astyanax mexicanus and Danio rerio, and more distantly from Oryzias latipes (Figure 6B). Based on the phylogentic analysis, all teleost ER stress–sensing genes (perk, ire-1α, and atf-6α) formed an independent cluster, whereas amphibian and mammalian corresponding proteins formed another cluster, clustering the yellow catfish most closely with Astyanax mexicanus and Danio rerio, and more distantly from Oryzias latipes (Figure 6, C–E).
Figure 6.
Phylogenetic trees based on the amino acid sequences of grp78 (A), crt (B), perk (C), ire-1α (D), and atf-6α (E) from yellow catfish and the other vertebrate species constructed by neighbor-joining method in MEGA 5.0 (Tamura et al. 2011) based on the JTT+G model (Jones et al. 1992) with 1000 bootstrap replicates. Accession numbers are shown next to each species. Numbers above branches indicate bootstrap support percentage over 50% in 1000 replicates.
mRNA tissue expression of grp78, crt, perk, ire-1α, and atf-6α
The grp78, crt, perk, ire-1α, and atf-6α genes from P. fulvidraco were detected in all sampled tissues, but their mRNA abundance varied (Figure 7). grp78 mRNA levels were the highest in liver, followed by heart, spleen, brain, head kidney, mid-intestine, and ovary, and were lowest in testis, gill, and skeletal muscle. crt mRNA levels were the highest in liver, followed by brain, heart, spleen, testis, and head kidney, and were the lowest in ovary, mid-intestine, gill, and skeletal muscle. perk mRNA levels were the highest in liver and heart, followed by spleen, brain, head kidney, ovary, testis, and mid-intestine, and were the lowest in skeletal muscle and gill. ire-1α mRNA expression is the highest in the liver, followed by heart, spleen, mid-intestine, brain, head kidney, and testis, and were the lowest in skeletal muscle and gill. atf-6α mRNA levels were the highest in liver and heart, followed by brain, spleen, head kidney, mid-intestine, and gill, and were the lowest in ovary, testis, and skeletal muscle.
Figure 7.
qPCR analysis for grp78, crt, perk, ire-1α, and atf-6α across skeletal muscle (M), gill (G), head kidney (K), mid-intestine (I), brain (B), spleen (S), liver (L), heart (H), testis (T), and ovary (O). Data (mean± SEM, n = 6) were normalized to housekeeping gene (β-actin and gapdh) expressed as a ratio of the control (value at skeletal muscle = 1). Bars that do not share a common letter are significantly different among the different tissues (P < 0.05).
Effect of dietary Cu deficiency and excess on expression of grp78, crt, perk, eif2α, ire-1α, xbp-1, and atf-6α
The effect of dietary Cu deficiency and excess on expression of grp78, crt, perk, eif2α, ire-1α, xbp-1, and atf-6α mRNAs in the liver and muscle of P. fulvidraco was shown in Figure 8. In liver, compared to dietary adequate Cu group, dietary Cu excess significantly upregulated mRNA expression levels of grp78, crt, perk, eif2α, ire-1α, and xbp-1, and dietary Cu deficiency only significantly upregulated mRNA expression levels of crt but showed no significant effects on other tested genes’ expressions. In muscle, compared to the dietary adequate Cu group, dietary Cu excess significantly upregulated mRNA levels of grp78, crt, and xbp-1, but showed no significant effects on mRNA expression of perk, eif2α, ire-1α, xbp-1, and atf-6α. mRNA levels of all seven tested genes showed no significant differences between dietary Cu deficiency group and dietary adequate Cu groups in skeletal muscle.
Figure 8.
Effects of dietary Cu levels on the expression of grp78, crt, perk, ire-1α, and atf-6α mRNAs in the liver and muscle of yellow catfish after 8 wk were quantified by qPCR. Data (mean ± SEM, n = 3 replicate tanks, three fish were sampled for each tank) were normalized to housekeeping gene (β-actin and gapdh) expressed as a ratio of the control (value at adequate Cu = 1). Different letters indicate significant differences among the treatments (P < 0.05).
Discussion
To date, the five full-length grp78, crt, perk, ire-1α, and atf-6α cDNAs have only been cloned in limited fish species, such as grass carp Ctenopharyngodon idella (grp78) (Zhu et al. 2013), rainbow trout Oncorhynchus mykiss (crt) (Kales et al. 2004), Asian seabass Lates calcarifer (crt) (Bai et al. 2012), and medaka Oryzias latipes (atf6) (Ishikawa et al. 2011). In this study, for the first time, we identified the full-length cDNA sequence of all five genes and explored their tissue distribution profiles from yellow catfish, and also indicated that the majority of their mRNA expression levels were differentially regulated by dietary Cu deficiency and excess, which would contribute to our understanding of the molecular basis of ER stress and would also greatly extend our understanding of the nutrition of Cu in fish.
In the present study, the ORF amino acid sequences of yellow catfish grp78, crt, perk, ire-1α, and atf-6α shared the high identity in the range of 91.9–94.9%, 69.9–84.8%, 56.8–74.4%, 65.1–81.7%, and 40.2–65.0% to those of other species, in agreement with other reports (Harding et al. 1999; Higashino et al. 2006; Ishikawa et al. 2011; Lenartowski et al. 2014; Das et al. 2015). The predicted amino acid sequences for the yellow catfish ER molecular chaperones (grp78 and crt) revealed that the peptide contained all of the structural features that are characteristic of ER molecular chaperones in other species, containing 16- and 18-residue signal peptide sequences and the KDEL motif (Kales et al. 2004; Higashino et al. 2006; Bai et al. 2012; Das et al. 2015). The signal peptide at the N terminus and the consensus tetrapeptide KDEL at the C terminus of the amino acid sequence ensure the protein targeting to the ER. KDEL is a retrieval motif essential for the precise sorting of ER resident proteins along the secretory pathway. It is also known that grp78 has two major domains, the N-terminal domain containing the ATPase catalytic site and the C-terminal substrate-binding domain (Gaut and Hendershot 1993; Gething 1999; Das et al. 2015), which are also conserved in the grp78 of yellow catfish. Structural predictions of crt indicated that the protein had at least three domains, i.e., N, P, and C domains. The P-domain of crt comprises a proline-rich sequence with three repeats of the amino acid sequence PXXIXDPDAXKPEDWDE followed by three repeats of the sequence GXWXPPXIXNPXYX. Similar results were also obtained in Japanese monkey (Macaca fuscata) (Higashino et al. 2006). The P-domain binds Ca2+ with high affinity, and the repeats may be essential for the high affinity of Ca2+ binding of crt (Fliegel et al. 1989; Ellgaard et al. 2001). The predicted amino acid sequence of the yellow catfish perk revealed that the peptide contained the structural features that are characteristic of perk, such as a signal peptide, a transmembrane domain, a cytosolic Ser/Thr kinase domain, sensor domain, and effector domain (Cui et al. 2011). ire-1α from mammals are characterized by an amino-terminal ER lumenal domain, a transmembrane region, and a carboxy-terminal domain that contains both kinase and RNase catalytic activities (Ishikawa et al. 2011), which are also conserved in the ire-1α of yellow catfish. The predicted amino acid sequence of atf-6α from yellow catfish and other species are characterized by a cytoplasmic N-terminus that contains a bZIP motif, which functions as a transcription factor following regulated intramembrane proteolysis in ER-stressed cells (Ishikawa et al. 2011). Phylogenetic analysis suggested that yellow catfish grp78, crt, perk, ire-1α, and atf-6α peptides were closely related to other fish with similar taxonomic classes, similar to those reported in other fish (Kales et al. 2004).
The analysis of tissue distribution profiles for genes is useful to better understand their physiological roles. At present, in fish several studies have been involved in mRNA expression of grp78 (Hori et al. 2010; Zhu et al. 2013; Rebl et al. 2014; Das et al. 2015) and crt (Kales et al. 2004; Bai et al. 2012), and other reports in mRNA expression profiles of ER stress-relevant transcripts in fish (Chen et al. 2014; Komoike and Matsuoka 2013). However, the mRNA expression profiles of perk, ire-1α, and atf-6α were still absent in fish. The present study indicated that grp78 mRNA levels were the highest in liver, followed by heart and brain. The noninduced level of expression of grp78 indicated that it might play multiple functions in yellow catfish as its homologs reported in other living organisms (Xi and Ma 2013). Zhu et al. (2013) indicated that the expression of grp78 in the liver of grass carp Ctenopharyngodon idella was higher than other tissues. Other studies also indicated higher Grp78 accumulation in brain (Higashino et al. 2006; Xi and Ma 2013) and heart tissue in mammals (Oh-hashi et al. 2003). In the present study, crt mRNA was expressed ubiquitously in every tissue, with the highest in liver and the lowest in ovary, mid-intestine, gill, and skeletal muscle. Kales et al. (2004) reported the ubiquitous expression of crt mRNA in all tissues tested, with highest expression in liver. In Asian seabass, Bai et al. (2012) observed that the highest expression was seen in the liver, followed by that in the brain, spleen, and kidney. In Japanese monkey, Higashino et al. (2006) pointed out that Crt mRNA was expressed ubiquitously in every tissue except the thymus and skeletal muscle, which had low expression. The present study also detected the low crt mRNA expression in the intestine, in agreement with the reports by Kales et al. (2004). The present study indicated the Crt mRNA levels in testis. crt has been identified in the sperm of several mammals (Ho and Suarez 2003; Naaby-Hansen et al. 2001) and appears to be involved in regulating sperm capacitation, motility, and acrosome reaction (Naaby-Hansen et al. 2001; Ho and Suarez 2003). Like mammals (Koyabu et al. 1994) and rainbow trout (Kales et al. 2007), yellow catfish crt may play only a minor role in skeletal muscle, conserving its role in calcium modulation for smooth muscle such as the heart (Waisman et al. 1986). The present study reported the ubiquitous mRNA expression of the three ER stress sensors (perk, ire-1α, and atf-6α) in all tested tissues, with the highest expression in liver and the lowest in skeletal muscle and gill. At present, mRNA expression profiles of these genes in fish were very scarce. Studies pointed out that the expression level of ER stress proteins was expected to be high in secretory cells, as these cells are rich in ER (Palade 1975). Their commonly high expression in the liver, spleen, and head kidney is consistent with this hypothesis. The present study also indicated the high expression of ER stress–related genes was found in the heart. Studies suggested that ER stress might protect the heart and even foster hypertrophic growth of the myocardium in mammals (Okada et al. 2004; Mao et al. 2006).
In mammals, lipid droplet accumulation is often induced by ER stress (Ozcan et al. 2004; Oyadomari et al. 2008). Studies indicated Cu could act as a modifier in lipid metabolism (Burkhead and Lutsenko 2013) and the variations in dietary Cu have a profound effect on lipid deposition in fish (Berntssen et al. 1999; Tan et al. 2011; Shaw and Handy 2006). Recently, we have indicated that dietary deficiency and excess could influence the key lipid metabolism–related genes expression (i.e., 6pgd, g6pd, fas, accα, pparγ, lxr, hsl, pparα, and atgl) and enzyme activities (i.e., 6pgd, G6pd, and Fas) in yellow catfish (Chen et al. 2015). To shed light on the molecular mechanisms underlying the change of lipid storage in yellow catfish, we analyzed the changes of mRNA expression of these ER-related genes known to play an important molecular role in ER stress–induced changes of lipid deposition. The present study, for the first time, found dietary Cu excess activated ER stress and the UPR signaling in a tissue-specific manner in yellow catfish. First, the chaperones and folding sensors in ER, including grp78 and crt, were significantly upregulated in liver and skeletal muscle of P. fulvidraco in dietary Cu excess groups. Upregulation of grp78 and crt often acts as an index of ER stress (Lee 2005). Studies reported marked upregulation of ER chaperones such as grp78 and crt in response to metal ions (Liu et al. 2006; Shinkai et al. 2010; Zhu et al. 2013). Second, we found that two UPR branches, the perk–eif2α pathway and the ire1–xbp1 pathway, were activated by dietary Cu excess in liver of P. fulvidraco. Studies also suggested that under ER stress, UPR pathways, i.e., perk–eif2α and the ire1–xbp1, are activated to reduce the ER load of the unfolded proteins (Ron and Walter 2007). In LLC-PK1 cells, Liu et al. (2006) found eif2α mRNA levels were elevated in response to CdCl2 exposure. The activation of UPR pathways will in turn influence lipid metabolism because these pathways control hepatic lipogenesis, as suggested by several studies (Lee et al. 2008; Oyadomari et al. 2008; Lee and Glimcher 2009). atf6 also plays an important role in regulating hepatic lipid homeostasis (Yamamoto et al. 2007; Rutkowski et al. 2008). In the present study, dietary Cu excess did not significantly influence atf6α expression, which showed that the atf6α pathway was not the main branch for the Cu-induced ER stress. Considering the lack of change in ER stress–related genes’ expressions in skeletal muscle following dietary Cu treatments, we believe this lack of response in these genes’ expression levels suggests a different mode of regulation or function in fish skeletal muscle. Because the ER stress plays important roles in lipid homeostasis, the ER stress and UPR may be the fundamental reaction to the dietary Cu exposure. Thus, ER stress may be the unrecognized mechanism underlying the changes of lipid deposition that are common to dietary Cu levels.
In summary, we cloned the full-length sequences of five key genes (grp78, crt, perk, ire-1α, and atf-6α) involved in ER stress response from yellow catfish and investigated their molecular characterization, which will facilitate further studies on the regulation of ER stress response at the molecular level. mRNA expression patterns of these genes in different tissues also provided a basis for further exploring the differential regulatory patterns of ER stress response in different tissues for the fish species. Dietary Cu excess activated ER stress and the UPR signaling in a tissue-specific manner in yellow catfish, which will help us to understand the effects of dietary Cu levels on the upstream pathway of lipid metabolism (the ER) and thus provide some novel insights on the nutrition of Cu in fish.
Acknowledgments
The authors thank the staff of Aquatic Animal Nutrition and Feed Laboratory of Huazhong Agricultural University for their assistance with excellent sample analysis. This work was supported by the National Natural Science Foundation of China (grant no. 31422056), the Fundamental Research Funds for the Central Universities, China (grant nos. 2662015PY017, 2014JQ002, 2013PY073), and by the PhD Candidate Research Innovation Project of Huazhong Agricultural University (Program No. 2014bs36).
Footnotes
Supporting information is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.115.019950/-/DC1
Communicating editor: W. S. Davidson
Literature Cited
- Bai Z. Y., Zhu Z. Y., Wang C. M., Xia J. H., He X. P., et al. , 2012. Cloning and characterization of the calreticulin gene in Asian seabass (Lates calcarifer). Animal 6: 887–893. [DOI] [PubMed] [Google Scholar]
- Berntssen M. H. G., Hylland K., Wendelaar Bonga S. E., Maage A., 1999. Toxic levels of dietary copper in Atlantic salmon (Salmo salar L.) parr. Aquat. Toxicol. 46: 87–99. [Google Scholar]
- Bertolotti A., Zhang Y., Hendershot L. M., Harding H. P., Ron D., 2000. Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat. Cell Biol. 2: 326–332. [DOI] [PubMed] [Google Scholar]
- Borgese N., Francolini M., Snapp E., 2006. Endoplasmic reticulum architecture: structures in flux. Curr. Opin. Cell Biol. 18: 358–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhead J. L., Lutsenko S., 2013. The role of copper as a modifier of lipid metabolism, pp. 39–61 in Lipid Metabolism, InTech Open Access Publisher, edited by Baez R. V. [Google Scholar]
- Caspersen C., Pedersen P. S., Treiman M., 2000. The sarco/endoplasmic reticulum calcium-ATPase 2b is an endoplasmic reticulum stress-inducible protein. J. Biol. Chem. 275: 22363–22372. [DOI] [PubMed] [Google Scholar]
- Chen Q. L., Luo Z., Song Y. F., Wu K., Huang C., et al. , 2014. Hormonesensitive lipase in yellow catfish Pelteobagrus fulvidraco: molecular characterization, mRNA tissue expression and transcriptional regulation by leptin in vivo and in vitro. Gen. Comp. Endocrinol. 206: 130–138. [DOI] [PubMed] [Google Scholar]
- Chen Q. L., Luo Z., Wu K., Huang C., Zhuo M. Q., et al. , 2015. Differential effects of dietary copper deficiency and excess on lipid metabolism in yellow catfish Pelteobagrus fulvidraco. Comp. Biochem. Physiol. 184: B19–B28. [DOI] [PubMed] [Google Scholar]
- Cui W., Li J., Ron D., Sha B., 2011. The structure of the PERK kinase domain suggests the mechanism for its activation. Acta Crystallogr. D Biol. Crystallogr. 67: 423–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S., Mohapatra A., Sahoo P. K., 2015. Expression analysis of heat shock protein genes during Aeromonas hydrophila infection in rohu, Labeo rohita, with special reference to molecular characterization of Grp78. Cell Stress Chaperones 20: 73–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellgaard L., Riek R., Herrmann T., Güntert P., Braun D., et al. , 2001. NMR structure of the calreticulin P-domain. Proc. Natl. Acad. Sci. USA 98: 3133–3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fliegel L., Burns K., Maclennan D. H., Reithmeier R. A., Michalak M., 1989. Molecular cloning of the high affinity calcium-binding protein (calreticulin) of skeletal muscle sarcoplasmic reticulum. J. Biol. Chem. 264: 21522–21528. [PubMed] [Google Scholar]
- Gatlin D. M., III, Wilson R. P., 1986. Dietary copper requirement of fingerling channel catfish. Aquaculture 54: 277–285. [Google Scholar]
- Gaut J. R., Hendershot L. M., 1993. Mutations within the nucleotide binding site of immunoglobulin-binding protein inhibit ATPase activity and interfere with release of immunoglobulin heavy chain. J. Biol. Chem. 268: 7248–7255. [PubMed] [Google Scholar]
- Gething M. J., 1999. Role and regulation of the ER chaperone BiP. In Seminars in cell & developmental biology. Semin. Cell Dev. Biol. 10: 465–472. [DOI] [PubMed] [Google Scholar]
- Gong Y., Luo Z., Zhu Q. L., Zheng J. L., Tan X. Y., et al. , 2013. Characterization and tissue distribution of leptin, leptin receptor and leptin receptor overlapping transcript genes in yellow catfish (Pelteobagrus fulvidraco). Gen. Comp. Endocrinol. 182: 1–6. [DOI] [PubMed] [Google Scholar]
- Gorlach A., Klappa P., Kietzmann T., 2006. The endoplasmic reticulum: folding, calcium homeostasis, signaling, and redox control. Antioxid. Redox Signal. 8: 1391–1418. [DOI] [PubMed] [Google Scholar]
- Harding H. P., Zhang Y., Ron D., 1999. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature 397: 271–274. [DOI] [PubMed] [Google Scholar]
- Higashino A., Fukuhara R., Tezuka T., Kageyama T., 2006. Molecular cloning and gene expression of endoplasmic reticulum stress proteins in Japanese monkey, Macaca fuscata. J. Med. Primatol. 35: 376–383. [DOI] [PubMed] [Google Scholar]
- Ho H. C., Suarez S. S., 2003. Characterization of the intracellular calcium store at the base of the sperm flagellum that regulates hyperactivated motility. Biol. Reprod. 68: 1590–1596. [DOI] [PubMed] [Google Scholar]
- Hori T. S., Gamperl A. K., Afonso L. O., Johnson S. C., S. Hubert et al, 2010. Heat-shock responsive genes identified and validated in Atlantic cod (Gadus morhua) liver, head kidney and skeletal muscle using genomic techniques. BMC Genomics 11: 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa T., Taniguchi Y., Okada T., Takeda S., Mori K., 2011. Vertebrate unfolded protein response: mammalian signaling pathways are conserved in medaka fish. Cell Struct. Funct. 36: 247–259. [DOI] [PubMed] [Google Scholar]
- Jones D. T., Taylor W. R., Thornton J. M., 1992. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8: 275–282. [DOI] [PubMed] [Google Scholar]
- Kales S., Fujiki K., Dixon B., 2004. Molecular cloning and characterization of calreticulin from rainbow trout (Oncorhynchus mykiss). Immunogenetics 55: 717–723. [DOI] [PubMed] [Google Scholar]
- Kales S. C., Bols N. C., Dixon B., 2007. Calreticulin in rainbow trout: a limited response to Endoplasmic reticulum (ER) stress. Comp. Biochem. Physiol. 147: B607–B615. [DOI] [PubMed] [Google Scholar]
- Kammoun H. L., Chabanon H., Hainault I., Luquet S., Magnan C., et al. , 2009. GRP78 expression inhibits insulin and ER stress–induced SREBP-1c activation and reduces hepatic steatosis in mice. J. Clin. Invest. 119: 1201–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Misawa K., Kuma K. I., Miyata T., 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30: 3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komoike Y., Matsuoka M., 2013. Exposure to tributyltin induces endoplasmic reticulum stress and the unfolded protein response in zebrafish. Aquat. Toxicol. 142: 221–229. [DOI] [PubMed] [Google Scholar]
- Koyabu S., Imanaka-Yoshida K., Ioshii S. O., Nakano T., Yoshida T., 1994. Switching of the dominant calcium sequestering protein during skeletal muscle differentiation. Cell Motil. Cytoskeleton 29: 259–270. [DOI] [PubMed] [Google Scholar]
- Kozutsumi Y., Segal M., Normington K., Gething M. J., Sambrook J., 1988. The presence of malfolded proteins in the endoplasmic reticulum signals the induction of glucose-regulated proteins. Nature 332: 462–464. [DOI] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F., 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157: 105–132. [DOI] [PubMed] [Google Scholar]
- Lee A. S., 2005. The ER chaperone and signaling regulator GRP78/BiP as a monitor of endoplasmic reticulum stress. Methods 35: 373–381. [DOI] [PubMed] [Google Scholar]
- Lee A. H., Glimcher L. H., 2009. Intersection of the unfolded protein response and hepatic lipid metabolism. Cell. Mol. Life Sci. 66: 2835–2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A. H., Scapa E. F., Cohen D. E., Glimcher L. H., 2008. Regulation of hepatic lipogenesis by the transcription factor XBP1. Science 320: 1492–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenartowski R., Suwińska A., Prusińska J., Gumowski K., Lenartowska M., 2014. Molecular cloning and transcriptional activity of a new Petunia calreticulin gene involved in pistil transmitting tract maturation, progamic phase, and double fertilization. Planta 239: 437–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z. H., Li P., 2015. Evaluation of tributyltin toxicity in Chinese rare minnow larvae by abnormal behavior, energy metabolism and endoplasmic reticulum stress. Chem. Biol. Interact. 227: 32–36. [DOI] [PubMed] [Google Scholar]
- Lin Y. H., Shie Y. Y., Shiau S. Y. Dietary copper requirements of juvenile grouper, Epinephelus malaaricus. Aquaculture 274: 161–165. [Google Scholar]
- Liu F., Inageda K., Nishitai G., Matsuoka M., 2006. Cadmium induces the expression of Grp78, an endoplasmic reticulum molecular chaperone, in LLC-PK1 renal epithelial cells. Environ. Health Perspect. 114: 859–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D., 2001. Analysis of relative gene expression data using realtime quantitative PCR and the 2 –ΔΔCT method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Lundebye A. K., Berntssen M. H. G., Wendelaar Bonga S. E., Maage A., 1999. Biochemical and physiological responses in Atlantic salmon (Salmo salar) following dietary exposure to copper and cadmium. Mar. Pollut. Bull. 39: 137–144. [Google Scholar]
- Mao C., Tai W. C., Bai Y., Poizat C., Lee A. S., 2006. In vivo regulation of Grp78/BiP transcription in the embryonic heart: role of the endoplasmic reticulum stress response element and GATA-4. J. Biol. Chem. 281: 8877–8887. [DOI] [PubMed] [Google Scholar]
- Naaby-Hansen S., Wolkowicz M. J., Klotz K., Bush L. A., Westbrook V. A., et al. , 2001. Co-localization of the inositol 1, 4, 5-trisphosphate receptor and calreticulin in the equatorial segment and in membrane bounded vesicles in the cytoplasmic droplet of human spermatozoa. Mol. Hum. Reprod. 7: 923–933. [DOI] [PubMed] [Google Scholar]
- National Research Council (NRC) , 2011. Nutrient Requirement of Fish and Shrimp, National Academic Press, Washington, DC. [Google Scholar]
- Oh-hashi K., Naruse Y., Amaya F., Shimosato G., Tanaka M., 2003. Cloning and characterization of a novel GRP78-binding protein in the rat brain. J. Biol. Chem. 278: 10531–10537. [DOI] [PubMed] [Google Scholar]
- Oikawa D., Kimata Y., Kohno K., Iwawaki T., 2009. Activation of mammalian IRE1 alpha upon ER stress depends on dissociation of BiP rather than on direct interaction with unfolded proteins. Exp. Cell Res. 315: 2496–2504. [DOI] [PubMed] [Google Scholar]
- Ojima N., Yamashita M., Watabe S., 2005. Quantitative mRNA expression profiling of heat-shock protein families in rainbow trout cells. Biochem. Biophys. Res. Commun. 329: 51–57. [DOI] [PubMed] [Google Scholar]
- Okada K., Minamino T., Tsukamoto Y., Liao Y., Tsukamoto O., et al. , 2004. Prolonged endoplasmic reticulum stress in hypertrophic and failing heart after aortic constriction: possible contribution of endoplasmic reticulum stress to cardiac myocyte apoptosis. Circulation 110: 705–712. [DOI] [PubMed] [Google Scholar]
- Oyadomari S., Harding H. P., Zhang Y., Oyadomari M., Ron D., 2008. Dephosphorylation of translation initiation factor 2alpha enhances glucose tolerance and attenuates hepatosteatosis in mice. Cell Metab. 7: 520–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozcan U., Cao Q., Yilmaz E., Lee A. H., Iwakoshi N. N., et al. , 2004. Endoplasmic reticulum stress links obesity, insulin action, and type 2 diabetes. Science 306: 457–461. [DOI] [PubMed] [Google Scholar]
- Palade G., 1975. Intracellular aspects of the process of protein synthesis. Science 189: 347–358. [DOI] [PubMed] [Google Scholar]
- Penn O., Privman E., Ashkenazy H., Landan G., Graur D., et al. , 2010. GUIDANCE: a web server for assessing alignment confidence scores. Nucleic Acids Res. 38: W23–W28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebl A., Brietzke A., Goldammer T., Seyfert H. M., 2014. GRP94 is encoded by two differentially expressed genes during development of rainbow trout (Oncorhynchus mykiss). Fish Physiol. Biochem. 40: 1917–1926. [DOI] [PubMed] [Google Scholar]
- Ron D., Walter P., 2007. Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol. 8: 519–529. [DOI] [PubMed] [Google Scholar]
- Rutkowski D. T., Wu J., Backetal S. H., 2008. UPR pathways combine to prevent hepatic steatosis caused by ER stressmediated suppression of transcriptional master regulators. Dev. Cell 15: 829–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröder M., Kaufman R. J., 2005. The mammalian unfolded protein response. Annu. Rev. Biochem. 74: 739–789. [DOI] [PubMed] [Google Scholar]
- Shaw B. J., Handy R. D., 2006. Dietary copper exposure and recovery in Nile tilapia, Oreochromis niloticus. Aquat. Toxicol. 76: 111–121. [DOI] [PubMed] [Google Scholar]
- Shiau S. Y., Ning Y. C., 2003. Estimation of dietary copper requirements of juvenile tilapia, Oreochromis niloticus × O. aureus. Anim. Sci. 77: 287–292. [Google Scholar]
- Shinkai Y., Yamamoto C., Kaji T., 2010. Lead induces the expression of endoplasmic reticulum chaperones GRP78 and GRP94 in vascular endothelial cells via the JNK-AP-1 pathway. Toxicol. Sci. 114: 378–386. [DOI] [PubMed] [Google Scholar]
- Song Y. F., Luo Z., Pan Y. X., Zhang L. H., Chen Q. L., et al. , 2014. Three unsaturated fatty acid biosynthesis-related genes in yellow catfish Pelteobagrus fulvidraco: Molecular characterization, tissue expression and transcriptional regulation by leptin. Gene 563: 1–9. [DOI] [PubMed] [Google Scholar]
- Sriburi R., Jackowski S., Mori K., Brewer J. W., 2004. XBP1 a link between the unfolded protein response, lipid biosynthesis, and biogenesis of the endoplasmic reticulum. J. Cell Biol. 167: 35–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., et al. , 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan X. Y., Luo Z., Liu X., Xie C. X., 2011. Dietary copper requirement of juvenile yellow catfish Pelteobagrus fulvidraco. Aquacult. Nutr. 17: 170–176. [Google Scholar]
- Tan X. Y., Luo Z., Xie P., Li X. D., Liu X. J., Xi W. Q., 2010. Effect of dietary conjugated linoleic acid (CLA) on growth performance, body composition and hepatic intermediary metabolism in juvenile yellow catfish Pelteobagrus fulvidraco. Aquaculture 310: 186–191. [Google Scholar]
- Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., et al. , 2002. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3: research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waisman D., Khanna N., Tokuda M., 1986. Identification of a major bovine heart Ca2+ binding protein. Biochem. Biophys. Res. Commun. 139: 596–603. [DOI] [PubMed] [Google Scholar]
- Watanabe T., Kiron V., Satoh S., 1997. Trace minerals in fish nutrition. Aquaculture 151: 185–207. [Google Scholar]
- Xi X. Z., Ma K. S., 2013. Molecular cloning and expression analysis of glucose-regulated protein78 (GRP78) gene in silkworm Bombyx mori. Biologia 68: 559–564. [Google Scholar]
- Yamamoto K., Sato T., Matsui T., Sato M., Okada T., et al. , 2007. Transcriptional induction of mammalian ER quality control proteins is mediated by single or combined action of ATF6alpha and XBP1. Dev. Cell 13: 365–376. [DOI] [PubMed] [Google Scholar]
- Zhao Y., Gul Y., Li S., Wang W., 2011. Cloning, identification and accurate normalization expression analysis of PPARαgene by GeNorm in Megalobrama amblycephala. Fish Shellfish Immunol. 31: 462–468. [DOI] [PubMed] [Google Scholar]
- Zheng J. L., Luo Z., Liu C. X., Chen Q. L., Tan X. Y., et al. , 2013a Differential effects of acute and chronic zinc (Zn) exposure on hepatic lipid deposition and metabolism in yellow catfish Pelteobagrus fulvidraco. Aquat. Toxicol. 132: 173–181. [DOI] [PubMed] [Google Scholar]
- Zheng J. L., Luo Z., Zhu Q. L., Tan X. Y., Chen Q. L., et al. , 2013b Molecular cloning and expression pattern of 11 genes involved in lipid metabolism in yellow catfish Pelteobagrus fulvidraco. Gene 531: 53–63. [DOI] [PubMed] [Google Scholar]
- Zhu Y., Fan Q., Mao H., Liu Y., Hu C., 2013. GRP78 from grass carp (Ctenopharyngodon idella) provides cytoplasm protection against thermal and Pb2+ stress. Fish Shellfish Immunol. 34: 617–622. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
File S1 contains detailed data about tissue expression mRNA of grp78, crt, perk, ire-1α, and atf-6α. File S2 contains detailed about effect of dietary Cu deficiency and excess on epression of grp78, crt, perk, eif2α, ire-1α, xbp-1α, and atf-6α.