Abstract
Three major DNA polymerases replicate the linear eukaryotic chromosomes. DNA polymerase α-primase (Pol α) and DNA polymerase δ (Pol δ) replicate the lagging-strand and Pol α and DNA polymerase ε (Pol ε) the leading-strand. To identify factors affecting coordination of DNA replication, we have performed genome-wide quantitative fitness analyses of budding yeast cells containing defective polymerases. We combined temperature-sensitive mutations affecting the three replicative polymerases, Pol α, Pol δ, and Pol ε with genome-wide collections of null and reduced function mutations. We identify large numbers of genetic interactions that inform about the roles that specific genes play to help Pol α, Pol δ, and Pol ε function. Surprisingly, the overlap between the genetic networks affecting the three DNA polymerases does not represent the majority of the genetic interactions identified. Instead our data support a model for division of labor between the different DNA polymerases during DNA replication. For example, our genetic interaction data are consistent with biochemical data showing that Pol ε is more important to the Pre-Loading complex than either Pol α or Pol δ. We also observed distinct patterns of genetic interactions between leading- and lagging-strand DNA polymerases, with particular genes being important for coupling proliferating cell nuclear antigen loading/unloading (Ctf18, Elg1) with nucleosome assembly (chromatin assembly factor 1, histone regulatory HIR complex). Overall our data reveal specialized genetic networks that affect different aspects of leading- and lagging-strand DNA replication. To help others to engage with these data we have generated two novel, interactive visualization tools, DIXY and Profilyzer.
Keywords: DNA replication, DNA polymerase, Profilyzer, DIXY, quantitative fitness analyses (QFA), Saccharomyces cerevisiae
DNA replication requires the coordinated action of numerous proteins and nucleic acids to ensure accurate genome duplication. At the core of the replicative machinery in eukaryotes are three B-family DNA polymerases. The three polymerases, Pol α, Pol δ, and Pol ε, work in concert with many other accessory proteins. For example, the six-subunit complex, the origin recognition complex, binds at replication origins throughout the cell cycle and recruits replication proteins to initiate replication. The origin recognition complex helps recruit the hexameric helicase MCM in G1, along with Cdc45 and GINS (CMG for Cdc45-MCM-GINS) in S-phase to initiate replication and unwind DNA at the front of the replication fork. The five-polypeptide complex, replication factor C (RFC), the sliding clamp loader, loads the trimeric sliding clamp, proliferating cell nuclear antigen (PCNA), onto DNA. PCNA helps tether polymerases to DNA to improve processivity during strand elongation (Waga and Stillman 1998).
DNA polymerase α-primase (Pol α) initiates replication of both leading and lagging strands (Georgescu et al. 2015). To perform this task, Pol α synthesizes a short RNA primer that is extended first by Pol α DNA polymerase activity before other DNA polymerases take over. The most widely accepted current view is that DNA polymerase δ (Pol δ) completes lagging-strand synthesis, whereas DNA polymerase ε (Pol ε) completes leading-strand synthesis (Garg and Burgers 2005). An alternative model is that Pol δ is the major polymerase for both leading and lagging strands and Pol ε is a repair polymerase (Johnson et al. 2015; Waga and Stillman 1994). Pol α has limited processivity and lacks intrinsic 3′ exonuclease activity for proofreading functions (Kunkel et al. 1989). The current evidence indicates that Pol α synthesizes about 1.5% of the genome (Reijns et al. 2015). In budding yeast, all four subunits of Pol α, Pri1, Pri2, Pol1, and Pol12, are essential for cell viability.
Pol δ principally completes synthesis of the lagging strand. In budding yeast Pol δ consists of three subunits: Pol3, Pol31, and Pol32. Pol3 is the catalytic subunit, and it has an associated 3′ exonuclease activity that can remove misincorporated nucleotides. Pol31, like Pol3, is essential in budding yeast. Pol32 is a nonessential subunit of Pol δ that help stabilize the other two subunits. The subunits of Pol δ play additional roles since, for example, Pol31 and Pol32 also physically interact with Rev3 and by this criterion are subunits of the error-prone DNA polymerase zeta (Pol ζ), which catalyses synthesis opposite DNA lesions (Kochenova et al. 2015).
Pol ε principally completes synthesis of the leading-strand. Pol ε is a four-subunit complex consisting of Pol2, Dpb2, Dpb3, and Dpb4. Pol2, the catalytic subunit, and Dpb2 are essential proteins in yeast, whereas Dpb3 and Dpb4 can be deleted. Pol2 DNA polymerase and 3′ exonuclease activities are located in the N-terminal domain while its C-terminal domain carries conserved zinc-finger motifs (Supporting Information, Figure S1). Interestingly, the C-terminus of Pol2 is essential for yeast viability, whereas the N-terminal polymerase domain of Pol2 is not (Dua et al. 1999; Feng and D’Urso 2001; Kesti et al. 1999). The current model to explain the fact that yeast cells can survive in the absence of the leading-strand DNA polymerase function is that the lagging-strand DNA polymerase Pol δ can, if necessary, replace Pol ε polymerase to replicate the leading strand (Daigaku et al. 2015). Additionally, Pol δ can replace Pol ε to proofread errors created by defective Pol ε and can complete DNA synthesis on the leading strand (Flood et al. 2015). It is thought that the most important (essential) function of Pol ε is to help initiate DNA replication as part of the preloading complex (pre-LC), working in concert with the GINS, Sld2, and Dpb11 proteins (Sengupta et al. 2013). Dpb3 and Dpb4, the nonessential subunits affect Pol ε binding to DNA and its processivity (Aksenova et al. 2010; Araki et al. 1991; Tsubota et al. 2003).
In eukaryotes, three multi-subunit polymerases interact to complete DNA replication, whereas in bacteria a single DNA polymerase is sufficient. The reasons for this difference are unclear but may reflect that fact that in eukaryotes, unlike prokaryotes, numerous origins are used to replicate each chromosome and eukaryotic chromosomes are linear, bringing an additional “end replication problem.” At each eukaryotic replication origin, a pair of replisomes travels in opposite directions from the origin (Masai et al. 2010) and will encounter replisomes coming in the opposite direction as well as other obstacles such as nucleosomes, DNA barrier elements, transcription factors, transcribing RNA polymerases, and damaged DNA, as well as chromosome ends (Tourriere and Pasero 2007). Accordingly, a large number of mechanisms have evolved to help the DNA replication machinery to deal with such challenges (Yeeles et al. 2013). Interestingly, it seems that distinct mechanisms have evolved to regulate the stability of leading- and lagging-strand machinery (Kurth and O’Donnell 2013).
Genetic suppressor and enhancer screens are a powerful way to explore how complex biochemical processes work in vivo and in particular how processes are controlled and interact with other cellular components or pathways. We have previously performed suppressor and enhancer analysis of telomere defective strains to identify pathways that are important for maintaining functional chromosome ends in budding yeast (Addinall et al. 2008, 2011). Here, we applied quantitative fitness analysis (QFA) to identify factors that affect fitness of strains defective in DNA replication. We identify numerous genes that affect general as well as specific aspects of eukaryotic DNA replication. To help others engage with this resource we also provide web-based tools that permit interaction with these and similar data sets.
Materials and Methods
Creation of the double-mutant libraries by synthetic genetic array (SGA)
Genotypes of the strains used in this study are described in Table S1. The thermosensitive (ts) alleles have been resequenced and mutations are shown in Figure S1. The SGA technique (Tong and Boone 2006; Tong et al. 2001) was used to combine the yeast genome knock-out (yfgΔ) and DAmP [decreased abundance by mRNA perturbation, which affects the function of essential genes (Schuldiner et al. 2005), (yfg-d)] collections with the lyp1Δ, pol1-4, pol2-12, cdc2-2, and cdc13-1 mutations. The recessive, ts-mutated alleles from (Li et al. 2011b) were flanked by the selectable LEU2 and HphMX markers in strains bearing can1::STE2pr-SpHis5 and lyp1::NatMX. Diploids were selected on rich media containing geneticin, hygromycin B, and nourseothricin. Diploids were sporulated on solid medium, and meiotic haploid MATa double-mutant progeny were isolated on SC medium containing canavanine, thialysine, geneticin, hygromycin B, and nourseothricin while lacking leucine, lysine, histidine, and arginine. Selection was based on haploid selection markers (can1::STE2pr-SpHis5), double-mutant selection markers (KanMX, HphMX, NatMX, and LEU2), and thialysine (lyp1::NatMX). Solid agar to solid agar pinning was performed on a Biomatrix BM3-SC robot (S&P Robotics Inc., Toronto, Canada) at 20° as previously described in Addinall et al. (2011).
Spotting yeast cultures for QFA
Double mutants obtained from SGA were inoculated into liquid (final media: SC-ARG, −HIS, −LYS, −LEU, +canavanine, +thialysine, +geneticin, +hygromycin, and +nourseothricin) at 20° in 96-wells plates and grown to saturation. Cultures were spotted on solid agar-plates in 384-format using a Biomek FX robot [Beckman Coulter (UK) Limited, High Wycombe, UK], and growth assays carried out as in Addinall et al. (2011), except we used concentrated cultures, without diluting in water, for lyp1Δ (control), pol1-4, pol2-12, and cdc2-2 experiments. Plates were imaged automatically, usually every 4 hr. For each genotype, we performed four independent genetic crosses and analyzed two growth curves from each cross (eight growth curves in total). In some cases, however, we examined more than eight cultures (i.e., up to 144 for his3Δ, repeated across several plates) and for others less than eight (i.e., down to four because of to technical errors). The specific number of growth curves analyzed for each double mutant can be found in File S1.
QFA fitness estimates
Cell-density estimates are generated from time-course photographs using Colonyzer2 [(Lawless et al. 2010), http://research.ncl.ac.uk/colonyzer/]. Strain fitness was quantified by first fitting the generalized logistic model to growth curves:
where t is time since inoculation, x is cell density, r is a growth rate parameter, K is carrying capacity, and ν is a parameter controlling symmetry of the growth curve. This differential equation has an analytical solution:
where x0 is the cell density at inoculation (assumed constant for all spots on all plates). We derive expressions for maximum doubling rate (MDR) and maximum doubling potential (MDP) for this generalized logistic model, equivalent to those presented by Addinall et al. (2011) for the logistic model [see qfa R package (qfa_0.0-39, http://qfa.r-forge.r-project.org/) for details]:
Following Addinall et al. (2011), we define fitness as the product of MDR and MDP.
QFA genetic interaction strength estimates
To estimate the strength and significance of genetic interaction with a query background (xyz), we compared the fitness of each strain in our collection (yfgΔ-d) in that background (xyz yfgΔ) with the equivalent fitness in a control background (lyp1Δ yfgΔ-d) under the same temperature and on the same growth media. We first fit a regression, equivalent to Fisher’s multiplicative model of genetic independence [see (Addinall et al. 2011) for details], to mean fitnesses across the collection, using the lm function in R:
where square brackets denote fitness and tildes denote average (mean) values for each strain. Assuming that most strains in the collection do not interact with the query background, estimating the slope m allows us to predict the fitnesses of individual strains in the query background, given fitness observed in the control background:
If the mean of predicted fitnesses for a strain is significantly different from the mean of observed fitnesses for that strain that suggests that the strain interacts with the query background. We carried out unpaired, two-tailed tests of the null hypothesis that is equal to using the t.test function in R. We corrected for multiple testing by applying a false discovery rate correction to the vector of P-values for the strain collection using the p.adjust function in R. This gave us a vector of corrected q-values. We defined interactions with q < 0.05 as significant. We defined genetic interaction strength as the difference between the mean of the observed fitnesses and the mean of the predicted fitnesses.
Stripping genes from QFA analysis
Genes within 20 kb of query mutations were stripped from analysis because of genetic linkage (Tong and Boone 2006). In addition, strains that did not complete SGA, such as genes required for mating and the biosynthesis of histidine, lysine, arginine, and leucine, were excluded as well as genes that failed the transition between SGA and QFA (probably due to prolonged storage on plates at 4°). Lists of genes stripped in each screen are available in the File S2 and in the GISReports folder on the following Web site: https://github.com/lwlss/profilyzer_Dubarry2015.
Gene ontology (GO) terms analysis
A list of enriched biological processes and biological components was downloaded from the Gene Ontology enRIchement anaLysis and visuaLizAtion tool (GOrilla: http://cbl-gorilla.cs.technion.ac.il/) using a p-value threshold of 10−3 (Eden et al. 2009). Significant negative or positive genetic interactions for each screen were compared with all genes screened as a background gene set to identify enriched GO terms. The lists were filtered to include only terms that annotated between two and 250 genes. Final lists were ranked by “enrichment” score (Table S3, Table S4, and Table S5) and filtered to include only the smallest terms of any branch ranked at the eighth level, or below, of the GO hierarchy (see Figure S3). p-value is not corrected for multiple testing. The q-value is adjusted to control for the false discovery rate (< 0.05) (Klipper-Aurbach et al. 1995). The dates when enrichments were assessed are indicated in each table.
Dynamic interactive X-Y (DIXY) plots
DIXY is an online data visualization tool for browsing QFA fitness plots, which show evidence for genetic interaction by comparing fitnesses of strains in a query screen with fitnesses in a control screen (Addinall et al. 2011). QFA screen data presented in this paper along with data from (Andrew et al. 2013) can be visualized here: http://bsu-srv.ncl.ac.uk/dixy-pol/viz/. Note that the DAmP alleles are missing from the data obtained in Andrew et al. (2013). The DIXY Web site includes documentation to help users interact with plots. DIXY uses the Python web-framework Django (https://www.djangoproject.com/) to serve dynamic web pages powered by the javascript libraries D3 (http://d3js.org/) and jQuery (http://jquery.com/). The tool requires genetic interaction strength (*GIS.txt) files as input, as generated by the R package for QFA: http://qfa.r-forge.r-project.org/.
Profilyzer
Profilyzer is an online tool for visualizing and interrogating the results of multiple QFA screens at once. QFA screen data presented in this paper, along with data from Andrew et al. (2013), can be visualized here: http://research.ncl.ac.uk/qfa/Dubarry2015/. Note that the DAmP alleles are missing from the data obtained in Andrew et al. (2013). The Profilyzer Web site includes documentation to help users interact with plots. Profilyzer is built using the Shiny framework (http://shiny.rstudio.com/) as a wrapper around a set of custom-built R functions. Source code and data underlying this instance can be found on GitHub: https://github.com/lwlss/profilyzer_Dubarry2015.
Data availability
All strains and materials are available upon request. File S1 and File S2 contain the raw data from the screens performed in this study. These information are also available through GitHub: https://github.com/lwlss/profilyzer_Dubarry2015 with description of the fitness estimates (generated by the qfa R package http://qfa.r-forge.r-project.org/) and description of the Profilyzer tool. Source code for the DIXY visualization tool can be found at https://github.com/Bioinformatics-Support-Unit/dixy. Table S1 contains the list of strains used in the study. Table S2 summarizes the positive and negative interactions identified (q < 0.005). Table S3, Table S4 and Table S5 show enriched gene ontology terms for the DNA polymerase mutants. Table S6 provides a description of all the negative interactions affecting the DNA polymerase mutants.
Results
We performed QFA to uncover genetic interactions affecting the fitness of strains containing hypomorphic, ts alleles affecting each of the catalytic subunits of the three major replicative polymerases. We examined pol1-4 affecting Pol α function, pol2-12 affecting Pol ε function (Budd and Campbell 1993), and cdc2-2 affecting Pol δ function (Hartwell et al. 1973) (Figure S1).
First, at permissive temperature, we crossed polymerase-defective strains to a genome-wide loss of function library using the SGA method (Tong et al. 2001). The library contained nonessential gene disruptions (yfgΔ, replaced with the KanMX marker) and DAmP alleles affecting essential genes [yfg-d, DAmP, which can partially reduce mRNA stability by inserting the KanMX marker into the 3′UTR of essential genes (Breslow et al. 2008)]. We indicate the combined library as yfgΔ-d. Fitness of strains was deduced from growth curves of cultures spotted on solid agar. Individual cultures were monitored by time course photography and curves fitted to the data allowing us to estimate MDR and MDP for each (see Figure S2 for a small set of examples). Generally, for each genotype, eight curves were analyzed. Fitness was defined as the product of the MDR and MDP, as before (Addinall et al. 2011). We performed QFA at temperatures that reduced fitness of strains with ts alleles to approximately 40–60% of wild-type fitness. In total, approximately 230,000 growth curves were generated (File S1).
DNA polymerase screens
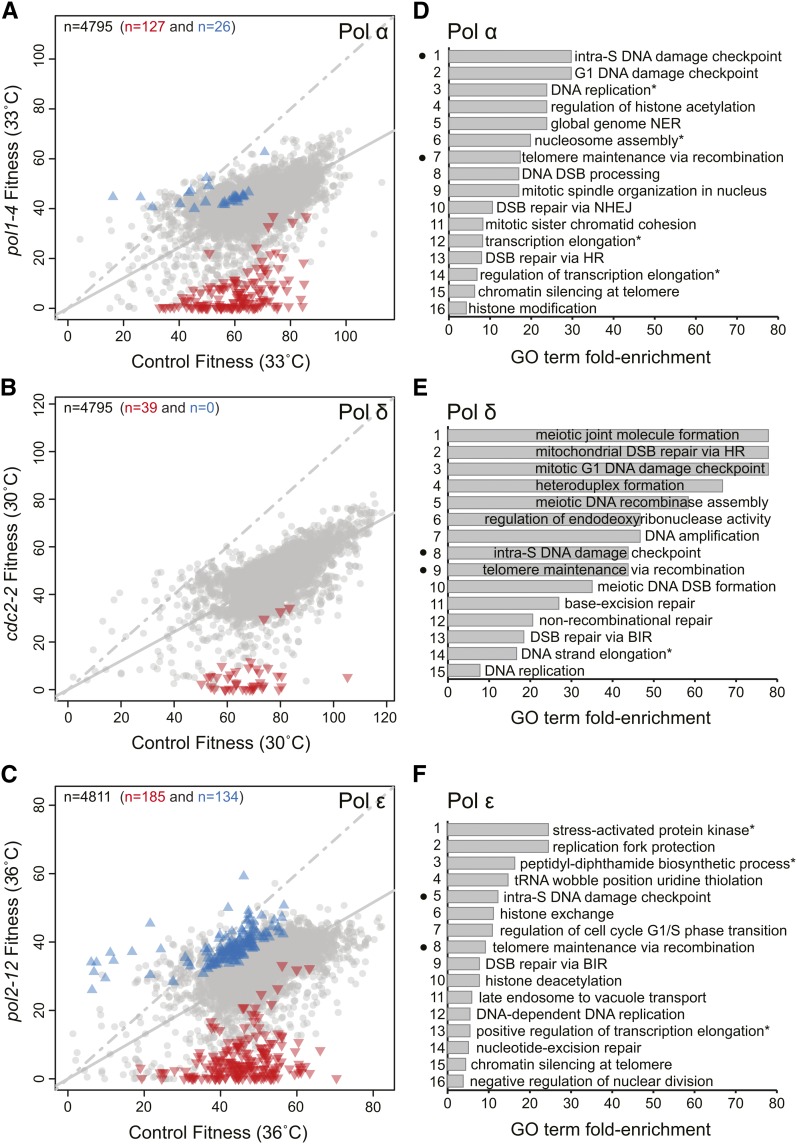
Pol α-primase initiates DNA replication of both leading and lagging strands. Figure 1A shows the effects of about 4800 library mutations combined with either a Pol α mutation (pol1-4; y-axis) or a control mutation (lyp1Δ, x-axis). It is clear that at 33°, pol1-4 yfgΔ-d double-mutants grew on average more poorly than lyp1Δ yfgΔ-d mutants. We observed 127 double-mutants that grew significantly more poorly than expected (inverted red triangles), reflecting negative genetic interactions with the Pol α mutation. 26 positive genetic interactions (blue upward triangles) also were identified (Table S2).
Figure 1.
Genetic interactions that affect DNA polymerase functions. (A) Fitnesses of yeast strains with yfgΔ-d mutations in pol1-4 or lyp1Δ genetic backgrounds and cultured at 33°. Each symbol shows the effect of a single yfgΔ-d in the two contexts, and the total number of yfgΔ-d genes shown is indicated by the number in top left corner of the plot. The solid gray line is a linear regression through all points, and the dashed line is the line of equal fitness (if there were no fitness cost caused by pol1-4 at 33°). Statistically significant positive genetic interactions with pol1-4 are represented by blue triangles, negative genetic interactions by red inverted triangles, and the remaining observations by gray dots. The numbers of significant interactions are indicated by the colored numbers across the top of the plot. (B) Same as in (A) but for DNA polymerase δ (Pol δ) mutation (cdc2-2) at 30°. (C) Same as in (A) but for the DNA polymerase ε (Pol ε) mutation (pol2-12) at 36°. (D) A ranked list of enriched biological process Gene Ontology (GO) terms across the negative genetic interactions identified for pol1-4. The complete list of enriched terms GO terms and classifications are found in Table S3. Black dots correspond to the common terms found enriched in the DNA polymerase α-primase (Pol α), Pol δ, and Pol ε screens. The asterisk indicates that the GO term has been abbreviated from Table S3. (E) Same as in (D) but for Pol δ mutation (cdc2-2) at 30°. See Table S4 for the complete list of enriched terms GO terms. (F) Same as in (D) but for the Pol ε mutation (pol2-12) at 36°. See Table S5 for the complete list of enriched terms GO terms.
We next examined the results obtained with the Pol δ mutation. Pol δ primarily contributes to lagging-strand synthesis but is sometimes engaged in leading-strand synthesis (Daigaku et al. 2015; Flood et al. 2015). Interestingly we only identified 39 statistically significant negative genetic interactions with the cdc2-2 mutation and no positive interactions (Figure 1B and Table S2).
Finally, we focused on Pol ε, which is the major leading strand DNA polymerase and in addition plays an important role in the assembly of the CMG helicase at origins of replication (Muramatsu et al. 2010). With pol2-12, we saw the largest number of genetic interactions, identifying 134 positive interactions and 185 negative interactions (Figure 1C and Table S2).
We interrogated the biological significance of the genetic interactions identified in the three screens by searching for enriched GO terms in the positive and negative interactions (Figure 1, D−F, and Figure S3). Negative genetic interactions gave enriched GO terms for Pol α, Pol δ, and Pol ε mutants, whereas positive genetic interactions only gave enriched GO terms with Pol ε mutants (Table S3, Table S4, and Table S5). The two enriched GO terms found from pol2-12 yfgΔ-d−positive interactions are related to cytoplasmic translation and ubiquitin-dependent protein catabolic process. The proteolytic and ubiquitin systems clearly contribute to DNA replication, but the many potential mechanisms of action remain to be fully understood (Finley et al. 2012).
The enriched GO terms found among the negative interactions with the different DNA polymerase mutants were enriched for numerous pathways functioning in DNA replication, DNA damage checkpoint regulation, and various DNA repair pathways (Figure 1, D−F). Interestingly, only two of 47 GO terms: the intra-S DNA damage checkpoint and telomere maintenance via recombination, were found in all three Pol α, Pol δ, and Pol ε screens. We conclude that defects in any of three DNA polymerases causes cells to rely on genes important for DNA checkpoint pathways and telomere integrity. However, the more specific nature of the most of the genetic interactions we have uncovered suggest that polymerase-specific interactions allow us to distinguish the division of labor between the three replicative DNA polymerases. If so, these data are a valuable resource to help to understand the numerous, complex, biochemical interactions combine together to result in efficient DNA replication.
DIXY plots: an interactive data visualization tool
To better facilitate the appreciation of and interaction with these data and to compare the effects of specific genes in the context of different polymerase mutations, we have developed an online data visualization tool called DIXY (http://bsu-srv.ncl.ac.uk/dixy-pol). DIXY connects a series of fitness plots like the three shown in Figure 1. It is possible to highlight genes based on their location in one plot, their name, GO function, or presence in known complexes/pathways and determine where the chosen genes lie across the fitness plots. To help highlight different genetic interactions, the points (yfgΔ-d mutations) move positions as the user switches between datasets (i.e., all points in Figure 1A move to different positions in Figure 1, B or C).
With DIXY, it is possible to compare the effects of mutations across datasets to identify factors that affect DNA replication in different ways. For example the rrm3Δ mutation, which encodes a DNA helicase that helps replisome progression in some regions of the genome (Ivessa et al. 2002, 2003), seems to cause particularly poor growth in the Pol ε mutants but less so in Pol α or Pol δ mutants (Figure S4). In contrast, the sgs1Δ mutation, which affects a different DNA helicase of the RecQ family (homologous to human BLM) that is involved in the suppression of illegitimate recombination (Myung et al. 2001; Watt et al. 1996), shows the opposite phenotype. Both Rrm3 and Sgs1 limit genome instability but this pattern shows that clear differences in the roles played by these helicases during regulation of DNA replication. Such differences in genetic interactions are informative about DNA replication processes and can be explored using DIXY.
Specificity of genetic interactions affecting DNA replication
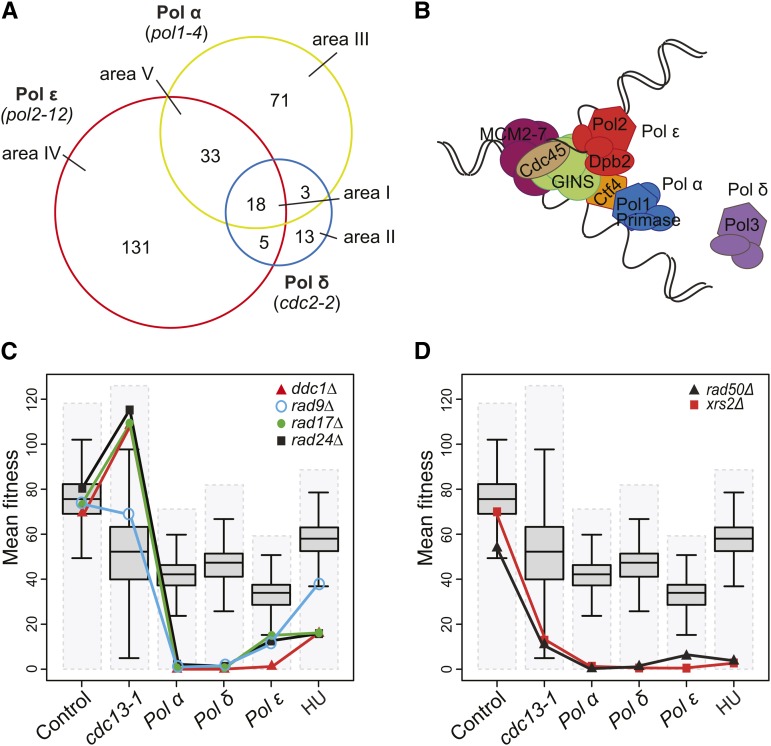
To explore the differences between the genetic networks affecting the three DNA polymerase mutants, we analyzed the distribution of specific negative genetic interactions over the three screens (Figure 2A). The interactions can be considered in the context of a current view of the eukaryotic DNA replication fork (Figure 2B). Only 18 gene deletions/DAmP mutations reduced fitness of all three DNA polymerase mutants (area I, Figure 2A, Table 1, Table S6). This small number is consistent with the fact that only two GO terms, the intra S DNA damage checkpoint and telomere maintenance via recombination, were found in all three screens (Figure 1). Much larger numbers of genes were found in other areas of the plot e.g., 131 gene deletions specifically interacted with the Pol ε mutation (area IV, Figure 2A). Thus, these data reinforce the view that there are many more genetic interactions that are specific to single polymerases than interactions that affect fitness of strains defective in all three polymerases.
Figure 2.
Specificity of genetic interactions. (A) Venn diagram showing overlaps between negative genetic interactions that affect cells with a DNA polymerase α-primase (Pol α) mutation (pol1-4), DNA polymerase δ (Pol δ) mutation (cdc2-2), or DNA polymerase ε (Pol ε) mutation (pol2-12) (Table 1 and Table S6). (B) A simplified view of the replication fork based on Sengupta et al. (2013). Pol ε binds to origins via Dpb2-GINS interaction. Pol ε remains associated with the CMG helicase (Cdc45-MCM2-7-GINS) during the leading-strand synthesis. Pol α-primase is coupled to the CMG helicase via Ctf4 and initiates the DNA synthesis on both strands. Pol α is replaced by Pol δ on the lagging-strand to extend the nascent strand. Note: for simplicity, numerous essential replication factors have been omitted (i.e., PCNA, RFC, Dpb11 etc.) (C) Fitness profiles of gene deletions affecting the DNA damage checkpoint (ddc1Δ, rad9Δ, rad17Δ, and rad24Δ) in combination with lyp1Δ (control), cdc13‐1, pol1-4, pol2-12, and cdc2-2 mutations and in presence of 100 mM hydroxyurea (HU). Dashed gray boxes represent the fitness range of the double-mutants in each screen. Box plots show 50% range, the whiskers represent 1.5-fold the 50% range from the box, and the horizontal black line is the median fitness. (D) Fitness profiles of gene deletions affecting the Mre11 complex (rad50Δ and xrs2Δ) as in C).
Table 1. Enhancers affecting defective DNA polymerase strains (q = 0.05).
| Defective DNA Polymerase(s) | Genes |
|---|---|
| Pol α, Pol δ, and Pol ε | BRE1 CAF40 DDC1 HTZ1 IPP1 LEA1 NCB2 POL32 RAD9 RAD17 RAD24 RAD50 RAD52 RAD57 SHR3 SMI1 XRS2 YLR339C |
| Pol ε and Pol δ | INP52 LGE1 SAD1 |
| Pol α and Pol δ | BMH1 NMD2 SPE1 TEN1 YDR269C |
| Pol α and Pol ε | ASF1 CSM3 CTF18 CTF8 GIM4 HDA3 HPC2 IES5 LSM7 MRC1 MTC1 MTC4 NHP10 NST1 PAT1 PPH21 RRD2 RTF1 SOK2 SPN1 SRS2 SWC5 SWD1 SWD3 TCO89 TIF5 TIM50 UBA4 VMS1 VPS63 YGL042C YNL235C YPL205C |
| Pol α | AIM29 ALT1 APQ12 ATG3 BDF2 BUD21 BUD28 CHD1 CIK1 CLA4 CTF4 DOT1 EAP1 ELC1 ELP6 FRS2 GPB2 HIR3 IMP2’ LSM6 MF(ALPHA)1 MTC6 MUP1 NHP2 PBY1 PET20 PSY1 PUS7 REI1 RIF1 RPB7 RPC11 RPL14A RPL31B RPS0B RPS1B RPS20 RPS4A RPS9B RRD1 RSC8 RTT10 SGS1 SIS2 SKO1 SOK1 SPT2 STI1 SUI2 TIM10 TPK3 TPT1 TRM11 UPF3 YBR099C YBR100W YJR087W YKE2 YKL069W YKL075C YLR374C YMD8 YMR166C YMR245W YNL171C YOL134C YPD1 YPR050C YPR153W YPT6:::MRC1 |
| Pol δ | CFD1 GPI15 INO2 PKR1 PMR1 PRE8 PRI1 RAD51 RAD54 RAD55 RFC5 RMI1 YNL011C |
| Pol ε | AIM4 ARC18 ARP4 ARP6 ATG21 ATO2 AZF1 BEM1 BET3 BFA1 BPH1 BRE2 BUB2 BUD27 CAT5 CDC21 CDC33 CHK1 CHS5 CKB1 CKB2 CLB2 CLB5 DCC1 DCR2 DEG1 DFG16 DIA2 DPB11 DPB2 DPB3 DPH2 DPH5 DPH6 DST1 ELF1 ELP3 EPS1 ERG3 ESBP6 ETP1 GCS1 GET3 GIM3 GPB1 GYP1 HCR1 HDA1 HIR2 HOS2 ILM1 IPK1 JJJ3 LAP2 LSM1 LSP1 LTE1 MID1 MIT1 MMS22 MRL1 MRN1 NGL2 NOP4 NPR3 NUP133 NUP188 PCP1 PER1 PET130 PKP1 PRE9 PSF1 PSF2 RIM101 RIM13 RIM21 RIM8 RIM9 RRM3 RRN9 RXT3 SAC3 SAF1 SAP190 SET3 SGF29 SHE1 SHE4 SIF2 SIR1 SIW14 SLT2 SNT1 SNX3 SOH1 SRB2 SRN2 SSO2 SWF1 SWR1 TRR2 TRS20 TRS85 TUM1 UME6 URM1 VAC8 VAM10 VHS3 VID22 VIP1 VPS13 VPS17 VPS21 VPS29 VPS30 VPS5 VPS55 VPS60 VPS71 VPS8 XRN1 YDR090C YGL046W YGR122W YGR237C YJL169W YOL050C YSC83 YSP1 |
Pol α, DNA polymerase α-primase; Pol δ, DNA polymerase δ; Pol ε, DNA polymerase ε.
Profilyzer: a tool for assessing genetic interactions in different contexts
To better understand the relevance of the genetic interactions in the three different DNA polymerase mutants, we created Profilyzer, an interactive, fitness-profiling web-tool (http://research.ncl.ac.uk/qfa/Dubarry2015/). This allows us to view the fitness profile of specific mutations across the three DNA polymerase screens and in other contexts. We also assessed the effects mutations in the context of cdc13-1, causing telomere defects, since “telomere maintenance via recombination” was one of only two GO terms enriched in all three polymerase screens and in the presence of the S-phase poison hydroxyurea (HU) [Figure S5, A−B, (Andrew et al. 2013)]. Profilyzer can be used to see fitness profiles of individual genes or groups of genes and to identify genes with similar profiles.
We first assessed the profiles of the 18 gene deletions that negatively affect all three polymerase mutants (area I, Figure 2A, Table 1). These mutations affect proteins that are important for sensing and responding to many replication defects and/or contribute to the stability or function of the DNA replication fork (Table S6). For example, DNA damage checkpoint mutations, including rad9Δ, rad17Δ, ddc1Δ, and rad24Δ (Table 1), which were all previously reported to reduce fitness of DNA polymerase mutants (Weinert 1992), were found in this group. In Figure 2C, we observe that strains with checkpoint mutations (rad9Δ, rad17Δ, ddc1Δ, and rad24Δ) are close to average fitness in control conditions, very fit when combined with cdc13-1, but very unfit when combined with all polymerase mutations or grown on HU. In contrast, other genes found in area I, like RAD50 and XRS2 (affecting two of three components of the Mre11 complex), showed a different profile. rad50Δ and xrs2Δ interacted negatively with all three polymerases and HU and also interacted negatively with cdc13-1 (Figure 2D). Thus, Profilyzer allows us to see that although all six genes affect fitness of DNA polymerase mutants similarly, rad9Δ, rad17Δ, ddc1Δ,and rad24Δ can be distinguished from rad50Δ and xrs2Δ by their effects in cdc13-1 strains. Such comparisons will be useful for helping to clarify the roles played by specific genes during DNA replication.
rad50Δ and xrs2Δ, along with mre11Δ, affect the three components of the MRX complex but MRE11 was not in central area I of Figure 2A. To investigate the reason why, we generated profiles of all three genes (Figure S5C). Reassuringly it is clear the all three genes behave very similarly, apart from in the control genetic background. Therefore, the reason why mre11Δ was not classified, like rad50Δ and xrs2Δ, as being in area I, seems to be that in the control experiment mre11Δ strains were unusually sick, probably because of a technical error during the SGA procedure. Thus, Profilyzer complements statistical cut off−based approaches to explore genome wide genetic interactions.
Profilyzer also permits the search for genes that cluster similarly to any particular query mutation. When we used this function to identify the nearest fitness profiles to RAD24 (Figure S5D), we found RAD17, DDC1, and RAD9 were the top three fitness profiles like RAD24. Furthermore, the ranking and distance of these three genes from RAD24 reflect the fact that RAD24, RAD17, and DDC1, encoding components of the checkpoint sliding clamp and its loader, share many functions that are distinct to RAD9, encoding a checkpoint mediator protein (Ngo et al. 2014). Profilyzer can be customized to explore subsets of data. For example, if we search for the closest profiles to mre11Δ and choose to ignore the control data then xrs2Δ and rad50Δ are among the top 12 most similar profiles to mre11Δ.
The other 12 genes in area I also could be classified based on their fitness in the context of the cdc13-1 mutation. Nine gene deletions including rad52Δ, bre1Δ, and pol32Δ, behaved liked rad50Δ and xrs2Δ mutants and were unfit in all contexts (Figure S5E), whereas rad57Δ, smi1Δ and lea1Δ differed because they were comparatively fit in combination with cdc13-1 (Figure S5F). Profilyzer therefore allows us to identify genes that are important in different contexts, in the specific examples highlighted, in polymerase mutants and telomere defective mutants.
Genes that affect specific polymerase functions
Regulators that work in concert with Pol δ (area II):
We next wanted to understand those genes that seemed to affect cells with defects in specific DNA polymerases. We first examined the Pol δ−specific interactors (13 genes, area II, Figure 2A and Table 1). We found that rad51Δ, rad54Δ, and rad55Δ, from the RAD52 epistasis group involved in HR repair, were in this group (Figure S6A). Other, similarly functioning HR genes pol32Δ, rad50Δ, rad52Δ, and rad57Δ were among those genes that affected all three polymerases (area I, discussed previously in Profilyzer: a tool for assessing genetic interactions in different contexts). The fitness profiles of rad51Δ, rad54Δ, and rad55Δ are somewhat difficult to distinguish from pol32Δ, rad50Δ, rad52Δ, and rad57Δ (Figure S6A vs. Figure S5, E−F). The major difference is that rad51Δ, rad54Δ, and rad55Δ are slightly fitter in Pol α and Pol ε mutants. Clearly the boundaries between the areas of the Venn diagram in Figure 2A are indistinct and areas I and II contain genes with similar roles. Other genes in area II of the Venn diagram seemed to affect the fitness of all three polymerase mutants somewhat similarly to those in area I (i.e., rfc5-d, rmi1Δ, and pkr1Δ). In summary, analysis using Profilyzer shows that most gene deletion/DAmP mutations found to interact with Pol δ alone (area II) also interact with Pol α or Pol ε and therefore, by this criterion, are general regulators of DNA replication.
Among the Pol δ−specific interactors in area II was pri1-d, affecting an essential DNA Pol α-primase subunit. This is curious, because we imagined that Pri1 would be most important in mutants defective in Pol α. Interestingly when we examined the effects of pri2-d, affecting another subunit of Pol α-primase, we saw a similar-shaped profile of genetic interactions across the screens (Figure S6B). Pri1 and Pri2 are each essential genes disrupted by DAmP alleles, and it has been estimated that less than half of the DAmP alleles affect gene function (Breslow et al. 2008). Inefficiency of the DAmP allele could explain why the effects of pri2-d are so mild in all cases. Irrespective, it seems that based on the fitness profiles, Pri1 is particularly important in Pol δ, Pol α, and cdc13-1 mutants and less important to Pol ε mutants. This finding suggests, perhaps, that Pri1, with the help of the accessory protein Pri2, plays critical roles in the switch from Pol α to Pol δ DNA polymerase activity during lagging strand synthesis and at telomeres.
Regulators that work in concert with Pol α (area III):
From area III, we highlight two particularly relevant genes, CTF4 and RSC8, that specifically interact with Pol α (area III, Figure 2A) in Figure S6C. CTF4 encodes a core component of the replication fork and is important for coupling the CMG helicase with Pol α (see Figure 2B) (Gambus et al. 2009). Strong interactions of ctf4Δ with Pol α suggest that the most critical role for Ctf4 is tethering Pol α at the replication fork. Perhaps as expected, ctf4Δ mutants also were unfit when combined with the other polymerase mutations and very unfit when combined with the telomere-defective cdc13-1 allele. The rsc8-d mutation causes a defect in an essential subunit of the RSC chromatin remodeling complex (Cairns et al. 1996), suggesting that interactions between Pol α and the RSC complex are critical for coordination of replication with chromatin structure.
Regulators that work in concert with Pol ε (area IV):
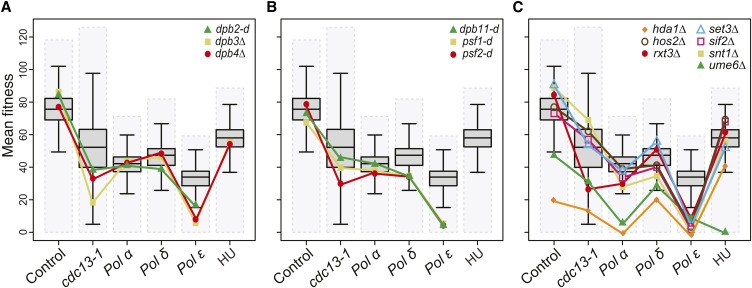
The largest number of specific genetic interactors was seen with Pol ε (area IV, Figure 2A). As expected, we found other components of Pol ε: dpb2-d and dpb3Δ (dpb4Δ was not significant but shows a similar profile) in this area (Figure 3A). It is interesting that defects in any of Pol ε subunits affect Pol ε whereas, in contrast, defects in Pol α-primase subunits affect Pol δ function more (compare Figure 3A with Figure S6B). Pol ε has at least two major functions during DNA replication, one as the leading strand polymerase and two, as part of the pre-LC containing GINS (Sld5, Psf1, Psf2, Psf3), Sld2, and Dpb11 that forms off chromatin and is recruited to origins before replication initiation (Muramatsu et al. 2010). Interestingly psf1-d, psf2-d, and dpb11-d were also Pol ε enhancers (Figure 3B). These data are consistent with the idea that GINS-Pol ε interactions being crucial for loading of Pol ε to the replisome (in the pre-LC) and/or for maintaining Pol ε at the replication fork after replication initiation (Langston et al. 2014; Sengupta et al. 2013) (see Figure 2B).
Figure 3.
Fitness profiling of genes affecting DNA polymerase ε (Pol ε). (A) Fitness profiles of dpb2-d, dpb3Δ, dpb4Δ mutations as in Figure 2C. (B) Fitness profiles of components of the preloading complex: dpb11-d and psf1-d, psf-2-d (GINS) as in Figure 2C. (C) Fitness profiles of hda1Δ, hos2Δ, rtx3Δ, set3Δ, sif2Δ, snt1Δ, and ume6Δ mutations (Rpd3L-expanded complex) as in Figure 2C. Note: Note that the DAmP alleles are missing from the hydroxyurea (HU) experiments (Andrew et al. 2013).
DNA replication initiation is influenced by chromatin structure, for example, histone acetylation. Additionally, after DNA replication is completed, epigenetic chromatin marks are maintained on the replicated sister chromatids. Histone deacetylases (HDACs), particularly Rpd3, delay replication initiation (Knott et al. 2009; Vogelauer et al. 2002). Interestingly we identified genes affecting seven HDAC components as negatively interacting with pol2-12 (Figure 3C). This was a surprise because we might expect that reduction of HDAC activity would improve the fitness of Pol ε mutants if DNA replication is easier to initiate when chromatin is more acetylated (Unnikrishnan et al. 2010). Therefore, this somewhat surprising result leads us to hypothesize instead that loss of HDACs, combined with defective Pol ε, leads to failure to properly replicate and maintain heterochromatic regions of the genome. Consistent with this hypothesis, in Schizosaccharomyces pombe, chromatin-modifying complexes travel with Pol ε to control modification of newly replicated chromatin at the fork (Li et al. 2011a).
Genes that differently interact with Pol α and Pol ε vs. Pol δ (area V):
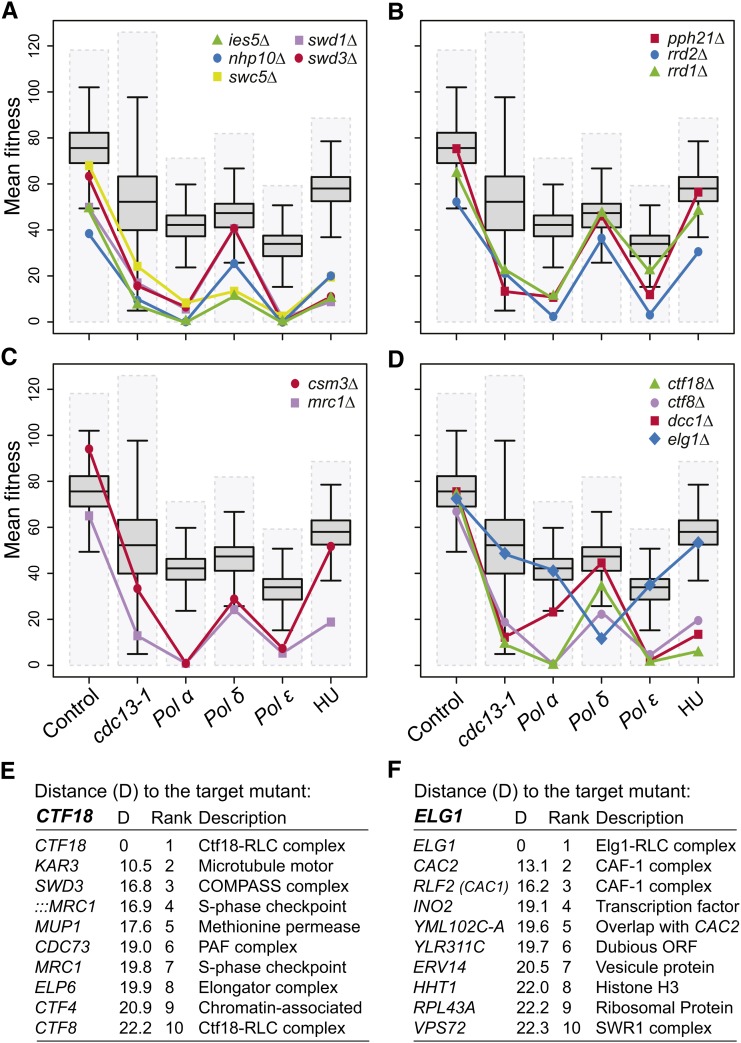
In area V, we found genes affecting protein complexes that modify chromatin, like the INO80-type complex (ies5Δ, nhp10Δ, swc5Δ) and the Set1/COMPASS complex (swd1Δ and swd3Δ) (Figure 4A). Both of these complexes have been shown to promote recovery of stalled replication forks (Rizzardi et al. 2012; Shimada et al. 2008) and perhaps, therefore, this is the reason why they were in area V.
Figure 4.
Fitness profiling genes associated with DNA polymerase α-primase (Pol α) and DNA polymerase ε (Pol ε). (A) Fitness profiles of components of the Set1-COMPASS complex: swd1Δ and swd3Δ and the INO80-type complex: ies5Δ, nhp10Δ and swc5Δ as in Figure 2C. (B) Fitness profiles of pph21Δ, rrd1Δ, and rrd2Δ mutations (Protein Phosphatase 2A complex) as in Figure 2C. (C) Fitness profiles of csm3Δ and mrc1Δ mutations (S-phase checkpoint) as in Figure 2C. (D) Fitness profiles of ctf18Δ, ctf8Δ, and dcc1Δ mutations (Ctf18-RLC) and elg1Δ mutation (Elg1-RLC) as in Figure 2C. (E) List of the top 10 gene deletions/DAmP mutations that show similar profiles to ctf18Δ when combined with control (lyp1Δ), cdc13‐1, pol1-4, pol2-12, and cdc2-2 mutations and in presence of 100 mM hydroxyurea (HU). (F) As in (E) but show similar profiles to elg1Δ mutation.
In area V we also identified two mutations (rrd2Δ and pph21Δ) affecting the PP2A complex. The rrd1Δ mutation affecting another subunit was in the Pol α−specific area of the Venn diagram (area III, Figure 2A) but its pattern of interactions is very similar to rrd2Δ and pph21Δ (Figure 4B). PP2A plays a critical role in the G1/S transition possibly by helping the recruitment of Cdc45, a key component of CMG complex (Chou et al. 2002; Lin et al. 1998). The recruitment of Cdc45 is necessary for CMG helicase activation and for Pol ε and Pol α recruitment to the fork (Yeeles et al. 2015). It is clear that pph21Δ, rrd1Δ, and rrd2Δ have comparatively small effects on the fitness of Pol δ mutants, suggesting that PP2A specifically affects the recruitment of the pre-LC and/or maintains stability of the core replication fork components. Pol δ is distinct to Pol α and Pol ε because it is recruited by Pol α and PCNA, rather than the CMG complex, to replication forks (Georgescu et al. 2015) (see Figure 2B).
Other enriched components in area V correspond to S-phase checkpoint components csm3Δ and mrc1Δ (Figure 4C) (Alcasabas et al. 2001; Bando et al. 2009). It is interesting that, like the pph21Δ, rrd1Δ, and rrd2Δ mutations discussed previously, the S-phase checkpoint genes have comparatively small effects in Pol δ mutants. Perhaps this is because these S phase checkpoint proteins and PP2A are particularly important for the stability of the replication fork (Figure 2B).
Ctf8 and Ctf18, which with Dcc1, help form the Ctf18-RFC-like complex (RLC) and load PCNA onto chromatin (Bermudez et al. 2003), were found in area V. Although dcc1Δ was not identified in area V, when we examined its pattern in different mutant backgrounds, it is very similar to ctf8Δ and ctf18Δ (Figure 4D). Rfc1 along with Rfc2, Rfc3, Rfc4, and Rfc5 forms the main RFC complex and the Ctf18-RFC complex complements RFC1 in loading PCNA during replication. Interestingly when we examined the effect of the elg1Δ mutation, part of an alternative RLC, we saw a very different (opposite) pattern of interactions to those seem by ctf8Δ, ctf18Δ, and dcc1Δ mutations. elg1Δ shows a strong negative genetic interaction with the Pol δ mutation but much weaker interactions with Pol α or Pol ε mutations (Figure 4D). The opposite patterns of genetic interactions seen for elg1Δ and ctf18Δ may reflect the fact that Elg1-RLC unloads PCNA whereas Ctf18-RLC loads PCNA during replication (Kubota et al. 2013; Yu et al. 2014), and PCNA-Pol δ interactions are more critical to efficient DNA binding than PCNA-Pol ε interactions, because Pol ε seems to be tethered directly by CMG (Sengupta et al. 2013).
Coordination of PCNA (un)loading with histone deposition
Figure 4D, in particular, illustrates how different profiles of fitness measurements in yeast strains carrying related mutations (elg1Δ vs. ctf18Δ) can draw attention to different cellular functions. Therefore, we next used fitness profiling to identify the nearest fitness profiles to ctf18Δ. The top 10 nearest fitness profiles to ctf18Δ, of more than 4800 profiles created, are shown in Figure 4E and Figure S7A. Importantly, as positive controls for this approach, we identified CTF8 as the ninth closest gene and DCC1 was 14th closest (Figure 4E and data not shown). Other known relationships are with CTF4, which as we have discussed previously (in Regulators that work in concert with Pol α, area III), couples Pol α with CMG at the replication fork. In addition Ctf4 and Ctf18 are involved in sister chromatid cohesion (Gambus et al. 2009; Hanna et al. 2001). MRC1 (affected by both :::MRC1/YCL060C and MRC1/YCL061C disruptions) and CTF18 are also important for the S-phase checkpoint pathway (Crabbe et al. 2010). This profiling approach found similar profiles for SWD3 affecting the COMPASS complex, CDC73 a member of the Paf1 complex required for gene expression, modification of histones, and telomere maintenance, ELP6, part of the Elongator complex, KAR3 and MUP1. Further experiments will be necessary to investigate the biochemical mechanisms underlying these related patterns.
We next investigated the closest fitness profiles to elg1Δ, which behaved in a very different way to ctf18Δ (Figure 4, D and F and Figure S7B). Interestingly, the 3/4 of the closest genes to ELG1 affect the chromatin assembly factor 1 (CAF-1) (RLF2 (CAC1)/YPR018W; CAC2/YML102W and CAC2/YML102C-A), responsible for histone deposition during DNA replication. We also found VPS72 encoding, an Htz1p-binding component of the SWR1 complex, and HHT1 that encodes for the histone H3. This leads us to hypothesize that Elg1 and CAF-1 orchestrate replication fork progression by coupling PCNA unloading to nucleosome assembly on lagging-strand. The HIR complex also loads histones, but this activity was defined as being independent of DNA replication in mammalian cells (Ray-Gallet et al. 2002). However, in yeast, it is not clear whether the role of the HIR complex is restrained to outside of S-phase. The fact that yeast mutants lacking CAF-1 subunits are viable suggests redundant pathways for histone incorporation during S-phase. On the basis of our data (Figure S7C), we suggest that the mirrored profiles of CAF-1 and HIR complexes reflect distinct activities of histone deposition during DNA replication.
Discussion
Genome replication is perhaps the most critical task that faces any cell. Replication is a complex task, depending on numerous different proteins and nucleic acids that function together in what has been likened to a machine (Alberts 1984). A powerful combination of biochemical and genetic approaches has succeeded in identifying the core components of the replisome. The replication machinery interfaces with the cell cycle, chromatin, and transcription machinery to ensure that the processes of DNA replication are coordinated within the cell cycle and with other cellular processes. Replication machinery can falter, for example, if it encounters DNA damage, and therefore a complex series of error correction pathways help overcome difficult situations. Although the core components of eukaryotic DNA replication have been identified our understanding how these replication components are regulated by the cell cycle, or respond to error, remains far from complete. In this paper we have performed genome-wide suppressor/enhancer interaction screens to identify genes that affect the fitness of replication defective strains. We examined strains defective in the each of the three major DNA polymerases (Pol α, Pol δ, and Pol ε).
Our unbiased genome-wide experiments have identified many genetic interactions consistent with known connections between DNA replication, repair, recombination, telomere maintenance, chromatin modification, and transcription. Interestingly, most of the genetic interactions we identified were specific to individual polymerases, rather than shared across the three polymerases, which indicates that most interactions affect specific aspects of the eukaryotic DNA replication process and this pattern likely reflects the different roles of Pol α, Pol δ, and Pol ε during DNA replication. Pol ε plays a major structural role during DNA replication, as part of the Pre-LC that is so critical for initiating DNA replication and, with Pol α, a part of the core replication fork machinery. Pol δ is less structurally important (Figure 2B). We also identified interactions of Pol ε with histone deacetylases. Histone acetylation is an important aspect of chromatin replication and assembly, and histone deacetylation has been found to occur after histone deposition to allow the newly assembled chromatin to acquire higher order structure (Annunziato 2013). How or why histone deacetylation is particularly important for leading-strand synthesis will require further investigation.
Elg1 is part of an alternative RFC complex and was recently shown to be involved in the removal of PCNA from DNA (Kubota et al. 2013; Yu et al. 2014). Removal of PCNA is particularly important for lagging-strand replication because each Okazaki fragment uses a single PCNA trimer. Consistent with these recent observations, we found that the elg1Δ mutation particularly affects fitness of Pol δ mutants. Furthermore, elg1Δ caused very similar effects to cac1Δ and cac2Δ mutations in all polymerase-defective strains (Figure 4 and Figure S7). Cac1 and Cac2 encode subunits of CAF-1, involved in chromatin reassembly after DNA replication (Kaufman et al. 1997). Recently, it was shown that nucleosome assembly helps regulate termination of Okazaki fragment synthesis in yeast (Smith and Whitehouse 2012) and defects in histone deposition delays PCNA unloading in human cells (Mejlvang et al. 2014). Our results lead us to suggest that Elg1 and CAF-1 orchestrate replication fork progression by coupling PCNA unloading to nucleosome assembly. We envisage that this function is specific to CAF-1 because hir2Δ and hir3Δ mutations affecting HIRA, another nucleosome assembly complex, show an “opposite” profile to cac1Δ and cac2Δ when combined with the different DNA polymerase mutants. However, the finding that hir2Δ and hir3Δ show interactions with polymerase mutations and are sensitive to the S-phase poison HU strongly suggests that in yeast HIRA plays a role in histone deposition during the S-phase. These types of insights, built on unbiased, genome-wide genetic interaction studies, lead us to believe that our data will be a valuable resource for understanding the processes of DNA replication in the context related processes such as cell-cycle progression, transcription, histone deposition, and epigenetic inheritance.
In conclusion, we report thousands of genetic interactions that are informative about how the three major eukaryotic DNA polymerases coordinate their activities to replicate DNA.
Acknowledgments
We thank Charles Boone for providing thermosensitive strains, Simon Whitehall for constructive comments and critical reading of the manuscript, and the members of the Lydall lab for helpful discussion on the manuscript. M.D. was funded by an EMBO Long Term Fellowship (ALTF 218-2012). This work was supported by Biotechnology and Biological Sciences Research Council (BB/M002314/1), Cancer Research UK (C23629), and Wellcome Trust (WT093088MA) grants (to D.L.).
Footnotes
Supporting information is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.115.021493/-/DC1
Communicating editor: R. A. Sclafani
Literature Cited
- Addinall S. G., Downey M., Yu M., Zubko M. K., Dewar J., et al. , 2008. A genomewide suppressor and enhancer analysis of cdc13–1 reveals varied cellular processes influencing telomere capping in Saccharomyces cerevisiae. Genetics 180: 2251–2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Addinall S. G., Holstein E. M., Lawless C., Yu M., Chapman K., et al. , 2011. Quantitative fitness analysis shows that NMD proteins and many other protein complexes suppress or enhance distinct telomere cap defects. PLoS Genet. 7: e1001362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aksenova A., Volkov K., Maceluch J., Pursell Z. F., Rogozin I. B., et al. , 2010. Mismatch repair-independent increase in spontaneous mutagenesis in yeast lacking non-essential subunits of DNA polymerase epsilon. PLoS Genet. 6: e1001209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberts B. M., 1984. The DNA enzymology of protein machines. Cold Spring Harb. Symp. Quant. Biol. 49: 1–12. [DOI] [PubMed] [Google Scholar]
- Alcasabas A. A., Osborn A. J., Bachant J., Hu F., Werler P. J., et al. , 2001. Mrc1 transduces signals of DNA replication stress to activate Rad53. Nat. Cell Biol. 3: 958–965. [DOI] [PubMed] [Google Scholar]
- Andrew E. J., Merchan S., Lawless C., Banks A. P., Wilkinson D. J., et al. , 2013. Pentose phosphate pathway function affects tolerance to the G-quadruplex binder TMPyP4. PLoS One 8: e66242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annunziato A. T., 2013. Assembling chromatin: the long and winding road. Biochim. Biophys. Acta 1819: 196–210. [DOI] [PubMed] [Google Scholar]
- Araki H., Hamatake R. K., Morrison A., Johnson A. L., Johnston L. H., et al. , 1991. Cloning DPB3, the gene encoding the third subunit of DNA polymerase II of Saccharomyces cerevisiae. Nucleic Acids Res. 19: 4867–4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bando M., Katou Y., Komata M., Tanaka H., Itoh T., et al. , 2009. Csm3, Tof1, and Mrc1 form a heterotrimeric mediator complex that associates with DNA replication forks. J. Biol. Chem. 284: 34355–34365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermudez V. P., Maniwa Y., Tappin I., Ozato K., Yokomori K., et al. , 2003. The alternative Ctf18-Dcc1-Ctf8-replication factor C complex required for sister chromatid cohesion loads proliferating cell nuclear antigen onto DNA. Proc. Natl. Acad. Sci. USA 100: 10237–10242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breslow D. K., Cameron D. M., Collins S. R., Schuldiner M., Stewart-Ornstein J., et al. , 2008. A comprehensive strategy enabling high-resolution functional analysis of the yeast genome. Nat. Methods 5: 711–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budd M. E., Campbell J. L., 1993. DNA polymerases delta and epsilon are required for chromosomal replication in Saccharomyces cerevisiae. Mol. Cell. Biol. 13: 496–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns B. R., Lorch Y., Li Y., Zhang M., Lacomis L., et al. , 1996. RSC, an essential, abundant chromatin-remodeling complex. Cell 87: 1249–1260. [DOI] [PubMed] [Google Scholar]
- Chou D. M., Petersen P., Walter J. C., Walter G., 2002. Protein phosphatase 2A regulates binding of Cdc45 to the prereplication complex. J. Biol. Chem. 277: 40520–40527. [DOI] [PubMed] [Google Scholar]
- Crabbe L., Thomas A., Pantesco V., De Vos J., Pasero P., et al. , 2010. Analysis of replication profiles reveals key role of RFC-Ctf18 in yeast replication stress response. Nat. Struct. Mol. Biol. 17: 1391–1397. [DOI] [PubMed] [Google Scholar]
- Daigaku Y., Keszthelyi A., Muller C. A., Miyabe I., Brooks T., et al. , 2015. A global profile of replicative polymerase usage. Nat. Struct. Mol. Biol. 22: 192–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dua R., Levy D. L., Campbell J. L., 1999. Analysis of the essential functions of the C-terminal protein/protein interaction domain of Saccharomyces cerevisiae pol epsilon and its unexpected ability to support growth in the absence of the DNA polymerase domain. J. Biol. Chem. 274: 22283–22288. [DOI] [PubMed] [Google Scholar]
- Eden E., Navon R., Steinfeld I., Lipson D., Yakhini Z., 2009. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics 10: 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng W., D’Urso G., 2001. Schizosaccharomyces pombe cells lacking the amino-terminal catalytic domains of DNA polymerase epsilon are viable but require the DNA damage checkpoint control. Mol. Cell. Biol. 21: 4495–4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley D., Ulrich H. D., Sommer T., Kaiser P., 2012. The ubiquitin-proteasome system of Saccharomyces cerevisiae. Genetics 192: 319–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flood C. L., Rodriguez G. P., Bao G., Shockley A. H., Kow Y. W., et al. , 2015. Replicative DNA polymerase delta but not epsilon proofreads errors in Cis and in Trans. PLoS Genet. 11: e1005049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambus A., van Deursen F., Polychronopoulos D., Foltman M., Jones R. C., et al. , 2009. A key role for Ctf4 in coupling the MCM2–7 helicase to DNA polymerase alpha within the eukaryotic replisome. EMBO J. 28: 2992–3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg P., Burgers P. M., 2005. DNA polymerases that propagate the eukaryotic DNA replication fork. Crit. Rev. Biochem. Mol. Biol. 40: 115–128. [DOI] [PubMed] [Google Scholar]
- Georgescu R. E., Schauer G. D., Yao N. Y., Langston L. D., Yurieva O., et al. , 2015. Reconstitution of a eukaryotic replisome reveals suppression mechanisms that define leading/lagging strand operation. eLife 10.7554/eLife.04988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna J. S., Kroll E. S., Lundblad V., Spencer F. A., 2001. Saccharomyces cerevisiae CTF18 and CTF4 are required for sister chromatid cohesion. Mol. Cell. Biol. 21: 3144–3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell L. H., Mortimer R. K., Culotti J., Culotti M., 1973. Genetic control of the cell division cycle in yeast: V. Genetic analysis of cdc mutants. Genetics 74: 267–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivessa A. S., Zhou J. Q., Schulz V. P., Monson E. K., Zakian V. A., 2002. Saccharomyces Rrm3p, a 5′ to 3′ DNA helicase that promotes replication fork progression through telomeric and subtelomeric DNA. Genes Dev. 16: 1383–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivessa A. S., Lenzmeier B. A., Bessler J. B., Goudsouzian L. K., Schnakenberg S. L., et al. , 2003. The Saccharomyces cerevisiae helicase Rrm3p facilitates replication past nonhistone protein-DNA complexes. Mol. Cell 12: 1525–1536. [DOI] [PubMed] [Google Scholar]
- Johnson R. E., Klassen R., Prakash L., Prakash S., 2015. A major role of DNA polymerase delta in replication of both the leading and lagging DNA strands. Mol. Cell 59: 163–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman P. D., Kobayashi R., Stillman B., 1997. Ultraviolet radiation sensitivity and reduction of telomeric silencing in Saccharomyces cerevisiae cells lacking chromatin assembly factor I. Genes Dev. 11: 345–357. [DOI] [PubMed] [Google Scholar]
- Kesti T., Flick K., Keranen S., Syvaoja J. E., Wittenberg C., 1999. DNA polymerase epsilon catalytic domains are dispensable for DNA replication, DNA repair, and cell viability. Mol. Cell 3: 679–685. [DOI] [PubMed] [Google Scholar]
- Klipper-Aurbach Y., Wasserman M., Braunspiegel-Weintrob N., Borstein D., Peleg S., et al. , 1995. Mathematical formulae for the prediction of the residual beta cell function during the first two years of disease in children and adolescents with insulin-dependent diabetes mellitus. Med. Hypotheses 45: 486–490. [DOI] [PubMed] [Google Scholar]
- Knott S. R., Viggiani C. J., Tavare S., Aparicio O. M., 2009. Genome-wide replication profiles indicate an expansive role for Rpd3L in regulating replication initiation timing or efficiency, and reveal genomic loci of Rpd3 function in Saccharomyces cerevisiae. Genes Dev. 23: 1077–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochenova O. V., Daee D. L., Mertz T. M., Shcherbakova P. V., 2015. DNA polymerase zeta-dependent lesion bypass in Saccharomyces cerevisiae is accompanied by error-prone copying of long stretches of adjacent DNA. PLoS Genet. 11: e1005110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota T., Nishimura K., Kanemaki M. T., Donaldson A. D., 2013. The Elg1 replication factor C-like complex functions in PCNA unloading during DNA replication. Mol. Cell 50: 273–280. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A., Hamatake R. K., Motto-Fox J., Fitzgerald M. P., Sugino A., 1989. Fidelity of DNA polymerase I and the DNA polymerase I-DNA primase complex from Saccharomyces cerevisiae. Mol. Cell. Biol. 9: 4447–4458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurth I., O’Donnell M., 2013. New insights into replisome fluidity during chromosome replication. Trends Biochem. Sci. 38: 195–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langston L. D., Zhang D., Yurieva O., Georgescu R. E., Finkelstein J., et al. , 2014. CMG helicase and DNA polymerase epsilon form a functional 15-subunit holoenzyme for eukaryotic leading-strand DNA replication. Proc. Natl. Acad. Sci. USA 111: 15390–15395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawless C., Wilkinson D. J., Young A., Addinall S. G., Lydall D. A., 2010. Colonyzer: automated quantification of micro-organism growth characteristics on solid agar. BMC Bioinformatics 11: 287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Martienssen R., Cande W. Z., 2011a Coordination of DNA replication and histone modification by the Rik1-Dos2 complex. Nature 475: 244–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Vizeacoumar F. J., Bahr S., Li J., Warringer J., et al. , 2011b Systematic exploration of essential yeast gene function with temperature-sensitive mutants. Nat. Biotechnol. 29: 361–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X. H., Walter J., Scheidtmann K., Ohst K., Newport J., et al. , 1998. Protein phosphatase 2A is required for the initiation of chromosomal DNA replication. Proc. Natl. Acad. Sci. USA 95: 14693–14698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masai H., Matsumoto S., You Z., Yoshizawa-Sugata N., Oda M., 2010. Eukaryotic chromosome DNA replication: where, when, and how? Annu. Rev. Biochem. 79: 89–130. [DOI] [PubMed] [Google Scholar]
- Mejlvang J., Feng Y., Alabert C., Neelsen K. J., Jasencakova Z., et al. , 2014. New histone supply regulates replication fork speed and PCNA unloading. J. Cell Biol. 204: 29–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramatsu S., Hirai K., Tak Y. S., Kamimura Y., Araki H., 2010. CDK-dependent complex formation between replication proteins Dpb11, Sld2, Pol (epsilon}, and GINS in budding yeast. Genes Dev. 24: 602–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myung K., Datta A., Chen C., Kolodner R. D., 2001. SGS1, the Saccharomyces cerevisiae homologue of BLM and WRN, suppresses genome instability and homeologous recombination. Nat. Genet. 27: 113–116. [DOI] [PubMed] [Google Scholar]
- Ngo G. H., Balakrishnan L., Dubarry M., Campbell J. L., Lydall D., 2014. The 9–1-1 checkpoint clamp stimulates DNA resection by Dna2-Sgs1 and Exo1. Nucleic Acids Res. 42: 10516–10528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray-Gallet D., Quivy J. P., Scamps C., Martini E. M., Lipinski M., et al. , 2002. HIRA is critical for a nucleosome assembly pathway independent of DNA synthesis. Mol. Cell 9: 1091–1100. [DOI] [PubMed] [Google Scholar]
- Reijns M. A., Kemp H., Ding J., de Proce S. M., Jackson A. P., et al. , 2015. Lagging-strand replication shapes the mutational landscape of the genome. Nature 518: 502–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzardi L. F., Dorn E. S., Strahl B. D., Cook J. G., 2012. DNA replication origin function is promoted by H3K4 di-methylation in Saccharomyces cerevisiae. Genetics 192: 371–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuldiner M., Collins S. R., Thompson N. J., Denic V., Bhamidipati A., et al. , 2005. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell 123: 507–519. [DOI] [PubMed] [Google Scholar]
- Sengupta S., van Deursen F., de Piccoli G., Labib K., 2013. Dpb2 integrates the leading-strand DNA polymerase into the eukaryotic replisome. Curr. Biol. 23: 543–552. [DOI] [PubMed] [Google Scholar]
- Shimada K., Oma Y., Schleker T., Kugou K., Ohta K., et al. , 2008. Ino80 chromatin remodeling complex promotes recovery of stalled replication forks. Curr. Biol. 18: 566–575. [DOI] [PubMed] [Google Scholar]
- Smith D. J., Whitehouse I., 2012. Intrinsic coupling of lagging-strand synthesis to chromatin assembly. Nature 483: 434–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong A. H., Boone C., 2006. Synthetic genetic array analysis in Saccharomyces cerevisiae. Methods Mol. Biol. 313: 171–192. [DOI] [PubMed] [Google Scholar]
- Tong A. H., Evangelista M., Parsons A. B., Xu H., Bader G. D., et al. , 2001. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 294: 2364–2368. [DOI] [PubMed] [Google Scholar]
- Tourriere H., Pasero P., 2007. Maintenance of fork integrity at damaged DNA and natural pause sites. DNA Repair (Amst.) 6: 900–913. [DOI] [PubMed] [Google Scholar]
- Tsubota T., Maki S., Kubota H., Sugino A., Maki H., 2003. Double-stranded DNA binding properties of Saccharomyces cerevisiae DNA polymerase epsilon and of the Dpb3p-Dpb4p subassembly. Genes Cells 8: 873–888. [DOI] [PubMed] [Google Scholar]
- Unnikrishnan A., Gafken P. R., Tsukiyama T., 2010. Dynamic changes in histone acetylation regulate origins of DNA replication. Nat. Struct. Mol. Biol. 17: 430–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelauer M., Rubbi L., Lucas I., Brewer B. J., Grunstein M., 2002. Histone acetylation regulates the time of replication origin firing. Mol. Cell 10: 1223–1233. [DOI] [PubMed] [Google Scholar]
- Waga S., Stillman B., 1994. Anatomy of a DNA replication fork revealed by reconstitution of SV40 DNA replication in vitro. Nature 369: 207–212. [DOI] [PubMed] [Google Scholar]
- Waga S., Stillman B., 1998. The DNA replication fork in eukaryotic cells. Annu. Rev. Biochem. 67: 721–755. [DOI] [PubMed] [Google Scholar]
- Watt P. M., Hickson I. D., Borts R. H., Louis E. J., 1996. SGS1, a homologue of the Bloom’s and Werner’s syndrome genes, is required for maintenance of genome stability in Saccharomyces cerevisiae. Genetics 144: 935–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert T. A., 1992. Dual cell cycle checkpoints sensitive to chromosome replication and DNA damage in the budding yeast Saccharomyces cerevisiae. Radiat. Res. 132: 141–143. [PubMed] [Google Scholar]
- Yeeles J. T., Poli J., Marians K. J., Pasero P., 2013. Rescuing stalled or damaged replication forks. Cold Spring Harb. Perspect. Biol. 5: a012815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeeles J. T., Deegan T. D., Janska A., Early A., Diffley J. F., 2015. Regulated eukaryotic DNA replication origin firing with purified proteins. Nature 519: 431–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C., Gan H., Han J., Zhou Z. X., Jia S., et al. , 2014. Strand-specific analysis shows protein binding at replication forks and PCNA unloading from lagging strands when forks stall. Mol. Cell 56: 551–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All strains and materials are available upon request. File S1 and File S2 contain the raw data from the screens performed in this study. These information are also available through GitHub: https://github.com/lwlss/profilyzer_Dubarry2015 with description of the fitness estimates (generated by the qfa R package http://qfa.r-forge.r-project.org/) and description of the Profilyzer tool. Source code for the DIXY visualization tool can be found at https://github.com/Bioinformatics-Support-Unit/dixy. Table S1 contains the list of strains used in the study. Table S2 summarizes the positive and negative interactions identified (q < 0.005). Table S3, Table S4 and Table S5 show enriched gene ontology terms for the DNA polymerase mutants. Table S6 provides a description of all the negative interactions affecting the DNA polymerase mutants.