Significance
The arginine-glycine-rich (RGG) box is an abundant RNA-binding motif present in many proteins. The RGG motif of the Fragile Mental Retardation Protein (FMRP) specifically binds guanine-quadruplex-containing RNA. Using X-ray crystallography, we uncovered molecular principles that account for specificity and affinity of the interactions between the RGG motif and guanine-quadruplex-containing RNA. The structure-based biochemical assays revealed that specific recognition of the RNA extends to the duplex region, thus identifying a broad network of important RNA–protein interactions and suggesting a common RNA binding principle for other RGG motif-containing proteins.
Keywords: fragile X syndrome, RNA structure, RGG box, FMRP, G-quadruplex
Abstract
Fragile X Mental Retardation Protein (FMRP) is a regulatory RNA binding protein that plays a central role in the development of several human disorders including Fragile X Syndrome (FXS) and autism. FMRP uses an arginine-glycine-rich (RGG) motif for specific interactions with guanine (G)-quadruplexes, mRNA elements implicated in the disease-associated regulation of specific mRNAs. Here we report the 2.8-Å crystal structure of the complex between the human FMRP RGG peptide bound to the in vitro selected G-rich RNA. In this model system, the RNA adopts an intramolecular K+-stabilized G-quadruplex structure composed of three G-quartets and a mixed tetrad connected to an RNA duplex. The RGG peptide specifically binds to the duplex–quadruplex junction, the mixed tetrad, and the duplex region of the RNA through shape complementarity, cation–π interactions, and multiple hydrogen bonds. Many of these interactions critically depend on a type I β-turn, a secondary structure element whose formation was not previously recognized in the RGG motif of FMRP. RNA mutagenesis and footprinting experiments indicate that interactions of the peptide with the duplex–quadruplex junction and the duplex of RNA are equally important for affinity and specificity of the RGG–RNA complex formation. These results suggest that specific binding of cellular RNAs by FMRP may involve hydrogen bonding with RNA duplexes and that RNA duplex recognition can be a characteristic RNA binding feature for RGG motifs in other proteins.
RNA-binding proteins (RBPs) control all aspects of RNA metabolism and are fundamental to core cellular processes. ∼14% of identified human RBPs are implicated in a broad spectrum of human pathologies, including neurodegenerative and muscular diseases, metabolic disorders, and cancers (1, 2). Fragile X Mental Retardation Protein (FMRP) is among the most important RBPs because of its central role in several human diseases (3). Loss of FMRP function due to CGG triplet repeat expansion-associated transcriptional silencing or missense mutations in the protein (4, 5) lead to fragile X syndrome (FXS), the most common cause of inherited intellectual disability. Mutations in FMRP are also the leading monogenic cause of autism (6, 7). Intermediate length repeat expansions in the FMR1 gene are linked to fragile X-associated tremor ataxia syndrome (8) and fragile X-associated primary ovarian insufficiency (9).
FMRP contains four canonical nucleic acid-binding motifs, three KH domains and one arginine-glycine-rich (RGG) box, which mediate interactions with RNAs in mRNA transport, storage, stability, and regulation of translation (Fig. 1A) (3, 10). Each KH domain has been reported to have a mutation either causing FXS or found in patients with intellectual disability (4, 5, 11). In neurons, FMRP associates with a subset of mRNAs and represses their translation both in the cell body and near synapses (12). Loss of repression of these mRNAs is associated with alterations in synaptic plasticity and dendritic spine dynamics thought to underlie the manifestation of FXS (13). The mechanisms of the FMRP-dependent translational repression have been suggested to include stalling of ribosomes by direct interactions of FMRP with mRNA (12) involving the RGG box (14) and KH2 domain (15). Individual contribution of each domain to RNA binding and translational repression and their functional relationship remain to be understood.
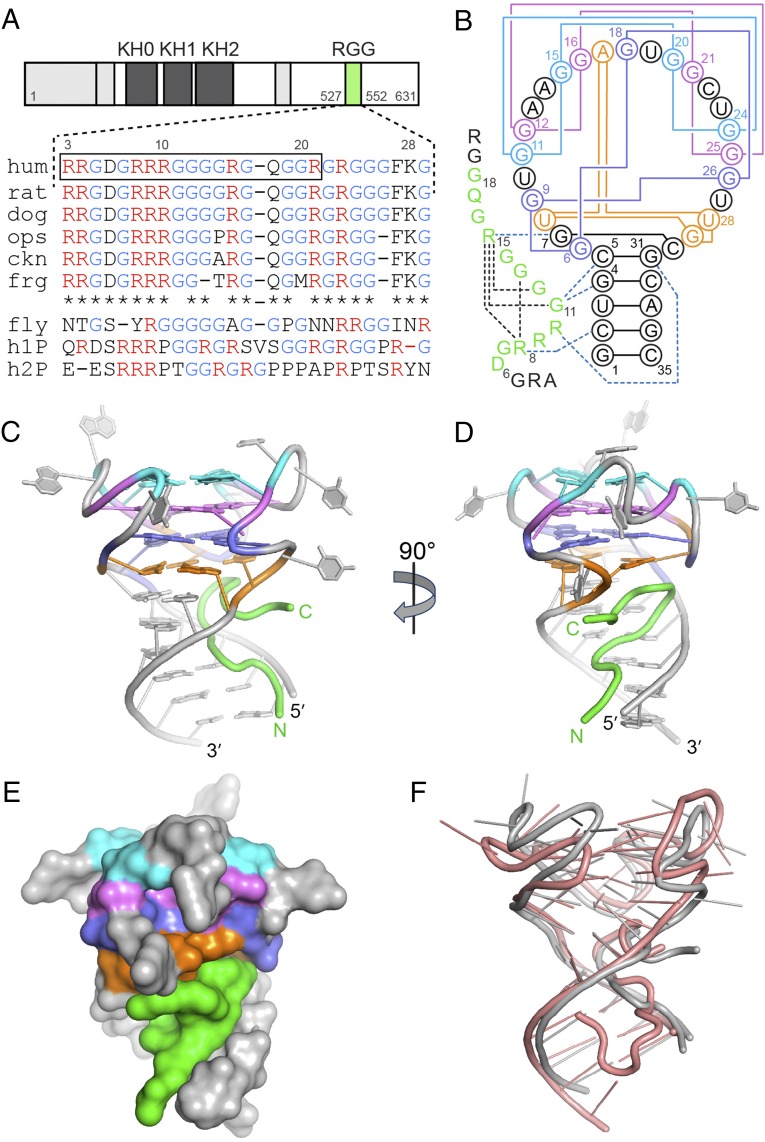
Fig. 1.
Crystal structure of the complex between the FMRP RGG box and sc1 RNA. (A) FMRP schematic and the RGG motif sequence alignment for FMRP and its autosomal paralogs. FMRPs were from opossum (ops), chicken (ckn), Xenopus (frg), and Drosophila (fly). h1P, human FXR1P; h2P, human FXR2P. The boxed sequence was used for the design of the crystallization construct. (B) Secondary structure schematic and interactions in the RGG–sc1 complex. RNA is in gray color with guanine quartets in cyan, violet and blue, and the mixed tetrad in orange. Pairing alignments in RNA are shown by solid lines. The well defined region of the RGG peptide is in green and intramolecular hydrogen bonds are depicted by black dashed lines. Blue dashed lines show base-specific intermolecular interactions. (C–D) Overall view of the complex in front (C) and side (D) views. (E) A surface view of the complex. (F) Superposition of the X-ray (current study, gray color) and NMR (salmon color) (37) structures.
A major research priority has been the identification of mRNA targets of FMRP and characterization of the FMRP–RNA interactions (3, 12, 16–23). RNA immunoprecipitation followed by microarray analysis (19), in vitro RNA selection (21), and in vitro binding assays (24) identified guanine (G)-rich RNAs as FMRP targets. Some of these RNAs harbor motifs that may form G-quartets and G-quadruplexes in vivo (21, 25). Several mRNAs contain regions shown to form G-quadruplexes implicated in FMRP binding in vitro (21, 24, 26–29). Notably, binding of FMRP to G-rich RNAs in vitro requires only the RGG motif, which specifically interacts with natural and in vitro selected G-quadruplex-containing RNAs such as a 35-nucleotide sc1 RNA (21, 26–29). Recent studies showed that G-quadruplexes facilitate mRNA interactions with ribosome-bound FMRP (30), whereas the RGG motif, in addition to mRNA binding, contributes to association with ribosomes and proteins and translational control (14, 31–34). The RGG motif is well conserved in FMRP of vertebrates but differs significantly from the motifs in FXR1P and FXR2P, brain-expressed autosomal paralogs of FMRP (35), which do not appear to compensate for the loss of FMRP function in FXS patients and Drosophila FXS model (36) (Fig. 1A and see SI Appendix, Fig. S1). Therefore, characterization of RGG–RNA interactions is important for understanding the mechanism of FMRP function and development of FXS.
Naturally occurring G-quadruplexes targeted by FMRP adopt multiple conformations and are challenging to study by structural methods (27, 28). Therefore, detailed structural information about RGG recognition of G-quadruplexes has been mostly obtained from studies on sc1 RNA (21), the RNA selected against the entire human FMRP but shown to require only the RGG motif of the protein for high affinity and specific binding (29, 37, 38). Previously, we determined the NMR solution structure of the RGG–sc1 complex that revealed an RNA fold composed of a three-layered G-quadruplex linked to a duplex RNA via a mixed junctional tetrad (37). The RGG peptide was found in the duplex–quadruplex junction, where it adopted a sharp turn specifically bound to guanines from two consecutive G-C base pairs. However, experimental limitations of the NMR study precluded us from fully defining the structure of the complex, identifying all RNA–peptide interactions, and revealing major specificity determinants of the FMRP-G-quadruplex recognition.
In this work, we determined the X-ray structure of the FMRP RGG peptide–sc1 RNA complex, to our knowledge, the first crystal structure of the RNA–protein complex bearing a component from FMRP. This structure and accompanying biochemical experiments uncovered previously unidentified determinants for the complex formation and clarified previously unexplained observations. Our data provides a comprehensive picture of important interactions between FMRP and G-quadruplex-containing RNA, with implications for specific RNA recognition by other RGG-motif-bearing proteins.
Results
Determination of the RGG Peptide–sc1 RNA Complex Structure.
Crystals of RGG–sc1 complexes were obtained with several RNA variants and peptides centered on the Arg10-to-Arg15 turn previously reported to contain determinants for sc1 RNA binding (37). The largest crystals, diffracted at 3.4 Å resolution, were grown with a 18-mer peptide (residues 3–20, numbering from ref. 37) and 35-mer RNA (Fig. 1 A and B). R1A mutation improved the diffraction limit to 2.8 Å resolution. Molecular replacement efforts using NMR structures (37) as search models did not yield a correct solution and therefore the X-ray structure was solved de novo by single wavelength anomalous dispersion (SAD) with iridium hexamine-soaked crystal (Materials and Methods and SI Appendix, Table S1). The structure contained two RNA-peptide complexes in the asymmetric unit, with all nucleotides and 13 amino acids of peptide chain present in the electron density map.
sc1 RNA Adopts a Conformation Exhibiting Structural Plasticity.
In the crystal structure of the sc1-RGG complex, sc1 RNA forms a three-layered intramolecular G-quadruplex connected to the helical stem via a mixed base tetrad stacked on the bottom G-quartet (Fig. 1 B–D). The well defined 13 amino acids (residues 6–18; ref. 37) of the RGG peptide snugly fit into the major groove of the RNA duplex and the junction between G-quadruplex and RNA duplex (Fig. 1E). The N terminus of the peptide is positioned along the 5′ RNA strand and rises toward G-quadruplex. The middle part of the peptide makes a sharp turn after reaching the mixed base tetrad so that the peptide occupies a large cavity predominantly formed by the 5′ region of RNA and the junctional tetrad.
Two RGG–sc1 complexes from the asymmetric unit display structural differences between the RNA molecules. Although the duplexes and G-quadruplex cores form practically identical structures, the loops, especially A13, A14, U23, and U27, adopt distinct conformations, as evidenced by ∼3.1-Å r.m.s.d. between the G-quadruplex regions (SI Appendix, Fig. S2 A and B and Table S2). These conformational differences are in line with poorly defined loops in the NMR structure (37) and high B-factors in some nucleotides of the loops, such as U10 and U19, in the crystal structure (SI Appendix, Fig. S3). These results suggest that loops are dynamic in solution but are trapped in the crystal in diverse conformations likely facilitated by crystal packing interactions (SI Appendix, Fig. S2 C–I). Further comparison of G-quadruplexes in the crystal and NMR structures revealed shifts between residues of G-quartets, which together with differences in loops account for high r.m.s.d. 2.5–3.9 Å between the structures (Fig. 1F and SI Appendix, Fig. S2B and Tables S3 and S4). Thus, despite good overall resemblance of RNA folds in the crystal and NMR structures, the G-quadruplexes have certain structural plasticity and display conformational dynamics associated primarily with loop regions.
Three K+ Cations Stabilize the G-Quadruplex Structure.
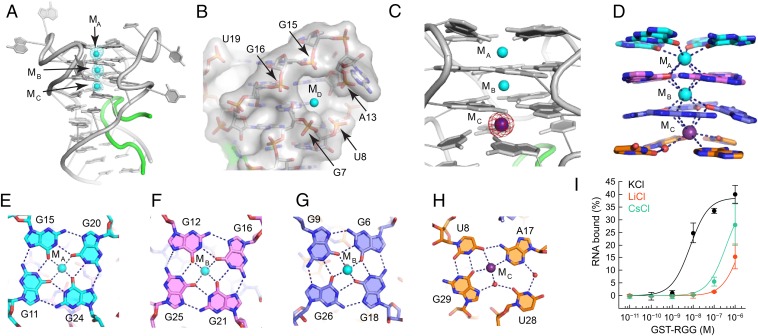
Our crystal structure provides the molecular basis for previous biochemical (21) and NMR (37) observations that formation of the sc1 RNA structure and RGG peptide binding critically depend on K+ cations. Each RGG–sc1 complex contains three K+ cations (MA, MB, and MC) separated by 3.5–3.9 Å distance and located along the central axis of the G-quadruplex in between planes of four tetrads (Fig. 2A). A fourth K+ cation MD was found in the cavity formed by phosphates of G7, U8, A13, G15 and G16 in one of the monomers (Fig. 2B).
Fig. 2.
Formation of the G-quadruplex. (A) The RGG–sc1 structure shown with K+ cations (cyan spheres) and the Fo − Fc electron density map contoured at the 5 σ level (cyan mesh). The map was generated before addition of metal cations to the refined RNA model and therefore is unbiased for cations. (B) General cation-binding site shown in surface representation with bound K+ cation in the MD position. (C) Anomalous difference Fourier map contoured at the 6 σ level (magenta) indicates replacement of K+ cation by Cs+ (purple sphere). (D) Coordination of K+ cations by base tetrads. Coordination bonds are depicted with dashed lines. (E–H) Nucleotide tetrad alignments and cation binding. Dashed lines show coordination and hydrogen bonds. Note that each cation binds two layers of the quadruplex and this figure does not depict all interactions. (I) Binding curves of the RGG peptide and sc1 RNA in the presence of various cations.
Two K+ cations MA and MB bind three G-quartets stably and specifically and are not replaced by similar Cs+ cations in a crystal grown in the mixture of KCl and CsCl (SI Appendix, Fig. S4 A–D). The K+ cation MC can be replaced, at least partially, by a Cs+ cation, as indicated by the ∼1.7 fold increase of the anomalous signal (Fig. 2C and SI Appendix, Fig. S4D). In addition to K+, the MD site can accommodate Cs+ and the iridium hexamine cation (SI Appendix, Fig. S4 E and F). Therefore, this site is not specific for K+ and can be considered as a general cation-binding site. MA and MB are octacoordinated with O6 atoms of guanine bases from the top and middle G-quartets as viewed in Fig. 2 D–H. MC directly coordinates the bottom G-quartet and U8, A17 and G29 from the mixed tetrad (Fig. 2H) as well as interacts with U28 and G29 via a water molecule observed in the better quality “Cs+ structure”. Thus, each K+ cation holds in place eight bases from two adjacent tetrads.
Despite differences in loop conformations, the X-ray and NMR structures agree on the nucleotide alignments within the G-quadruplex core. Typically for G-quartets, guanines are arranged in four-base circles stabilized by two hydrogen bonds between Watson–Crick and Hoogsteen edges (Fig. 2 E–H). The G11•G15•G20•G24 and G12•G16•G21•G25 quartets and the mixed tetrad have the same hydrogen bond directionality, whereas the third G-quartet flips over resulting in opposite directionality. Although the (U8•G29)•A17 triple alignment in the mixed tetrad is similar in NMR and crystal structures, in the X-ray structure, U28 was located in the middle between two positions proposed by the NMR study (37). This residue binds to the nucleotides of the mixed tetrad through water-mediated hydrogen bonds and is therefore weakly bound and likely responsible for the K+-to-Cs+ exchange in the MC site.
To test whether Cs+ cations support G-quadruplex formation, we compared binding affinities of sc1 RNA to RGG peptide with K+, Cs+ and Li+ ions by Differential Radial Capillary Action of Ligand Assay (DRaCALA) (39) (see below). In the presence of K+ cations, RGG motif binds RNA with KD = 10.8 ± 4.3 nM (Fig. 2I) similar to the previously determined values of 3.8 and 10.0 nM (21, 37). Cs+ cations decrease binding affinity ∼30 fold to KD ∼ 360 nM, the value comparable with KD measured in the presence of G-quadruplex-disrupting Li+ cations (21). Similar low binding affinities with Cs+ and Li+ indicate that peptide binding strongly depends on the presence of K+ cations, which favor G-quadruplex folding. Given that Li+ cations preclude formation of the sc1 G-quadruplex, we concluded that Cs+ alone also does not support the G-quadruplex formation despite its ability to bind in MC site between two bottom tetrads.
RGG Peptide Adopts a β-Turn Conformation.
The N-terminal amino acids of the peptide are poorly defined in both crystal and NMR structures in agreement with the observation that only 10 residues in the middle of the peptide facilitate the G-quadruplex formation (37). The rest of the peptide, except the last two residues, is well ordered and similar in both monomers of the crystal structure (SI Appendix, Fig. S2B). These data suggest that the most important intermolecular interactions between RGG motif and sc1 RNA involve residues 6–18 of the peptide.
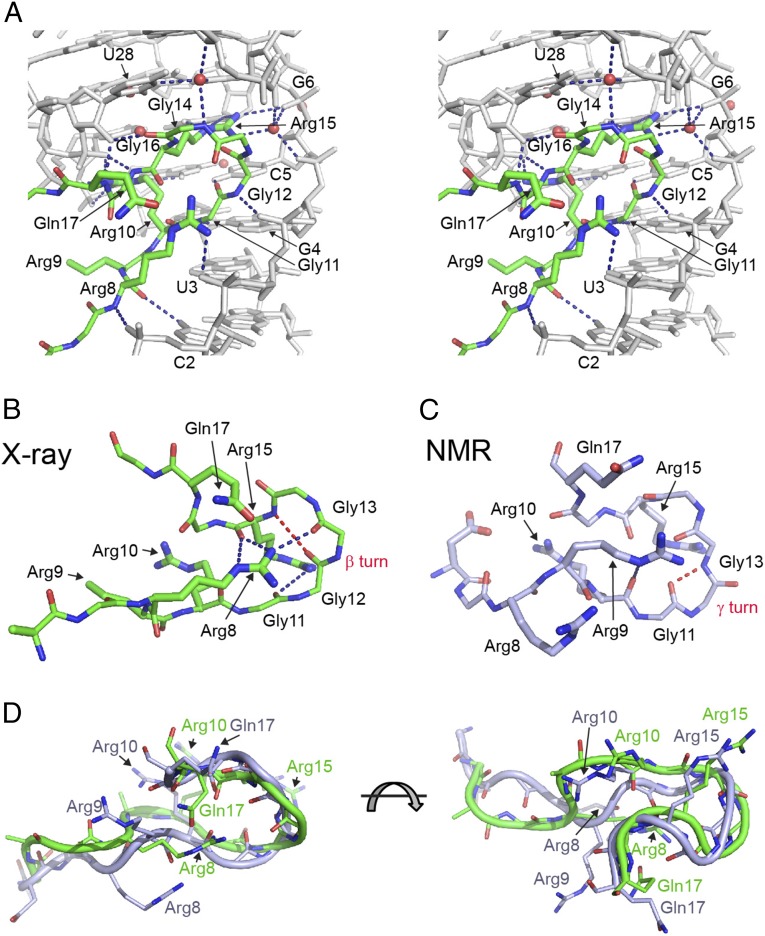
The prominent feature of the peptide is a reverse turn in the middle region (Fig. 3 A and B) that positions the well defined N-terminal part of the peptide alongside the 5′ RNA strand and straightens out the C terminus underneath the mixed tetrad. The turn adopts a type I β-turn conformation characterized by an intramolecular hydrogen bond between main chain groups of Gly12 and Arg15 (Fig. 3B). The guanidinium group of Arg8 stabilizes the turn by hydrogen bonding and electrostatic interactions with carbonyls of Gly13, Arg15, and Gln17. The X-ray structure resolves some violations observed in the NMR structure and shows several differences in the peptide conformation, accounting for 1.9–2.6 Å r.m.s.d. between peptide atoms (SI Appendix, Table S3). Most importantly, the X-ray structure unambiguously demonstrates the β-turn in the RGG motif, an element that was not previously recognized in the NMR structure because of the lack of sufficient constraints. Instead of the β-turn, the NMR structure suggested formation of a γ-turn characterized by an intramolecular hydrogen bond between Gly11 and Gly13 (37) (Fig. 3C). In the X-ray structure, Gly11 is positioned for an intermolecular contact with RNA. Additionally, in the X-ray structure, side chain functionalities of Arg8 are engaged in several intra- and intermolecular interactions. In contrast to the stretched conformation in the X-ray structure, in the majority of NMR models (37), the side chain of Arg8 adopts a more compact conformations so that its guanidinium group interacts with phosphates of C2 and U3 (Fig. 3D and SI Appendix, Fig. S5 C and D). The position of Arg8 in the NMR structure was defined by several intermolecular NOEs with the sugar of G1 and the base of C2 (37). These NOEs are incompatible with the X-ray structure, suggesting that they likely reflect certain flexibility of Arg8 and report on minor interactions of Arg8 with the RNA. Another prominent difference between the X-ray and NMR structures is the position of Arg9. In the NMR structure, conformation of this amino acid is poorly defined because of the lack of intermolecular NOEs. In the majority of the NMR models, Arg9 occupies the position of Arg8 in the X-ray structure, whereas in some NMR models the side chain of Arg9 reverses its orientation. In the X-ray structure, Arg9 is oriented inwards to the interior of the structure and is defined better although is missing the density for the guanidinium group.
Fig. 3.
Protein–RNA interactions in the RGG–sc1 complex. Putative hydrogen bonds are shown as blue dashed lines. (A) Stereoview of hydrogen bonding between the RGG peptide (green) and sc1 RNA (gray). (B and C) Comparison of the RGG peptides from the X-ray (B, current study) and NMR (C) (37) structures. Hydrogen bonds characteristic for peptide turns are in red dashed lines. (D) Superposition of the RGG peptide from the X-ray (green) and NMR (light blue) structures.
Crystal Structure Reveals a Dense Network of Intermolecular Interactions.
The crystal structure identified multiple RNA-peptide contacts that form three sets of interactions along the Arg8-to-Gln17 region of the peptide (SI Appendix, Table S5). These interactions include direct hydrogen bonding of eight evolutionarily conserved amino acids with ten nucleotides, several inferred water-mediated hydrogen bonds, three cation–π interactions, ionic interactions, and van der Waals contacts (Figs. 3A and 4 and SI Appendix, Fig. S5).
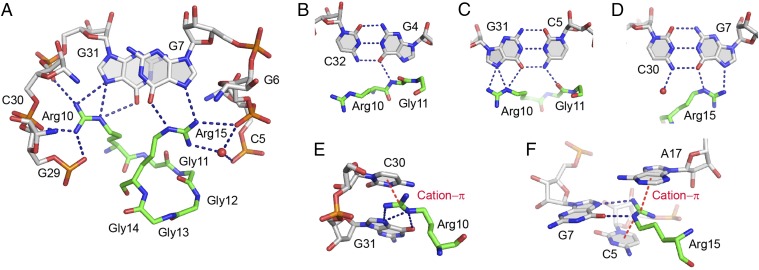
Fig. 4.
Details of protein–RNA interactions in the RGG–sc1 complex. (A) Specific recognition of RNA by Arg10 and Arg15 of the RGG peptide. (B–D) Base-specific recognition of the RNA base pairs in the upper part of the RNA duplex. (E and F) Cation–π interactions (red dashed lines) involving guanidinium groups of arginines and nucleotide bases.
The first set of contacts specifies the major groove recognition of RNA bases and phosphates in the C2U3G4 region. Phosphate recognition involves hydrogen bonding between nonbridging oxygen atoms of all three nucleotides with Arg8 and Gly12 (Fig. 3A). The base-specific contacts are formed between main chain atoms of amino acids and include Arg8•C2, Gly11•G4 and Arg10•U3 hydrogen bonding (Fig. 3A). In addition to binding affinity, these base-specific interactions are positioned to contribute to specificity of RNA-peptide recognition, although they are not formed by amino acid side chains.
The second set of contacts involves the major groove binding of the Arg10-to-Gly16 region of the peptide to C5-G31 and G7-C30 base pairs adjacent to the mixed tetrad (Fig. 4). This set of interactions includes a single hydrogen bond between the main chain of Gly11 and the base of C5 (Fig. 4C) and several base-specific hydrogen bonds involving side chains of Arg10 and Arg15. Guanidinium groups of these side chains point in opposite directions and align with Hoogsteen edges of guanines G7 and G31 to form hydrogen bonds with O6 and N7 atoms (Fig. 4 A–D). Because Arg10 and Arg15 flank the β-turn, their interactions with RNA define the peptide conformation and contribute to specificity of RNA recognition, as suggested earlier (37). In addition, guanidinium groups of Arg10 and Arg15 make several direct and water-mediated hydrogen bonds with nonbridging oxygen atoms of phosphate moieties of G5, G6, G29, C30 and G31 (Fig. 4A) as well as cation–π interactions with C30, C5 and A17 (Fig. 4 E and F). These interactions most likely hold together the RNA strands in the duplex–quadruplex junction and stabilize the mixed tetrad and G-quadruplex above it.
The last set of contacts includes hydrogen bonds between sugar-phosphate backbone of U28 and main chain atoms of Gly14, Gly16, and Gln17. These three amino acids further strengthen peptide binding to the RNA duplex and stabilize the mixed tetrad (Fig. 3A).
Binding Studies Identify Determinants of Specific Interactions.
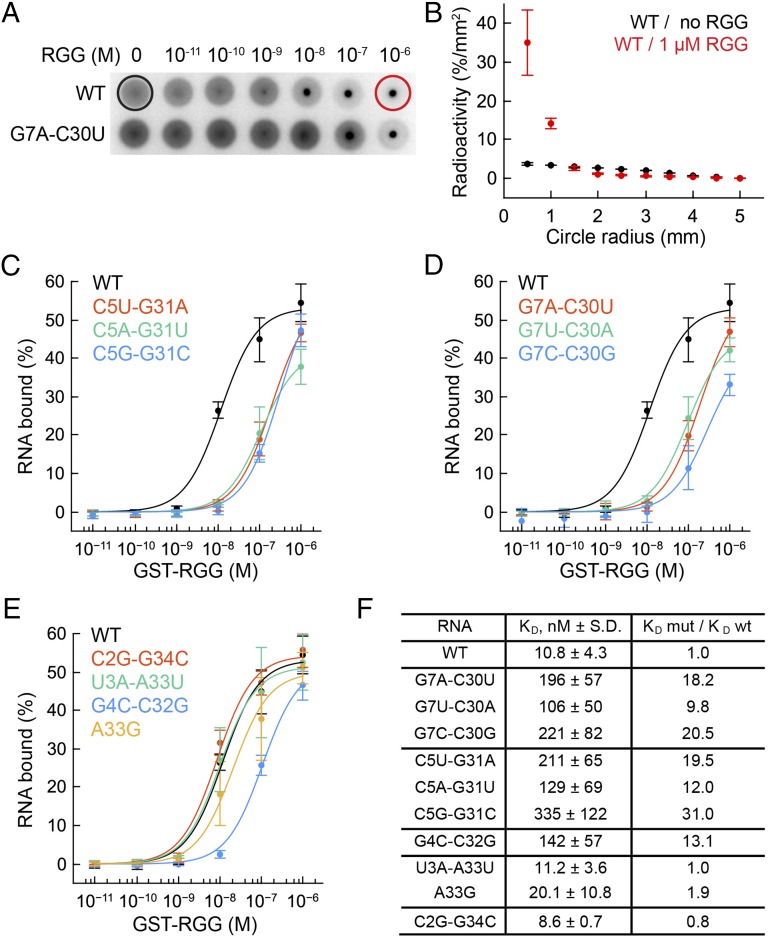
The RGG–sc1 crystal structure revealed base-specific contacts with five nucleotides. To identify which of these interactions contribute to specificity of RNA recognition, we conducted structure-guided mutagenesis of RNA and determined binding affinity of the RNA mutants to the GST-tagged RGG peptide by DRaCALA (39). This technique is based on different diffusion rates of radioactively labeled RNA and RGG–RNA complex on nitrocellulose membranes. Free RNA diffuses radially, whereas protein-bound RNA remains stationary (Fig. 5 A and B).
Fig. 5.
DRaCALA binding experiments with the GST-RGG protein and sc1 RNA mutants. KDs are average values from at least three independent experiments. Error bars are SDs. (A) Diffusion of radioactively labeled wild type (Upper) and G7A-C30U mutant (Lower) RNAs on nitrocellulose filters in the DRaCALA assay. Black and red circles show maximal diffusion limits used for quantification of binding affinity. (B) Quantification of the wild type RNA diffusion in the absence and presence of the protein. (C–E) Binding curves of the RNA variants. Mutated base pairs are shown in insets. (F) Dissociation constants and effects of mutations expressed as a ratio between KDs for wild-type and mutant RNAs.
Nucleotides G7 and G31 from adjacent G7-C30 and C5-G31 base pairs are the primary candidates for specific determinants because these residues make base-specific double hydrogen bonds with Arg10 and Arg15. To test the contribution of G31 to peptide binding, we replaced the C5-G31 base pair by U-A, A-U, or G-C base pairs and determined binding affinity of the mutants. All mutations strongly reduced binding affinity by 12- to 31-fold (Fig. 5 C and F) because of unfavorable placement of functional groups that disrupted hydrogen bonding. Similar effects were observed when G7-C30 was replaced by C-G, A-U, and U-A base pairs (Fig. 5 D and F).
To evaluate the significance of observed Arg8•C2, Gly11•G4, and Arg10•U3 and potential Arg9•A33 (with 3.6 Å distance) base-specific contacts for RGG–sc1 binding, we tested several mutants of the corresponding base pairs. Only the G4C-C32G mutant decreased peptide binding considerably (Fig. 5 E and F), whereas substitutions of C2-G34 and U3-A33 base pairs did not affect the binding. The 13.1-fold affinity reduction of the G4C-C32G mutant is comparable to the effects of mutating G7 and G31, therefore the close alignment of the main chain of Gly11 and the G4 base is as important for RNA-peptide binding as the juxtaposition of arginine side chains with G7 and G31.
Nuclease Cleavage Suggests Similar Conformations of Wild-Type and Mutant sc1 RNAs.
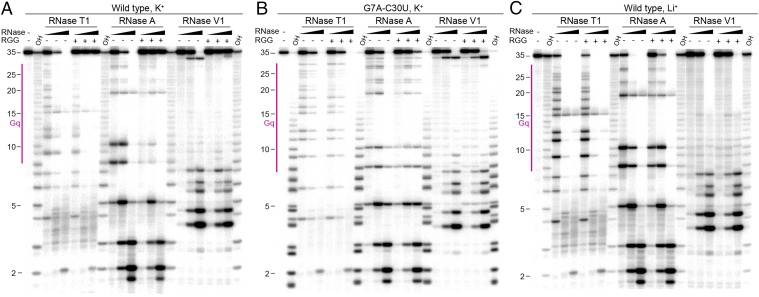
To rule out alternative folding of sc1 RNA mutants, we performed structure probing and RGG peptide footprinting using ribonuclease T1, A, and V1 which specifically cut unpaired guanosines, unpaired pyrimidines, and RNA helices, respectively. Previously, reduction of ribonuclease T1 cleavage in guanosines of G-rich sequences was associated with the formation of G-quartets and G-quadruplexes (21, 24). Because the G7-C30 base pair stabilizes the mixed tetrad, the G7A-C30U mutation is the most likely candidate for disrupting the RNA structure and is therefore representative of mutant RNAs used for binding assays. To obtain a comprehensive picture of nuclease cleavage and resistance to cleavage upon peptide binding, we compared probing and footprinting patterns using three nuclease concentrations (Fig. 6 and SI Appendix, Fig. S6).
Fig. 6.
Representative gels with ribonuclease T1, A, and V1 footprinting of the RGG–sc1 complex. Enzyme concentrations were: RNase T1, 0.01, 0.1, and 1.0 U/µL; RNase A, 0.01, 0.1 and 1.0 ng/µL; RNase V1, 0.001, 0.01, and 0.1 U/µL. OH, alkaline ladder. Gq, G-quadruplex region. (A) Wild-type RNA in K+. (B) G7A-C30U mutant RNA in K+. (C) Wild-type RNA in Li+.
RNase T1 treatment of sc1 RNA at low enzyme concentration in the presence of K+ showed that the majority of guanosines are susceptible to weak cleavage (Fig. 6A). Digestions with medium and high RNase T1 concentrations resulted in progressive reduction of the full-length RNA and appearance of smaller RNA species. Addition of the RGG peptide protected the RNA region from residue 9–35 at the low RNase concentration and eliminated specific cleavages at medium and high concentrations. Experiments with RNase A revealed similar tendencies although cleavages around mixed tetrad (U8) and loops (U10 and U19) were more efficient and peptide protections at low enzyme concentration were more pronounced. RNase V1 made several cleavages in the duplex region that were strongly (C32) or weakly (G4-to-U8 region) protected by the RGG peptide. These data demonstrate that the G-quadruplex is less stable in the absence of peptide and that peptide binding stabilizes G-quadruplex and directly shields and/or strengthens the duplex region.
Nuclease treatment of the G7A-C30U RNA demonstrated cleavage patterns similar to ones of the wild-type RNA, with only slightly less efficient RNase A cleavage at U8 and U10 (Fig. 6B). These data suggest that G7A-C30U mutation does not unfold RNA and that the mutant and wild-type RNAs adopt similar conformations in the ligand-unbound state. In agreement with reduced binding, effects of peptide addition were weak and mostly observed as cleavage reduction in RNase A patterns.
As a control for unfolded G-quadruplex, wild-type sc1 RNA and RNA-peptide complex samples were prepared and digested by RNases in the presence of Li+ cations (Fig. 6C) previously shown to disrupt G-quadruplexes (21, 24, 37). As expected, RNase T1 and A cleave RNA more efficiently in the presence of Li+ cations than in the presence of K+ cations, especially in the G-quadruplex part, suggestive of unfolding of the G-quadruplex. The loss of RNase V1 cut at C32 also indicates disruption or reduced stability of the duplex stem. In agreement with significantly reduced binding, addition of the RGG peptide does not affect RNase T1 and V1 patterns and only slightly protects RNA from RNase A cleavage. These results confirm that the G7A-C30U RNA and likely other mutants in our study adopt conformations similar to the wild-type RNA structure in the presence of K+ cations.
Discussion
The lack of a complementary strand in RNA, compared with DNA, makes G-rich sequences more likely to form quadruplexes in vivo, therefore, many G-quadruplex-forming sequences predicted in the human genome (40) may adopt G-quadruplexes in the encoded RNA transcriptome. An increasing number of studies identifies G-quadruplexes in natural RNAs and implicates these structures in transcription, mRNA processing, translation and other cellular processes (reviewed in ref. 41). RNA G-quadruplexes are also implicated in sequestration of RNA-binding proteins that may lead to amyotrophic lateral sclerosis and possibly other neurodegenerative pathologies (42). Despite the abundance of functions suggested for RNA G-quadruplexes and the growing list of G-quadruplex-binding proteins, 3D structures were determined for only several G-quadruplex-containing RNAs. However, the majority of these structures are built by short RNAs and only two RNA structures, Spinach and sc1 (37, 43, 44), embed G-quadruplexes into long sequences. In these two structures, only the NMR structure of the RGG–sc1 complex (37) contains a protein component bound near the G-quadruplex, whereas Spinach contains a binding site for a small ligand. Hence, the current study reports on the first X-ray crystal structure of a G-quadruplex-containing RNA bound to a biologically relevant peptide.
The crystal structure of the RGG–sc1 complex reveals a K+ and peptide-stabilized intricate architecture of the G-quadruplex RNA composed of three G-quartets connected to a duplex via a stacked mixed tetrad. This architecture is in good agreement with the NMR structure of the RGG–sc1 complex (37) and can be paralleled with chromophore-bound Spinach RNA that also contains a G-quadruplex connected to a duplex through a mixed tetrad (43, 44) (SI Appendix, Fig. S7). However, aside from this marginal similarity, sc1 and Spinach bear little in common. The RNAs have different topology and the G-quadruplex of sc1 is connected to a single RNA duplex unlike the G-quadruplex of Spinach that is situated between two helical regions.
Comparison of two sc1 RNA monomers in the crystal structure and NMR models revealed a certain degree of structural plasticity in the G-quadruplex parts. The structural differences are mostly associated with highly dynamic loops and are rather substantial if compared, for instance, with G-quadruplexes from the independently determined structures of Spinach RNAs (43, 44), which have 2–3-fold lower r.m.s.d. Although the nucleotide identity in loop regions is not important for sc1 G-quadruplex recognition by the RGG motif (37), other G-quadruplexes may exploit loops for specific protein binding. Therefore, strategies to design and study G-quadruplex binders should take into account the dynamic nature of loop regions and a possibility of conformational shifts even in seemingly rigid G-quadruplexes.
The RGG–sc1 structures show that two out of four K+-binding sites cannot accommodate similar Cs+ cations and are therefore K+-specific. The other two sites are less specific, as indicated by Cs+ binding, and are not sufficient to induce G-quadruplex folding, as suggested by reduced peptide binding in the presence of Cs+ cations. One of these sites is located between quadruplex-closing tetrads and could facilitate preformation of structure(s) capable of K+ and protein recognition. Such partially folded intermittent structure(s) could be folded on the basis of short RNA duplexes observed by our nuclease probing studies as well as NMR studies on sc1 RNA and natural G-quadruplex mRNAs targeted by FMRP (29, 37). Formation of the intermittent structure(s) could accelerate K+-dependent quadruplex folding and FMRP binding when intracellular conditions are favorable, for instance, when FMRP is present in the vicinity of mRNA.
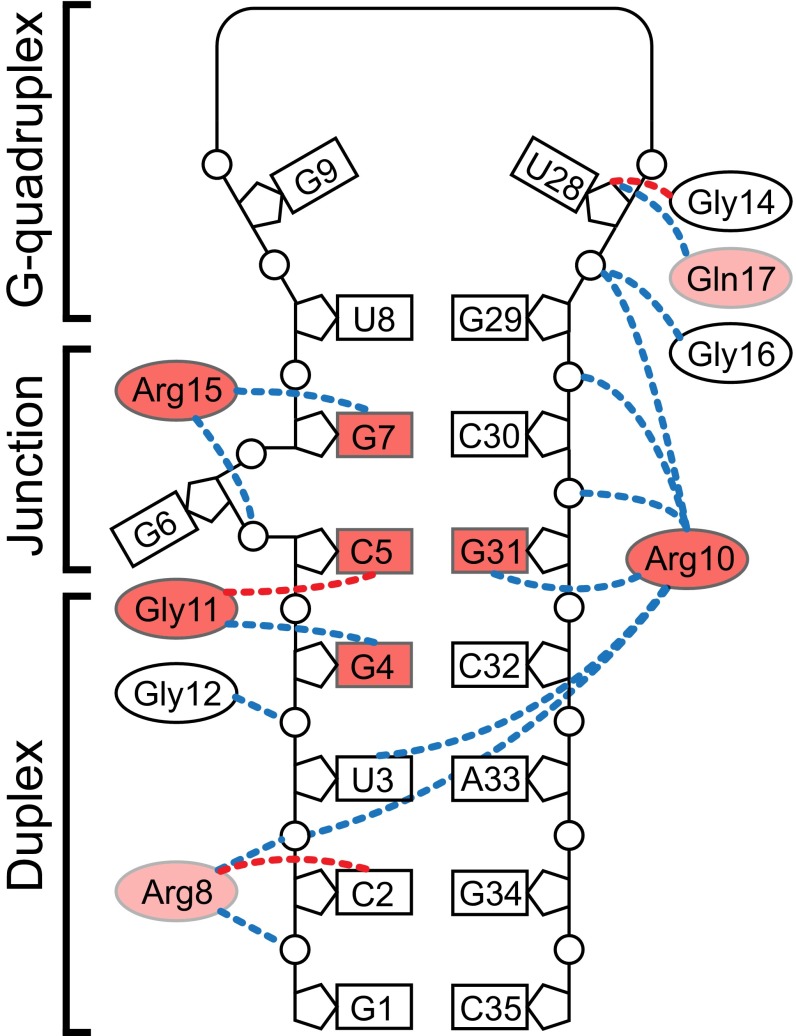
The crystal structure shows that the RGG peptide creates a dense network of various interactions with sc1 RNA. Although the peptide does not bind to G-quartets directly, it stabilizes the mixed base tetrad that in turn facilitates formation of the G-quadruplex. As anticipated from our previous study (37), the key contacts are formed by residues around RGG’s β-turn, in particular side chains of Arg10 and Arg15, which make base-specific interactions with two top base pairs C5-G31 and G7-C30 of the duplex in the quadruplex–duplex junction (Fig. 7). However, the base-specific contacts of Arg10 and Arg15 are not the only essential sequence-specific interactions. Surprisingly, our data identified Gly11-bound G4 from the G4-C32 base pair of the duplex as another major specificity determinant. Therefore, sequence-specific recognition of sc1 RNA by the FMRP RGG motif extends from the quadruplex–duplex junction to the duplex region.
Fig. 7.
Molecular determinants of RGG–sc1 recognition. Effects of selected mutations (current study and ref. 37) are color-coded: red and light red are >10 and 2- to 10-fold reduction of binding affinity, respectively. Putative intermolecular hydrogen bonds involving C-O and N-H main chain functionalities of the peptide are depicted by dashed lines of red and blue colors, respectively.
In addition to pinpointing major determinants for RGG–sc1 binding, the structure explains the effects of peptide mutations that were difficult to rationalize based on the NMR structure. For instance, the outward direction of Gln17 in the NMR structure could not explain a 3.5-fold reduction in binding affinity of the Q17N mutant (37). These observations are consistent with the formation of a stabilizing contact with Arg8, observed in the crystal structure, and its loss in the mutant. Strong ninefold reduction in RNA binding of R8K mutant (37) would be easier to explain by the loss of stabilizing contacts within the important β-turn, as suggested by the X-ray structure, than by the loss of two nonessential hydrogen bonds, according to the NMR structure.
An exciting finding of our study is the identification of type I β-turn in the RNA-bound RGG box of FMRP. This finding raises a question about formation of the β-turn in the free RGG peptide. Previous NMR (29, 37) and circular dichroism (29) studies did not identify any secondary structure in the RGG motif. In contrast, circular dichroism and infrared spectroscopy suggested such turns in the RGG box of nucleolin at moderate KCl concentrations (45); however, presented data did not demonstrate all spectral features characteristic for β-turns (46). Although a solid evidence for the formation of β-turns in the unbound RGG box is missing, our structural data clearly shows feasibility of the turn formation in the RGG motif. Our results indicate that the RGG motif does not necessarily interact with other molecules in the flexible linear conformation but can form a secondary structure element that reverses directionality of the protein chain and may position the termini of the motif in close proximity to each other. This property of the RGG motif should be taken into consideration when analyzing RGG box binding to its partner proteins and nucleic acids. Sequence alignment demonstrates that the β-turn-forming sequence GlyGlyGlyArg is shared by the FMRP RGG motifs from some mammals but not from all vertebrates (Fig. 1A). Because β-turns favor a Gly-Gly sequence in the middle (47), an ArgGlyGly(X) quadropeptide could be an alternative β-turn-forming sequence in other FMRPs as well as RGG box proteins, such as Ewing sarcoma protein, EBNA1, and nucleolin (48–50), also known to bind G-quartets. What could be the physiological consequences of β-turn formation in the RGG box? The turn can increase specificity of the RGG box binding and limit the number of RNA targets for the peptide by reducing the flexibility of the peptide, and providing a specific spatial pattern of functional groups for intermolecular interactions. Additionally, the RGG box in a rigid conformation bound to a compact site can better stabilize the adjacent structural elements of RNA, which could serve as signals for cellular activities. In line with this suggestion, G-quadruplexes have been suggested to function as neurite localization signals for neuronal mRNAs (25).
Although Arg10 and Arg15 are crucial for sc1 RNA binding, our previous data showed that Arg10 is not required for specific recognition of some G-quadruplex-forming RNAs, whereas mutations of Arg15 strongly impact RNA binding (37). These observations imply that the RGG box of FMRP retains some but not all recognition features for binding to G-quadruplex RNAs. Such adaptability of the RGG motif could be critical for high-affinity recognition of natural mRNAs that contain variable duplexes and G-quadruplexes. On the other hand, commonalities in the RNA binding are beneficial for global regulation of protein–RNA interactions or switching binding partners as a result of, for instance, arginine methylation frequently observed in RGG boxes. This side chain modification disfavors RNA binding but facilitates protein interactions with Tudor family of proteins (reviewed in ref. 51). It is intriguing that Arg10 and Arg15, involved in polyribosome binding (14), are among four arginines targeted for methylation in FMRP and their methylation decreases RNA binding affinity (14, 52).
In addition to positively charged arginines that are naturally adept to bind negatively charged RNA, our results established critical contributions of glycines to the specific binding of RNA. This involvement may not be limited to RGG boxes and related motifs, which are well known RNA binding domains found in hundreds of proteins (51, 53). In addition to the RGG motif, recent interactome capture experiments identified the YGG (tyrosine-glycine-glycine) sequence as another frequent motif in RNA-binding architectures (2). The function of this motif is unknown; nevertheless it could use a similar mechanism of RNA binding as RGG boxes by using the tyrosine side chain for stacking and hydrogen bonding with nucleobases and glycines for formation of local structural elements and RNA binding. Glycine-specific recognition of groove functionalities in the RNA helix could be envisioned as a shared feature in many biological systems using RGG boxes because RNA duplexes are the most common secondary structural elements of RNA. Thus, our structural and biochemical data on the binding of FMRP RGG box and G-quadruplex RNA illustrate structural principles that may be applicable to many other proteins that contain RGG box and similar motifs. These general recognition principles could also be common to proteins that contain other motifs enriched in arginine and glycines. For example, the NMR structures of a complex between a bovine immunodeficiency virus (BIV) transactivation response element (TAR) and a peptide from a transactivating protein (Tat) showed that the peptide adopts a β-turn conformation and sits in the major grove of the RNA, where it forms base-specific contacts with RNA using glycines and arginines (54, 55).
Two important features of the RGG peptide binding to sc1 RNA, the accommodation of the RGG peptide in the distorted helical stem and the lack of direct contacts with the G-quadruplex, emphasize that RNA binding of the RGG box may not be limited to specific G-quadruplex-containing RNAs. In the sc1-RGG complex structure, the G-quadruplex expands the peptide binding surface by forming the “roof” of the binding pocket. This additional binding surface, created by the mixed base tetrad, does not form base-specific interactions with the peptide and can involve any kind of tetrad, thus possibly explaining why the RGG box can interact with mRNAs that contain seemingly different duplexes and quadruplexes (19, 21). Furthermore, RGG binding RNAs could be principally different from G-quadruplex-containing RNAs. Such RNAs may be composed from a duplex distorted by connection to a planar nucleotide arrangement, which is stabilized by stacking interactions with an RNA element distinct from a G-quadruplex.
A major unanswered question about the pathogenesis of Fragile X Syndrome is whether the highly homologous paralogs of FMRP, FXR1P, and FXR2P, can compensate for the loss of FMRP function. Most of the functionally annotated domains of this family are highly conserved among the three proteins, including the nuclear localization and export signals, two Tudor and three KH domains, and the phosphorylation domain. However, the G-quadruplex-binding RGG box is unique to FMRP and is not shared by human FXR1P and FXR2P and Drosophila dFMRP, which contain RGG-rich regions incapable of binding to sc1 RNA and significantly divergent from FMRP (35). In light of the apparent failure of FXR1P and FXR2P to rescue loss of FMRP both in models and in the human disease, understanding the specialized role of the RGG box is imperative. Although G-quadruplex RNA motifs definitively bound by FMRP have not been identified in vivo yet, such RNA structural motifs clearly exist and their importance has been documented. These motifs probably do not crosslink with high efficiency using HITS-CLIP and PAR-CLIP protocols, or they are present in noncoding RNAs such as lincRNA or rRNA that has not yet been analyzed for these interactions. Nonetheless, the structural and biochemical data we present here should aid in unraveling the complex interactions that FMRP and its multiple RNA binding domains have with key RNA targets once these technical limitations have been addressed. Given that mRNA binding and translational repression are the most important functions of FMRP, our results will help to decipher the regulatory circuits underlying development of FXS.
Materials and Methods
Preparation of RNA and Proteins.
Wild-type and mutated 35-mer sc1 RNAs were transcribed in vitro with T7 RNA polymerase (37). All RNAs were purified by denaturing polyacrylamide gel electrophoresis (PAGE).
The GST-RGG protein containing 28-mer RGG peptide fused to GST was produced in Escherichia coli BL21(DE3)CodonPlusRIL strain bearing the pGEX6P-FMRP-RGG plasmid (37). The cells were resuspended in lysis buffer (50 mM Tris⋅HCl pH 7.5, 150 mM NaCl, 1 mM EDTA, 1 mM DTT) supplemented with 1× EDTA-free protease inhibitor mixture (Roche) and disrupted by EmulsiFlex C-5 (Avestine). The protein was purified by affinity chromatography on GSTrap (GE Healthcare), dialyzed against a low-salt buffer (50 mM Tris⋅HCl pH 7.5, 0.1 mM EDTA, and 1 mM DTT), and further purified by cation-exchange chromatography on HiTrap SP column (GE Healthcare). The protein eluted by the NaCl gradient was extensively dialyzed to remove traces of K+ cations, concentrated, frozen in liquid nitrogen, and stored −80 °C. Before RNA binding assay, the protein was thawed on ice, diluted with 50 mM Tris⋅HCl pH 7.5, 5 mM DTT, 0.1 mg/mL BSA, 50% (vol/vol) glycerol to concentrations 10−10 to 10−5 M, and stored at −20 °C.
Crystallization of the RGG–sc1 Complex and Structure Determination.
To prepare the complex, chemically synthesized 35-mer sc1 RNA and 18-mer RGG peptide (residues 527–544, Uniprot ID: Q06787-1; Arg1Ala mutation) were mixed at 0.2 mM concentration in 50 mM K-Acetate (pH 6.7), heated at 95 °C for 2 min, and chilled on ice as previously described for the NMR studies. Before crystallization, the complex was concentrated twofold by air drying to the final concentration of ∼0.4 mM. Crystals of the elongated cube shape and ∼0.15 × 0.05 × 0.05 mm in size were grown by hanging-drop vapor diffusion after mixing 1 µL of complex with 1 µL of reservoir solution at 20 °C. The reservoir solution contained 0.2 M ammonium acetate, 0.1 M Na-Citrate (pH 5.6), and 18% (wt/vol) PEG4000. To obtain Cs+-containing crystals, crystals were grown with the K+-containing RNA-protein complex sample in 0.1 M CsCl, 0.1 M ammonium acetate, 0.1 M Na-Citrate (pH 5.6), and 18% (wt/vol) PEG4000. To obtain a heavy atom derivative, native crystals were incubated with 10 mM [Ir(NH3)6]Cl3 for 2 h. For data collection, crystals were passed through a reservoir solution supplemented with 25% (vol/vol) glycerol and flash-frozen in liquid nitrogen. X-ray diffraction data were collected at 100 K at 24-ID-C beamline at the Argonne National Laboratory. The data were reduced with XDS (56) and corrected for anisotropy with Diffraction Anisotropy Server at services.mbi.ucla.edu/anisoscale/ (SI Appendix, Table S1) to significantly improve the quality of the experimental map. The structure was phased by SAD using PHENIX (57). The experimental SAD electron density map was of excellent quality and model tracing was completed manually in Coot (58) (SI Appendix, Fig. S5). The structures were refined with PHENIX (57) and Refmac (59) using the data corrected for anisotropy. Figures were prepared with PyMol (pymol.org/).
RNA Binding Assay.
The affinity of RNA-binding interactions was determined by DRaCALA (39) using GST-RGG protein and 5ʹ [32P]-labeled 36-mer sc1 RNA variants (37). Labeled RNAs were diluted to 1 nM concentration in 10 mM Tris⋅HCl pH 7.5, 100 mM K-Acetate (KCl, LiCl, or CsCl when needed), and 5% glycerol. RNA samples were folded by heating at 95 °C for 2 min and cooling down to room temperature over 20 min. Nine-microliter RNA samples were mixed with 1-μL aliquot of GST-RGG to yield final protein concentrations 10−11 to 10−6 M. After 10-min incubation, 4-μL aliquot from each sample was spotted onto nitrocellulose membrane (Bio-Rad) and allowed to diffuse and dry on air for 30 min before exposing on the phosphorimager screen and scanning on Typhoon scanner (GE Healthcare). Radioactive signal was measured using OptiQuant software (PerkinElmer), the bound RNA was quantified as described in (39), and the binding curves were fitted to 1:1 binding model with QtiPlot (www.qtiplot.com). Approximately half of the RNA sample did not fold correctly and did not interact with the peptide.
Ribonuclease Footprinting.
5ʹ [32P]-labeled sc1 RNAs were diluted to 0.1 μM in a buffer containing 10 mM Tris⋅HCl (pH 7.5), 100 mM K-acetate (or Li-Acetate), 10 mM Na-acetate, 1 mM Mg-acetate, and 0.1% Triton X-100. RNA samples were folded by heating–cooling with and without addition of the chemically synthesized 28-mer FMRP RGG peptide at 1 μM final concentration. Ten-microliter RNA samples were treated with RNases T1, A, and V1 for 10 min at room temperature. Reactions were quenched by mixing with 10 μL of urea loading buffer (10 M urea, 2% β-mercaptoethanol, 10 mM EDTA pH 8.0, 0.05% xylene cyanol FF, 0.05% bromophenol blue) and heating at 75 °C for 30 s. Five-microliter samples were separated by denaturing 20% PAGE. After the run, gels were placed onto filter paper, dried, and analyzed by phosphorimager.
Supplementary Material
Acknowledgments
We thank Dr. Anh Tuan Phan for assistance in preparing RNA–protein complexes and the staff of beamlines 24-ID (Advanced Photon Source, Argonne National laboratory), supported by NIH Grant P41 GM103403 and Department of Energy funds, for assistance in data collection. This work was supported by New York University start-up funds; NIH Grant R21MH103655; Whitehead fellowship; Edward Mallinckrodt, Jr. Foundation award; and Irma T. Hirschl Career Scientist Award (to A.S.), NIH Grant R01HD040647 (to J.C.D.), and NIH Grant TR01 GM104962 (to D.J.P.). D.J.P. acknowledges support by the Memorial Sloan Kettering Cancer Center Support Grant/Core Grant (P30 CA008748).
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 5DE5, 5DE8, and 5DEA).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1515737112/-/DCSupplemental.
References
- 1.Baltz AG, et al. The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol Cell. 2012;46(5):674–690. doi: 10.1016/j.molcel.2012.05.021. [DOI] [PubMed] [Google Scholar]
- 2.Castello A, et al. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell. 2012;149(6):1393–1406. doi: 10.1016/j.cell.2012.04.031. [DOI] [PubMed] [Google Scholar]
- 3.Ashley CT, Jr, Wilkinson KD, Reines D, Warren ST. FMR1 protein: Conserved RNP family domains and selective RNA binding. Science. 1993;262(5133):563–566. doi: 10.1126/science.7692601. [DOI] [PubMed] [Google Scholar]
- 4.De Boulle K, et al. A point mutation in the FMR-1 gene associated with fragile X mental retardation. Nat Genet. 1993;3(1):31–35. doi: 10.1038/ng0193-31. [DOI] [PubMed] [Google Scholar]
- 5.Myrick LK, et al. Fragile X syndrome due to a missense mutation. Eur J Hum Genet. 2014;22(10):1185–1189. doi: 10.1038/ejhg.2013.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bassell GJ, Warren ST. Fragile X syndrome: Loss of local mRNA regulation alters synaptic development and function. Neuron. 2008;60(2):201–214. doi: 10.1016/j.neuron.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hernandez RN, et al. Autism spectrum disorder in fragile X syndrome: A longitudinal evaluation. Am J Med Genet A. 2009;149A(6):1125–1137. doi: 10.1002/ajmg.a.32848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jacquemont S, et al. Penetrance of the fragile X-associated tremor/ataxia syndrome in a premutation carrier population. JAMA. 2004;291(4):460–469. doi: 10.1001/jama.291.4.460. [DOI] [PubMed] [Google Scholar]
- 9.Sullivan SD, Welt C, Sherman S. FMR1 and the continuum of primary ovarian insufficiency. Semin Reprod Med. 2011;29(4):299–307. doi: 10.1055/s-0031-1280915. [DOI] [PubMed] [Google Scholar]
- 10.Myrick LK, Hashimoto H, Cheng X, Warren ST. Human FMRP contains an integral tandem Agenet (Tudor) and KH motif in the amino terminal domain. Hum Mol Genet. 2015;24(6):1733–1740. doi: 10.1093/hmg/ddu586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collins SC, et al. Identification of novel FMR1 variants by massively parallel sequencing in developmentally delayed males. Am J Med Genet A. 2010;152A(10):2512–2520. doi: 10.1002/ajmg.a.33626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Darnell JC, et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011;146(2):247–261. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Portera-Cailliau C. Which comes first in fragile X syndrome, dendritic spine dysgenesis or defects in circuit plasticity? Neuroscientist. 2012;18(1):28–44. doi: 10.1177/1073858410395322. [DOI] [PubMed] [Google Scholar]
- 14.Blackwell E, Zhang X, Ceman S. Arginines of the RGG box regulate FMRP association with polyribosomes and mRNA. Hum Mol Genet. 2010;19(7):1314–1323. doi: 10.1093/hmg/ddq007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Darnell JC, et al. Kissing complex RNAs mediate interaction between the Fragile-X mental retardation protein KH2 domain and brain polyribosomes. Genes Dev. 2005;19(8):903–918. doi: 10.1101/gad.1276805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zou K, et al. Identification of FMRP-associated mRNAs using yeast three-hybrid system. Am J Med Genet B Neuropsychiatr Genet. 2008;147B(6):769–777. doi: 10.1002/ajmg.b.30678. [DOI] [PubMed] [Google Scholar]
- 17.Dolzhanskaya N, Sung YJ, Conti J, Currie JR, Denman RB. The fragile X mental retardation protein interacts with U-rich RNAs in a yeast three-hybrid system. Biochem Biophys Res Commun. 2003;305(2):434–441. doi: 10.1016/s0006-291x(03)00766-6. [DOI] [PubMed] [Google Scholar]
- 18.Sung YJ, Conti J, Currie JR, Brown WT, Denman RB. RNAs that interact with the fragile X syndrome RNA binding protein FMRP. Biochem Biophys Res Commun. 2000;275(3):973–980. doi: 10.1006/bbrc.2000.3405. [DOI] [PubMed] [Google Scholar]
- 19.Brown V, et al. Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell. 2001;107(4):477–487. doi: 10.1016/s0092-8674(01)00568-2. [DOI] [PubMed] [Google Scholar]
- 20.Brown V, et al. Purified recombinant Fmrp exhibits selective RNA binding as an intrinsic property of the fragile X mental retardation protein. J Biol Chem. 1998;273(25):15521–15527. doi: 10.1074/jbc.273.25.15521. [DOI] [PubMed] [Google Scholar]
- 21.Darnell JC, et al. Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell. 2001;107(4):489–499. doi: 10.1016/s0092-8674(01)00566-9. [DOI] [PubMed] [Google Scholar]
- 22.Suhl JA, Chopra P, Anderson BR, Bassell GJ, Warren ST. Analysis of FMRP mRNA target datasets reveals highly associated mRNAs mediated by G-quadruplex structures formed via clustered WGGA sequences. Hum Mol Genet. 2014;23(20):5479–5491. doi: 10.1093/hmg/ddu272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ascano M, Jr, et al. FMRP targets distinct mRNA sequence elements to regulate protein expression. Nature. 2012;492(7429):382–386. doi: 10.1038/nature11737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schaeffer C, et al. The fragile X mental retardation protein binds specifically to its mRNA via a purine quartet motif. EMBO J. 2001;20(17):4803–4813. doi: 10.1093/emboj/20.17.4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Subramanian M, et al. G-quadruplex RNA structure as a signal for neurite mRNA targeting. EMBO Rep. 2011;12(7):697–704. doi: 10.1038/embor.2011.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bole M, Menon L, Mihailescu MR. Fragile X mental retardation protein recognition of G quadruplex structure per se is sufficient for high affinity binding to RNA. Mol Biosyst. 2008;4(12):1212–1219. doi: 10.1039/b812537f. [DOI] [PubMed] [Google Scholar]
- 27.Menon L, Mihailescu MR. Interactions of the G quartet forming semaphorin 3F RNA with the RGG box domain of the fragile X protein family. Nucleic Acids Res. 2007;35(16):5379–5392. doi: 10.1093/nar/gkm581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Menon L, Mader SA, Mihailescu MR. Fragile X mental retardation protein interactions with the microtubule associated protein 1B RNA. RNA. 2008;14(8):1644–1655. doi: 10.1261/rna.1100708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramos A, Hollingworth D, Pastore A. G-quartet-dependent recognition between the FMRP RGG box and RNA. RNA. 2003;9(10):1198–1207. doi: 10.1261/rna.5960503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen E, Sharma MR, Shi X, Agrawal RK, Joseph S. Fragile X mental retardation protein regulates translation by binding directly to the ribosome. Mol Cell. 2014;54(3):407–417. doi: 10.1016/j.molcel.2014.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blackwell E, Ceman S. A new regulatory function of the region proximal to the RGG box in the fragile X mental retardation protein. J Cell Sci. 2011;124(Pt 18):3060–3065. doi: 10.1242/jcs.086751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mazroui R, et al. Trapping of messenger RNA by Fragile X Mental Retardation protein into cytoplasmic granules induces translation repression. Hum Mol Genet. 2002;11(24):3007–3017. doi: 10.1093/hmg/11.24.3007. [DOI] [PubMed] [Google Scholar]
- 33.Pfeiffer BE, et al. Fragile X mental retardation protein is required for synapse elimination by the activity-dependent transcription factor MEF2. Neuron. 2010;66(2):191–197. doi: 10.1016/j.neuron.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Taha MS, et al. Subcellular fractionation and localization studies reveal a direct interaction of the fragile X mental retardation protein (FMRP) with nucleolin. PLoS One. 2014;9(3):e91465. doi: 10.1371/journal.pone.0091465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Darnell JC, Fraser CE, Mostovetsky O, Darnell RB. Discrimination of common and unique RNA-binding activities among Fragile X mental retardation protein paralogs. Hum Mol Genet. 2009;18(17):3164–3177. doi: 10.1093/hmg/ddp255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Coffee RL, Jr, Tessier CR, Woodruff EA, 3rd, Broadie K. Fragile X mental retardation protein has a unique, evolutionarily conserved neuronal function not shared with FXR1P or FXR2P. Dis Model Mech. 2010;3(7-8):471–485. doi: 10.1242/dmm.004598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Phan AT, et al. Structure-function studies of FMRP RGG peptide recognition of an RNA duplex-quadruplex junction. Nat Struct Mol Biol. 2011;18(7):796–804. doi: 10.1038/nsmb.2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zanotti KJ, Lackey PE, Evans GL, Mihailescu MR. Thermodynamics of the fragile X mental retardation protein RGG box interactions with G quartet forming RNA. Biochemistry. 2006;45(27):8319–8330. doi: 10.1021/bi060209a. [DOI] [PubMed] [Google Scholar]
- 39.Roelofs KG, Wang J, Sintim HO, Lee VT. Differential radial capillary action of ligand assay for high-throughput detection of protein-metabolite interactions. Proc Natl Acad Sci USA. 2011;108(37):15528–15533. doi: 10.1073/pnas.1018949108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huppert JL, Balasubramanian S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005;33(9):2908–2916. doi: 10.1093/nar/gki609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Millevoi S, Moine H, Vagner S. G-quadruplexes in RNA biology. Wiley Interdiscip Rev RNA. 2012;3(4):495–507. doi: 10.1002/wrna.1113. [DOI] [PubMed] [Google Scholar]
- 42.Haeusler AR, et al. C9orf72 nucleotide repeat structures initiate molecular cascades of disease. Nature. 2014;507(7491):195–200. doi: 10.1038/nature13124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Warner KD, et al. Structural basis for activity of highly efficient RNA mimics of green fluorescent protein. Nat Struct Mol Biol. 2014;21(8):658–663. doi: 10.1038/nsmb.2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang H, et al. A G-quadruplex-containing RNA activates fluorescence in a GFP-like fluorophore. Nat Chem Biol. 2014;10(8):686–691. doi: 10.1038/nchembio.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ghisolfi L, Joseph G, Amalric F, Erard M. The glycine-rich domain of nucleolin has an unusual supersecondary structure responsible for its RNA-helix-destabilizing properties. J Biol Chem. 1992;267(5):2955–2959. [PubMed] [Google Scholar]
- 46.Gao F, et al. β-turn formation by a six-residue linear peptide in solution. J Pept Res. 2002;60(2):75–80. doi: 10.1034/j.1399-3011.2002.02982.x. [DOI] [PubMed] [Google Scholar]
- 47.Sibanda BL, Blundell TL, Thornton JM. Conformation of beta-hairpins in protein structures. A systematic classification with applications to modelling by homology, electron density fitting and protein engineering. J Mol Biol. 1989;206(4):759–777. doi: 10.1016/0022-2836(89)90583-4. [DOI] [PubMed] [Google Scholar]
- 48.Hanakahi LA, Sun H, Maizels N. High affinity interactions of nucleolin with G-G-paired rDNA. J Biol Chem. 1999;274(22):15908–15912. doi: 10.1074/jbc.274.22.15908. [DOI] [PubMed] [Google Scholar]
- 49.Norseen J, Johnson FB, Lieberman PM. Role for G-quadruplex RNA binding by Epstein-Barr virus nuclear antigen 1 in DNA replication and metaphase chromosome attachment. J Virol. 2009;83(20):10336–10346. doi: 10.1128/JVI.00747-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Takahama K, Kino K, Arai S, Kurokawa R, Oyoshi T. Identification of Ewing’s sarcoma protein as a G-quadruplex DNA- and RNA-binding protein. FEBS J. 2011;278(6):988–998. doi: 10.1111/j.1742-4658.2011.08020.x. [DOI] [PubMed] [Google Scholar]
- 51.Thandapani P, O’Connor TR, Bailey TL, Richard S. Defining the RGG/RG motif. Mol Cell. 2013;50(5):613–623. doi: 10.1016/j.molcel.2013.05.021. [DOI] [PubMed] [Google Scholar]
- 52.Stetler A, et al. Identification and characterization of the methyl arginines in the fragile X mental retardation protein Fmrp. Hum Mol Genet. 2006;15(1):87–96. doi: 10.1093/hmg/ddi429. [DOI] [PubMed] [Google Scholar]
- 53.Rajyaguru P, Parker R. RGG motif proteins: Modulators of mRNA functional states. Cell Cycle. 2012;11(14):2594–2599. doi: 10.4161/cc.20716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ye X, Kumar RA, Patel DJ. Molecular recognition in the bovine immunodeficiency virus Tat peptide-TAR RNA complex. Chem Biol. 1995;2(12):827–840. doi: 10.1016/1074-5521(95)90089-6. [DOI] [PubMed] [Google Scholar]
- 55.Puglisi JD, Chen L, Blanchard S, Frankel AD. Solution structure of a bovine immunodeficiency virus Tat-TAR peptide-RNA complex. Science. 1995;270(5239):1200–1203. doi: 10.1126/science.270.5239.1200. [DOI] [PubMed] [Google Scholar]
- 56.Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Adams PD, et al. PHENIX: Building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58(Pt 11):1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 58.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53(Pt 3):240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.