Significance
Plant cells constantly survey their environment to fine-tune their internal processes. Plant receptor-like kinases (RLKs) of the Catharanthus roseus RLK1-like (CrRLK1L) subfamily have been implicated in many signaling processes, including the coordination of the intracellular growth machinery with the performance of the extracellular matrix, i.e. the cell wall (CW). To avoid loss of integrity and growth and to adapt their CWs to developmental and environmental perturbations, growing plant cells have developed complex sensing mechanisms. The CrRLK1Ls ANXUR1 and ANXUR2 and their closest homolog FERONIA control CW integrity in the tip-growing pollen tubes and root hairs, respectively. Here, we identify and characterize the receptor-like cytoplasmic kinase MARIS as a novel downstream component of the CrRLK1L-dependent signaling cascade that controls CW integrity in tip-growing cells.
Keywords: receptor-like kinase signaling, cell wall integrity, pollen tube, root hair, Arabidopsis
Abstract
Growing plant cells need to rigorously coordinate external signals with internal processes. For instance, the maintenance of cell wall (CW) integrity requires the coordination of CW sensing with CW remodeling and biosynthesis to avoid growth arrest or integrity loss. Despite the involvement of receptor-like kinases (RLKs) of the Catharanthus roseus RLK1-like (CrRLK1L) subfamily and the reactive oxygen species-producing NADPH oxidases, it remains largely unknown how this coordination is achieved. ANXUR1 (ANX1) and ANX2, two redundant members of the CrRLK1L subfamily, are required for tip growth of the pollen tube (PT), and their closest homolog, FERONIA, controls root-hair tip growth. Previously, we showed that ANX1 overexpression mildly inhibits PT growth by oversecretion of CW material, whereas pollen tubes of anx1 anx2 double mutants burst spontaneously after germination. Here, we report the identification of suppressor mutants with improved fertility caused by the rescue of anx1 anx2 pollen tube bursting. Mapping of one these mutants revealed an R240C nonsynonymous substitution in the activation loop of a receptor-like cytoplasmic kinase (RLCK), which we named MARIS (MRI). We show that MRI is a plasma membrane-localized member of the RLCK-VIII subfamily and is preferentially expressed in both PTs and root hairs. Interestingly, mri-knockout mutants display spontaneous PT and root-hair bursting. Moreover, expression of the MRIR240C mutant, but not its wild-type form, partially rescues the bursting phenotypes of anx1 anx2 PTs and fer root hairs but strongly inhibits wild-type tip growth. Thus, our findings identify a novel positive component of the CrRLK1L-dependent signaling cascade that coordinates CW integrity and tip growth.
Growing plant cells are in constant communication with their environment, monitoring external signals that lead to internal reactions. Because cellular growth depends on a tight coordination of external and internal processes, signaling between the extracellular matrix, i.e., the primary cell wall (CW), and the internal growth machinery plays a central role in its regulation. The turgor pressure-resisting CW that shields plant cells from a changing environment exhibits remarkable but seemingly contradictory properties: rigidity and extensibility. Growing cells must find a balance between loosening their CWs sufficiently to allow expansion but not so much that they lose CW integrity. Because any environmental perturbation affecting the properties of the CW can upset this fragile balance, plant cells have developed complex sensing mechanisms to coordinate the state of the CW with the internal growth machinery (1, 2). These CW sensing mechanisms must be particularly robust in fast-growing cells, such as the tip-growing root hairs and pollen tubes (PTs), the male gametophytes of flowering plants. PTs elongate rapidly and grow over long distances within female tissues to deliver the sperm cells to the female gametophytes, which are deeply embedded in the ovules. Considering the vast excess of pollen grains that germinate on a receptive stigma, there is strong competition, and PTs must grow as fast as possible to reach unfertilized ovules; otherwise they will not contribute to the next generation. However, PTs must ensure that, while elongating extremely rapidly, they do not lose their integrity. In PTs of the model plant Arabidopsis thaliana, the plasma membrane-localized ANXUR (ANX1 and ANX2) receptor-like kinases (RLKs) play a major role in controlling CW integrity and growth. On one hand, loss of function of the redundant RLKs ANX1 and ANX2 leads to precocious PT rupture shortly after germination, resulting in male sterility (3, 4). This phenotype also is seen when the ANX-RLKs are degraded by the endoplasmic reticulum-associated degradation pathway in the turan mutant affecting N-glycosylation (5). On the other hand, overexpression of ANX-RLKs inhibits pollen germination and PT elongation, most likely because of the oversecretion of CW material (6). The ANX-RLKs and their closest homolog FERONIA (FER) belong to the Catharanthus roseus RLK1-like (CrRLK1L) subfamily that has been investigated extensively as putative CW sensors involved in coordinating cell growth, cell–cell communication, defense against pathogens, and hormone signaling as well as CW remodeling and integrity (reviewed in refs. 7–9). FER, the most thoroughly studied member of this subfamily, controls many developmental processes, such as intercellular communication during fertilization (10–12), cell elongation (13), calcium and hormone signaling (10, 14, 15), mechanosensing (16), plant defense (17, 18), and growth control of root hairs (19). Despite the many reports describing the role of CrRLK1Ls in various signaling processes, the mechanistic basis of their function and their relationship with other pathways remain poorly understood. FER was recently found to mediate the inhibition of primary root growth by forming a receptor–ligand pair with the rapid alkalinization factor 1 (RALF1) peptide (20). Moreover, it was shown that the intracellular kinase domains of three CrRLK1Ls members are interchangeable and that the various CrRLK1Ls share downstream signaling components (21). This finding has been confirmed for at least one signaling component so far, namely the reactive oxygen species (ROS)-producing NADPH oxidases, also called “respiratory burst oxidase homologs” (Rboh). It was reported that RbohD/RbohF for THESEUS1 (THE1), RbohC for FER, and RbohH/RbohJ for ANX1/2 act downstream of the CrRLK1Ls in primary roots, root hairs, and PTs, respectively (6, 19, 22).
Here, we report, based on an anx1 anx2 male sterility suppressor screen, the identification of the receptor-like cytoplasmic kinase (RLCK) MARIS (MRI), a novel component of the CrRLK1L-mediated signaling pathway in tip-growing cells. MRI is expressed preferentially in PTs and root hairs, and its disruption triggers spontaneous bursting of PTs and root hairs. The suppressor mutant carries a R240C amino acid substitution in the activation loop of MRI. Expression of MRIR240C partially rescues the bursting phenotypes of anx1 anx2 and rbohH rbohJ PTs as well as fer root hairs while strongly inhibiting WT tip growth, indicating that this mutant form of MRI overactivates the CrRLK1L-dependent pathway downstream of the NADPH oxidases.
Results
An anx1 anx2 Sterility-Suppressor Screen Identifies Putative Components of the ANX1/2 Pathway.
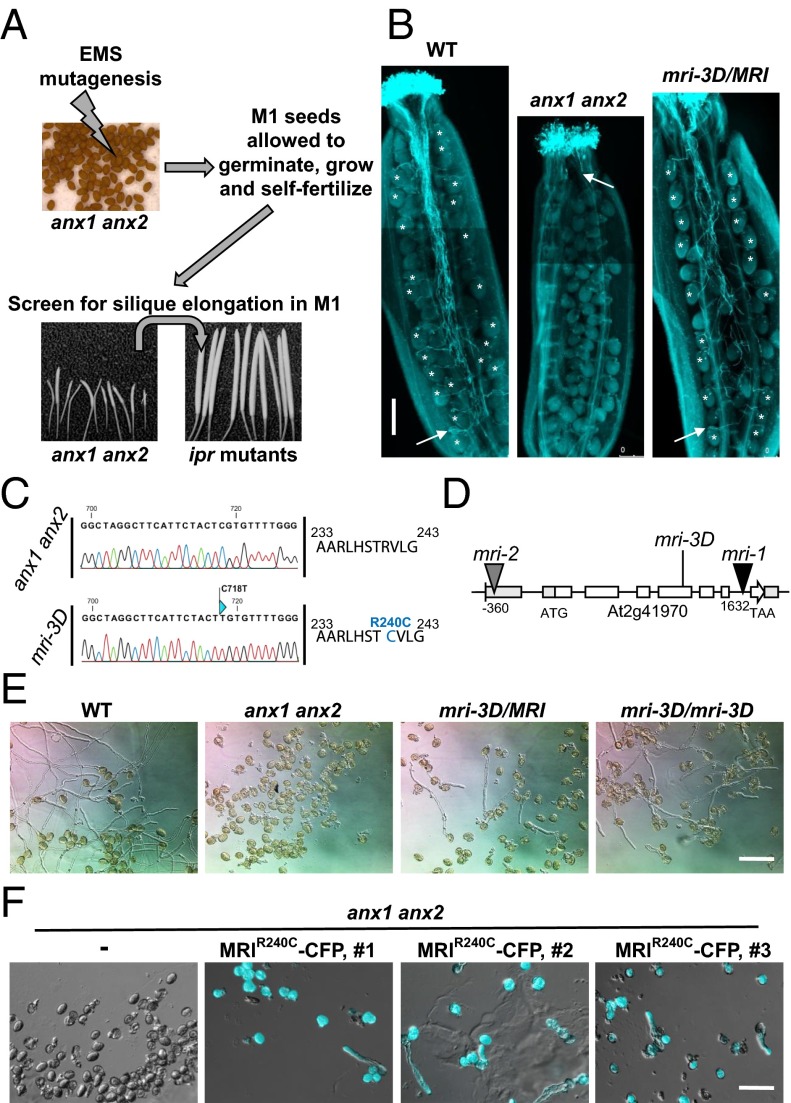
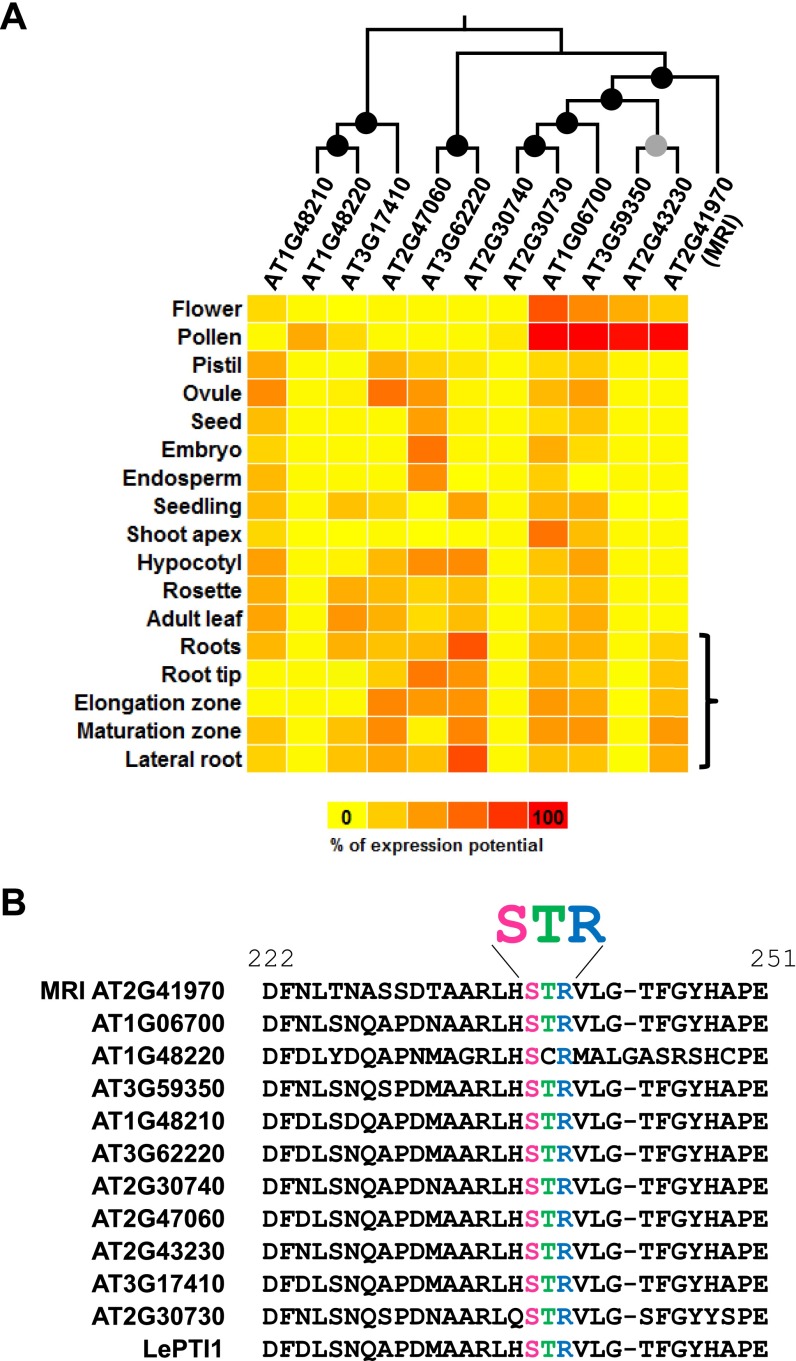
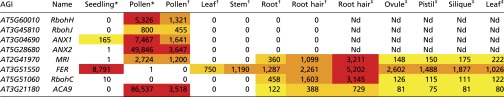
To identify putative downstream components of the ANX1/2 pathway, a forward genetic screen to identify suppressor mutations of the anx1 anx2 PT-bursting phenotype was carried out (Fig. 1A). We anticipated that rescue of anx1 anx2 PT bursting in vivo by a second site mutation would lead to improved plant fertility characterized by elongated siliques, an easily scorable phenotype. Approximately 7,000 ethyl-methyl sulfonate (EMS)-treated anx1-2 anx2-2 M1 plants were screened for silique elongation, resulting in the identification of ∼30 suppressor mutants, all of which were confirmed to be true double homozygous anx1-2 anx2-2 plants. In all these plants, aniline-blue staining showed rescue of anx1-2 anx2-2 PT growth in vivo as well as normal targeting of unfertilized ovules (Fig. 1B). Backcrosses of the M1 mutants as pollen donor on the stigmas of the original anx1-2 anx2-2 plants systematically led to elongated siliques, showing that the rescue of PT growth was caused by mutation(s) in the haploid male gametophyte, thus defining the impotence rescued (ipr) class of mutants. Here, we report the identification of one of these ipr mutants, which we named “maris” (mri-3D; see below) after the Etruscan god of fertility and agriculture (23). The mri-3D suppressor mutation restores in vivo growth of anx1 anx2 PTs, resulting in filled siliques (43 ± 5.4 seeds for mri-3D compared with less than one seed for anx1 anx2 siliques) (Fig. 1B). To identify the EMS-generated causative lesion in mri-3D, likely a single nucleotide polymorphism (SNP), an SNP-ratio mapping (SRM) approach (24), was carried out with modifications (SI Material and Methods). Briefly, the SNPs called with Freebayes were fed to a hidden Markov chain-based algorithm that estimates the probability that the SNP is the causative SNP, based on the read data and linkage between the SNPs within each chromosome. For mri-3D/MRI, the top candidate exonic SNP had a ratio of alternate to reference reads of 0.476, very close to the theoretically expected ratio of 0.500 (Datasets S1 and S2) (24). The affected gene AT2G41970 encodes an uncharacterized RLCK, henceforth referred to as “MARIS” (MRI) (see below). MRI belongs to the family of RLCK-VIII homologs of the tomato Pto-interacting protein 1 (Pti1) and is highly expressed in pollen as well as in roots and root hairs according to publicly available microarray data and previous transcriptomic studies (Fig. S1A and Table S1).
Fig. 1.
Identification of the ipr mutants, suppressors of anx1 anx2 male sterility, and characterization of maris-3D (mri-3D). (A) Scheme for the anx1 anx2 male sterility-suppressor screen. Reproduced from ref. 6. (B) Aniline blue staining reveals that mri-3D/MRI PTs are able to elongate in vivo and fertilize female gametophytes, unlike anx1 anx2 PTs, which burst and stop growth early in the style. White arrows indicate the tip of the longest PT. Asterisks indicate female gametophytes normally targeted by PTs. (Scale bar: 200 μm.) (C) The mri-3D mutant carries a C718T SNP in AT2G41970 that corresponds to an R240C mutation at the protein level. Shown are spectra from the Sanger sequencing reactions of the AT2G41970 cDNA amplified from anx1 anx2 (Upper) or mri-3D anx1 anx2 (Lower) flowers. (D) AT2G41970 locus with introns, exons, and positions of the mutant alleles. (E) In vitro PT growth assays of WT, anx1 anx2, heterozygous mri-3D/MRI anx1 anx2, and homozygous mri-3D/mri-3D anx1 anx2. Note how the mri-3D mutation partially rescues the anx1 anx2 pollen-bursting phenotype and allows PT growth. See Fig. S2C for corresponding quantification. (Scale bar: 100 μm.) (F) In vitro PT growth assays of untransformed anx1 anx2 and three independent anx1 anx2 transgenic lines expressing MRIR240C-CFP. DIC and CFP channels were overlaid for the transgenic lines. (Scale bar: 80 μm.)
Fig. S1.
Sequence alignment and phylogenic tree of the Arabidopsis RLCK-VIII homologs of the tomato Pto-interacting protein 1 to which MRI belongs. (A) Multiple alignments of Arabidopsis RLCK-VIII proteins were performed with ClustalW 2.0, and the phylogenetic tree was reconstructed with MEGA6 using the protein sequence parsimony method (bootstrap test, 1,000 replicates). Black and gray circles at nodes indicate bootstrap values of more than 900 and between 800 and 900, respectively. Then the tree was combined with the relative gene expression of Arabidopsis RLCK-VIII family members in various plant tissues according to the Genevestigator microarray database using the Meta-Profile Analysis tool, Anatomy Profile (45). Note the strong and preferential expression of MRI in pollen and roots (bracket). See Table S1, for more detailed gene-expression data in pollen and root hairs. (B) Multiple alignments of Arabidopsis RLCK-VIII protein sequences around the conserved STR motifs were performed with ClustalW 2.0.
Table S1.
Expression data from previous studies based on RNA-seq (46) or Affymetrix Arabidopsis ATH1 arrays (47, 48) for CrRLK1L-related pathways in tip-growing cells
 |
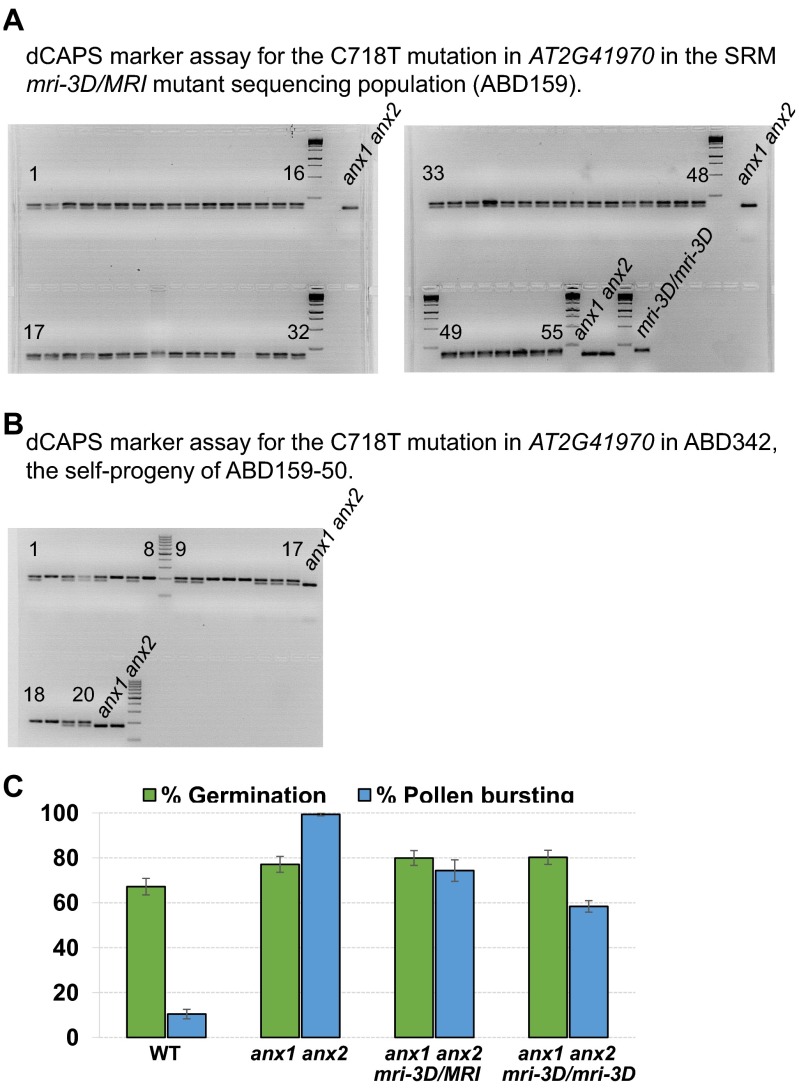
The putative causative SNP in mri-3D is a C-to-T transition at position 718 of the coding sequence of MRI that leads to a nonsynonymous substitution of a perfectly conserved arginine into a cysteine (R240C) in the activation loop of the kinase (Fig. 1C and Fig. S1B). All mutant plants used for the SRM approach were heterozygous for this SNP (Fig. S2A). mri-3D/MRI also displayed partial rescue of the anx1 anx2 PT-bursting phenotype in vitro (Fig. 1E and Fig. S2C). All the 20 progenies of selfed mri-3D/MRI were either heterozygous (n = 12), or homozygous (n = 8) for the causative SNP in MRI, suggesting that this mutation confers a reproductive advantage relative to the WT allele in the anx1 anx2 background (Fig. S2B). As expected, in mri-3D/mri-3D plants, roughly twice as many PTs were rescued from bursting in anx1 anx2 plants as in mri-3D/MRI plants (Fig. 1E and Fig. S2C).
Fig. S2.
(A) dCAPS marker assay for the C718T mutation in AT2G41970 in the mri-3D/MRI mutant sequencing population (ABD159). (B) dCAPS marker assay for the C718T mutation in AT2G41970 in population ABD342, the self-progeny of ABD159-50. Note that the gDNA of plant ABD159-13 was used as a control and labeled mri-3D/mri-3D in A. (C) Quantification of in vitro germination and bursting rates for WT, anx1 anx2, mri-3D/MRI anx1 anx2, and mri-3D/mri-3D anx1 anx2 plants. Data are mean ± SEM of three independent experiments with more than 150 pollen grains per genotype and experiment.
To confirm that the mutant form MRIR240C was indeed responsible for the suppression of PT bursting in anx1 anx2 plants, anx1/anx1 anx2/ANX2 plants were transformed with MRIR240C-CFP and MRI-CFP fusion constructs under the control of the pollen-specific promoter Lat52 (25) and were compared with previous complementation experiments with ANX1-YFP and ANX2-YFP (6). In the progeny of selfed anx1/anx1 anx2/ANX2 plants with high MRI-CFP expression levels, no homozygous anx1 anx2 individuals could be recovered (n = 85) (Table S2). In contrast, homozygous anx1 anx2 individuals could be identified in the self-progeny of anx1/anx1 anx2/ANX2 plants with high MRIR240C-CFP expression levels (n = 13 of 88), similar to the ANX1-YFP and ANX2-YFP fusion proteins (n = 13 of 87) (Table S2). Moreover, seed set for three independent MRIR240C-CFP transgenic lines homozygous for anx1 anx2 was 14.6 ± 4.4, 14.2 ± 2.8, and 14.6 ± 3.2 on average, as opposed to less than one seed from untransformed anx1 anx2 plants. These results indicate that MRIR240C-CFP, but not MRI-CFP, can partially rescue anx1 anx2 male sterility in vivo. Furthermore, unlike untransformed anx1 anx2 plants, pollen of homozygous anx1 anx2 plants expressing MRIR240C-CFP was able to produce some PTs in vitro, albeit at a lower rate than by the original mri-3D/MRI mutant (Fig. 1F, three independent lines). These results demonstrate that the MRIR240C mutant protein is indeed responsible for the partial rescue of the anx1 anx2 PT-bursting phenotype observed in the original mri-3D mutant and that MRIR240C exerts a dominant effect over the WT form.
Table S2.
Segregation analysis by PCR-based genotyping of the self-progeny of anx1/anx1 anx2/ANX2 transformed with pLAT52-MRI-CFP, pLAT52-MRIR240C-CFP, pACA9-ANX1-YFP, and pACA9-ANX2-YFP or untransformed
MRI Is a Positive Component of the ANX1/2 Pathway.
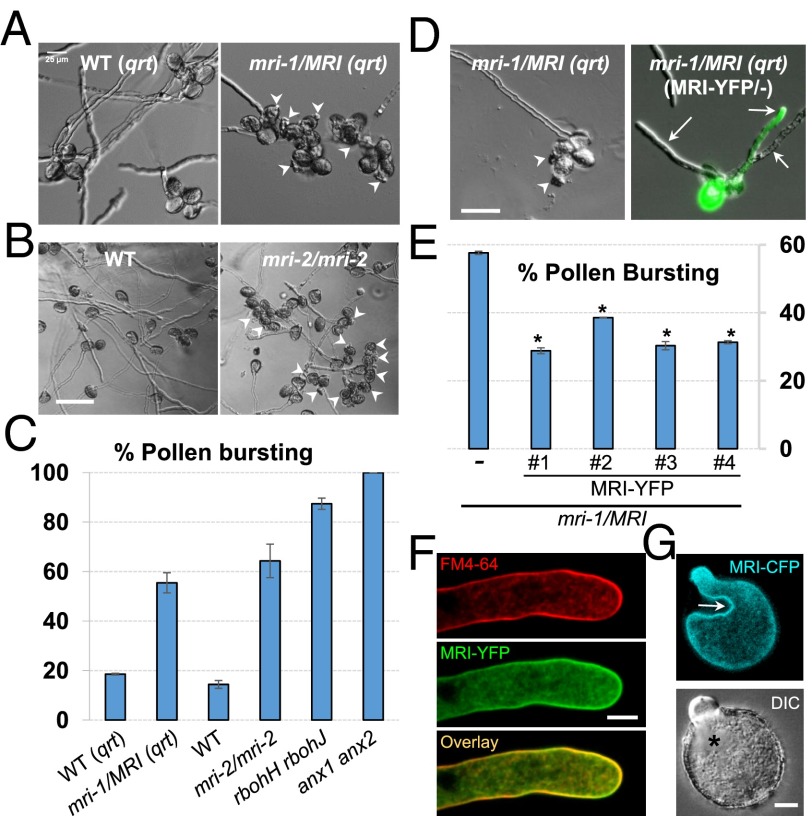
To understand the role of MRI further, we looked for insertion lines that potentially could disrupt MRI function. Two lines were recovered, CSHL_GT21229 (gene trap line, Ds transposon, Ler), renamed “mri-1,” and GABI_820D05 (T-DNA insertion, Col-0), renamed “mri-2” (Fig. 1D). Although homozygous mri-2 individuals could be retrieved from selfed mri-2/MRI plants, albeit less often than expected, no homozygous mri-1 individuals were identified (n = 353; Table S3). Moreover, both distortion of the segregation ratio and analysis of transmission efficiency (TE) showed that both mri-1 and mri-2 were normally transmitted through the female gametophyte but showed no or reduced transmission, respectively, through the male gametophyte (Table S4). To understand how the mri-1 and mri-2 alleles affect pollen development, in vitro pollen germination assays were carried out. Because mri-1/MRI was originally in the Ler accession that is recalcitrant to in vitro pollen germination assays, mri-1/MRI was introgressed into the Col-0 background carrying the quartet (qrt) mutation (26) (SI Materials and Methods). Unlike WT segregants, germinated PTs from mri-1/MRI quartets either burst (n = 194) or grew normally (n = 160) in a 1:1 ratio, as expected for the segregating mri-1 allele (two-tailed Fisher’s exact test, P = 0.2286) (Fig. 2A). Moreover, 64.3% of PTs from mri-2/mri-2 plants (vs. 14.6% of PTs from WT plants) lost integrity and burst in germination assays, similar to the percentages of bursting PTs from homozygous rbohH rbohJ (87.4%) and anx1 anx2 (100%) plants (Fig. 2 B and C). Next, we transformed mri-1/MRI plants with pLat52-MRI-YFP and measured PT bursting in four independent mri-1/MRI transgenic T1 lines hemizygous for MRI-YFP. All the lines displayed significantly less PT bursting (30–39%) than untransformed mri-1/MRI plants (56%) (P < 0.001 for all lines, Student's t test) (Fig. 2 D and E). Moreover, out of 90 plants 29 homozygous mri-1/mri-1 individuals were identified in the subsequent generation, showing that MRI-YFP is functional.
Table S3.
Segregation analysis of mri alleles by scoring herbicide/antibiotic resistance or PCR-based genotyping in the progeny derived from self-fertilization
| Genotype | MRI/MRI | mri/MRI | mri/mri | Ratios |
| mri-1/MRI | 183 | 170 | 0 | 1:0.93:0** |
| mri-2/MRI | 128 | 60 | 11 | 1:0.47:0.09** |
Statistically significant difference from the expected 1:2:1 ratio; P < 0.0001 (two-tailed χ2 test).
Table S4.
Segregation analysis of mri alleles by PCR-based genotyping or scoring herbicide resistance of the progeny resulting from reciprocal crosses with the wild type (Col-0)
| Female × male | MRI/MRI (a) | mri/MRI (b) | TE, % |
| mri-1/MRI × Col-0 | 92 | 89 | 96.7 |
| Col-0 × mri-1/MRI | 1,005 | 0 | 0** |
| mri-2/MRI × Col-0 | 50 | 46 | 92 |
| Col-0 × mri-2/MRI | 152 | 82 | 53.9* |
TE, transmission efficiency: TE = (b/a) × 100%. Asterisks denote a significant difference from the expected 1:1 ratio for normal Mendelian transmission: ** P < 0.001, * P = 0.0014 (two-tailed exact Fisher’s test).
Fig. 2.
MARIS is a positive component of the CW-integrity signaling pathway. (A) In vitro PT growth assays of WT and mri-1/MRI in the qrt background. Arrowheads point to rupturing PTs discharging cytoplasm into the medium. (Scale bar: 25 μm.) (B) In vitro PT growth assays of WT and mri-2/mri-2; arrowheads point to cytoplasmic discharge. (Scale bar: 100 μm.) (C) Quantification of PT rupture in vitro for WT (qrt), mri-1/MRI (qrt), WT, mri-2, rbohH rbohJ, and anx1 anx2. (D) Pollen tetrad of untransformed mri-1/MRI (qrt) and mri-1/MRI (qrt) hemizygous for pLAT52-MRI-YFP. Arrowheads and arrows point to cytoplasmic discharge and PT, respectively. DIC and YFP channels were overlaid for the transgenic line. (Scale bar: 50 μm.) (E) Four independent T1 lines of mri-1/MRI (qrt) hemizygous for pLAT52-MRI-YFP display significantly less bursting than untransformed mri-1/MRI (qrt) (-). Asterisks indicate significant differences from the untransformed control (P < 0.001, Student's t test). (F) Median plane of a normally growing PT expressing MRI-YFP. Before imaging, PTs were treated with liquid germination medium containing FM4-64 (2 µM) for 5 min. (Scale bar: 5 µm.) (G) An early arrested PT overexpressing MRI-CFP showing membrane invagination (arrow) and overaccumulation of CW material (asterisk). (Scale bar: 5 µm.)
Although MRI does not possess a transmembrane domain, MRI-YFP localized to the plasma membrane of growing PTs, possibly through palmitoylation (Fig. 2F). However, unlike plasma membrane localization of ANX1/2 or RbohH, which shows enrichment at the PT tip (6), MRI-YFP was localized uniformly in the plasma membrane, similar to other pollen-expressed RLCKs (27). Taken together, our results indicate that mri-1, mri-2, and mri-3D are amorphic, hypomorphic, and hypermorphic alleles of MRI, respectively, and that disruption of MRI leads to a loss of CW integrity during PT growth. Thus, MRI is a positive component of the CW integrity pathway.
Overexpression of the MRIR240C Mutant Form Strongly Inhibits WT Pollen Germination.
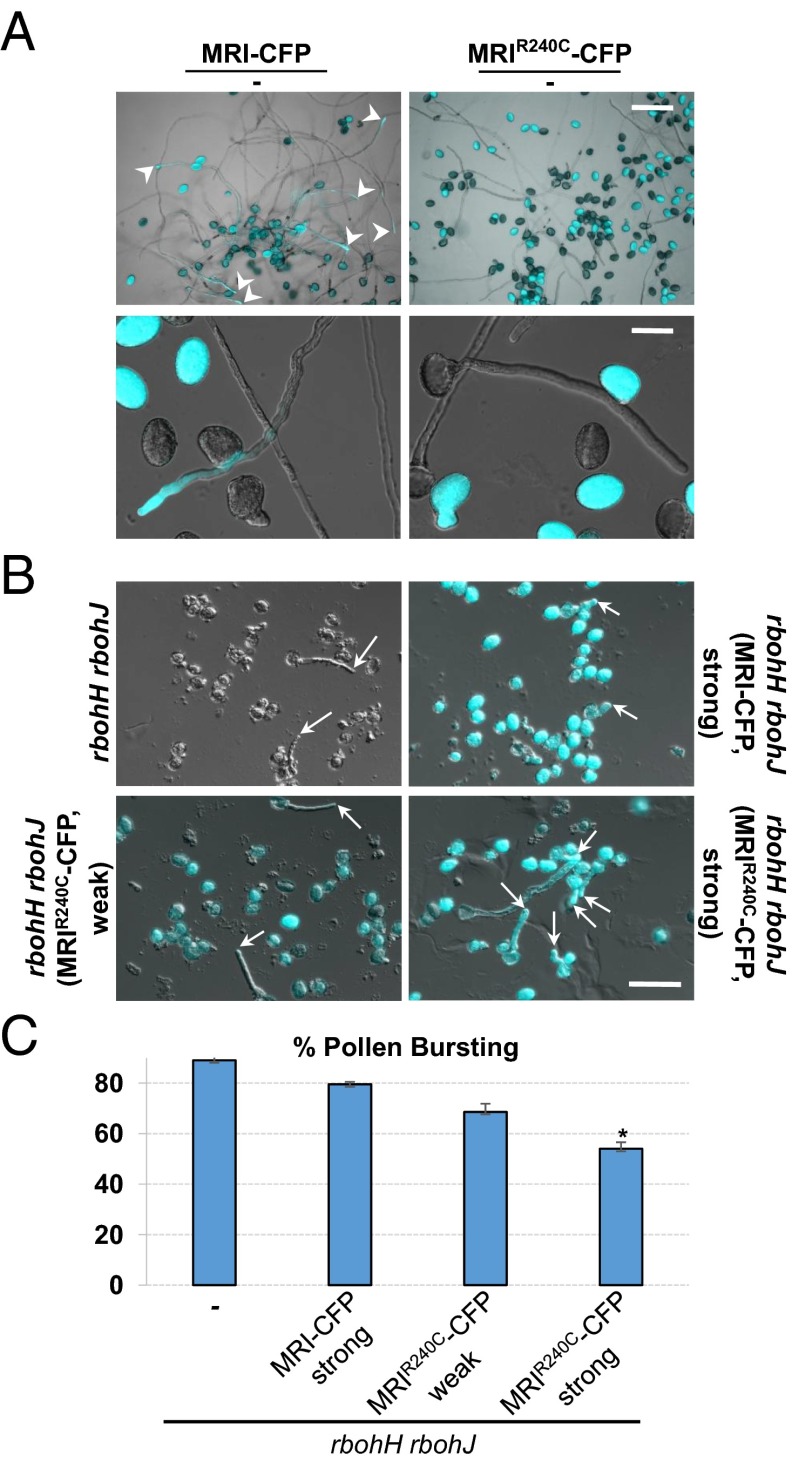
To study the effect of the R240C mutant form further, WT plants were transformed with both MRI-CFP and MRIR240C-CFP. Previously, we reported that overexpression of ANX1 mildly inhibits pollen germination and PT growth, most likely by triggering overaccumulation of secreted CW material (6). In transgenic lines with strong expression of MRI-CFP, only a few pollen grains displayed membrane invaginations and CW accumulation (n = 31 of 444 fluorescent PTs) (Fig. 2G). The occurrence of this phenotype was similar in lines strongly expressing GFP-RbohH (6) but was much lower than in the ANX1-OX lines (n = 65 of 133). This result suggests that the levels of MRI (and RbohH) do not constitute a rate-limiting step in the pathway. Furthermore, 32 homozygous individuals could be identified among 102 Basta-resistant T2 progenies expressing MRI-CFP (three independent transgenic lines). In contrast, no homozygous individual could be retrieved among 106 Basta-resistant T2 progenies expressing the MRIR240C-CFP fusion (three independent transgenic lines), suggesting that MRIR240C-CFP PTs cannot effect fertilization. To investigate this hypothesis further, we compared in vitro germination and growth of pollen from transgenic lines hemizygous for MRIR240C-CFP and MRI-CFP. For the latter, MRI-CFP–expressing and nonfluorescent WT PTs were observed in a 1:1.64 ratio, which is significantly different from the expected 1:1 ratio for plants hemizygous for a PT-expressed marker (P = 0.0022, two-tailed Fisher’s exact test, n = 330) ( Fig. 3A), suggesting that MRI-CFP partially inhibits pollen germination. Remarkably, only two very small fluorescent bulges were observed for plants hemizygous for MRIR240C-CFP (P < 0.0001, two-tailed Fisher’s exact test, n = 240) (Fig. 3A). This finding was confirmed further by the decreased or almost absent male TE of the MRI-CFP and MRIR240C-CFP transgenes, respectively (TEmale for MRI-CFP = 55.7%, n = 503; TEmale for MRIR240C-CFP = 5.7%, n = 258). Our results show that, in a WT background, overexpression of MRI and MRIR240C slightly or strongly inhibits pollen germination, respectively, and that they are detrimental to fertilization. These data further indicate that MRIR240C is more active than WT MRI, at least in rescuing anx1 anx2 sterility and inhibiting PT emergence in a WT background.
Fig. 3.
MRIR240C-CFP strongly inhibits WT pollen germination and partially rescues the rbohH rbohJ bursting phenotype. (A) In vitro growth assays with WT pollen hemizygous for either MRI-CFP (Left) or MRIR240C-CFP (Right). DIC and CFP channels were overlaid. (Upper) An overview of the growing PTs. Note that, unlike MRIR240C-CFP–expressing PTs (Right), MRI-CFP expressing PTs (Left) are observed frequently (arrowheads). (Lower) Close-up pictures of growing pollen expressing MRI-CFP (Left) and very rarely germinating pollen expressing MRIR240C-CFP (Right). (Scale bars: 100 μm, Upper; 30 μm, Lower.) (B) In vitro growth assays for untransformed rbohH rbohJ pollen (Upper Left) and transgenic rbohH rbohJ pollen strongly expressing MRI-CFP (Upper Right) or weakly (Lower Left) or strongly (Lower Right) expressing MRIR240C-CFP. DIC and CFP channels were overlaid. Strongly expressed MRIR240C-CFP significantly rescued the rbohH rbohJ bursting phenotype in vitro. (Scale bar: 30 μm.) (C) Quantification of the bursting phenotype relative to B. The asterisk denotes a significant difference compared to untransformed rbohH rbohJ PTs (−) (P < 0.001, two-tailed unpaired Student's t test).
MRI Functions Downstream of ROS-Producing NADPH Oxidases.
To position MRI in the ANX1/2 signaling pathway, we tested whether overexpression of MRI and MRIR240C could rescue partially male-sterile rbohH rbohJ mutant plants (6). All 32 independent T1 lines of rbohH rbohJ with MRI-CFP fluorescence remained partially sterile (e.g., for three independent T1 lines with high CFP expression, seed set was 8.5 ± 3.1, 8.8 ± 3.9, and 9.4 ± 4.2) similar to untransformed rbohH rbohJ plants (7.2 ± 2.1). In contrast, all of the 32 independent T1 lines of rbohH rbohJ with MRIR240C-CFP fluorescence produced elongated siliques with many more seeds (e.g., for three independent T1 lines, seed set was 23 ± 3.5, 28.6 ± 4.1, and 30.2 ± 4.9). In the next generation, we recovered a weak and a strong expressor of MRIR240C-CFP, both of which were fertile despite the rbohH rbohJ mutations (seed set of 25.6 ± 3.9 and 32.2 ± 3.7, respectively), and we compared their in vitro PT bursting rates with that of a strong MRI-CFP expressor (seed set of 4.6 ± 3.9). PTs from rbohH rbohJ plants homozygous for MRI-CFP or untransformed controls burst more frequently in vitro than PTs from rbohH rbohJ fertile plants homozygous for MRIR240C–CFP, although only the bursting rate of the strong MRIR240C–CFP line was significantly lower (P < 0.001, two-tailed unpaired Student's t test) (Fig. 3 B and C). These results show that hyperactive MRIR240C, but not WT MRI, can partially rescue rbohH rbohJ male sterility in vivo and in vitro. Thus, MRI acts downstream of both the ANX-RLKs and the NADPH oxidases to control CW integrity in PTs.
MRI Functions Downstream of FER in Root-Hair Growth Control.
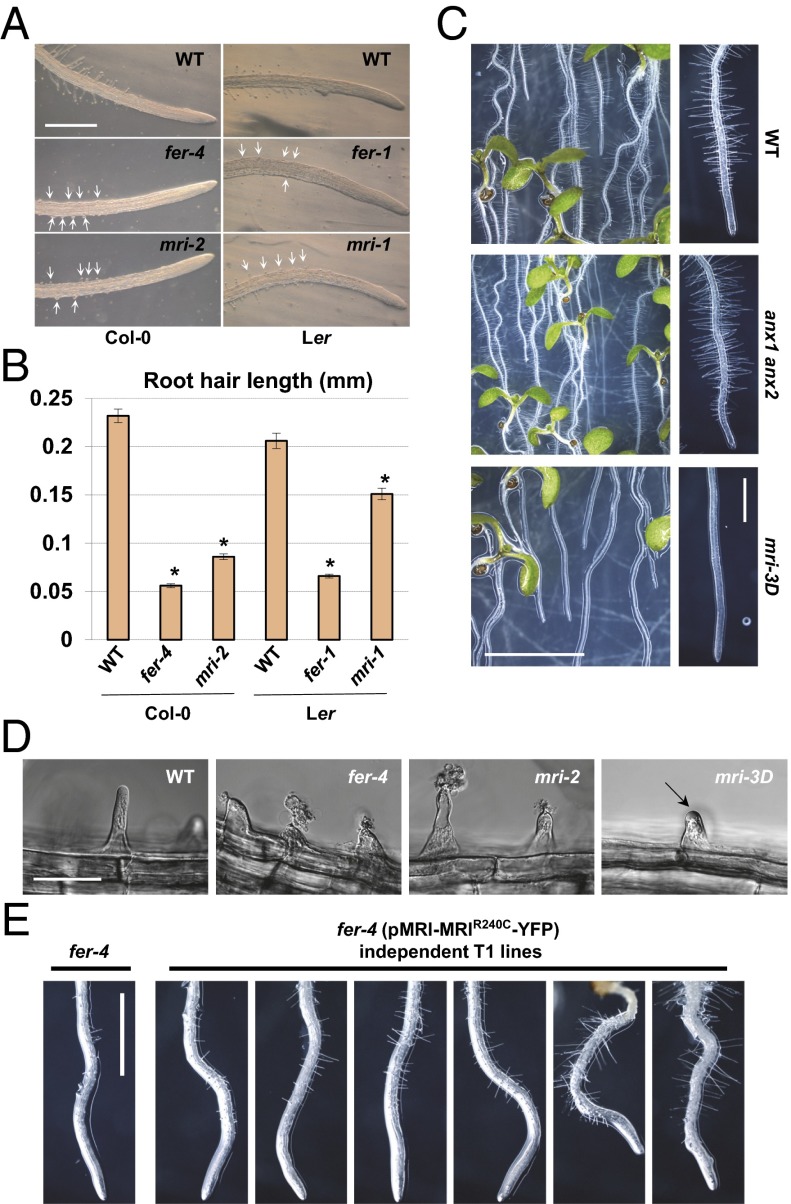
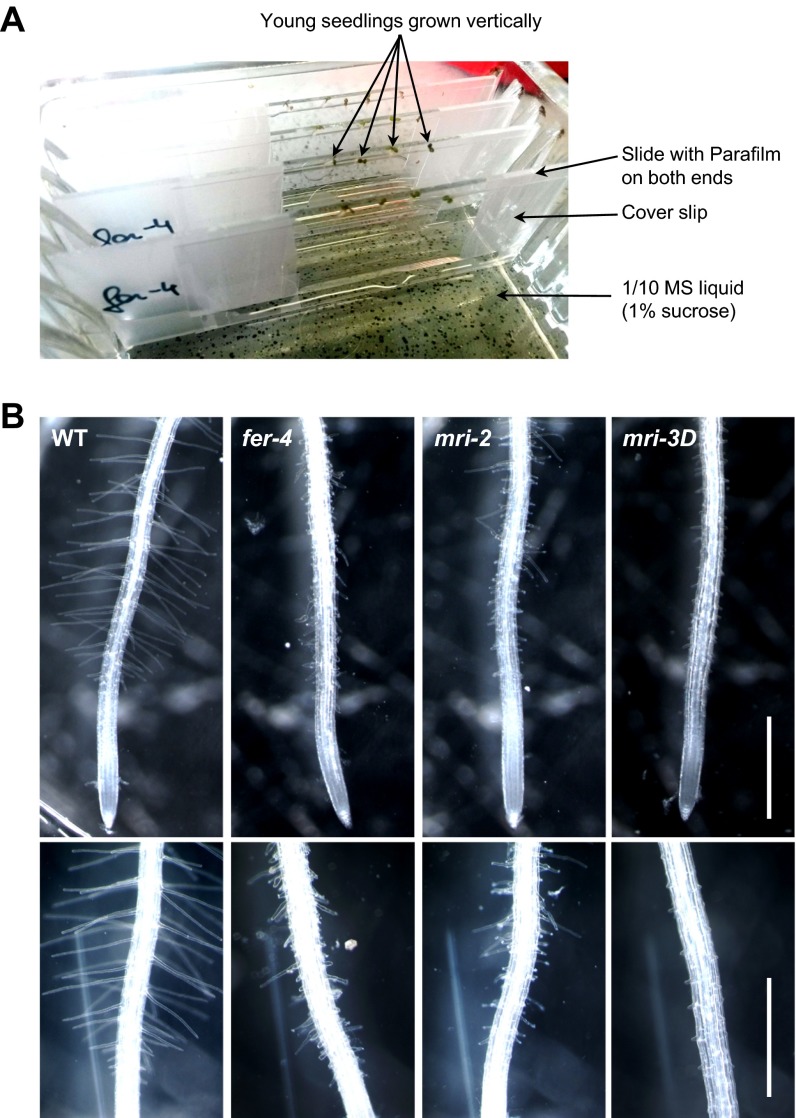
Unlike ANX1/2 and RbohH/J, MRI displays significant expression in root hairs, another plant cell type with tip growth (Table S1). Because it was reported that FER and RbohC (analogous to ANX1/2 and RbohH/J) functions to maintain root-hair integrity during growth (19), we tested whether MRI could also play a role in root-hair growth. First, we measured root-hair length in 5-d-old seedlings grown in agar and homozygous for mri-2 (vs. WT Col-0) and in seedlings homozygous for mri-1 whose male sterility was complemented with the pollen-preferential expression of pACA9-MRI-YFP (vs. WT Ler; see SI Materials and Methods). Both the mri-1 and mri-2 alleles showed much shorter root hairs than their respective WT controls (Fig. 4 A and B), similar to, but not as short as, the root hairs of plants homozygous for the fer-1 (Ler) (11) and fer-4 (Col-0) (19) alleles. Surprisingly, the strong mri-1 allele displayed a weaker phenotype in root hairs than the weak mri-2 allele. However, the pACA9 promoter also drives weak expression in root hairs (Table S1). It is possible that weak, undetectable expression of MRI-YFP in mri-1 root hairs may partially complement the short-root-hair phenotype. Moreover, in contrast to anx1 anx2 and WT seedlings grown on microagar, mri-3D seedlings expressing the MRIR240C protein in the anx1 anx2 background did not have any elongated root hairs (Fig. 4C). Microscopic observations of root hairs grown in liquid medium showed that, like fer-4 root hairs, mri-2 root hairs were bursting, whereas mri-3D short root hairs displayed overaccumulation of pectinaceous CW material (Fig. 4D, Figs. S3 and S4, and Movie S1). Taken together, our results show that MRI is essential for PT and root-hair CW integrity, whereas MRIR240C strongly inhibits the growth of both PTs and root hairs. Finally, independent T1 fer-4 seedlings expressing pMRI-MRIR240C-YFP displayed partial rescue of fer-4 root-hair phenotype (Fig. 4E), indicating that, in root hairs, MRI is a downstream component of FER signaling.
Fig. 4.
As in FERONIA, MARIS is required to sustain root-hair growth. (A) Main roots with root hairs of 5-d-old mri-2, fer-4, and WT (Col-0 accession) seedlings (Left) and mri-1, fer-1, and WT (Ler accession) seedlings (Right) grown in agar. Arrows point to short, defective root hairs. (Scale bar: 1 mm.) (B) Quantification of root-hair lengths relative to A. Asterisks denote significant differences versus the appropriate control (P < 0.001, two-tailed unpaired Student's t test). (C, Left) WT, anx1 anx2, and mri-3D anx1 anx2 seedlings growing on the surface of a microagar plate. (Right) Pictures of primary roots and their root hairs. (Scale bars: 5 mm, Left; 1 mm, Right.) (D) Young root hairs in the root elongation zone of WT, mri-2, and mri-3D anx1 anx2 seedlings grown in liquid medium. Note the loss of CW integrity or CW overaccumulation (arrow) for mri-2 and mri-3D root hairs, respectively. (Scale bar: 20 μm.) See also Figs. S3 and S4 and Movie S1. (E) Main root of untransformed fer-4 seedling and fer-4 T1s expressing pMRI-MRIR240C-YFP. (Scale bar: 1 mm.)
Fig. S3.
(A) Experimental set-up for root-hair growth assays in liquid medium. Four-day-old seedlings grown vertically on half-strength MS microagar plates were sandwiched between a slide and a coverslip containing 1/10th strength MS liquid medium (1% sucrose). Young root hairs in the elongation zone were observed 48 h later. (B) Representative roots of WT, fer-4, mri-2, and mri-3D anx1 anx2 seedlings grown in liquid medium relative to Fig. 4D. (Scale bar: 2 mm.)
Fig. S4.
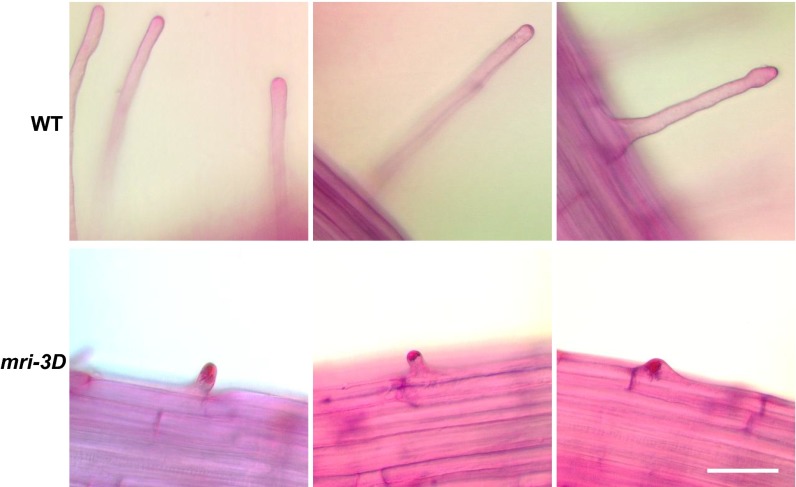
Ruthenium red staining reveals overaccumulation of cell wall material in mri-3D short root hairs. Roots of 4-d-old WT (Upper) and mri-3D anx1 anx2 (Lower) seedlings were allowed to grow for 48 h in liquid medium before being treated for 1 min with 0.01% Ruthenium red that stains for acidic pectin. Note that staining is restricted to the tip surface of WT root hairs but accumulates within the short root hairs of mri-3D anx1 anx2 plants. (Scale bar: 100 µm.)
SI Materials and Methods
Plant Material, Growth Conditions, and Genotyping of Mutants.
Plant growth conditions and transformation were as reported previously (1). All plants used in this study are from the Columbia (Col-0) accession except when otherwise stated. All primers used in this study are listed in Table S6. The anx1-2 anx2-2 mutant and the pACA9-ANX1-YFP binary vector were described previously (3). The rbohH-3 rbohJ-3 mutant was described previously (6). Genotyping PCR reactions were performed with primer pairs ABD681/ABD682 and ABD682/ABD747 for mri-1 and ABD735/ABD736 and ABD735/ABD614 for mri-2. To confirm and follow the causative SNP (C718T in AT2G41970) in mri-3D/MRI and subsequent generations, the derived cleaved amplified polymorphic sequence (dCAPS) primer pair ABD724/ABD725 was used. After XhoI digestion, PCR amplicons from WT, mri-3D/mri-3D, and mri-3D/MRI plants give rise to a 172-bp-long fragment, a 200-bp-long fragment, and both fragments, respectively. The original mutant mri-1/MRI (Ler) was outcrossed as a female recipient four consecutive times with pollen from WT Col-0 harboring qrt mutation. Then MRI/MRI and mri-1/MRI segregants were maintained separately for further pollen germination assays or transformation. To obtain homozygous mri-1 plants for root-hair assays, mri-1/MRI (Ler, kanamycin-resistant) plants were transformed with pACA9-MRI-YFP (see below). Three independent mri-1/MRI T1 lines showing high MRI-YFP expression in pollen were selected for investigating the occurrence of homozygous mri-1/mri-1 plants in their T2 progenies. Out of 69 kanamycin-resistant progenies (>20 individuals per line), homozygous (n = 26) and heterozygous (n = 43) mri-1 plants were found in a 1:2 ratio, as expected for complementation of mri-1 in pollen by MRI-YFP (P = 0.722, two-tailed exact Fisher’s test). The progenies of mri-1/mri-1 homozygous for pACA9-MRI-YFP were used for the root-hair-length assays. Note that no YFP-derived fluorescence was observed in the root hairs. Seed sets reported in the main text are all with n > 12 siliques per plant or genotype, ± SD. Aniline blue staining was performed as previously described (3).
Table S6.
Oligonucleotides used in this study
| Name | Sequence (5′-3′) | Forward or Reverse | Purpose |
| ABD724 | GCATATGGAGCAGCCAAAGG | F | dCAPS primer for mri-3D |
| ABD725 | TGGTAGCCGAATGTTCCCAAAACTC | R | dCAPS primer for mri-3D |
| ABD681 | TTCGGCTACCACGCTCCAGA | F | Genotype mri-1 (CSHL_GT21229) |
| ABD682 | GGACCGGCCGGTTTAGAGTT | R | Genotype mri-1 (CSHL_GT21229) |
| ABD747 | TCCGTTCCGTTTTCGTTTTTTAC | Ds5-2 | Genotype mri-1 (CSHL_GT21229) |
| ABD687 | GTCGACATGTTTTGTTGCGGTGGTGC | F | To clone MRI SalI/SpeI in pJet1.2 |
| ABD688 | GTATATTGTTACTAGTCCGGACGTAGACTC | R | To clone MRI SalI/SpeI in pJet1.2, no stop |
| ABD735 | GTTCTATTCTTCGACCAAATGG | F | Genotype mri-2 (GABI_820D05) |
| ABD736 | CTGCATACTGGTTTGCGGG | R | Genotype mri-2 (GABI_820D05) |
| ABD614 | GGGCTACACTGAATTGGTAGCTC | LBgabi2 | Genotype mri-2 (GABI_820D05) |
| ABD737 | GGGGACAAGTTTGTACAAAAAAGCAGGCTTAATGTTTTGTTGCGGTGGTGC | F | To clone MRI and MRIR240C in pDONR207 |
| ABD738 | GGGGACCACTTTGTACAAGAAAGCTGGGTAGGACGTAGACTCAGGACCGG | R | To clone MRI and MRIR240C in pDONR207, no stop |
| ABD762 | GAAATAAGAGCTCATTAAATTCAAACGG | F | To clone pMRI in pABD34 |
| ABD763 | CACCACCGCAACTAGTCATATCTTCAG | R | To clone pMRI in pABD34 |
Underlined nucleotides indicate mutations. Bold nucleotides indicate restriction sites.
Identification of ipr Mutants.
Approximately 23,500 seeds of anx1-2 anx2-2 were treated with 0.25% (wt/vol) EMS (Sigma Aldrich) and were sown uniformly on a total of 49 trays filled with ready-to-use soil (ED73; Universal Erde) along with a tray of untreated anx1 anx2 seeds. There was ∼60% lethality among the mutagenized seeds. After 3 d at 4 °C, plants were grown in growth chambers at 22 °C, 60% humidity with a 16-h light/8-h dark photoperiod regime at ∼75 μmol⋅m−2⋅s−1. Plants were grown until they set seeds. Sterile plants were discarded, and those carrying elongated siliques were kept and used for several tests. First, they were genotyped to ascertain that they were true double anx1 anx2 homozygous plants. Second, they were used as male donors for backcrossing on untreated anx1 anx2 plants. Third, some of their elongated siliques were subjected to aniline blue staining to confirm the rescue of the PT bursting phenotype in vivo. Finally, their elongated-silique phenotype was monitored in the M2 generation.
SNP Ratio Mapping of the mri-3D Mutant.
To identify the EMS-generated causative mri-3D suppressor mutation or SNP, an SRM approach with modifications was carried out (24). Briefly, this method is based on the observation that, after two rounds of backcrossing from the M1 mutant to the parental line anx1 anx2, unlinked EMS-generated SNPs are either lost or maintained in a 1:3 ratio, whereas the causative SNP is conserved in a 1:1 ratio. The M1 mri-3D/MRI plant was backcrossed as a pollen donor twice successively on the original anx1 anx2 plant, and 65 F1 plants from the second backcross were grown. Of the 65 plants, 63 displayed the elongated-silique phenotype, thereby showing the strong beneficial effect of the mri-3D mutation in an anx1 anx2 background. Genomic DNA of 55 plants with elongated siliques was extracted with the DNeasy Plant Mini kit (Qiagen) and pooled. The Functional Genomic Center of the University of Zürich prepared the Illumina-adapted library and sequenced mri-3D/MRI with the pair-end Illumina HiSeq2000 platform. The raw sequencing data were mapped to the TAIR10 reference genome with NGM mapper (cibiv.github.io/NextGenMap/)(39). The unambiguously mapped reads with a mapping quality score greater than 20 were subject to duplicate removal (40) (samtools; www.htslib.org/) and overlap clipping (41) (bamtools; github.com/pezmaster31/bamtools). The SNPs were called with Freebayes (42) (https://github.com/ekg/freebayes) with the lower threshold of three alternative reads per locus. The obtained data were fed to a hidden Markov chain-based algorithm, which estimated the likelihood of the SNP given the read data, assuming linkage between the SNPs within each chromosome. The SNPs were classified into intronic, intergenic, coding (synonymous and nonsynonymous amino acid substitutions), and splice-site variants with Ensemble Variant Effect Predictor (www.ensembl.org/info/docs/tools/vep/index.html). Dataset S2 is the list of 259 filtered EMS-generated SNPs with more than seven alternative reads and a ratio of alternative-to-reference reads between 0.05 and 0.75, ranked by probability of being the causative SNP. Dataset S1 is the same list filtered for SNPs occurring only in exonic regions (a total of 115 SNPs).
Stable MRI-CFP, MRI-YFP, MRIR240C-CFP, and MRIR240C-YFP Protein Fusion Expression in Arabidopsis PTs and Root Hairs.
To generate the pACA9-MRI-YFP construct, full-length MRI was amplified from WT cDNA flowers with the primer pairs ABD687 (introducing the SalI site) and ABD688 (introducing the SpeI site and removing the stop codon) using the Phusion DNA polymerase (Finnzymes). The 1.1-kb-long fragment was ligated into pJet1.2 (Thermo Scientific), sequenced, and then cloned into the SalI/SpeI sites of the binary ps779 [ACA9-promoter-TAP2(YFP)] vector that confers hygromycin resistance (43). The Gateway-compatible pLAT52::GW:YFP (pABD34) and pLAT52::GW:CFP (pABD35) destination vectors are derived from pB7YWG2 and pB7CWG2 (conferring Basta resistance) (44), whose 35S promoters were replaced by the LAT52 promoter using the HindIII/SpeI sites. Then, to generate the pLAT52-MRI-CFP and pLAT52-MRIR240C-CFP constructs, the Gateway-compatible primer pair ABD737/ABD738 was used to amplify MRI without a stop codon from cDNA of mri-3D/MRI flowers. The Phusion-amplified 1.1-kb fragment was cloned into pDONR207. Of 16 sequenced clones, 6 were wild-type MRI, and 10 were MRIR240C. Then both MRI and MRIR240C were remobilized into pABD34 and pABD35. The 1.1-kb-long MRI promoter was amplified with primers ABD762 and ABD763 and cloned into pJet1.2. After sequencing, the promoter was cloned into SacI-SpeI to replace the LAT52 promoter in pABD34, thereby generating the pMRI::GW:YFP plasmid, pABD83. Then, MRIR240C in pDONR207 was remobilized in pABD83, generating the pMRI-MRIR240C-YFP binary vector (pABD85). Finally, fer-4 plants were transformed with pABD85. Young Basta-resistant T1 seedlings were transferred on microagar plates without selection and allowed to grow for 2 d along with untransformed fer-4 before root-hair observations.
Discussion
During growth, any chemical or physical perturbation of the CW caused by developmental signals, abiotic stresses, and interactions with neighboring cells or microorganisms can potentially cause a cell to stop growing or lose its integrity. For the fast-growing PTs, this growth cessation would lead to a failure in fertilization, necessitating a robust signal transduction network to maintain CW integrity during tip growth. The ANX1/2 RLKs of the CrRLK1L subfamily and their closest homolog FER have been implicated in the control of tip growth in PTs and root hairs, respectively, acting upstream of ROS-producing NADPH oxidases (3, 4, 19). In this study, we identified MRI, a novel, positive downstream component of these RLK signaling pathways. MRI belongs to the Arabidopsis RLCK-VIII subfamily that shares homology with the tomato Pti1 protein involved in the Pto-mediated hypersensitive response (28). In tomato, the Pto kinase interacts with and transphorylates Pti1 in the amino acid motif STR at threonine 233; the biological relevance of this interaction awaits further studies (29). In Arabidopsis, four members of the RLCK-VIII subfamily, namely Pti1-1 (AT1G06700), Pti1-2 (AT2G30740), Pti1-3 (AT3G59350), and Pti1-4 (AT2G47060), interact with the AGC2 kinase OXIDATIVE SIGNAL-INDUCIBLE1 (OXI1) (30, 31). However, only Pti1-1, Pti1-2, and Pti1-4 appear to be phosphorylated by OXI1 (30, 31). Interestingly, Pti1-2 and OXI1 kinase activities are induced by ROS-generating stresses (30, 32). The Arabidopsis oxi1 mutant is more susceptible to pathogens and displays mild root-hair growth defects and less lignin deposition after CW damage (22, 32–34). However, to date no mutant phenotype has been reported for any Arabidopsis gene encoding a RLCK-VIII protein.
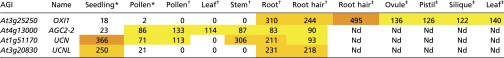
Here, we show that MRI, a member of the RLCK-VIII subfamily, is preferentially expressed in root hairs and PTs and localizes uniformly at the plasma membrane. Disruption of MRI leads to PT bursting similar to that observed in anx1 anx2 mutants and to root-hair bursting similar to that seen in fer homozygous plants. Furthermore, an R240C amino acid substitution in the activation loop of MRI is sufficient for partial rescue of both the anx1 anx2 and the rbohH rbohJ PT-bursting phenotypes and the fer-4 root-hair–bursting phenotype. Thus, although it is not known whether MRI kinase activity is activated by ROS similar to Pti1-2 (30), our data show that MRI controls CW integrity downstream of ANX1/2 and the NADPH oxidases in PTs and downstream of FER in root hairs. Because OXI1 also is involved in root-hair growth and can phosphorylate different members of the RLCK-VIII subfamily, OXI1 is likely to activate MRI during root-hair growth. In PTs, however, OXI1 expression levels are very low, and it seems more likely that either another member of this family, the AGC2 kinases (group VIII), or a combination of them fulfills this role (Table S5) (35–37). Whether CrRLK1Ls, NADPH oxidases, OXI1/AGC2 kinases, and MRI/Pti1-like proteins are all part of a linear pathway or, more likely, belong to different pathways that orchestrate complex cross-talk to sustain cell growth remains to be addressed. In this regard, the presence of a module composed of OXI/AGC2 kinases and MRI/Pti1-like RLCKs acting downstream of the CrRLK1Ls and NADPH oxidases could potentially allow the integration of inputs generated by biotic or abiotic stresses with the CW integrity pathway (36, 37).
Table S5.
Expression data from previous studies based on RNA-seq (46) or Affymetrix Arabidopsis ATH1 arrays (47, 48) for members of the AGC2 kinases subgroup VIII
 |
Interestingly, in all the different genetic backgrounds tested, MRIR240C is more efficient than the WT form in activating downstream responses. The question of how the R240C substitution makes MRI more active is intriguing, and several scenarios can be envisioned. First, this amino acid change in the core catalytic domain may enhance MRI kinase activity itself. Second, MRIR240C could be more resistant than MRI to either degradation or inactivation mechanisms. Third, the interaction of MRIR240C with upstream activators or downstream signaling components could be stronger than that of the WT form, resulting in more efficient activation of the downstream responses. In this respect it is noteworthy that, in tomato, Pto phosphorylates Pti1 at T233 in the conserved STR motif of RLCKs-VIII (29), but in Arabidopsis, OXI1 phosphorylates Pti1-2 at T238 (30). In MRI, the corresponding conserved threonine is located at position 239, just before R240, which is mutated in mri-3D (Fig. 1C and Fig. S1B). Therefore it is conceivable that the R240C substitution in MRI facilitates phosphorylation at the neighboring T239 or partially mimics a phosphorylated T239, enabling MRIR240C to be more efficient than MRI in activating downstream components. Future studies combining biochemistry and rescue experiments of mutants in these signaling pathways will be needed to distinguish which of these scenarios best explains the phenotypes triggered by MRIR240C expression. We anticipate that engineering and expressing this type of mutation in the conserved STR motif could be very useful for studying the other unexplored RLCK-VIII–mediated signaling pathways.
Materials and Methods
Plant Material, Growth Conditions, and Genotyping of Mutant Alleles.
Mutant lines mri-1 and mri-2 with insertions in the MRI gene (At2g41970) were obtained from Cold Spring Harbor Laboratory and the European Arabidopsis Stock Center and correspond to CSHL_GT21229 (Ds element inserted 1,632 bp downstream of the start codon, Ler accession, kanamycin resistance) and GABI_820D05 (T-DNA insertion located 360 bp upstream of the start codon, Col-0 accession, sulfadiazine resistance), respectively (Table S6 and SI Materials and Methods).
In Vitro Pollen Growth Assays.
In vitro pollen growth assays were carried out as described previously (38). Briefly, freshly opened flowers were incubated at 22 °C for 30 min in moisture incubation boxes and then were brushed on slides containing germination medium [0.01% boric acid, 5 mM CaCl2, 5 mM KCl, 1 mM MgSO4, 10% (wt/vol) sucrose (pH 7.5), 1.5% (wt/vol) low-melting agarose]. The boxes were preincubated for 35 min at 30 °C and then were returned to 22 °C for several hours. To determine the percentage of bursting PTs, germinating pollen grains and PTs were imaged with a Leica DM6000 microscope and were analyzed using the ImageJ 1.47d software (rsb.info.nih.gov/ij). All data presented here are the mean ± SEM of three independent experiments with more than 150 pollen grains (or 30 tetrads for the qrt background) scored per genotype and experiment.
Analysis of Root-Hair Length.
Root hairs located 1.5–3.5 mm from the primary root tip of 5-d-old seedlings grown in half-strength MS medium (1% sucrose) were observed with a Leica MZ16F stereomicroscope. The length of ∼100 root hairs in focus from at least 18 seedlings per genotype was measured with ImageJ 1.47d. Data are presented as the mean ± SEM of three independent experiments with more than 30 root hairs scored per genotype and experiment. Alternatively, roots from 7-d-old seedlings grown on microagar plates were imaged. For microscopic root-hair observations, 4-d-old seedlings grown on half-strength MS microagar plates were sandwiched between a slide and a coverslip containing 1/10th strength MS liquid medium supplemented with 1% sucrose (Fig. S3). Emerging root hairs were observed 48 h later with a Leica DM5500 equipped with differential interference contrast (DIC) optics.
Supplementary Material
Acknowledgments
We thank all members of the U.G. and Martin Hülskamp laboratories for help with mutant screening and enriching discussions, in particular Rita Galhano, Sharme Thirugnanarajah, Sucharita Roy, and the Plant Sex Club; the Functional Genomics Center Zurich and its staff for deep-sequencing of the mri-3D/MRI mutant; Sharon Kessler (University of Oklahoma), Michael R. Sussman, and Miyoshi Haruta (University of Wisconsin) for fer seeds; Yan Zhang (Shandong Agricultural University) for plasmids; and Boris Voigt (University of Bonn) for fruitful suggestions about live imaging of root hairs. This work was supported by the University of Zurich; by grants from the University of Zurich’s Research Priority Program “Functional Genomics/Systems Biology,” the European Union through the Marie Curie International Reintegration Grant FP7-PEOPLE-2009-RG (project 249247) and the PLANTFELLOWS Grant GA-2010-267243, and the Deutsche Forschungsgemeinschaft Grant BO 4470/1-1 (to A.B.-D.); as well as by Swiss National Science Foundation Grant 31003A-112489; an interdisciplinary PhD project from SystemsX.ch; and European Research Council Grant AdG 250358 (to U.G.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1512375112/-/DCSupplemental.
References
- 1.Engelsdorf T, Hamann T. An update on receptor-like kinase involvement in the maintenance of plant cell wall integrity. Ann Bot (Lond) 2014;114(6):1339–1347. doi: 10.1093/aob/mcu043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wolf S, Hématy K, Höfte H. Growth control and cell wall signaling in plants. Annu Rev Plant Biol. 2012;63:381–407. doi: 10.1146/annurev-arplant-042811-105449. [DOI] [PubMed] [Google Scholar]
- 3.Boisson-Dernier A, et al. Disruption of the pollen-expressed FERONIA homologs ANXUR1 and ANXUR2 triggers pollen tube discharge. Development. 2009;136(19):3279–3288. doi: 10.1242/dev.040071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miyazaki S, et al. ANXUR1 and 2, sister genes to FERONIA/SIRENE, are male factors for coordinated fertilization. Curr Biol. 2009;19(15):1327–1331. doi: 10.1016/j.cub.2009.06.064. [DOI] [PubMed] [Google Scholar]
- 5.Lindner H, et al. TURAN and EVAN mediate pollen tube reception in Arabidopsis synergids through protein glycosylation. PLoS Biol. 2015;13(4):e1002139. doi: 10.1371/journal.pbio.1002139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boisson-Dernier A, et al. ANXUR receptor-like kinases coordinate cell wall integrity with growth at the pollen tube tip via NADPH oxidases. PLoS Biol. 2013;11(11):e1001719. doi: 10.1371/journal.pbio.1001719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boisson-Dernier A, Kessler SA, Grossniklaus U. The walls have ears: The role of plant CrRLK1Ls in sensing and transducing extracellular signals. J Exp Bot. 2011;62(5):1581–1591. doi: 10.1093/jxb/erq445. [DOI] [PubMed] [Google Scholar]
- 8.Lindner H, Müller LM, Boisson-Dernier A, Grossniklaus U. CrRLK1L receptor-like kinases: Not just another brick in the wall. Curr Opin Plant Biol. 2012;15(6):659–669. doi: 10.1016/j.pbi.2012.07.003. [DOI] [PubMed] [Google Scholar]
- 9.Cheung AY, Wu H-M. THESEUS 1, FERONIA and relatives: A family of cell wall-sensing receptor kinases? Curr Opin Plant Biol. 2011;14(6):632–641. doi: 10.1016/j.pbi.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 10.Ngo QA, Vogler H, Lituiev DS, Nestorova A, Grossniklaus U. A calcium dialog mediated by the FERONIA signal transduction pathway controls plant sperm delivery. Dev Cell. 2014;29(4):491–500. doi: 10.1016/j.devcel.2014.04.008. [DOI] [PubMed] [Google Scholar]
- 11.Escobar-Restrepo J-M, et al. The FERONIA receptor-like kinase mediates male-female interactions during pollen tube reception. Science. 2007;317(5838):656–660. doi: 10.1126/science.1143562. [DOI] [PubMed] [Google Scholar]
- 12.Duan Q, et al. Reactive oxygen species mediate pollen tube rupture to release sperm for fertilization in Arabidopsis. Nat Commun. 2014;5:3129. doi: 10.1038/ncomms4129. [DOI] [PubMed] [Google Scholar]
- 13.Guo H, et al. Three related receptor-like kinases are required for optimal cell elongation in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2009;106(18):7648–7653. doi: 10.1073/pnas.0812346106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deslauriers SD, Larsen PB. FERONIA is a key modulator of brassinosteroid and ethylene responsiveness in Arabidopsis hypocotyls. Mol Plant. 2010;3(3):626–640. doi: 10.1093/mp/ssq015. [DOI] [PubMed] [Google Scholar]
- 15.Yu F, et al. FERONIA receptor kinase pathway suppresses abscisic acid signaling in Arabidopsis by activating ABI2 phosphatase. Proc Natl Acad Sci USA. 2012;109(36):14693–14698. doi: 10.1073/pnas.1212547109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shih H-W, Miller ND, Dai C, Spalding EP, Monshausen GB. The receptor-like kinase FERONIA is required for mechanical signal transduction in Arabidopsis seedlings. Curr Biol. 2014;24(16):1887–1892. doi: 10.1016/j.cub.2014.06.064. [DOI] [PubMed] [Google Scholar]
- 17.Kessler SA, et al. Conserved molecular components for pollen tube reception and fungal invasion. Science. 2010;330(6006):968–971. doi: 10.1126/science.1195211. [DOI] [PubMed] [Google Scholar]
- 18.Keinath NF, et al. PAMP (pathogen-associated molecular pattern)-induced changes in plasma membrane compartmentalization reveal novel components of plant immunity. J Biol Chem. 2010;285(50):39140–39149. doi: 10.1074/jbc.M110.160531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duan Q, Kita D, Li C, Cheung AY, Wu H-M. FERONIA receptor-like kinase regulates RHO GTPase signaling of root hair development. Proc Natl Acad Sci USA. 2010;107(41):17821–17826. doi: 10.1073/pnas.1005366107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haruta M, Sabat G, Stecker K, Minkoff BB, Sussman MR. A peptide hormone and its receptor protein kinase regulate plant cell expansion. Science. 2014;343(6169):408–411. doi: 10.1126/science.1244454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kessler SA, Lindner H, Jones DS, Grossniklaus U. Functional analysis of related CrRLK1L receptor-like kinases in pollen tube reception. EMBO Rep. 2015;16(1):107–115. doi: 10.15252/embr.201438801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Denness L, et al. Cell wall damage-induced lignin biosynthesis is regulated by a reactive oxygen species- and jasmonic acid-dependent process in Arabidopsis. Plant Physiol. 2011;156(3):1364–1374. doi: 10.1104/pp.111.175737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gerhard E. Über die Gottheiten der Etrusker. Druckerei der Königlichen Akademie der Wissenschaften; Berlin: 1847. [Google Scholar]
- 24.Lindner H, et al. SNP-Ratio Mapping (SRM): Identifying lethal alleles and mutations in complex genetic backgrounds by next-generation sequencing. Genetics. 2012;191(4):1381–1386. doi: 10.1534/genetics.112.141341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Twell D, Yamaguchi J, Wing RA, Ushiba J, McCormick S. Promoter analysis of genes that are coordinately expressed during pollen development reveals pollen-specific enhancer sequences and shared regulatory elements. Genes Dev. 1991;5(3):496–507. doi: 10.1101/gad.5.3.496. [DOI] [PubMed] [Google Scholar]
- 26.Preuss D, Rhee SY, Davis RW. Tetrad analysis possible in Arabidopsis with mutation of the QUARTET (QRT) genes. Science. 1994;264(5164):1458–1460. doi: 10.1126/science.8197459. [DOI] [PubMed] [Google Scholar]
- 27.Liu J, et al. Membrane-bound RLCKs LIP1 and LIP2 are essential male factors controlling male-female attraction in Arabidopsis. Curr Biol. 2013;23(11):993–998. doi: 10.1016/j.cub.2013.04.043. [DOI] [PubMed] [Google Scholar]
- 28.Zhou J, Loh Y-T, Bressan RA, Martin GB. The tomato gene Pti1 encodes a serine/threonine kinase that is phosphorylated by Pto and is involved in the hypersensitive response. Cell. 1995;83(6):925–935. doi: 10.1016/0092-8674(95)90208-2. [DOI] [PubMed] [Google Scholar]
- 29.Sessa G, D’ascenzo M, Martin GB. The major site of the Pti1 kinase phosphorylated by the Pto kinase is located in the activation domain and is required for Pto-Pti1 physical interaction. Eur J Biochem. 2000;267(1):171–178. doi: 10.1046/j.1432-1327.2000.00979.x. [DOI] [PubMed] [Google Scholar]
- 30.Anthony RG, Khan S, Costa J, Pais MS, Bögre L. The Arabidopsis protein kinase PTI1-2 is activated by convergent phosphatidic acid and oxidative stress signaling pathways downstream of PDK1 and OXI1. J Biol Chem. 2006;281(49):37536–37546. doi: 10.1074/jbc.M607341200. [DOI] [PubMed] [Google Scholar]
- 31.Forzani C, et al. The Arabidopsis protein kinase Pto-interacting 1-4 is a common target of the oxidative signal-inducible 1 and mitogen-activated protein kinases. FEBS J. 2011;278(7):1126–1136. doi: 10.1111/j.1742-4658.2011.08033.x. [DOI] [PubMed] [Google Scholar]
- 32.Rentel MC, et al. OXI1 kinase is necessary for oxidative burst-mediated signalling in Arabidopsis. Nature. 2004;427(6977):858–861. doi: 10.1038/nature02353. [DOI] [PubMed] [Google Scholar]
- 33.Anthony RG, et al. A protein kinase target of a PDK1 signalling pathway is involved in root hair growth in Arabidopsis. EMBO J. 2004;23(3):572–581. doi: 10.1038/sj.emboj.7600068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Petersen LN, Ingle RA, Knight MR, Denby KJ. OXI1 protein kinase is required for plant immunity against Pseudomonas syringae in Arabidopsis. J Exp Bot. 2009;60(13):3727–3735. doi: 10.1093/jxb/erp219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Enugutti B, et al. Regulation of planar growth by the Arabidopsis AGC protein kinase UNICORN. Proc Natl Acad Sci USA. 2012;109(37):15060–15065. doi: 10.1073/pnas.1205089109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rademacher EH, Offringa R. Evolutionary adaptations of plant AGC kinases: From light signaling to cell polarity regulation. Front Plant Sci. 2012;3:250. doi: 10.3389/fpls.2012.00250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Garcia AV, Al-Yousif M, Hirt H. Role of AGC kinases in plant growth and stress responses. Cell Mol Life Sci. 2012;69(19):3259–3267. doi: 10.1007/s00018-012-1093-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boavida LC, McCormick S. Temperature as a determinant factor for increased and reproducible in vitro pollen germination in Arabidopsis thaliana. Plant J. 2007;52(3):570–582. doi: 10.1111/j.1365-313X.2007.03248.x. [DOI] [PubMed] [Google Scholar]
- 39.Sedlazeck FJ, Rescheneder P, von Haeseler A. NextGenMap: Fast and accurate read mapping in highly polymorphic genomes. Bioinformatics. 2013;29(21):2790–2791. doi: 10.1093/bioinformatics/btt468. [DOI] [PubMed] [Google Scholar]
- 40.Li H, et al. 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barnett DW, Garrison EK, Quinlan AR, Strömberg MP, Marth GT. BamTools: A C++ API and toolkit for analyzing and managing BAM files. Bioinformatics. 2011;27(12):1691–1692. doi: 10.1093/bioinformatics/btr174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Erik G, Gabor M. 2012 Haplotype-based variant detection from short-read sequencing. Available at arxiv.org/pdf/1207.3907.pdf. Accessed May 14, 2015.
- 43.Myers C, et al. Calcium-dependent protein kinases regulate polarized tip growth in pollen tubes. Plant J. 2009;59(4):528–539. doi: 10.1111/j.1365-313X.2009.03894.x. [DOI] [PubMed] [Google Scholar]
- 44.Karimi M, De Meyer B, Hilson P. Modular cloning in plant cells. Trends Plant Sci. 2005;10(3):103–105. doi: 10.1016/j.tplants.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 45.Hruz T, et al. Genevestigator v3: A reference expression database for the meta-analysis of transcriptomes. Adv Bioinforma. 2008;2008:420747. doi: 10.1155/2008/420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Loraine AE, McCormick S, Estrada A, Patel K, Qin P. RNA-seq of Arabidopsis pollen uncovers novel transcription and alternative splicing. Plant Physiol. 2013;162(2):1092–1109. doi: 10.1104/pp.112.211441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Honys D, Twell D. Transcriptome analysis of haploid male gametophyte development in Arabidopsis. Genome Biol. 2004;5(11):R85. doi: 10.1186/gb-2004-5-11-r85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Becker JD, Takeda S, Borges F, Dolan L, Feijó JA. Transcriptional profiling of Arabidopsis root hairs and pollen defines an apical cell growth signature. BMC Plant Biol. 2014;14:197. doi: 10.1186/s12870-014-0197-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.