Significance
Compound Activity Mapping provides an alternative approach to natural products drug discovery by integrating high-content biological screening and untargeted metabolomics to directly reveal the identities and biological functions of individual bioactive compounds in complex natural product libraries. This accelerated discovery pipeline addresses many of the issues that have contributed to the decline of natural products research in some areas by improving dereplication and lead prioritization strategies. The detailed structural and functional annotation offered by this tool may help to improve the integration of natural products with modern high-content, high-throughput screening and provide an additional strategy for the discovery of the next generation of natural product-inspired drug leads and chemical probes.
Keywords: metabolomics, natural products, image-based screening, bioactive small molecules, informatics
Abstract
Traditional natural products discovery using a combination of live/dead screening followed by iterative bioassay-guided fractionation affords no information about compound structure or mode of action until late in the discovery process. This leads to high rates of rediscovery and low probabilities of finding compounds with unique biological and/or chemical properties. By integrating image-based phenotypic screening in HeLa cells with high-resolution untargeted metabolomics analysis, we have developed a new platform, termed Compound Activity Mapping, that is capable of directly predicting the identities and modes of action of bioactive constituents for any complex natural product extract library. This new tool can be used to rapidly identify novel bioactive constituents and provide predictions of compound modes of action directly from primary screening data. This approach inverts the natural products discovery process from the existing ‟grind and find” model to a targeted, hypothesis-driven discovery model where the chemical features and biological function of bioactive metabolites are known early in the screening workflow, and lead compounds can be rationally selected based on biological and/or chemical novelty. We demonstrate the utility of the Compound Activity Mapping platform by combining 10,977 mass spectral features and 58,032 biological measurements from a library of 234 natural products extracts and integrating these two datasets to identify 13 clusters of fractions containing 11 known compound families and four new compounds. Using Compound Activity Mapping we discovered the quinocinnolinomycins, a new family of natural products with a unique carbon skeleton that cause endoplasmic reticulum stress.
Notwithstanding the historical importance of natural products in drug discovery (1) the field continues to face a number of challenges that affect the relevance of natural products research in modern biomedical science (2). Among these are the increasing rates of rediscovery of known classes of natural products (3–6) and the high rates of attrition of bioactive natural products in secondary assays due to limited information about compound modes of action in primary whole-cell assays (7). Although pharmaceutical companies recognize that natural products are an important component of drug discovery programs because of the different pharmacologies of natural products and synthetic compounds (8), there is a reluctance to return to “grind and find” discovery methods (9). Therefore, there is a strong need for technologies that address these issues and provide new strategies for the prioritization of lead compounds with unique structural and/or biological properties (10).
Natural product drug discovery is challenging in any assay system because extract libraries are typically complex mixtures of small molecules in varying titers, making it difficult to distinguish biological outcomes (11). This is compounded by issues of additive effects of multiple bioactive compounds and the presence of nuisance compounds that cause false positives in assay systems (12). To address these issues, our laboratory has recently developed several image-based screening platforms that are optimized for natural product discovery (13–16). The cytological profiling platform optimized by Schulze and coworkers characterizes the biological activities of extracts using untargeted phenotypic profiling. These phenotypic profiles are compared with natural products extracts and a training set of compounds with known modes of action to characterize the bioactivity landscape of the screening library (17, 18). This cytological profiling tool forms the basis of the biological characterization component of the Compound Activity Mapping platform, as described below.
In the area of chemical characterization of natural product libraries, untargeted metabolomics is gaining attention as a method for evaluating chemical constitution (3, 19–22). Modern “genes-to-molecules” and untargeted metabolomics approaches taking advantage of principal component analysis and MS2 spectral comparisons have also been developed to quickly dereplicate complex extracts and distinguish noise and nuisance compounds from new molecules (23–27). Unfortunately, although these techniques are well suited to the discovery of new chemical scaffolds, they are unable to describe the function or biological activities of the compounds they identify. Therefore, there is still a need for new approaches to systematically identify novel bioactive scaffolds from complex mixtures.
To overcome some of these outstanding challenges we have developed the Compound Activity Mapping platform to integrate phenotypic screening information from the cytological profiling assay with untargeted metabolomics data from the extract library (Fig. 1). By correlating individual mass signals with specific phenotypes from the high-content cell-based screen (Fig. 2), Compound Activity Mapping allows the prediction of the identities and modes of action of biologically active molecules directly from complex mixtures, providing a mechanism for rational lead selection based on desirable biological and/or chemical properties. To evaluate this platform for natural products discovery we examined a 234-member extract library, from which we derived 58,032 biological measurements (Fig. 1C) and 10,977 mass spectral features (Fig. 1A). By integrating and visualizing these data we created a Compound Activity Map for this library composed of 13 clusters containing 16 compounds from 11 compound classes (Fig. 3). This integrated data network enabled the discovery of four new compounds, quinocinnolinomycins A–D (1–4, Fig. 4), which are the first examples to our knowledge of microbial natural products containing the unusual cinnoline core (Fig. 5). Clustering the cytological profiles of the quinocinnolinomycins with those of the Enzo library training set suggests that these compounds induce endoplasmic reticulum (ER) stress and the protein unfolding response.
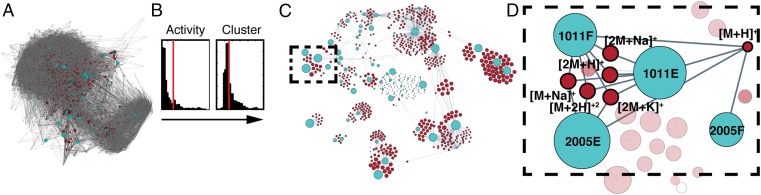
Fig. 1.
Overview of Compound Activity Mapping. (A) Representation of the chemical space in the tested extract library. The network displays extracts (light blue) connected by edges to all m/z features (red) observed from the metabolomics analysis, illustrating the chemical complexity of even small natural product libraries. (B) Histograms of activity and cluster scores for all m/z features with cutoffs indicated as red lines (for full-size histograms see SI Appendix, Fig. S5). (C) Compound Activity Map, with the network displaying only the m/z features predicted to be associated with consistent bioactivity, and their connectivity to extracts within the library. (D) Expansion of the staurosporine cluster (dotted box in C) with extract numbers and relevant m/z features labeled.
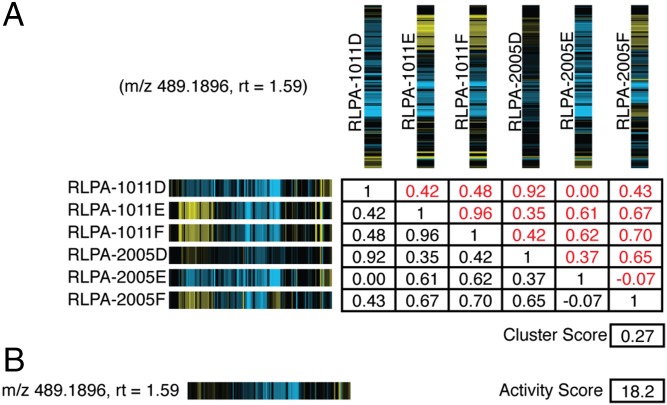
Fig. 2.
Determination of synthetic fingerprints and cluster and activity scores. (A) Table of Pearson correlations for the cytological profiles between all extracts containing a specific m/z feature (m/z of 489.1896, rt of 1.59). In each cytological profile, yellow stripes correspond to positive perturbations in the observed cytological attribute and blue stripes correspond to negatively perturbed attributes. The cluster score is determined by calculating the average of the Pearson correlation scores for all relevant extracts. (B) Calculated synthetic fingerprint and activity score for feature (m/z of 489.1896, rt of 1.59). Synthetic fingerprints are calculated as the averages of the values for each cytological attribute to give a predicted cytological profile for each bioactive m/z feature in the screening set.
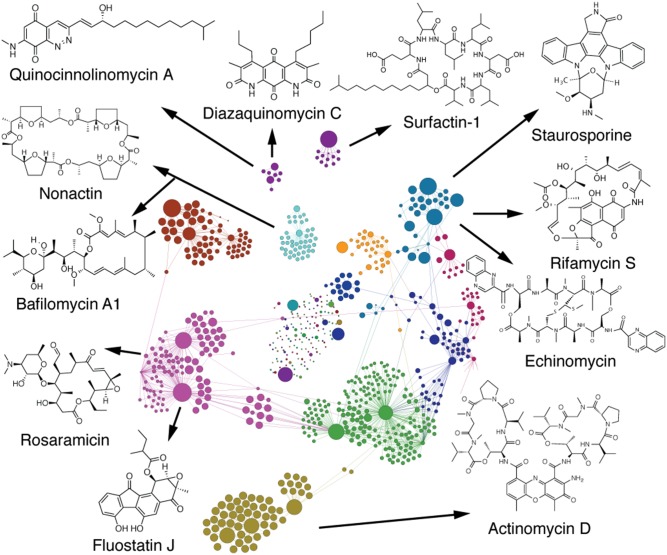
Fig. 3.
Annotated Compound Activity Map. An expanded view of the Compound Activity Map from Fig. 1C, with the extracts and m/z features separated into subclusters and colored coded using the Gephi modularity function. Each bioactive subcluster is composed of extracts containing a family of compounds with a defined biological activity. The Compound Activity Map is annotated with a representative molecule from each of the families of compounds that have been independently confirmed by purification and chemical analysis.
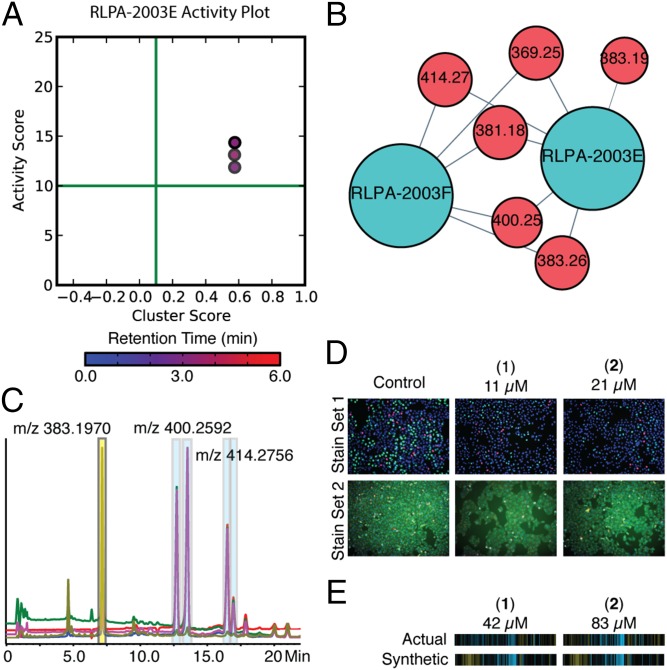
Fig. 4.
The prioritization, isolation, and confirmation of the quinocinnolinomycins A–D (1–4). (A) Bioactive m/z features plotted on a graph of activity score vs. cluster score. The color of the dot corresponds to the retention time of the m/z feature with the color bar and scale below in minutes. (B) Isolated cluster from Fig. 1C and Fig. 3 containing both the relevant extracts (blue) and bioactive m/z features (red). (C) HPLC trace of extract RLPA-2003E and the isolation of quinocinnolinomycins A–D (highlighted with blue boxes on HPLC trace). (D) Cell images of pure compounds screened as a twofold dilution series for quinocinnolinomycins A and B in both stain sets compared with images of vehicle (DMSO) wells. (E) Comparison of the synthetic and actual cytological fingerprints of the pure compounds is presented below the relevant images, demonstrating the relationship between experimental and calculated cytological profiles for these two metabolites.
Fig. 5.
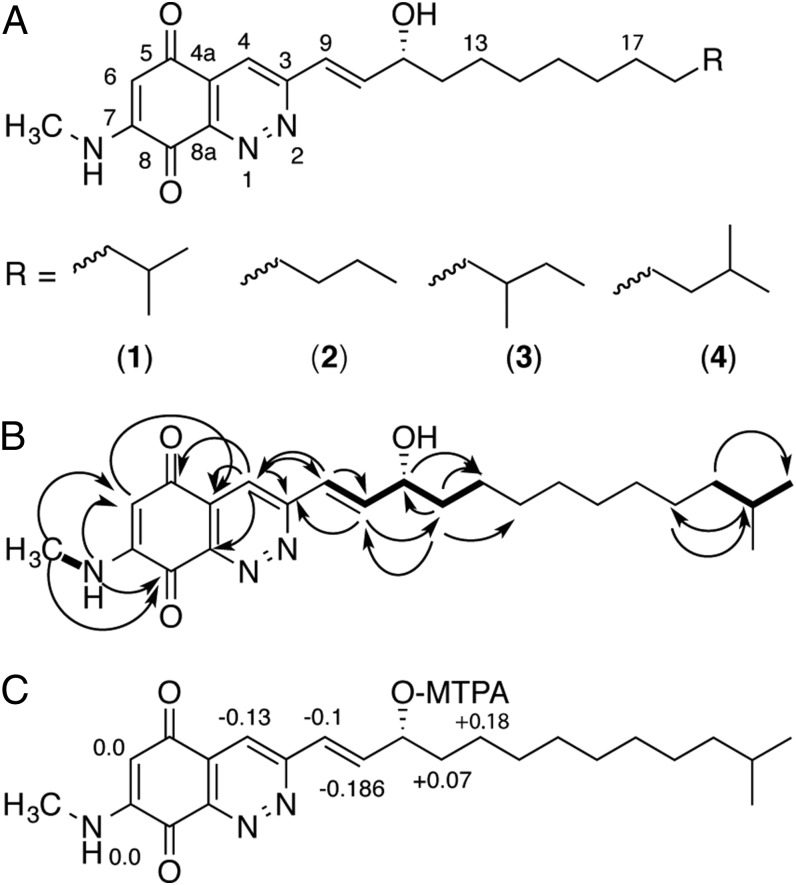
Structure elucidation of quinocinnolinomycins A–D (1–4). (A) Structures of quinocinnolinomycins A–D. (B) Key NMR correlations used in the structure elucidation of quinocinnolinomycin A. COSY correlations are indicated by bold lines. Heteronuclear multiple-bond correlations are indicated by curved arrows. (C) ∆δSR values for the Mosher’s α-methoxy-α-trifluoromethylphenylacetic acid (MTPA) ester analysis of the secondary alcohol in quinocinnolinomycin A (1) to assign the absolute configuration at position C11.
Experimental Procedures
Library Preparation.
Cell culture, extraction, library preparation, and crude extract fractionation were performed as described by Schulze et al. (15). Briefly, bacterial strains were isolated from marine sediment collected by hand using SCUBA from coastal areas of Panama at depths of 5–30 m. Bacterial isolates were grown under standard fermentation conditions (16), extracted with 1:1 methanol/dichloromethane, and fractionated on a reverse-phase C18 column with an eleutropic series of water and methanol [20, 40, 60, 80, and 100% (vol/vol) methanol in water, and an ethyl acetate wash]. These extracts were concentrated to dryness in vacuo and resuspended in 1 mL of DMSO. DMSO stock solutions were diluted 1:5 in DMSO for cytological profiling and 1:25,000 in 50% (vol/vol) methanol/water into Corning V bottom 96-well plates for metabolomic analysis.
Chemical Profiling.
Ultra performance liquid chromatography TOF-MS experiments were performed using an Agilent 1260 binary pump in low dwell volume mode, an Agilent column oven heated to 45 °C, and an Agilent 6230 time-of-flight mass spectrometer with a Jetstream ESI source. From the 1:25,000 fold dilution of the DMSO extract,1 µL of sample, dissolved in 50% (vol/vol) methanol/water, was injected onto a 1.8-µm particle size, 50 × 2.3 mm i.d., ZORBAX RRHT C18 column. Each sample was subjected to a water-acetonitrile gradient from 10 to 90% (vol/vol) acetonitrile over 4 min with a 1.5 min hold at 90% (vol/vol) acetonitrile before a 3 min reequilibration. The flow rate was maintained at 0.8 mL/min. Formic acid (200 µL of acid per liter of solvent) was added to both the water and the acetonitrile. Water (1 mL of water per liter) was added to the acetonitrile. Mass spectrometry acquisitions and peak selection were performed using standard instrument settings for small molecules (SI Appendix).
MS Data Alignment.
Comparison of selected peaks between injections of the same sample were performed using high-resolution mass (parts per million), retention time, and an isotope pattern matching method adapted from Pluskal et al. (28). After initial data acquisition, processing, and CEF file (peak list) output, peaks from MeOH media blanks were removed from the extract peak lists using 20 ppm, 0.4 min, and 0.5 isotopic score difference windows. Detector ringing was removed by eliminating all peaks within 0.4 min and 1 mass unit of the most abundant peak in saturated data. We developed a decision tree to align m/z retention time (rt) pairs between extended dynamic range (2-GHz) and high-resolution (4-GHz) detector modes to select the most accurate data between 2- and 4-GHz modes from both positive and negative ESI experiments (SI Appendix, Fig. S2). The blank, media, and saturation filtered peaks were aligned between 4-GHz and 2-GHz modes with 7 or 20 ppm, 0.4 min, 0.5 isotopic score difference windows. Each aligned peak was assigned a tag that indicated whether or not the peak was present and not saturated or present and saturated in both 4-GHz and 2-GHz modes (SI Appendix, Fig. S2). To ensure the highest accuracy data were stored, m/z values that were not saturated from the 4-GHz data were selected preferentially, with 2-GHz data being substituted in instances where the 4-GHz data were saturated. We stored the postvalidated peak list in a SQLite database for rapid indexing during incorporation with biological data.
Basketing.
To compare and align peaks from different extracts in the database, we performed 2D binning based on m/z and rt values using the same cutoffs of 7 ppm and 0.4 min. These baskets, referred to as m/z features, include the m/z, rt, and mass intensity data, as well as a list of each extract from which peaks in the basket were detected. For each m/z feature, this extract set was used to generate the integrated biological profiling metrics activity score and cluster score.
Cytological Profile Screening and Image Analysis.
Methods for cell culture and staining were used as previously reported (15, 18). HeLa cells were plated into two clear-bottom 384-well plates at a target density of 2,500 cells per well. The plates were incubated for 24 h under 5% CO2 at 37 °C, 150 nL of extract was pinned into the culture plates, and the plates were incubated for 19 h under 5% CO2 at 37 °C. The plates were then fixed and stained with either cell cycle or cytoskeletal stain sets, which report on the number of cells in S-phase or mitosis and amount and distribution of tubulin and actin, respectively (15, 18). Both stain sets contained a nuclear stain (Hoechst), which was used to count the number of cells and segment the image. The plates were imaged with a 10× objective lens acquiring four images per well. For each extract, 248 different parameters were measured from the images of each plate. Together these values report on a diverse range of size and shape features including, for example, those representing the total area and shape of the nuclei or the number of mitotic cells. Comparing extract-treated wells with DMSO-treated wells and reduction of these cell-by-cell metrics to population values for each well using our in-house data management pipeline afforded a 248-parameter fingerprint for each extract displaying the positive (yellow) or negative (blue) perturbations for each attribute with values between −1 and 1 (Fig. 2).
Death Dilutions.
Before submitting each screening plate for image analysis, the raw imaging data were used to count the number of cells in each well. In some instances treatment with extracts resulted in significant cell death, precluding the determination of accurate cytological profiles. The extracts that caused a reduction in cell count outside of three SDs of control wells were submitted for serial dilution and rescreened. For extracts that elicited a response with acceptable cell counts, the journaled cytological profiles were used for data integration and clustering. For the extracts that caused a three-SD reduction in the number of cells, the cytological profile of the first dilution with a cell count within three SDs of the mean control cell count was used for clustering and integration.
Integrating Untargeted Metabolomics Data and Cytological Profiling Data.
To integrate the cytological profiling and metabolomics datasets, each m/z feature stored in the database is ascribed a synthetic fingerprint, an activity score, and a cluster score, which together predict the biological activity of each feature. A visual representation of the calculations performed on an example compound is displayed in Fig. 2.
Synthetic Fingerprints.
The synthetic fingerprint of an m/z feature is the average of each cytological attribute value for the set extracts in which the m/z feature is detected. This calculated or “synthetic fingerprint” represents the predicted cytological profile for each m/z feature in the sample set (Fig. 2B).
| [1] |
Eq. 1 is the calculation of the synthetic fingerprint. For the set of extracts E in which the m/z feature was detected, extracts 1 to j, F is the set of each extract’s cytological profile, fingerprints f1 to fj, where each fingerprint, fk, contains attributes a1 to an. The synthetic fingerprint is the set containing the average of each attribute of the fingerprints in F.
Activity Score.
From a cytological profile fk or a synthetic fingerprint, the activity score is defined as the sum of the square each attribute. This singular value can be used to assess whether or not each m/z feature is predicted to perturb cell development above baseline values.
| [2] |
Eq. 2, the activity score for a fingerprint, is the sum of the square of each attribute.
Cluster Score.
The cluster score of an m/z feature is the average of the cube of the Pearson correlations between all combinations of two different cytological profile fingerprints in set F. The Pearson correlation values are cubed to reduce the emphasis of low values caused by the presence of one or more extracts containing a given m/z feature at a concentration high enough to be detected by the mass spectrometer, but too low to cause a significant impact on cell development. This single value is used to assess whether or not each m/z feature is correlated with a specific phenotype within the biological dataset.
| [3] |
Using Eq. 3, the cluster score is calculated as the average of the cube the Pearson correlations of all combinations of two fingerprints in F where j is the number of extracts. The number of extracts j is subtracted from the numerator and denominator to remove Pearson correlations between fi and fi.
Network Visualization.
We use NetworkX in Python to create and edit networks and Gephi to visualize and analyze networked data. In general, blue nodes represent extracts and are connected to red nodes representing the m/z features detected in those extracts (Fig. 1D). Using Gephi, we display the relative bioactivities of each node by making the diameter of each node proportional to extract activity score or m/z feature activity score. Distinct clusters (represented by different colors in Fig. 3) are identified using network modularity with weighted edges and a resolution of one. We use Gephi’s built-in Force Atlas 2 algorithm to distribute nodes with default parameters except the following: approximate repulsion of 0.2, scaling of 10, gravity of 2, and “prevent overlap.”
Results
Cytological Profiling.
Preliminary screening generated cytological profiles for all 234 extracts, of which 50 were serially diluted and rescreened based on low cell count (Experimental Procedures). After these samples were diluted, 57 of the 234 profiles had activity scores greater than 10, with 13 discrete clusters with Pearson correlations <0.875 (SI Appendix, Figs. S3 and S4).
Metabolomics.
Features (n = 10,977) were stored into the mass feature sequel database after media and blank subtraction. Of these, 346 were eliminated because they appeared in greater than 10% of extracts and 5,310 singletons were removed, affording 5,321 filtered features for network analysis. The removal of singletons must be considered carefully, because it eliminates these features from further consideration as bioactive constituents. This is a particular challenge for small extract sets, such as the one used in this initial study, where the probability of metabolites being observed only once is relatively high. The long-term objective of our laboratory is to build a database containing profiles from many thousands of extracts. This will reduce the number of instances where singletons are encountered because of the inherent redundancy of the occurrence of individual compounds in natural product extract libraries and permit the integration of singleton features into the Compound Activity Mapping workflow. For this initial study, incorporation of singletons was impractical because of the large number of m/z features appearing only once. However, because the 13 clusters derived from the cytological profiling screen all contained multiple extracts, we were able to exclude singleton features in this instance based on the hypothesis that the bioactive constituents should be present in more than one extract in each case. Examination of the final network reveals validated bioactive compounds for all clusters, supporting this assumption.
Data Integration.
To integrate the biological and chemical datasets synthetic fingerprints, cluster scores, and activity scores were generated for each m/z feature in the database. These results were used to generate activity plots for each extract (for an example activity plot, see Fig. 4A), displaying m/z features as points on the graph, with the activity score on the y axis, the cluster score on the x axis, and the color of each point corresponding to the retention time of that m/z feature.
The activity and cluster score metrics were used to filter the m/z feature database to select for features that were correlated with strong and consistent phenotypes. Cutoffs were selected so that only m/z features with activity scores greater than 10 and cluster scores greater than 0.10 were retained for subsequent network generation (SI Appendix, Fig. S5). These cutoffs were selected to include all m/z features with “nonnegative” scores for both values. This was accomplished by plotting a rank order of values for every m/z feature for each metric (SI Appendix, Fig. S5 A and C) and selecting cutoffs below the vertex of each curve to eliminate all features in the lower pseudolinear region of each plot. These initial cutoff values were then manually adjusted using the pan-specific kinase inhibitor staurosporine as a positive control compound within the dataset to obtain networks containing all relevant connections between the m/z features for staurosporine and their associated extracts. For reference, the average activity score in this study was 4.66 with an SD of 5.53, and the average cluster score was 0.13 with an SD of 0.14.
After applying these filters 634 features remained that represented the m/z features predicted to be responsible for the observed bioactivities. A network was then generated from these 634 features with extract nodes connected to their corresponding m/z features by edges (unweighted). The size of the node is defined by the activity score of the extract or half the activity score of the m/z feature for easy visualization. Subclusters could then be assigned using the modularity feature of Gephi based on connectivities between extract and m/z feature nodes. From this we were able to observe 13 unique clusters, each of which contained mass spectral features for the natural products predicted to be responsible for the bioactivity of the extracts.
Discussion
Compound Activity Mapping provides a new approach to the characterization of natural product libraries that is complementary to existing discovery methods. The main advantage of this approach is that it provides a mechanism for the early prediction of the identities and biological behaviors of bioactive compounds from complex mixtures, permitting hypothesis-driven lead selection and a streamlined discovery workflow. Data acquisition can be completed in under 2 wk for each 384-well plate (including both metabolomics analysis and cytological profiling), making this method suitable for medium-throughput natural product discovery efforts.
Of the 234 extracts examined in this study, 57 had cytological profiles with activity scores above 10. All of these extracts possessed associated m/z features from the metabolomic analysis predicted to be responsible for the observed activities, suggesting that Compound Activity Mapping is suitable for the systematic characterization of complex screening libraries. In general, these active clusters fall into one of three classes: clusters where the activity is caused by a single known natural product class, clusters where the activity is caused by the presence of multiple classes of known bioactives, and clusters where the activity is caused by bioactives that have no matches to available databases of microbially-derived natural products.
Clusters Containing Single Bioactives.
One example of a cluster driven by the presence of a single known bioactive class is the cluster containing extracts RLPA2008C, E, and F (Fig. 3, olive-green cluster). It is clear from the network that the chemical constitutions of RLPA2008C, E and F are distinct from the rest of the library. Three of the m/z features in this cluster were consistent with the [M + H]+, [M-H]−, and [M+Na]+ adducts of a compound with the molecular formula C62H86N12O16. Searching the AntiMarin database (a comprehensive database of microbial and marine-derived natural products) identified actinomycin D as a match with the molecular formula (29). Clustering and visualizing the synthetic fingerprints of these features with the cytological profiles of the Enzo compound library using Cluster 3.0 and Java TreeView (30, 31) strongly supported this result (SI Appendix, Fig. S7), with extracts RLPA2008C, E and F also clustering closely with the pure actinomycin D standard. The identification was confirmed by coinjection with a commercial standard of actinomycin D, which possessed the same m/z features and retention time as the predicted hits from the extract.
A second example of clustering driven by the presence of a single bioactive compound is the cluster containing extracts RLPA1011E, RLPA1011F, RLPA2005E, and RLPA2005F (Fig. 3, cyan cluster). In this case the activity plot for RLPA1011F reveals seven m/z features consistent with the single molecular formula C28H26N4O3. Comparison of this formula to the AntiMarin database reveals a match to the pan-specific kinase inhibitor staurosporine. This assignment was confirmed by coinjection with an authentic standard of staurosporine, which had a retention time and high-resolution MS signals that matched those for the bioactive components in these extracts.
Clusters Containing Multiple Bioactives.
Although extracts 1011E and F were correctly predicted to contain staurosporine, examination of the Compound Activity Map and activity plots for extracts RLPA2005E and F revealed a second set of two m/z features predicted to contribute strongly to the observed biological activities of these extracts. These new m/z features were consistent with a compound with the molecular formula C51H64N12O12S2, which corresponded to the DNA intercalator echinomycin. Presence of this second bioactive metabolite was also confirmed by coinjection with a standard.
Importantly, although these two situations (staurosporine only, staurosporine and echinomycin) are connected in one “supercluster” because they are related by the extracts in which they are found, they resolve into individual subclusters based on the interconnectivities of the extract nodes and m/z features. This demonstrates that this approach is able to resolve convoluted situations involving mixtures of compounds with different biological mechanisms of action and provide useful characterization of bioactive metabolites even in situations in which mixtures of bioactives cause phenotypic responses that are not closely related to either compound individually. The synthetic fingerprints of the m/z features corresponding to staurosporine cluster closely with the pure compound from the Enzo library and are readily distinguishable from those of the echinomycin (SI Appendix, Fig. S8).
A second example of clusters containing multiple bioactive metabolites is provided by the cluster containing extracts RLPA2021C, E, and F (Fig. 3, purple cluster). In this instance the cluster contains a large number of candidate m/z features, many of which are consistent with different members of two separate classes of natural products: the fluorenone-containing fluostatins and the macrolide antibiotic rosaramicins. This situation is significantly more complex than the previous example, with multiple members of two separate bioactive compound classes contributing to the overall phenotypes observed for these extracts. Isolation and NMR evaluation of representative members of these two compound classes (fluostatins C, D, and J and rosaramicin) confirmed their initial assignments and permitted the evaluation of each compound class as pure compounds in the cytological profiling assay. The fluostatins all clustered closely with kinase inhibitors (32), whereas rosaramicins induced only a very weak phenotype that is consistent with their previous annotation as antibiotics and not cytotoxic agents (SI Appendix, Figs. S6 and S7) (33). Compound Activity Mapping was able to identify the fluostatins as the correct bioactive constituent, but because the fluostatins and the rosaramicins always appeared together, the macrolides were called as a false positive. This limitation of the platform can be resolved by analyzing larger libraries of extracts from similar organisms, which will reduce the probability that two compounds are always coexpressed. Once each constituent appears individually in the dataset, inactive compounds will display lower activity and cluster scores, eventually excluding them from the network.
Discovery and Structure Determination of Quinocinnolinomycins A–D.
In addition to the annotation of known bioactive metabolites, Compound Activity Mapping is well suited to the discovery of novel compounds and the characterization of their biological attributes. Within this set of extracts we observed a number of unique clusters with high activity scores and no matches in the AntiMarin database. One such cluster, containing extracts RLPA2003E and F, was prioritized because it contained just five m/z features that were common to only those extracts (Fig. 4B). Examination of the activity plot (Fig. 4A) highlighted one m/z feature with high activity and cluster scores (m/z of 400.2590, rt of 3.50 min, activity score 13.12, cluster score 0.57) that was prioritized for chemical analysis. Liquid chromatography–MS analysis of this extract revealed the presence of two peaks with m/z features at 400.2590 amu and similar UV profiles, as well as two additional peaks that possessed the same UV profiles but had m/z values of 414.2756, suggestive of the presence of a family of related compounds (Fig. 4C and SI Appendix, Fig. S11).
The molecular formulae C23H33N3O3 and C24H35N3O3 were determined based on the strong consensus between the [M + H]+ and [M + Na]+ m/z features for each set of two constitutional isomers. The earliest eluting compound with the molecular formula C23H33N3O3 was solved by NMR analysis, using a combination of 1H, 13C, gCOSY, gHSQC, gHMBC, and 1D-TOCSY spectra (Fig. 5; for full structure determination description see SI Appendix). The R stereochemistry of quinocinnolinomycin A was determined using Mosher’s ester method (Fig. 5D), and this assignment extended to quinocinnolinomycins B–D based on their common biosynthetic origin (SI Appendix, Fig. S12).
Mechanism of Action of the Quinocinnolinomycins.
Purified quinocinnolinomycins A–D were rescreened as twofold dilution series (166.7 µM–2.5 nM) in the cytological profiling assay (SI Appendix, Fig. S9). Clustering these cytological profiles with those of the Enzo compound library training set revealed a distinct cluster containing all four analogs over a range of concentrations between 0.3 and 83.3 μM along with the known compounds thapsigargin (calcium ATPase inhibitor) (34), tunicamycin (N-linked protein glycosylation inhibitor) (35), lycorine (ribosome inhibitor) (36), and brefeldin A (ARF GTPase inhibitor) (37). Although the precise molecular targets of these compounds differ, they are all mechanistically related because they affect the function of different components of the ER and result in ER stress and the induction of the protein unfolding response (38–40). Active concentrations of quinocinnolinomycins A–D are present within this cluster with Pearson correlations to the other training set compounds on the order 0.6–0.7, indicating close matches between these cytological profiling fingerprints; these data suggest that the quinocinnolinomycins have a mode of action that causes ER stress. Moderate ER stress may be mitigated by macroautophagy (autophagy) in mammalian cells and can lead to cell death or survival depending on the context; this is an active area of research for future cancer therapies (39–42). Further studies to elucidate the precise molecular target of the quinocinnolinomycins will expand our understanding of the cellular processes involved with ER stress, the unfolded protein response, and autophagy with direct implications for human disease.
Conclusion
By predicting the identity and mode of action of all detectable metabolites from complex extracts Compound Activity Mapping aims to expedite the discovery process by changing the traditional “blind” discovery model to a hypothesis-driven approach to novel bioactive compound discovery. This approach reduces the time required to go from a hit in an assay to a lead molecule by minimizing iterative bioassay-guided fractionation and screening steps and allows hypothesis-driven exploration of natural product libraries by providing a global view of compound diversity and activity across any library. In this study, analysis of the 234-member library revealed 13 unique clusters based on chemical and biological similarities. We were able to confirm the identities of 16 compounds from these clusters using a combination of analytical approaches, providing a detailed molecular picture of the bioactivity landscape for this extract library in this biological assay.
The discovery of quinocinnolinomycins A–D highlights the utility of this platform for novel compound discovery and mode of action characterization. The cluster containing extracts RLPA2003E and F is distinct in the Compound Activity Map and contained m/z features suggesting the presence of unique compounds correlated with a strong and distinct phenotype. These data strongly suggested that these mass features should be prioritized for structure elucidation, leading to the discovery of this new structural class of natural products with accurately predefined biological activities.
Supplementary Material
Acknowledgments
We thank R. S. Lokey and W. Bray for assistance with cytological profiling screening. This work was funded by NIH Grant TW006634.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1507743112/-/DCSupplemental.
References
- 1.Newman DJ, Cragg GM. Natural products as sources of new drugs over the 30 years from 1981 to 2010. J Nat Prod. 2012;75(3):311–335. doi: 10.1021/np200906s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Koehn FE, Carter GT. The evolving role of natural products in drug discovery. Nat Rev Drug Discov. 2005;4(3):206–220. doi: 10.1038/nrd1657. [DOI] [PubMed] [Google Scholar]
- 3.Nielsen KF, Månsson M, Rank C, Frisvad JC, Larsen TO. Dereplication of microbial natural products by LC-DAD-TOFMS. J Nat Prod. 2011;74(11):2338–2348. doi: 10.1021/np200254t. [DOI] [PubMed] [Google Scholar]
- 4.Yang JY, et al. Molecular networking as a dereplication strategy. J Nat Prod. 2013;76(9):1686–1699. doi: 10.1021/np400413s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Månsson M, et al. Explorative solid-phase extraction (E-SPE) for accelerated microbial natural product discovery, dereplication, and purification. J Nat Prod. 2010;73(6):1126–1132. doi: 10.1021/np100151y. [DOI] [PubMed] [Google Scholar]
- 6.Ibrahim A, et al. Dereplicating nonribosomal peptides using an informatic search algorithm for natural products (iSNAP) discovery. Proc Natl Acad Sci USA. 2012;109(47):19196–19201. doi: 10.1073/pnas.1206376109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gerwick WH, Moore BS. Lessons from the past and charting the future of marine natural products drug discovery and chemical biology. Chem Biol. 2012;19(1):85–98. doi: 10.1016/j.chembiol.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wassermann AM, et al. A screening pattern recognition method finds new and divergent targets for drugs and natural products. ACS Chem Biol. 2014;9(7):1622–1631. doi: 10.1021/cb5001839. [DOI] [PubMed] [Google Scholar]
- 9.Davies J. The garden of antimicrobial delights. F1000 Biol Rep. 2010;2(26):26. doi: 10.3410/B2-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Potterat O, Hamburger M. Concepts and technologies for tracking bioactive compounds in natural product extracts: Generation of libraries, and hyphenation of analytical processes with bioassays. Nat Prod Rep. 2013;30(4):546–564. doi: 10.1039/c3np20094a. [DOI] [PubMed] [Google Scholar]
- 11.Suffness M, Douros JD. Discovery of antitumor agents from natural sources. Trends Pharmacol Sci. 1981;2:307–310. [Google Scholar]
- 12.Rishton GM. Natural products as a robust source of new drugs and drug leads: Past successes and present day issues. Am J Cardiol. 2008;101(10A):43D–49D. doi: 10.1016/j.amjcard.2008.02.007. [DOI] [PubMed] [Google Scholar]
- 13.Navarro G, et al. Image-based 384-well high-throughput screening method for the discovery of skyllamycins A to C as biofilm inhibitors and inducers of biofilm detachment in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2014;58(2):1092–1099. doi: 10.1128/AAC.01781-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peach KC, Bray WM, Winslow D, Linington PF, Linington RG. Mechanism of action-based classification of antibiotics using high-content bacterial image analysis. Mol Biosyst. 2013;9(7):1837–1848. doi: 10.1039/c3mb70027e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schulze CJ, et al. “Function-first” lead discovery: Mode of action profiling of natural product libraries using image-based screening. Chem Biol. 2013;20(2):285–295. doi: 10.1016/j.chembiol.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wong WR, Oliver AG, Linington RG. Development of antibiotic activity profile screening for the classification and discovery of natural product antibiotics. Chem Biol. 2012;19(11):1483–1495. doi: 10.1016/j.chembiol.2012.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perlman ZE, et al. Multidimensional drug profiling by automated microscopy. Science. 2004;306(5699):1194–1198. doi: 10.1126/science.1100709. [DOI] [PubMed] [Google Scholar]
- 18.Woehrmann MH, et al. Large-scale cytological profiling for functional analysis of bioactive compounds. Mol Biosyst. 2013;9(11):2604–2617. doi: 10.1039/c3mb70245f. [DOI] [PubMed] [Google Scholar]
- 19.Duncan KR, et al. Molecular networking and pattern-based genome mining improves discovery of biosynthetic gene clusters and their products from Salinispora species. Chem Biol. 2015;22(4):460–471. doi: 10.1016/j.chembiol.2015.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoffmann T, Krug D, Hüttel S, Müller R. Improving natural products identification through targeted LC-MS/MS in an untargeted secondary metabolomics workflow. Anal Chem. 2014;86(21):10780–10788. doi: 10.1021/ac502805w. [DOI] [PubMed] [Google Scholar]
- 21.El-Elimat T, et al. High-resolution MS, MS/MS, and UV database of fungal secondary metabolites as a dereplication protocol for bioactive natural products. J Nat Prod. 2013;76(9):1709–1716. doi: 10.1021/np4004307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krug D, Müller R. Secondary metabolomics: The impact of mass spectrometry-based approaches on the discovery and characterization of microbial natural products. Nat Prod Rep. 2014;31(6):768–783. doi: 10.1039/c3np70127a. [DOI] [PubMed] [Google Scholar]
- 23.Cimermancic P, et al. Insights into secondary metabolism from a global analysis of prokaryotic biosynthetic gene clusters. Cell. 2014;158(2):412–421. doi: 10.1016/j.cell.2014.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krug D, et al. Discovering the hidden secondary metabolome of Myxococcus xanthus: A study of intraspecific diversity. Appl Environ Microbiol. 2008;74(10):3058–3068. doi: 10.1128/AEM.02863-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hou Y, et al. Microbial strain prioritization using metabolomics tools for the discovery of natural products. Anal Chem. 2012;84(10):4277–4283. doi: 10.1021/ac202623g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Watrous J, et al. Mass spectral molecular networking of living microbial colonies. Proc Natl Acad Sci USA. 2012;109(26):E1743–E1752. doi: 10.1073/pnas.1203689109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Doroghazi JR, et al. A roadmap for natural product discovery based on large-scale genomics and metabolomics. Nat Chem Biol. 2014;10(11):963–968. doi: 10.1038/nchembio.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pluskal T, Uehara T, Yanagida M. Highly accurate chemical formula prediction tool utilizing high-resolution mass spectra, MS/MS fragmentation, heuristic rules, and isotope pattern matching. Anal Chem. 2012;84(10):4396–4403. doi: 10.1021/ac3000418. [DOI] [PubMed] [Google Scholar]
- 29.Blunt JW, Munro M, Laatsch H. 2006. AntiMarin Database (University of Canterbury, Christchurch, New Zealand)
- 30.de Hoon MJL, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics. 2004;20(9):1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- 31.Saldanha AJ. Java Treeview--extensible visualization of microarray data. Bioinformatics. 2004;20(17):3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- 32.Baur S, et al. Novel members of the fluostatin family produced by Streptomyces strain Acta 1383. J Antibiot (Tokyo) 2006;59(5):293–297. doi: 10.1038/ja.2006.41. [DOI] [PubMed] [Google Scholar]
- 33.Lin CC, et al. Pharmacokinetics and metabolism of rosaramicin in humans. Antimicrob Agents Chemother. 1984;26(4):522–526. doi: 10.1128/aac.26.4.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Treiman M, Caspersen C, Christensen SB. A tool coming of age: Thapsigargin as an inhibitor of sarco-endoplasmic reticulum Ca(2+)-ATPases. Trends Pharmacol Sci. 1998;19(4):131–135. doi: 10.1016/s0165-6147(98)01184-5. [DOI] [PubMed] [Google Scholar]
- 35.Heifetz A, Keenan RW, Elbein AD. Mechanism of action of tunicamycin on the UDP-GlcNAc:dolichyl-phosphate Glc-NAc-1-phosphate transferase. Biochemistry. 1979;18(11):2186–2192. doi: 10.1021/bi00578a008. [DOI] [PubMed] [Google Scholar]
- 36.Garreau de Loubresse N, et al. Structural basis for the inhibition of the eukaryotic ribosome. Nature. 2014;513(7519):517–522. doi: 10.1038/nature13737. [DOI] [PubMed] [Google Scholar]
- 37.Donaldson JG, Finazzi D, Klausner RD. Brefeldin A inhibits Golgi membrane-catalysed exchange of guanine nucleotide onto ARF protein. Nature. 1992;360(6402):350–352. doi: 10.1038/360350a0. [DOI] [PubMed] [Google Scholar]
- 38.Samali A, Fitzgerald U, Deegan S, Gupta S. Methods for monitoring endoplasmic reticulum stress and the unfolded protein response. Int J Cell Biol. 2010;2010(11):830307. doi: 10.1155/2010/830307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ding W-X, et al. Differential effects of endoplasmic reticulum stress-induced autophagy on cell survival. J Biol Chem. 2007;282(7):4702–4710. doi: 10.1074/jbc.M609267200. [DOI] [PubMed] [Google Scholar]
- 40.Verfaillie T, Salazar M, Velasco G, Agostinis P. Linking ER stress to autophagy: Potential implications for cancer therapy. Int J Cell Biol. 2010;2010(12):930509. doi: 10.1155/2010/930509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hau AM, et al. Coibamide A induces mTOR-independent autophagy and cell death in human glioblastoma cells. PLoS One. 2013;8(6):e65250. doi: 10.1371/journal.pone.0065250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Høyer-Hansen M, Jäättelä M. Connecting endoplasmic reticulum stress to autophagy by unfolded protein response and calcium. Cell Death Differ. 2007;14(9):1576–1582. doi: 10.1038/sj.cdd.4402200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.