Significance
Propagation of protein aggregates throughout the nervous system is thought to play a significant role in the pathology of neurodegenerative diseases. Thus, understanding how this propagation is regulated is crucial for identifying potential therapeutic interventions. Here we use experimental advantages of Drosophila to investigate aggregate spreading. We demonstrate that huntingtin, containing an expanded polyglutamine tract of 138 repeats, but not an unexpanded form, aggregates and spreads rapidly throughout the brain, resulting in loss of vulnerable neurons. Aggregate spreading and neuronal loss are impaired by blocking synaptic exocytosis in releasing cells or endocytic uptake in target cells. These studies enable screens for genetic and pharmacological modifiers that could ameliorate the deleterious effects of spreading in neurodegenerative diseases.
Keywords: Huntington's disease, neurodegeneration, transmission, disease model, expanded triplet repeat
Abstract
A key feature of many neurodegenerative diseases is the accumulation and subsequent aggregation of misfolded proteins. Recent studies have highlighted the transcellular propagation of protein aggregates in several major neurodegenerative diseases, although the precise mechanisms underlying this spreading and how it relates to disease pathology remain unclear. Here we use a polyglutamine-expanded form of human huntingtin (Htt) with a fluorescent tag to monitor the spreading of aggregates in the Drosophila brain in a model of Huntington’s disease. Upon expression of this construct in a defined subset of neurons, we demonstrate that protein aggregates accumulate at synaptic terminals and progressively spread throughout the brain. These aggregates are internalized and accumulate within other neurons. We show that Htt aggregates cause non–cell-autonomous pathology, including loss of vulnerable neurons that can be prevented by inhibiting endocytosis in these neurons. Finally we show that the release of aggregates requires N-ethylmalemide–sensitive fusion protein 1, demonstrating that active release and uptake of Htt aggregates are important elements of spreading and disease progression.
Accumulation of protein aggregates is a key feature of many neurodegenerative diseases. Lesions in each of these diseases are initially limited to defined regions of selectively vulnerable neurons, but staging of pathology in Alzheimer’s disease (1), Parkinson’s disease (2, 3), amyotropic lateral sclerosis (4, 5), and Huntington’s disease (HD) (6) reveal broader deposition of pathological aggregates at more advanced stages of disease progression. The observation that the pathology appeared to progress into regions that were synaptically connected led to the idea that pathology was spreading through neuronal circuits (7, 8). Converging lines of evidence demonstrated that aggregates of disease-associated misfolded proteins, including α-synuclein, tau, and superoxide dismutase 1, are in fact transmissible from cell to cell and that this transmission propagates throughout the brain (9, 10). More recently, mutant huntingtin (Htt) aggregates were also shown to spread between neurons in vivo (11).
Although the cell-to-cell spreading of pathogenic proteins has been demonstrated in several neurodegenerative diseases, the mechanism by which this spreading occurs, and how it contributes to pathology and later stages of disease progression, remain unclear. To gain a better understanding of how this protein spreading contributes to disease pathology, we sought to study this phenomenon in Drosophila. Drosophila has been used to create useful models of many neurodegenerative diseases, including Parkinson’s disease (12) and the polyglutamine (polyQ) expansion diseases spinocerebellar ataxia type 1 (13) and type 3 (14) as well as Huntington’s disease (15–18). These models have proven to reproduce many of the key structural and functional deficits associated with disease pathology and provide insight into the underlying mechanisms. For example, a recent study demonstrated a “prion-like” spread of huntingtin aggregates into phagocytic glia, cells which carry out a protective clearance function but also potentially contribute to spreading itself (19). One advantage to studying protein aggregate spreading in Drosophila is the ability to independently label and manipulate separate populations of neurons simultaneously by using the yeast Gal4/Upstream Activating Sequence (UAS) (20) and bacterial LexA/LexA operator (LexAop) (21) binary expression systems. Additionally, the ability to rapidly identify and characterize genetic and chemical modifiers of this spreading phenomenon should help unravel mechanisms responsible for spreading.
In this study, we demonstrate that mutant huntingtin aggregates accumulate at synaptic terminals in the antennal lobe of the Drosophila central brain when expressed in olfactory receptor neurons (ORNs). Over time, these aggregates begin to spread to various regions of the brain, where they are internalized by other populations of neurons, resulting in some instances in loss of these neurons. This neuronal loss is prevented by blocking endocytosis, suggesting that spreading requires active internalization of the pathogenic protein. We observe unique spreading patterns when huntingtin is expressed in different populations of neurons, supporting the idea that nearby cells and neuronal circuits are likely targets of spreading. However, rapid accumulation of aggregates far from the original source also suggests that transmission is not limited to these circuits. The release of aggregates depends on N-ethylmaleimide–sensitive fusion protein 1 (NSF1), suggesting that soluble NSF attachment protein receptor (SNARE)-mediated fusion events are required for aggregate spreading. The extensive and efficient spread of huntingtin aggregates in the Drosophila brain that we report here provides a powerful experimental system for detailed genetic, molecular, and cellular analyses to dissect the underlying mechanisms and consequences.
Results
Transmission of Mutant Htt Throughout the Brain.
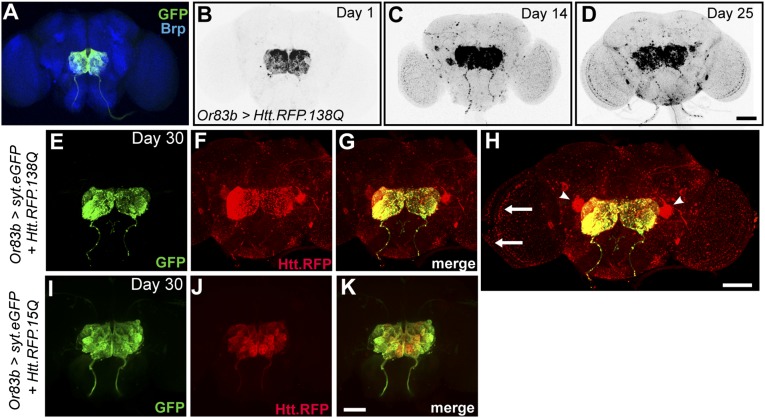
To visualize Htt aggregates in the brain, we expressed a 588-aa N-terminal fragment of the human Htt gene containing exons 1–12 with an expanded polyQ tract of 138 repeats fused to monomeric red fluorescent protein (mRFP) (UAS-mRFP.Htt.138Q) that was previously used to examine Htt aggregation kinetics in Drosophila (22). This fragment is a cleavage product formed by caspase-6 in HD (23) and is thus a biologically relevant fragment for use in a disease model. We expressed this construct with the Or83b-Gal4 driver to target expression in ORNs that project axons into the antennal lobe of the central brain (Fig. 1A). To prevent Htt aggregates from forming before adult flies emerged, we also used a temperature-sensitive Gal80 (tubulin-Gal80ts1) (24) to repress Gal4 in flies raised at 18 °C. At the adult stage, the flies were shifted to 29 °C to repress Gal80 and allow Gal4 activation. We found that Htt aggregates initially accumulated at ORN terminals within the antennal lobe (Fig. 1B). However, as flies aged these aggregates spread throughout the brain within 25 d (Fig. 1 C and D). To verify that Htt aggregates were spreading beyond the neurons in which mutant Htt was expressed, we simultaneously labeled ORN synaptic terminals with synaptotagmin-GFP (UAS-syt.eGFP). Although the GFP signal remained within the antennal lobe in 30-d-old flies, Htt.RFP aggregates were apparent throughout various brain regions (Fig. 1 E–G). Brain areas accumulating Htt aggregates include the optic lobe (Fig. 1H, arrows) as well as a pair of large neurons on the posterior side of the brain (Fig. 1H, arrowheads). To test whether this spreading phenomenon is unique to the expanded polyQ form of Htt, we used a construct with a much shorter nonpathogenic polyQ tract, (UAS-mRFP.Htt.15Q) (22), which does not form aggregates, as a control. In these flies, the Htt.RFP remained within the antennal lobe (Fig. 1 I–K), demonstrating that spreading of Htt aggregates is specific to the expanded polyQ form.
Fig. 1.
Htt aggregates spread throughout the Drosophila brain. (A) Expression pattern of or83b-Gal4 in the Drosophila brain labeling the antennal lobe. Neuropil is labeled by anti-Brp (blue) (B–D). Aggregates of Htt.RFP.138Q expressed in ORNs become more widely distributed throughout the brain as a function of age. (E–G) PolyQ-expanded Htt aggregates (red) spread far beyond ORN terminals marked by syt.eGFP (green). (H) Expanded view of G to illustrate Htt aggregates within large posterior neurons (arrowheads) and in the optic lobe (arrows). (I–K) Nonpathogenic Htt.RFP is confined to synaptic terminals in the antennal lobe. (Scale bar in D, 50 μm, also applies to A–C; scale bar in H, 50 μm; and scale bar in K, 50 μm, also applies to E–G, I, and J.)
Internalization of Htt Aggregates by Large Posterior Neurons.
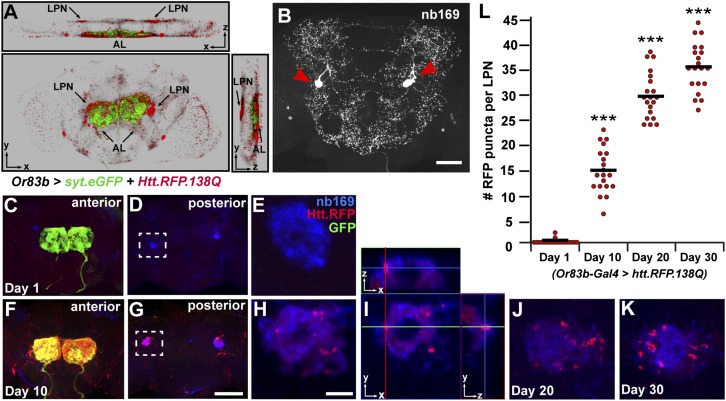
One prominent area of accumulation of Htt aggregates following expression in ORNs is a pair of large posterior neurons (LPNs) with cell bodies located in the posterior protocerebrum (Fig. 2A and Movie S1). These neurons appear identical to the large cells labeled by the monoclonal antibody nb169 from the Würzburg hybridoma library (25, 26) (Fig. 2B). To confirm the nonautonomous accumulation of Htt.RFP aggregates in these neurons, we expressed Htt.RFP and mCD8-GFP in ORNs and stained with nb169. Once again, we used tubulin-Gal80ts1 to repress Gal4 in flies raised at 18 °C to prevent Htt aggregates from forming before adult flies emerged. Adult flies were shifted to 29 °C to repress Gal80 and allow Gal4 activation. Although initially there was no RFP staining in these cells (Fig. 2 C–E), aggregates are seen within these cells by day 10 (Fig. 2 F–I) and continue to increase in number as the flies age (Fig. 2 J–L). The lack of GFP in the large posterior cells at both time points demonstrates that Or83b-Gal4 is not inappropriately driving expression of Htt.RFP in these neurons (Fig. 2 E and H). These results confirm that the Htt aggregates are spreading from ORNs and internalized by the large posterior neurons.
Fig. 2.
Htt aggregates are taken up by large posterior neurons. (A) Three-dimensional projection of the distribution of Htt.RFP aggregates on day 30, as shown from frontal (Bottom), top (Upper), and side (Right panel) views. AL, antennal lobe; LPN, large posterior neurons. (B) Central brain stained with monoclonal antibody nb169. Red arrowheads mark a pair of large posterior neurons in the posterior protocerebrum. (C–E) On day 1, Htt.RFP aggregates (red) have started to accumulate within ORN terminals (green) on the anterior side of the brain (C), but not within large posterior cells (blue) (D and E). (F–H) By day 10, Htt.RFP aggregates colocalize with GFP in ORN terminals (F) and are now present in posterior cells as well (G and H). E and H are enlarged areas marked by boxes within D and G, respectively. (I) Orthogonal view of a single optical slice from H, demonstrating that Htt.RFP aggregates are localized within large posterior cells. (J and K) Accumulation of Htt.RFP aggregates in large posterior neurons on day 20 (J) and day 30 (K). (L) The number of Htt.RFP aggregates within individual large posterior neurons is quantified at various time points. ***P < 0.001 using Student’s t test. Black bars represent mean values for each condition. (Scale bar in B, 50 μm; scale bar in G, 50 μm, also applies to C, D, and F; and scale bar in H, 5 μm, also applies to E, J, and K.)
Spreading of Htt Aggregates Causes Non–Cell-Autonomous Damage.
Aggregates clearly spread beyond the original expression pattern and accumulate in other neurons. However, it remained unclear whether this spreading resulted in non–cell-autonomous damage, which has been demonstrated in neurodegenerative diseases. Recently, non–cell-autonomous neurodegeneration was demonstrated in a Drosophila model of Parkinson’s disease, where expression of leucine-rich repeat kinase 2 (LRRK2-G2019S) in dopaminergic neurons caused cell death in photoreceptors (27). Additionally, human neurons integrated in Huntington’s disease mouse model brain slices developed abnormal morphology, including shorter and fewer primary and secondary neurites when bearing mutant huntingtin aggregates (11).
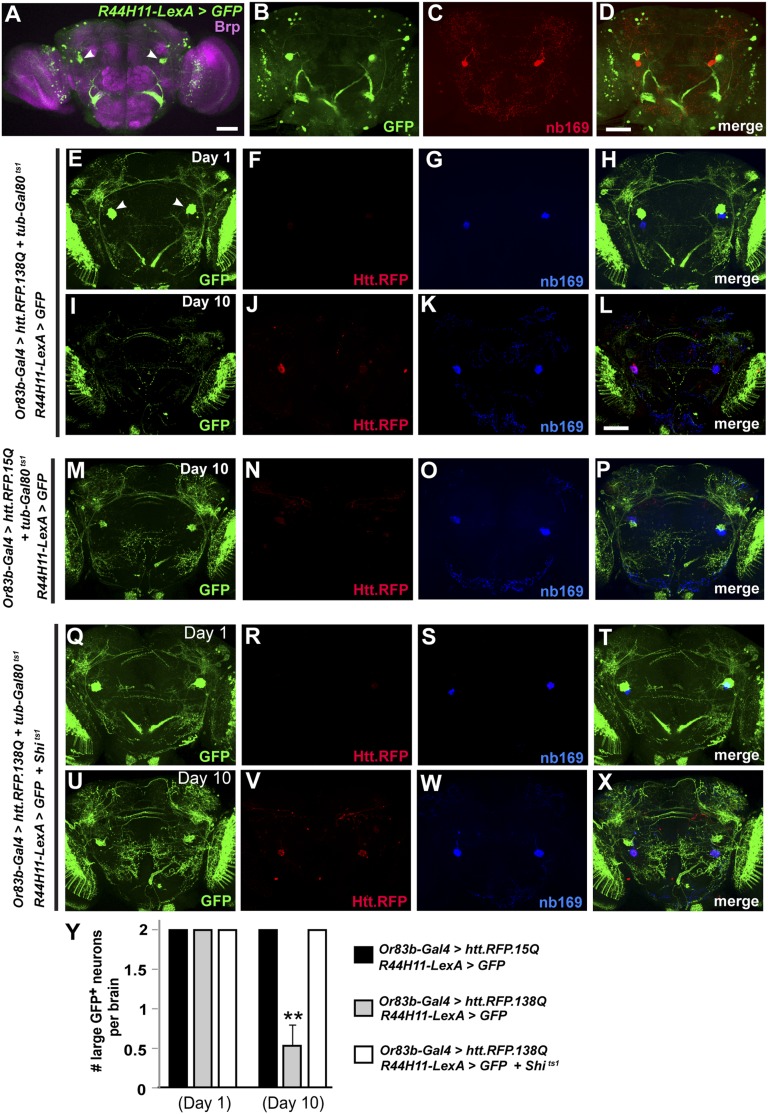
To examine occurrence of non–cell-autonomous pathology in our spreading model and to determine if blocking the uptake of aggregates would serve a neuroprotective function, we first searched through the Janelia Gal4 collection (28) to find GFP expression patterns that labeled the pair of large posterior cells so that they could be genetically manipulated independently of Gal4. We found one line (R44H11) that labeled a pair of large posterior neurons and also had an existing LexA line available. R44H11-LexA driving LexAop-GFP clearly labeled a pair of large neurons in the posterior protocerebrum (Fig. 3A). However, when we costained with GFP and nb169, we found that neurons labeled with R44H11-LexA > LexAop::GFP and those positive for nb169 were separate pairs of neurons (Fig. 3 B–D). When we drove expression of of Htt.RFP in ORNs using tubulin-Gal80ts1 to prevent Htt.RFP expression before eclosion and costained with nb169 and GFP, we observed a surprising result: Although we again saw aggregates of Htt.RFP in nb169-positive neurons, the large GFP-expressing cells were no longer detectable (Fig. 3 E–L). These neurons were not lost when using the nonpathogenic Htt.RFP.15Q controls (Fig. 3 M–P). These results suggest that expression of Htt.RFP in ORNs results in the loss of large posterior cells labeled by R44H11-LexA > LexAop::GFP within 10 d.
Fig. 3.
Blocking endocytosis with shibire mutant protects against neurodegeneration. (A) Expression pattern of R44H11-LexA in the adult central brain. Pair of large posterior neurons are marked by white arrowheads. Neuropil is labeled by anti-Brp (pink). (B–D) Comparison of large cells labeled by R44H11-LexA (B) and nb169 (C) showing that these labels mark distinct cells (D). (E–H) At day 1 following expression of Htt.RFP.138Q in ORNs, few HTT aggregates are observed and both R44H11-LexA > GFP-positive cells and nb169-positive cells are present. (I–L) At day 10, spreading of Htt aggregates is noticeable and the GFP-positive cells are no longer detectable, although the nb169-positive cells are still present. (M–P) Expression of Htt.RFP.15Q as a control, showing the presence of GFP-positive cells at day 10. (Q–X) Identical brain images as in E–L, but with endocytosis now blocked in R44H11-LexA > GFP-positive cells by coexpression of LexAop-Shits1. Note that at day 10 (U–X) under these conditions GFP-positive cells are still present. (Y) Average number of large posterior GFP-positive neurons labeled using R44H11-LexA in all conditions at day 1 and day 10. **P < 0.01 using Student’s t test. (Scale bar in A, 50 μm; scale bar in D, 50 μm, also applies to B and C; scale bar in L, 50 μm, also applies to E–K and M–X.)
If this loss of neurons was due to the uptake of mutant huntingtin, we hypothesized that blocking endocytosis in these neurons could be neuroprotective. To test this hypothesis, we expressed a temperature-sensitive form of dynamin, encoded by shibire (LexAop-shits1) (29), which acts in a dominant negative manner at restrictive temperatures. Flies were raised at 18 °C until eclosion to repress Gal4 expression and then shifted to 32 °C, the restrictive temperature for shibirets1. This same temperature also relieves Gal80ts1-induced repression of Gal4 to ensure that Htt aggregation would not begin before impairing endocytosis in the target cells. When endocytosis was blocked in the R44H11-LexA–expressing cells by coexpressing shibirets1, we found that these neurons were no longer lost (Fig. 3 Q–Y). These data demonstrate non–cell-autonomous neurodegeneration in our model and further indicate that blocking endocytosis is protective in otherwise vulnerable neurons.
Aggregate Spreading Patterns Vary Using Different Gal4 Drivers.
Recent studies have shown that the transmission of pathogenic proteins often spreads through neuronal circuits (7, 8). If propagation in our model were primarily through circuits, we would expect to see very different patterns of aggregate accumulation when mutant huntingtin is expressed in different regions of the brain. When Htt.RFP was expressed in ORNs, we saw widespread accumulation throughout the brain, with certain areas of high concentration such as the nb169-positive cells.
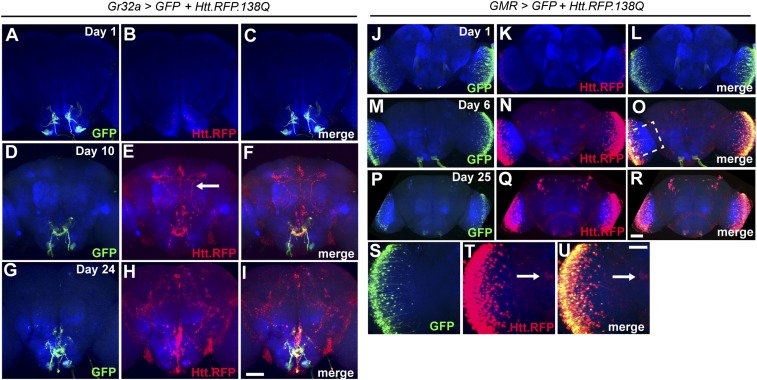
To test whether a similar type of spreading occurs in different populations of neurons, we expressed UAS-htt.RFP.138Q together with UAS-mCD8-GFP in various subsets of neurons in the Drosophila brain. First, we used Gr32a-Gal4 to drive expression in a subset of gustatory receptor neurons that send axonal projections to the subesophageal ganglion (30) (Fig. 4 A–C). In 10-d-old adults, Htt aggregates can be seen spreading far beyond the subesophageal ganglion (Fig. 4 D–F). In particular, aggregates were seen in prominent projections to dorsal areas of the central brain (Fig. 4E, arrow). By day 24, the aggregation pattern was much more diffuse. At this point, individual projections do not stand out as much, but rather the entire area of the central brain was covered (Fig. 4 G–I). Note that there was not much accumulation in the optic lobes as observed with Or83b-Gal4.
Fig. 4.
Expression of Htt.RFP.138Q using different Gal4 drivers results in distinct patterns of aggregate spreading. (A–I) Expression pattern of Gr32a-Gal4 (green) and distribution of Htt.RFP aggregates (red) in the adult brain at day 1 (A–C), day 10 (D–F), and day 24 (G–I). Arrow in E points to an axonal projection containing aggregates. Neuropil is labeled by anti-Brp (blue). (J–R) Expression pattern of GMR-Gal4 (green) in the adult brain and optic lobes and distribution of Htt.RFP aggregates (red) at day 1 (J–L), day 6 (M–O), and day 25 (P–R). (S–U) Enlarged view of the optic lobe from O. Arrow in T and U point to a cell body in the central brain that has taken up aggregates. (Scale bar in I, 50 μm, also applies to A–H; scale bar in R, 50 μm, also applies to J–Q; and scale bar in U, 20 μm, also applies to S and T.)
Next, we used GMR-Gal4 to drive expression in photoreceptors in the optic lobe (31) (Fig. 4 J–L). By day 6 after eclosion, we found Htt aggregates beyond the GFP expression pattern within the optic lobe, as well as within neurons located in the central brain (Fig. 4 M–O and S–U, arrows in T and U). By day 25 the spreading was again more diffuse, with aggregates present in regions throughout the entire brain (Fig. 4 P–R). These results reveal the capacity of protein aggregates to spread from various neurons in the Drosophila brain, with both local neurons and synaptic partners contributing to some degree to the unique patterns of accumulation depending on the initial expression pattern.
Spreading of Htt Aggregates Requires NSF1 and Dynamin.
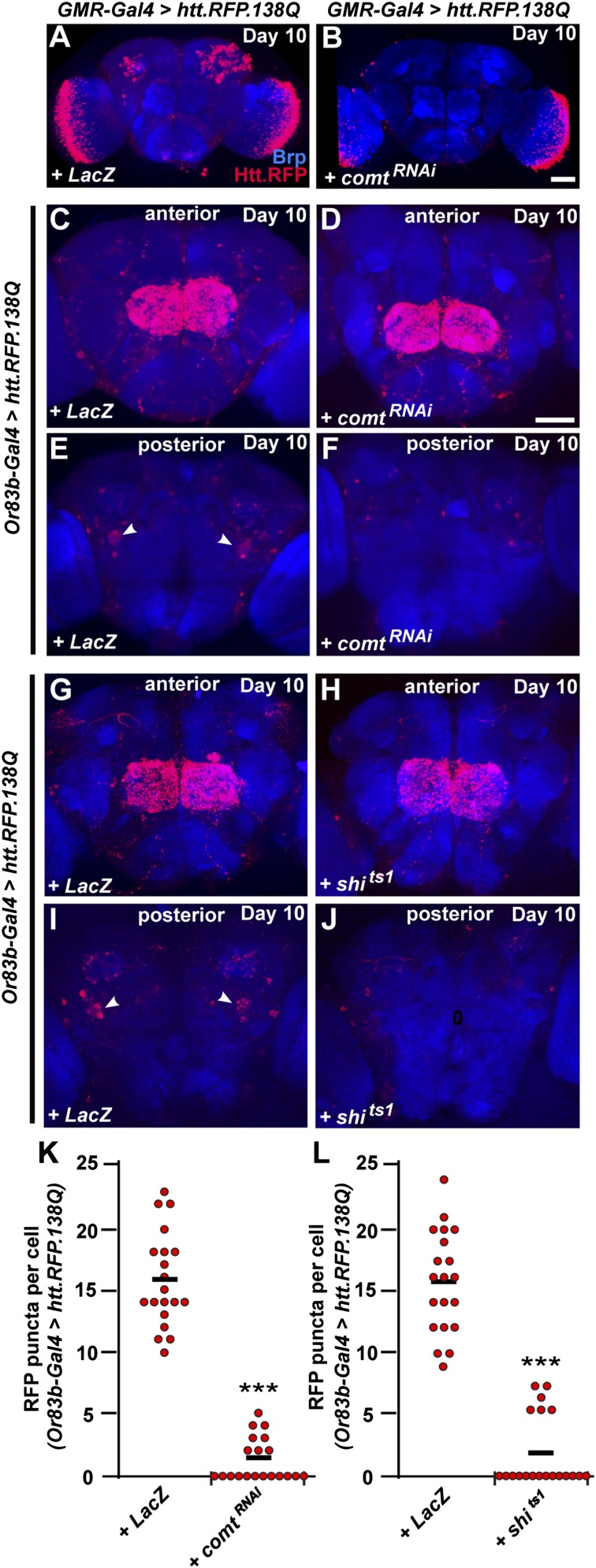
Previous work has shown that pharmacological application of tetanus toxins to inhibit the SNARE machinery prevents the spread of mutant huntingtin aggregates in cultured cells (11). To test the requirement of SNARE-mediated fusion events in the spread of Htt aggregates throughout the Drosophila brain, we knocked down expression of NSF1, encoded by comatose (comt). NSF1 is required for the disassembly and recycling of SNARE complexes involved in synaptic transmission (32–34) and is also required for fusion events involving lysosomal trafficking and autophagy (35). To inhibit NSF1 function in neurons expressing UAS-mRFP.Htt.138Q, we coexpressed UAS-comtRNAi (36). Although spreading of Htt aggregates from photoreceptors into the central brain is evident by day 10 in controls (Fig. 5A), the amount of spreading beyond photoreceptors is significantly diminished upon coexpression of UAS-comtRNAi (Fig. 5B). We found similar results when examining spreading from ORNs, where coexpression of UAS-comtRNAi resulted in less abundant spreading beyond the antennal lobe in anterior regions of the brain by day 10 (Fig. 5 C and D) as well as a lack of spreading to a pair of large cells on the posterior side of the brain (Fig. 5 E, F, and K). These results demonstrate that NSF1 and SNARE-mediated fusion events are required for the spread of HTT aggregates between neurons in the Drosophila brain. We found similar results by interfering with dynamin function in ORNs, where coexpression of UAS-shits1 resulted in a significant decrease in the number of aggregates present in large posterior neurons by day 10 (Fig. 5 G–J and L), further demonstrating that spreading of Htt aggregates requires the exocytotic machinery.
Fig. 5.
Spreading of Htt aggregates requires exocytosis. (A) Spreading pattern of Htt aggregates (red) into the central brain by day 10 when expressed in photoreceptors using GMR-Gal4. (B) Spreading pattern of Htt aggregates when UAS-comtRNAi is coexpressed to inhibit SNARE-mediated fusion. UAS-LacZ is coexpressed in A to standardize the number of transgenes expressed. (C and D) Anterior view of spreading pattern of Htt aggregates when expressed in ORNs using Or83b-Gal4 along with UAS-LacZ (C) or UAS-comtRNAi (D). (E and F) Posterior view of spreading pattern from C and D, respectively. Arrowheads in E mark large posterior cells with accumulated Htt aggregates in control, but not UAS-comtRNAi brains. (G and H) Anterior view of spreading pattern of Htt aggregates when expressed in ORNs using Or83b-Gal4 along with UAS-LacZ (G) or UAS-Shits1 (H). (I and J) Posterior view of spreading pattern from G and H, respectively. Arrowheads in I mark large posterior cells with accumulated Htt aggregates in control, but not UAS-Shits1 brains. Neuropil is labeled by anti-Brp (blue). (Scale bar in B, 50 μm, also applies to A; scale bar in D, 50 μm, also applies to C and E–J.) (K and L) The number of Htt.RFP aggregates found within large posterior neurons at day 10 in controls compared with UAS-comtRNAi (K) or UAS-Shits1 (L) brains. ***P < 0.001 using Student’s t test. Black bars represent mean values for each condition.
Transmission Is Not Observed in All PolyQ-Expanded Aggregate-Prone Proteins.
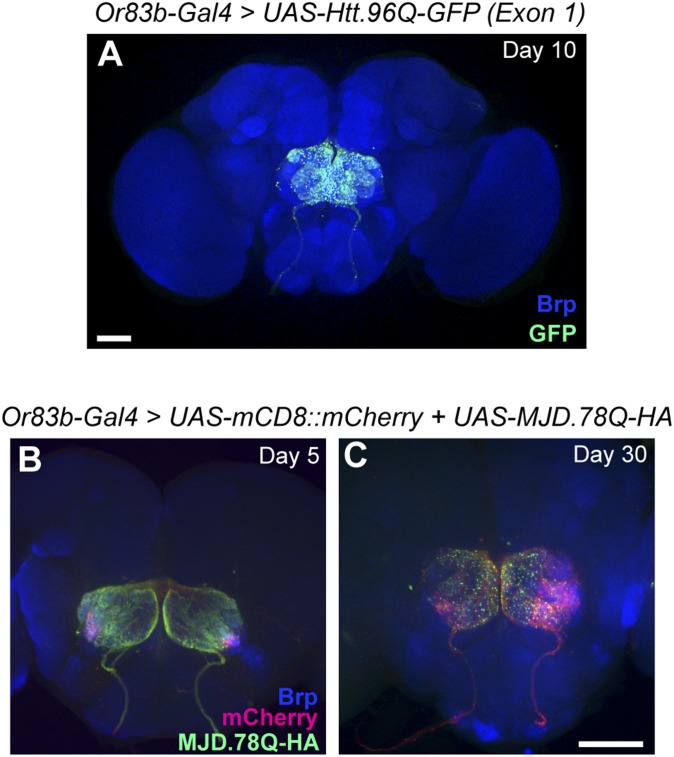
Is the pathogenic spreading observed by expression of mutant huntingtin common to all polyQ-expanded proteins? We first tested whether a different polyQ-expanded Htt construct would spread similarly to the 588-aa N-terminal fragment. Interestingly, we found that expression of a polyQ-expanded Htt exon 1 fragment (UAS-Htt.96Q-GFP exon 1) (22) resulted in accumulation of Htt.GFP aggregates in ORN that failed to spread beyond the antennal lobe by day 10 (Fig. S1A). This result suggests that aggregation of the polyQ-expanded exon 1 fragment is not sufficient to induce spreading. One possible reason for this difference in spreading behavior from the 588-aa N-terminal fragment is the presence of additional protein interaction sites included in the latter fragment (37).
Fig. S1.
Not all polyQ aggregates exhibit spreading in Drosophila brain. (A) Expression of polyQ-expanded GFP-tagged Htt exon 1 using or83b-Gal4. GFP-tagged aggregates do not spread beyond ORN terminals at day 10. (B and C) Expression of HA-tagged UAS-MJDtr-Q78 (green) together with UAS-mCD8-mCherry (red) to mark synaptic terminals in ORNs. HA-tagged aggregates do not spread beyond ORN terminals at day 5 (B) or day 30 (C). Neuropil is labeled by anti-Brp (blue). (Scale bar in A, 50 μm; scale bar in C, 50 μm, also applies to B.)
To test whether other pathogenic proteins with polyQ expansions show similar spreading behavior as the 588-aa Htt fragment, we expressed a truncated ataxin-3 construct with a pathogenic polyQ expansion (UAS-MJDtr-Q78) (14) with a hemagglutinin (HA) tag used to model spinocerebellar ataxia type 3 in Drosophila. Similar to Htt, we find that aggregates can be seen at ORN terminals in young flies (Fig. S1B). Although the number of aggregates increases by day 30, we do not observe widespread deposition of aggregates elsewhere in the brain as we did with the 588-aa Htt fragment (Fig. S1C). These results suggest that propagation of protein aggregates is not a feature of all polyglutamine-expanded proteins. It will be interesting to compare the properties of pathogenic proteins that spread versus those that do not to identify key mechanisms responsible for transmission.
Discussion
The ability of misfolded proteins to aggregate and spread throughout the brain has major implications for neurodegenerative diseases. However, there are still many unanswered questions regarding how spreading occurs and its consequences for disease progression. Here we demonstrate that mutant huntingtin aggregates spread throughout the Drosophila brain. Although aggregates initially accumulate at ORN synaptic terminals in the antennal lobe, over time these aggregates are distributed more broadly to the far posterior and lateral regions of the brain. After release from ORN terminals, we found that Htt aggregates become internalized in other populations of neurons. The most prominent accumulation we noticed was in a pair of large, possibly peptidergic neurons in the posterior protocerebrum.
Selective vulnerability of particular neurons is a common feature of many neurodegenerative diseases, including HD (38, 39). In HD there is a lack of correlation between neurons in which aggregates accumulate and neuronal loss. For example, striatal spiny projection neurons are particularly vulnerable in HD, yet these neurons accumulate far fewer aggregates than striatal interneurons (40). We observed a similar outcome in our model: neurons labeled with the nb169 monoclonal antibody accumulate Htt aggregates but they do not seem vulnerable to cell death. In contrast, neighboring neurons that express the R44H11-LexA driver are lost within 10 d after eclosion. One possible explanation for this discrepancy is that the most vulnerable neurons simply are not viable long enough to accumulate a quantity of Htt aggregates. Therefore, the only neurons where accumulation of aggregates can be seen in abundance are those that are most resistant to the toxic effects of the aggregates. Whereas the underlying cause of this selective vulnerability remains unknown, some leading ideas include differences in the microenvironment, metabolic activity, and translational machinery between neuronal populations (41, 42).
One striking result was that loss of the R44H11-LexA–expressing GFP+ neurons was prevented by blocking endocytosis in these cells. This result suggests that Htt.RFP protein is actively internalized by target neurons. Transmission of α-synuclein between cells in culture also depends on endocytosis (43), demonstrating that there may be some similarities between various pathogenic proteins in mechanism of transfer. Although we did not observe large aggregates in R44H11-LexA–expressing cells before loss of these neurons, it is possible that monomers or oligomers were transmitted, which would be difficult to detect. This possibility is also consistent with previous results, demonstrating that both aggregates and more soluble forms of the protein are likely pathogenic (22).
Understanding the cellular pathways involved in spreading of pathogenic proteins is an important next step because of its potential impact on therapeutic intervention. Although there is abundant evidence that spreading occurs through synaptic connections, other potential mechanisms include spreading between cells via exosomes (43, 44) or tunneling nanotubes (45). In the current study, we find unique patterns of spreading when mutant Htt is expressed in different subsets of neurons in the brain. This observation supports the idea that transcellular spreading is more likely to involve neurons in close proximity or within the same circuit as those containing aggregates. However, rapid accumulation of Htt aggregates throughout the brain when expressed in olfactory receptor neurons suggests that synaptic connections are not solely responsible for the spreading we observed. In addition to transneuronal spreading, mutant Htt aggregates have also recently been shown to spread to nearby phagocytic glia and are responsible for the prion-like conversion of soluble wild-type Htt (19). Although these glia provide a neuroprotective role through clearance of extracellular aggregates, they may also contribute to disease pathogenesis by spreading the aggregates themselves (19).
Our results demonstrate that release of Htt aggregates requires both NSF1 and dynamin, suggesting that SNARE-mediated fusion events play an important role in the spreading of pathology. This is consistent with previous data revealing that tetanus toxins targeting components of the synaptic vesicle fusion machinery block spreading of aggregates in culture (11). Although inhibition of NSF1 or dynamin significantly limited the spreading, it was not blocked completely. One possible reason for this is that normal protein function was not completely eliminated by our genetic manipulation. Alternatively, spreading of protein aggregates may also operate via additional mechanisms independent of SNARE-mediated fusion events such as release from dead or damaged cells. By use of a candidate gene approach as well as unbiased genetic screens in Drosophila, it should now be possible to identify additional modifiers that regulate spreading of Htt aggregates in vivo.
Our results demonstrate that whereas polyglutamine-expanded huntingtin aggregates can spread throughout the brain in Drosophila, polyglutamine-expanded ataxin-3 lacks this property. Furthermore, we established a distinction between the spreading capacities of both a 588-aa fragment of Htt and an 81-aa fragment containing only exon 1. The lack of spreading seen using the exon 1 fragment suggests that specific regions of the protein are required for transmission throughout the brain. These differences should help to identify properties of aggregate-prone proteins that influence the ability to spread and also highlight the need to consider specific forms of proteins used when modeling these diseases. Differences among various disease-associated, aggregate-prone protein in their ability to spread from cell to cell may depend on the type of aggregates they form or the cell type in which they are first expressed. By taking advantage of Drosophila to characterize spreading of other aggregate-prone proteins, it should now be possible to define the precise cellular and molecular mechanisms that are responsible and to determine why some proteins are more likely to undergo spreading.
Materials and Methods
Fly Strains.
Flies were raised on standard Drosophila medium at 25 °C unless otherwise noted. UAS-mRFP.Htt.138Q, UAS-mRFP.Htt.15Q, and UAS-Htt.96Q-GFP exon 1 (22) were obtained from Troy Littleton (Massachusetts Institute of Technology, Cambridge, MA). 13XLexAop2-IVS-Syn21-Shibirets1 (pJFRC104) and UAS-TTS-Shibirets1 (pJFRC100) (29) were provided by Gerald Rubin (Janelia Farm Research Campus, Ashburn, VA). The following lines were obtained from the Bloomington Drosophila Stock Center at Indiana University: Or83b-Gal4 (no. 23292) (46), UAS-syt.eGFP (47), GMR-Gal4 (31), Gr32a-Gal4 (30), R44H11-LexA (28, 48), UAS-mCD8-GFP, UAS-LacZ, LexAop-mCD8-GFP, Tubulin-Gal80ts1 (24), UAS-MJDtr-Q78 (14) UAS-mCD8-mCherry. UAS-comtRNAi (no. 105552) was obtained from the Vienna Drosophila RNAi Center.
Immunohistochemistry.
Immunohistochemistry was performed as previously described (35). Brains were dissected in PBS and fixed in 4% (vol/vol) formaldehyde at room temperature for 20 min. Brains were then incubated in blocking buffer (PBS with 0.2% Triton X-100 and 0.1% normal goat serum) for 1 h. Samples were then placed in primary antibodies overnight at 4 °C. After five washes in PBS, samples were incubated in secondary antibodies for 2 h at room temperature. Finally, brains were washed five times in PBS and mounted in Vectashield (Vector Laboratories). Primary antibodies include chicken anti-GFP (1:500) (Life Technologies), rabbit anti-DsRed (1:500) (Clontech), mouse anti-Bruchpilot (Brp) (1:50) (Developmental Studies Hybridoma Bank, University of Iowa), rat anti-HA (1:50) (Sigma-Aldrich). The mouse monoclonal antibody nb169 (1:20) (Würzburg Hybridoma Library) (25, 26) was provided by Erich Buchner (University of Würzburg, Würzburg, Germany). Species-specific Alexa Fluor conjugated secondary antibodies were used at 1:500 (Invitrogen), including Alexa Fluor 488, 568, and 633.
Imaging.
Images were acquired using a Zeiss LSM confocal microscope. Series of 1-μm z-stacks were taken for each image using a 20×/0.8 NA Plan-APOCHROMAT, 40×/1.3 NA Plan-NEOFLUAR, or 63×/1.4 NA Plan-APOCHROMAT objective. Image brightness and contrast were adjusted using ImageJ software (National Institutes of Health) and Adobe Photoshop CS5.
Statistical Analysis.
Aggregate number and cell viability were each measured and analyzed by use of the Student’s t test, with Bonferroni coffections applied in cases where multiple comparisons were made. Statistics were analyzed using SPSS software (IBM).
Supplementary Material
Acknowledgments
We thank Troy Littleton, Gerald Rubin, and Erich Buchner for fly stocks and reagents and David Wassarman, Grace Boekhoff-Falk, and all members of the B.G. laboratory for critical comments and helpful discussion of the manuscript. This work was supported by National Institutes of Health Grants F32 NS078958 (to D.T.B.) and R01 AG033620 and R01 NS15390 (to B.G.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1516217112/-/DCSupplemental.
References
- 1.Braak H, Braak E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 1991;82(4):239–259. doi: 10.1007/BF00308809. [DOI] [PubMed] [Google Scholar]
- 2.Braak H, et al. Staging of brain pathology related to sporadic Parkinson’s disease. Neurobiol Aging. 2003;24(2):197–211. doi: 10.1016/s0197-4580(02)00065-9. [DOI] [PubMed] [Google Scholar]
- 3.Braak H, Rüb U, Gai WP, Del Tredici K. Idiopathic Parkinson’s disease: Possible routes by which vulnerable neuronal types may be subject to neuroinvasion by an unknown pathogen. J Neural Transm. 2003;110(5):517–536. doi: 10.1007/s00702-002-0808-2. [DOI] [PubMed] [Google Scholar]
- 4.Ravits J, Laurie P, Fan Y, Moore DH. Implications of ALS focality: Rostral-caudal distribution of lower motor neuron loss postmortem. Neurology. 2007;68(19):1576–1582. doi: 10.1212/01.wnl.0000261045.57095.56. [DOI] [PubMed] [Google Scholar]
- 5.Ravits J, Paul P, Jorg C. Focality of upper and lower motor neuron degeneration at the clinical onset of ALS. Neurology. 2007;68(19):1571–1575. doi: 10.1212/01.wnl.0000260965.20021.47. [DOI] [PubMed] [Google Scholar]
- 6.Deng YP, et al. Differential loss of striatal projection systems in Huntington’s disease: A quantitative immunohistochemical study. J Chem Neuroanat. 2004;27(3):143–164. doi: 10.1016/j.jchemneu.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 7.Raj A, Kuceyeski A, Weiner M. A network diffusion model of disease progression in dementia. Neuron. 2012;73(6):1204–1215. doi: 10.1016/j.neuron.2011.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou J, Gennatas ED, Kramer JH, Miller BL, Seeley WW. Predicting regional neurodegeneration from the healthy brain functional connectome. Neuron. 2012;73(6):1216–1227. doi: 10.1016/j.neuron.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo JL, Lee VM. Cell-to-cell transmission of pathogenic proteins in neurodegenerative diseases. Nat Med. 2014;20(2):130–138. doi: 10.1038/nm.3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Soto C. Transmissible proteins: Expanding the prion heresy. Cell. 2012;149(5):968–977. doi: 10.1016/j.cell.2012.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pecho-Vrieseling E, et al. Transneuronal propagation of mutant huntingtin contributes to non-cell autonomous pathology in neurons. Nat Neurosci. 2014;17(8):1064–1072. doi: 10.1038/nn.3761. [DOI] [PubMed] [Google Scholar]
- 12.Feany MB, Bender WW. A Drosophila model of Parkinson’s disease. Nature. 2000;404(6776):394–398. doi: 10.1038/35006074. [DOI] [PubMed] [Google Scholar]
- 13.Fernandez-Funez P, et al. Identification of genes that modify ataxin-1-induced neurodegeneration. Nature. 2000;408(6808):101–106. doi: 10.1038/35040584. [DOI] [PubMed] [Google Scholar]
- 14.Warrick JM, et al. Expanded polyglutamine protein forms nuclear inclusions and causes neural degeneration in Drosophila. Cell. 1998;93(6):939–949. doi: 10.1016/s0092-8674(00)81200-3. [DOI] [PubMed] [Google Scholar]
- 15.Gunawardena S, et al. Disruption of axonal transport by loss of huntingtin or expression of pathogenic polyQ proteins in Drosophila. Neuron. 2003;40(1):25–40. doi: 10.1016/s0896-6273(03)00594-4. [DOI] [PubMed] [Google Scholar]
- 16.Jackson GR, et al. Polyglutamine-expanded human huntingtin transgenes induce degeneration of Drosophila photoreceptor neurons. Neuron. 1998;21(3):633–642. doi: 10.1016/s0896-6273(00)80573-5. [DOI] [PubMed] [Google Scholar]
- 17.Lee WC, Yoshihara M, Littleton JT. Cytoplasmic aggregates trap polyglutamine-containing proteins and block axonal transport in a Drosophila model of Huntington’s disease. Proc Natl Acad Sci USA. 2004;101(9):3224–3229. doi: 10.1073/pnas.0400243101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Steffan JS, et al. Histone deacetylase inhibitors arrest polyglutamine-dependent neurodegeneration in Drosophila. Nature. 2001;413(6857):739–743. doi: 10.1038/35099568. [DOI] [PubMed] [Google Scholar]
- 19.Pearce MM, Spartz EJ, Hong W, Luo L, Kopito RR. Prion-like transmission of neuronal huntingtin aggregates to phagocytic glia in the Drosophila brain. Nat Commun. 2015;6:6768. doi: 10.1038/ncomms7768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118(2):401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 21.Lai SL, Lee T. Genetic mosaic with dual binary transcriptional systems in Drosophila. Nat Neurosci. 2006;9(5):703–709. doi: 10.1038/nn1681. [DOI] [PubMed] [Google Scholar]
- 22.Weiss KR, Kimura Y, Lee WC, Littleton JT. Huntingtin aggregation kinetics and their pathological role in a Drosophila Huntington’s disease model. Genetics. 2012;190(2):581–600. doi: 10.1534/genetics.111.133710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Graham RK, et al. Cleavage at the caspase-6 site is required for neuronal dysfunction and degeneration due to mutant huntingtin. Cell. 2006;125(6):1179–1191. doi: 10.1016/j.cell.2006.04.026. [DOI] [PubMed] [Google Scholar]
- 24.McGuire SE, Le PT, Osborn AJ, Matsumoto K, Davis RL. Spatiotemporal rescue of memory dysfunction in Drosophila. Science. 2003;302(5651):1765–1768. doi: 10.1126/science.1089035. [DOI] [PubMed] [Google Scholar]
- 25.Blanco Redondo B, et al. Identification and structural characterization of interneurons of the Drosophila brain by monoclonal antibodies of the würzburg hybridoma library. PLoS One. 2013;8(9):e75420. doi: 10.1371/journal.pone.0075420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hofbauer A, et al. The Wuerzburg hybridoma library against Drosophila brain. J Neurogenet. 2009;23(1-2):78–91. doi: 10.1080/01677060802471627. [DOI] [PubMed] [Google Scholar]
- 27.Hindle SJ, Elliott CJ. Spread of neuronal degeneration in a dopaminergic, Lrrk-G2019S model of Parkinson disease. Autophagy. 2013;9(6):936–938. doi: 10.4161/auto.24397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jenett A, et al. A GAL4-driver line resource for Drosophila neurobiology. Cell Reports. 2012;2(4):991–1001. doi: 10.1016/j.celrep.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pfeiffer BD, Truman JW, Rubin GM. Using translational enhancers to increase transgene expression in Drosophila. Proc Natl Acad Sci USA. 2012;109(17):6626–6631. doi: 10.1073/pnas.1204520109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weiss LA, Dahanukar A, Kwon JY, Banerjee D, Carlson JR. The molecular and cellular basis of bitter taste in Drosophila. Neuron. 2011;69(2):258–272. doi: 10.1016/j.neuron.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Freeman M. Reiterative use of the EGF receptor triggers differentiation of all cell types in the Drosophila eye. Cell. 1996;87(4):651–660. doi: 10.1016/s0092-8674(00)81385-9. [DOI] [PubMed] [Google Scholar]
- 32.Littleton JT, et al. SNARE-complex disassembly by NSF follows synaptic-vesicle fusion. Proc Natl Acad Sci USA. 2001;98(21):12233–12238. doi: 10.1073/pnas.221450198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sanyal S, Tolar LA, Pallanck L, Krishnan KS. Genetic interaction between shibire and comatose mutations in Drosophila suggest a role for snap-receptor complex assembly and disassembly for maintenance of synaptic vesicle cycling. Neurosci Lett. 2001;311(1):21–24. doi: 10.1016/s0304-3940(01)02125-5. [DOI] [PubMed] [Google Scholar]
- 34.Tolar LA, Pallanck L. NSF function in neurotransmitter release involves rearrangement of the SNARE complex downstream of synaptic vesicle docking. J Neurosci. 1998;18(24):10250–10256. doi: 10.1523/JNEUROSCI.18-24-10250.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Babcock DT, Shen W, Ganetzky B. A neuroprotective function of NSF1 sustains autophagy and lysosomal trafficking in Drosophila. Genetics. 2015;199(2):511–522. doi: 10.1534/genetics.114.172403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dietzl G, et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448(7150):151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- 37.Harjes P, Wanker EE. The hunt for huntingtin function: Interaction partners tell many different stories. Trends Biochem Sci. 2003;28(8):425–433. doi: 10.1016/S0968-0004(03)00168-3. [DOI] [PubMed] [Google Scholar]
- 38.Cowan CM, Raymond LA. Selective neuronal degeneration in Huntington’s disease. Curr Top Dev Biol. 2006;75:25–71. doi: 10.1016/S0070-2153(06)75002-5. [DOI] [PubMed] [Google Scholar]
- 39.Han I, You Y, Kordower JH, Brady ST, Morfini GA. Differential vulnerability of neurons in Huntington’s disease: The role of cell type-specific features. J Neurochem. 2010;113(5):1073–1091. doi: 10.1111/j.1471-4159.2010.06672.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kuemmerle S, et al. Huntington aggregates may not predict neuronal death in Huntington’s disease. Ann Neurol. 1999;46(6):842–849. [PubMed] [Google Scholar]
- 41.Jackson WS. Selective vulnerability to neurodegenerative disease: the curious case of Prion Protein. Dis Model Mech. 2014;7(1):21–29. doi: 10.1242/dmm.012146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saxena S, Caroni P. Selective neuronal vulnerability in neurodegenerative diseases: From stressor thresholds to degeneration. Neuron. 2011;71(1):35–48. doi: 10.1016/j.neuron.2011.06.031. [DOI] [PubMed] [Google Scholar]
- 43.Desplats P, et al. Inclusion formation and neuronal cell death through neuron-to-neuron transmission of alpha-synuclein. Proc Natl Acad Sci USA. 2009;106(31):13010–13015. doi: 10.1073/pnas.0903691106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fraser KB, et al. LRRK2 secretion in exosomes is regulated by 14-3-3. Hum Mol Genet. 2013;22(24):4988–5000. doi: 10.1093/hmg/ddt346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Costanzo M, et al. Transfer of polyglutamine aggregates in neuronal cells occurs in tunneling nanotubes. J Cell Sci. 2013;126(Pt 16):3678–3685. doi: 10.1242/jcs.126086. [DOI] [PubMed] [Google Scholar]
- 46.Kreher SA, Kwon JY, Carlson JR. The molecular basis of odor coding in the Drosophila larva. Neuron. 2005;46(3):445–456. doi: 10.1016/j.neuron.2005.04.007. [DOI] [PubMed] [Google Scholar]
- 47.Zhang YQ, Rodesch CK, Broadie K. Living synaptic vesicle marker: Synaptotagmin-GFP. Genesis. 2002;34(1-2):142–145. doi: 10.1002/gene.10144. [DOI] [PubMed] [Google Scholar]
- 48.Pfeiffer BD, et al. Refinement of tools for targeted gene expression in Drosophila. Genetics. 2010;186(2):735–755. doi: 10.1534/genetics.110.119917. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.