Abstract
Introduction
Recent studies examining the association of Toll-like receptor 3 (TLR3) gene polymorphisms with the risk of developing various types of cancer have reported conflicting results. Clarifying this association could advance our knowledge of the influence of TLR3 single nucleotide polymorphisms (SNPs) on cancer risk.
Methods
We systematically reviewed studies that focused on a collection of 12 SNPs located in the TLR3 gene and the details by which these SNPs influenced cancer risk. Additionally, 14 case-control studies comprising a total of 7997 cases of cancer and 8699 controls were included in a meta-analysis of 4 highly studied SNPs (rs3775290, rs3775291, rs3775292, and rs5743312).
Results
The variant TLR3 genotype rs5743312 (C9948T, intron 3, C > T) was significantly associated with an increased cancer risk as compared with the wild-type allele (odds ratio [OR] = 1.11, 95 % confidence interval [CI] = 1.00–1.24, P = 0.047). No such association was observed with other TLR3 SNPs. In the stratified analysis, the rs3775290 (C13766T, C > T) variant genotype was found to be significantly associated with an increased cancer risk in Asian populations. Additionally, the rs3775291 (G13909A, G > A) variant genotype was significantly associated with an increased cancer risk in Asians, subgroup with hospital-based controls, and subgroup with a small sample size.
Conclusion
After data integration, our findings suggest that the TLR3 rs5743312 polymorphism may contribute to an increased cancer risk.
Keywords: The Toll-like receptor 3 (TLR3) gene, Single nucleotide polymorphism, Cancer risk, Meta-analysis, Systematic review
Background
Toll-like receptors (TLRs) are members of a membrane receptor protein family that recognize the antigenic determinants of viruses, bacteria, protozoa, and fungi, and are thus associated with immunity. There are two pathways associated with the immune deficiencies that lead to disease: the MyD88-IRAK4 pathway, involving all TLR family proteins except TLR3, and the TLR3-Unc93b-TRIF-TRAF3 pathway [1, 2]. Therefore, TLR3 can be considered a unique protein in the TLR family, and it has been further implicated in the development of tumors resulting from an activated immune system.
TLR3 single nucleotide polymorphisms (SNPs) could possibly affect cancer susceptibility and could therefore serve as potential biomarkers to evaluate cancer risk [3]. Thus, TLR3 polymorphisms that are associated with both heredity and environmental factors may be critical in bridging the relationship between genetic factors and external environmental conditions. TLR3 SNPs were originally identified by He et al. [4] in 2007. Since then, several studies have focused on examining the association of TLR3 SNPs with cancer risk. However, the first studied SNP site included 10 SNPs covering the entire TLR3 gene, and because each of these SNPs had several reported names, confusion arose in the literature. Furthermore, published results of the TLR3 gene have been conflicting. The TLR3 SNPs that exhibit the most variability are rs3775290 and rs3775291; however, their relationship with cancer risk remains unclear. For instance, although the majority of studies have reported that the rs3775290 variant T allele was associated with an increased cancer risk (odds ratio [OR] > 1), four studies testing this conclusion did not reach statistical significance (P > 0.05) [5–8] and only one additional study produced significant results (P < 0.05) [9]. Furthermore, another study reported the opposite result that the variant allele was associated with a decreased cancer risk (OR = 0.88) [4]. Thus, a comprehensive analysis integrating all studies on TLR3 SNPs and cancer risk is needed. In addition, no systematic review or meta-analysis for the TLR3 gene polymorphisms has been performed.
Here, we systematically reviewed published data and comprehensively analyzed and integrated all published studies on the relationship between TLR3 SNPs and cancer risk. We also contacted the authors of these studies to obtain any data that was omitted from published articles so as to enhance our comprehension of the details surrounding these SNPs. We included a meta-analysis for hotspot SNPs (rs3775290, rs3775291, rs3775292, and rs5743312) that have been described in at least three published studies to enhance the comprehensiveness of our assessment into the association between TLR3 SNPs and cancer risk.
Methods
Publication search
A systematic literature search was performed on the association between the TLR3 rs3775290, rs3775291, rs3775292, rs5743312 polymorphisms and cancer risk. Our final search concluded with literature published on or before October 5th, 2014. Two independent researchers (Ben-Gang Wang and De-Hui Yi) searched the PubMed, Chinese National Knowledge Infrastructure (CNKI), and Web of Science databases for the following keywords: “TLR3,” “cancer/carcinoma/tumor/neoplasm,” and “polymorphism”. The inclusion criteria were as follows: (1) case-control studies, (2) studies evaluating the association between TLR3 polymorphisms and cancer risk, and (3) the SNP was reported in at least 3 publications. The major exclusion criteria were as follows: (1) studies that presented duplicate data, (2) studies that included only cancer patients (i.e., no healthy controls), and (3) studies that investigated benign diseases compared with controls.
Data extraction
Two independent researchers (Ben-Gang Wang and De-Hui Yi) each extracted all data that were considered to be relevant, and in cases of inconsistent selection, a third author (Yong-Feng Liu) participated in data selection. In this way, a consensus was reached on which studies should be included for analysis. For each study, the following items were collected: first author’s name, publication year, country of origin, ethnicity, cancer type, source of control groups (population- or hospital-based), genotyping method, total numbers of cases and controls, and genotype distributions in cases and controls. When we considered the published data to be insufficient for our analyses, we contacted the authors to obtain the original data. Thus, the data included in this review were obtained from both published and unpublished studies.
Statistical analyses
The Chi-square test was used to determine the Hardy-Weinberg equilibrium (HWE) for the genotype frequencies of different TLR3 polymorphisms; a result of P < 0.05 was considered to indicate significant disequilibrium. The strength of the association between TLR3 polymorphisms and cancer risk was calculated using odds ratios (ORs) with 95 % confidence intervals (CIs). The heterogeneity between different studies was calculated using the Cochran’s Q test and quantified by the I2 (a significance level of P < 0.10). When heterogeneity did not exist, a fixed-effect model based on the Mantel-Haenszel method was used to assess the pooled OR of each study [10]. When heterogeneity did exist, a random-effect model based on the method developed by DerSimonian and Laird was employed [11]. Five comparison models were evaluated: heterozygote comparison (M1: Aa vs. AA), homozygote comparison (M2: aa vs. AA), dominant model (M3: Aa + aa vs. AA), recessive model (M4: aa vs. AA + Aa), and allelic model (M5: a vs. A). “A” indicates wild allele, and “a” indicates variant allele. OR1 and OR2 were calculated for the genotypes aa vs. AA (M2) and Aa vs. AA (M1), and were used to determine the most appropriate genetic model. According to the reference [12], a recessive model is recommended in cases where OR1 ≠ 1 and OR2 = 1, whereas a dominant model is suggested for cases in which OR1 = OR2 ≠ 1, and a codominant model is indicated if OR1 > OR2 > 1 or OR1 < OR2 < 1.
For studies with sufficient patients, we also performed stratification analyses on cancer type, ethnicity (Asian or Caucasian), sources of controls (population- or hospital-based study design), and sample size (total samples ≥1000 [large sample size] or <1000 [small sample size]). The Begg’s rank correlation and the Egger’s linear regression tests were used to evaluate publication bias [13, 14]. A value of P < 0.10 was considered statistically significant. All analyses were performed using STATA software, version 11.0 (STATA Corp., College Station, TX, USA).
Results
Characteristics of the included studies
Searches of the PubMed, CNKI, and Web of Science databases using different combinations of our keywords yielded a total of 155 records (after duplicates were removed). We excluded 50 studies based on the information presented in the title or abstract (17 were irrelevant articles, 16 were functional studies rather than polymorphism studies, and 17 were review articles) and 91 studies based on the information presented in the text (5 were not case-control studies, 62 were not about TLR3 gene polymorphisms, and 24 were not relevant to cancer). Thus, a total of 14 case-control studies that met our inclusion criteria were included in our systematic review and final meta-analysis, which consisted of 7997 cancer patients and 8699 cancer-free controls [4–9, 15–22] (Table 1).
Table 1.
The studies included in this systematic review for the association between Toll-like receptor 3 (TLR3) single nucleotide polymorphisms (SNPs) and the risk of developing cancer
| Publication year | Study | Country | Sample size | Source of controls | Genotyping method | Matched factors | Adjusted factors | |
|---|---|---|---|---|---|---|---|---|
| Cases | Controls | |||||||
| 2007 | He et al. [4] | China | 434 | 512 | Population-based | Sequencing | Sex and age matched | None |
| 2009 | Etokebe et al. [8] | Croatia | 130 | 101 | Population-based | qPCR | All females, age not matched | None |
| 2010 | Lei et al. [15] | China | 981 | 1221 | Population-based | SNP Stream | All females, age matched | Age and BMI |
| 2011 | Pandey et al. [7] | India | 200 | 200 | Not mentioned, only healthy controls | PCR-RFLP | All females, age matched | Age |
| Gast et al. [16] | Germany | 763 | 736 | Hospital-based | Two multiplex PCR | Sex not mentioned, age not matched | None | |
| 2012 | Mandal et al. [6] | India | 195 | 250 | Hospital-based | PCR-RFLP | All males, age matched | |
| Slattery et al.[17] | USA | 2309 | 2915 | Population-based | Multiplexed bead array | Sex and age matched | None | |
| 2013 | Singh et al. [5] | India | 200 | 200 | Hospital-based | PCR-RFLP | Sex and age matched | Age, sex, and smoking |
| Li et al. [9] | China | 466 | 482 | Population-based | PCR-RFLP | Sex and age matched | Sex and age | |
| Resler et al. [18] | USA | 840 | 800 | Population-based | Chip | All females, age matched | Age | |
| Zeljic et al. [19] | Yugoslavia | 93 | 104 | Not mentioned, only healthy controls | qPCR | Sex and age matched | Sex, age, smoking, and alcohol consumption | |
| Yeyeodu et al. [20] | USA | 102 | 72 | Hospital-based | SNP Stream | All females, age not mentioned | None | |
| Moumad et al. [21] | Germany | 472 | 362 | Hospital-based | KASPar | Sex and age matched | Sex and age | |
| 2014 | Zidi et al. [22] | Tunisia | 130 | 200 | Hospital-based | PCR-RFLP | All females, age not matched | None |
qPCR, quantitative polymerase chain reaction; PCR-RFLP, polymerase chain reaction-restriction fragment length polymorphism; KASPar, KASPar SNP genotyping system; BMI, body mass index.
Of the 14 enrolled studies, all but 4 were matched by age; 6 were sex-matched, 1 did not mention the sex-matching, and 7 that examined either breast, cervical, or prostate cancer did not need to match for sex. Six studies investigated Asians, 7 investigated Caucasians, and 1 was conducted in Africa. The controls were hospital-based in 6 studies, population-based in 6 studies, and not mentioned in 2 studies. Genotyping methods included polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP, 5 studies), quantitative polymerase chain reaction (qPCR, 2 studies), Chip (1 study), multiplexed bead array (1 study), two multiplex PCR (1 study), the KASPar SNP genotyping system (1 study), SNP Stream (2 studies), and sequencing (1 study). Eight studies checked genotypes for quality control [4–6, 9, 16–18, 21]. The genotype distribution of controls was consistent with the Hardy-Weinberg equilibrium model in all but four studies [9, 15, 17, 22].
In these 14 studies, 12 SNPs were reported. Table 2 lists the SNP locations, studied tumor types, and any associations between TLR3 SNPs and cancer risk. In summary, there are 10 types of cancer that have been evaluated in relation to TLR3 SNPs. The majority of the 12 SNPs located within an intronic region of the TLR3 gene, except for 2 that were found in the 3′ and 5′ regions of the gene and another 2 that located within exons. Only 4 SNPs (rs3775290, rs3775291, rs3775292, and rs5743312) were reported in at least 3 studies. For this reason, we defined them as hotspot SNPs which covered all 10 cancer types included in this study (Table 3).
Table 2.
The associations between TLR3 polymorphisms and cancer risk in the 14 selected studies
| TLR3 SNP | Location | Association(s) with tumor(s) |
|---|---|---|
| rs10025405 | 3′-near gene | Breast cancer, associated [20] |
| rs11721827 | Intron 1 | Nasopharyngeal carcinoma, associated [4] |
| Colorectal cancer, associated [17] | ||
| rs11730143 | Intron 1 | Melanoma, not associated [16] |
| rs13126816 | Intron 1 | Melanoma, not associated [16] |
| rs3775290 | Exon 4 | Nasopharyngeal carcinoma, not associated [4] |
| Bladder cancer, not associated [5] | ||
| Prostate cancer, not associated [6] | ||
| Cervical cancer, not associated [7] | ||
| Breast cancer, not associated [8] | ||
| Cervical cancer, associated [22] | ||
| rs3775291 | Exon 4, L412F | Nasopharyngeal carcinoma, not associated [4] |
| Hepatocellular carcinoma, associated [9] | ||
| Breast cancer, not associated [15] | ||
| Melanoma, not associated [16] | ||
| Colorectal cancer, not associated [17] | ||
| Breast cancer, not associated [18] | ||
| Oral cancer, associated [19] | ||
| Nasopharyngeal carcinoma, associated [21] | ||
| rs3775292 | Intron 3 | Melanoma, not associated [16] |
| Colorectal cancer, associated [17] | ||
| Breast cancer, not associated [18] | ||
| rs5743305 | 5′-near gene | Hepatocellular carcinoma, not associated [9] |
| Colorectal cancer, not associated [17] | ||
| rs5743312 | Intron 3 | Nasopharyngeal carcinoma, not associated [4] |
| Melanoma, not associated [16] | ||
| Oral cancer, associated with survival [19] | ||
| rs7657186 | Intron 1 | Melanoma, not associated [16] |
| Breast cancer, not associated [20] | ||
| rs7668666 | Intron 3 | Melanoma, not associated [16] |
Table 3.
Characteristics of the 14 studies included for this meta-analysis
| Study | Ethnicity | Cancer type | Sample size | Cases | Controls | P of HWE | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Case | Control | wt/wt | wt/var | var/var | wt/wt | wt/var | var/var | ||||
| rs3775290 (C13766T, C > T) | |||||||||||
| He et al. [4] | Asian | Nasopharyngeal carcinoma | 395 | 371 | 565* | 225* | 510* | 232a | |||
| Singh et al. [5] | Asian | Bladder cancer | 200 | 200 | 106 | 81 | 13 | 122 | 71 | 7 | 0.391 |
| Mandal et al. [6] | Asian | Prostate cancer | 195 | 250 | 115 | 68 | 12 | 157 | 84 | 9 | 0.585 |
| Pandey et al. [7] | Asian | Cervical cancer | 200 | 200 | 91 | 98 | 11 | 110 | 81 | 9 | 0.217 |
| Etokebe et al. [8] | Caucasian | Breast cancer | 130 | 101 | 58 | 56 | 14 | 46 | 42 | 8 | 0.713 |
| Zidi et al. [22] | Caucasian | Cervical cancer | 130 | 200 | 69 | 48 | 13 | 76 | 106 | 18 | 0.026 |
| rs3775291 (G13909A, L412F, G > A) | |||||||||||
| He et al. [4] | Asian | Nasopharyngeal carcinoma | 333 | 287 | 405* | 261* | 359a | 215a | |||
| Li et al. [9] | Asian | Hepatocellular carcinoma | 466 | 482 | 192 | 222 | 52 | 256 | 203 | 23 | 0.030 |
| Lei et al. [15] | Asian | Breast cancer | 981 | 1221 | 447 | 431 | 103 | 594 | 500 | 127 | 0.155 |
| Gast et al. [16] | Caucasian | Skin malignant melanoma | 732 | 668 | 379 | 291 | 62 | 332 | 284 | 52 | 0.415 |
| Slattery et al. [17] | Caucasian | Colon cancer | 1554 | 1955 | 748 | 653 | 153 | 947 | 817 | 191 | 0.446 |
| Rectal cancer | 754 | 959 | 363 | 332 | 59 | 478 | 381 | 100 | 0.066 | ||
| Resler et al. [18] | Caucasian | Breast cancer | 840 | 801 | 427 | 348 | 65 | 418 | 318 | 65 | 0.679 |
| Zeljic et al. [19] | Caucasian | Oral squamous cell carcinomas | 93 | 104 | 39 | 39 | 15 | 43 | 53 | 8 | 0.128 |
| Moumad et al. [21] | African | Nasopharyngeal carcinoma | 472 | 362 | 289 | 170 | 13 | 252 | 96 | 14 | 0.210 |
| rs3775292 (C/G, intron 3, C > G) | |||||||||||
| Gast et al. [16] | Caucasian | Skin malignant melanoma | 730 | 668 | 464 | 233 | 33 | 413 | 228 | 27 | 0.521 |
| Slattery et al. [17] | Caucasian | Colon cancer | 1554 | 1956 | 990 | 507 | 57 | 1253 | 601 | 102 | 0.008 |
| Rectal cancer | 754 | 959 | 494 | 222 | 38 | 586 | 335 | 38 | 0.247 | ||
| Resler et al. [18] | Caucasian | Breast cancer | 840 | 800 | 522 | 284 | 34 | 509 | 262 | 29 | 0.507 |
| rs5743312 (C9948T, intron 3, C > T) | |||||||||||
| He et al. [4] | Asian | Nasopharyngeal carcinoma | 405 | 200 | 640* | 170a | 316* | 84a | |||
| Lei et al. [15] | Asian | Breast cancer | 996 | 1245 | 582 | 348 | 66 | 770 | 433 | 42 | 0.044 |
| Gast et al. [16] | Caucasian | Skin malignant melanoma | 716 | 657 | 500 | 200 | 16 | 453 | 193 | 11 | 0.060 |
| Zeljic et al. [19] | Caucasian | Oral squamous cell carcinomas | 93 | 104 | 69 | 21 | 3 | 78 | 24 | 2 | 0.923 |
wt/wt, wild type/wild type, indicating wild genotype; wt/var, wild type/variant type, indicating heterozygote; var/var, variant type/variant type, indicating variant genotype; HWE, Hardy-Weinberg equilibrium. *Only the data of allelic model were available, and thus the studies were analyzed only when the allelic model was used. The bold means the genotype information of the rs3775291 and rs3775292 SNPs that readers could not found in [17], and these data were kindly provided by the contacted authors.
Unpublished data obtained from the original authors
Slattery et al. [17] detected TLR3 rs3775291 and rs3775292 SNPs in colorectal cancer patients and showed only the minimum allele frequency for these SNPs. We contacted the authors of this study to obtain the genotype information of these two SNPs that were found in the case and control groups, and we were especially interested in the results when dividing patients into colon cancer and rectal cancer groups. These results are presented in bold in Table 3.
Quantitative synthesis
The variant T allele of the rs5743312 SNP was significantly associated with an increased risk of cancer when compared with the wild C allele (OR = 1.11, 95 % CI = 1.00–1.24, P = 0.047) (Table 4, Fig. 1a). The OR1 and OR2 values of this rs5743312 SNP were 1.88 (P < 0.001) and 1.02 (P = 0.832), respectively. Thus, a codominant model (M2) was the most appropriate choice for rs5743312. Regardless of the genetic model, no other TLR3 SNPs (rs3775290, rs3775291, and rs3775292) were found to be associated with cancer risks.
Table 4.
Pooled ORs and 95 % CIs of TLR3 polymorphisms in this meta-analysis
| SNP | Number of studies | M1 | M2 | M3 | M4 | Number of studies | M5 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR (95 % CI) | P | I 2 (%) | OR (95 % CI) | P | I2 (%) | OR (95 % CI) | P | I2 (%) | OR (95 % CI) | P | I2 (%) | OR (95 % CI) | P | I2 (%) | |||
| rs3775290 (C13766T, C > T) | 5 | 1.04 | 0.854* | 70.0 | 1.38 | 0.110 | 0.0 | 1.09 | 0.640* | 69.4 | 1.41 | 0.082 | 0.0 | 6 | 1.06 | 0.550 | 59.3 |
| (0.72-1.49) | (0.93-2.06) | (0.77-1.53) | (0.96-2.08) | (0.87-1.30) | |||||||||||||
| rs3775291 (G13909A, L412F, G > A) | 8 | 1.12 | 0.056* | 53.9 | 1.13 | 0.335* | 66.5 | 1.12 | 0.054* | 58.6 | 1.08 | 0.507* | 65.2 | 9 | 1.09 | 0.064* | 59.5 |
| (1.00-1.26) | (0.88-1.45) | (1.00-1.26) | (0.86-1.37) | (1.00-1.19) | |||||||||||||
| rs3775292 (C/G, intron 3, C > G) | 4 | 0.96 | 0.554* | 54.5 | 0.93 | 0.500 | 34.7 | 0.97 | 0.476 | 23.1 | 0.94 | 0.554 | 49.9 | 4 | 0.97 | 0.416 | 0.0 |
| (0.83-1.11) | (0.75-1.15) | (0.89-1.06) | (0.76-1.16) | (0.90-1.05) | |||||||||||||
| rs5743312 (C9948T, intron 3, C > T) | 3 | 1.02 | 0.832 | 0.0 | 1.88 | <0.001 | 0.0 | 1.08 | 0.267 | 0.0 | 1.86 | <0.001 | 0.0 | 4 | 1.11 | 0.047 | 7.1 |
| (0.88-1.17) | (1.33-2.67) | (0.94-1.23) | (1.32-2.63) | (1.00-1.24) | |||||||||||||
OR, odds ratio; CI, confidence interval. *The heterogeneity exists; a random-effect model based on the DerSimonian and Laird method or a fixed-effect model based on the Mantel-Haenszel method was used. M1: Aa vs. AA, heterozygote comparison; M2: aa vs. AA, homozygote comparison; M3: Aa + aa vs. AA, dominant model; M4: aa vs. AA + Aa, recessive model; M5: a vs. A, allelic model (A indicates wild allele, a indicates variant allele). The bold means the significant results.
Fig. 1.
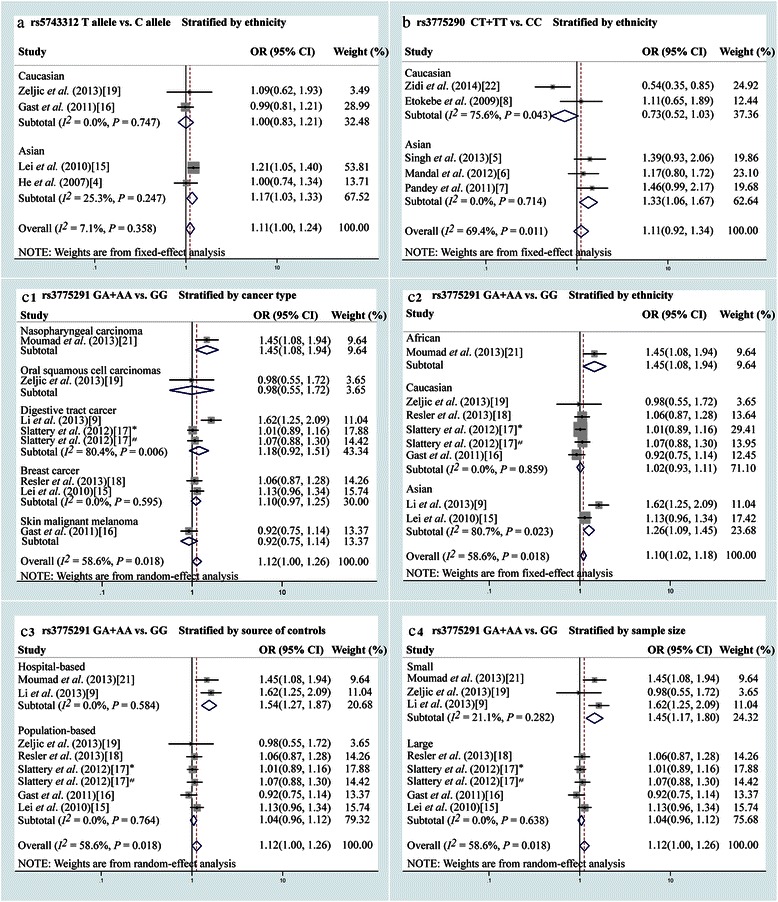
Forest plot of the odds ratios (ORs) for the association of Toll-like receptor 3 (TLR3) single nucleotide polymorphisms (SNPs) with cancer risk. a, the association of the TLR3 rs5743312 SNP with cancer risk when stratified by ethnicity (allelic model). b, the association of the TLR3 rs3775290 SNP with cancer risk when stratified by ethnicity (dominant model). c, The association of TLR3 rs3775291 SNP with cancer risk (dominant model) when stratified by cancer type (c1), ethnicity (c2), source of controls (c3), and sample size (c4). *The cancer type was colon cancer; #the cancer type was rectal cancer
In the stratified analysis, the rs3775290 variant genotype was significantly associated with an increased cancer risk in Asian populations in M1 (CT vs. CC: OR = 1.28, 95 % CI = 1.02–1.62, P = 0.036), M2 (TT vs. CC: OR = 1.79, 95 % CI = 1.05–3.05, P = 0.032), and M3 models (CT + TT vs. CC: OR = 1.33, 95 % CI = 1.06–1.67, P = 0.013) (Table 5, Fig. 1b). For the rs3775291 SNP, both the GA heterozygote and the GA + AA genotypes were consistently associated with an increased risk of nasopharyngeal cancer (GA vs. GG: OR = 1.54, 95 % CI = 1.14–2.09, P = 0.005; GA + AA vs. GG: OR = 1.45, 95 % CI = 1.09–1.94, P = 0.012) (Table 5, Fig. 1c1). When the data were stratified by ethnicity, the GA heterozygote was significantly associated with an increased cancer risk in the Asian subgroup (GA vs. GG: OR = 1.27, 95 % CI = 1.00–1.60, P = 0.048) (Table 5, Fig. 1c2). When the data were stratified by the source of controls, the hospital-based subgroup showed that the variant allele was significantly associated with an increased cancer risk (GA vs. GG: OR = 1.50, 95 % CI = 1.23–1.83, P < 0.001; GA + AA vs. GG: OR = 1.54, 95 % CI = 1.27–1.87, P < 0.001; A vs. G: OR = 1.43, 95 % CI = 1.23–1.67, P < 0.001) (Table 5, Fig. 1c3). Finally, when the data were stratified by sample size, the small sample size subgroup showed that the variant genotype was significantly associated with an increased cancer risk (AA vs. GG: OR = 2.77, 95 % CI = 1.75–4.39, P < 0.001; AA vs. GA + GG: OR = 2.46, 95 % CI = 1.58–3.83, P < 0.001) (Table 5, Fig. 1c4).
Table 5.
Pooled ORs and 95 % CIs of TLR3 polymorphisms in the stratified analysis
| Stratification | Number of studies | M1 | M2 | M3 | M4 | Number of studies | M5 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR (95 % CI) | P | I 2 (%) | OR (95 % CI) | P | I 2 (%) | OR (95 % CI) | P | I 2 (%) | OR (95 % CI) | P | I 2 (%) | OR (95 % CI) | P | I 2 (%) | |||
| rs3775290 (C13766T, C > T) | |||||||||||||||||
| Ethnicity | |||||||||||||||||
| Asian | 3 | 1.28 | 0.036 | 0 | 1.79 | 0.032 | 0 | 1.33 | 0.013 | 0 | 1.61 | 0.08 | 0 | 4 | 1.14 | 0.25a | 58.2 |
| (1.02-1.62) | (1.05-3.05) | (1.06-1.67) | (0.95-2.71) | (0.91-1.43) | |||||||||||||
| Caucasian | 2 | 0.72 | 0.372 | 75.5 | 1.00 | 0.996 | 0 | 0.76 | 0.45a | 75.6 | 1.35 | 0.52 | 0 | 2 | 0.89 | 0.60a | 63.6 |
| (0.34-1.49) | (0.55-1.82) | (0.38-1.54) | (0.54-3.36) | (0.58-1.38) | |||||||||||||
| Cancer type | |||||||||||||||||
| Genital system neoplasms | 4 | 0.97 | 0.892 | 75 | 1.26 | 0.3 | 0 | 1.04 | 0.73a | 73.9 | 1.33 | 0.2 | 0 | 1.07 | 0.64a | 58.8 | |
| (0.62-1.53) | (0.81-1.95) | (0.84-1.29) | (0.87-2.03) | (0.82-1.39) | |||||||||||||
| Source of controls | |||||||||||||||||
| Hospital-based | NA | NA | NA | NA | 5 | 1.12 | 0.32a | 54.7 | |||||||||
| (0.90-1.40) | |||||||||||||||||
| Population-based | NA | NA | NA | NA | 1 | 0.88 | 0.23 | ||||||||||
| (0.70-1.09) | |||||||||||||||||
| rs3775291 (G13909A, L412F, G > A) | |||||||||||||||||
| Cancer type | |||||||||||||||||
| Digestive tract cancer | 3 | 1.16 | 0.132a | 65.4 | 1.28 | 0.429 | 89.1 | 1.18 | 0.186 | 80.4 | 1.18 | 0.559a | 87.4 | 3 | 1.14 | 0.288a | 87.5 |
| (0.96-1.41) | (0.69-2.38) | (0.92-1.51) | (0.68-2.05) | (0.90-1.44) | |||||||||||||
| Breast cancer | 2 | 1.11 | 0.118 | 0 | 1.04 | 0.739 | 0 | 1.10 | 0.148 | 0 | 0.99 | 0.911 | 0 | 2 | 1.05 | 0.288 | 0 |
| (0.97-1.27) | (0.70-1.55) | (0.97-1.25) | (0.79-1.23) | (0.96-1.16) | |||||||||||||
| Nasopharyngeal carcinoma | 1 | 1.54 | 0.005 | 0.81 | 0.593 | 1.45 | 0.012 | 0.70 | 0.37 | 2 | 1.16 | 0.083 | 0 | ||||
| (1.14-2.09) | (0.37-1.76) | (1.09-1.94) | (0.33-1.52) | (0.98-1.37) | |||||||||||||
| Ethnicity | |||||||||||||||||
| Caucasian | 5 | 1.02 | 0.613 | 0 | 0.98 | 0.78 | 4.6 | 1.02 | 0.732 | 0 | 0.97 | 0.685 | 40.9 | 5 | 1.00 | 0.94 | 0 |
| (0.94-1.12) | (0.84-1.14) | (0.93-1.11) | (0.84-1.13) | (0.93-1.07) | |||||||||||||
| Asian | 2 | 1.27 | 0.048 a | 54 | 1.76 | 0.272 | 91.2 | 1.33 | 0.105 | 80.7 | 1.55 | 0.334 | 89.5 | 3 | 1.21 | 0.108a | 79.5 |
| (1.00-1.60) | (0.64-4.82) | (0.94-1.89) | (0.64-3.77) | (0.96-1.53) | |||||||||||||
| African | 1 | 1.23 | 0.005 | 0.81 | 0.593 | 1.45 | 0.012 | 0.70 | 0.37 | 1 | 1.27 | 0.062 | |||||
| (1.06-1.43) | (0.37-1.76) | (1.09-1.94) | (0.33-1.52) | (0.99-1.63) | |||||||||||||
| Source of controls | |||||||||||||||||
| Hospital-based | 2 | 1.50 | <0.001 | 0 | 1.61 | 0.466a | 86.8 | 1.54 | <0.001 | 0 | 1.37 | 0.616a | 86.3 | 2 | 1.43 | <0.001 | 32.6 |
| (1.23-1.83) | (0.45-5.84) | (1.27-1.87) | (0.40-4.76) | (1.23-1.67) | |||||||||||||
| Population-based | 6 | 1.05 | 0.259 | 0 | 1.00 | 0.999 | 0 | 1.04 | 0.338 | 0 | 0.98 | 0.748 | 26.9 | 7 | 1.02 | 0.465a | 0 |
| (0.97-1.14) | (0.87-1.15) | (0.96-1.12) | (0.86-1.12) | (0.97-1.08) | |||||||||||||
| Sample size | |||||||||||||||||
| Large | 5 | 1.05 | 0.218 | 0 | 0.98 | 0.823 | 0 | 1.04 | 0.328 | 0 | 0.96 | 0.545 | 0 | 5 | 1.02 | 0.629 | 0 |
| (0.97-1.14) | (0.86-1.13) | (0.96-1.12) | (0.84-1.10) | (0.96-1.08) | |||||||||||||
| Small | 3 | 1.41 | <0.001 | 45.6 | 1.76 | 0.184a | 73.7 | 1.47 | <0.001 | 21.1 | 1.63 | 0.242a | 73.9 | 4 | 1.30 | <0.001 | 47.9 |
| (1.16-1.70) | (0.77-4.05) | (1.23-1.76) | (0.72-3.65) | (1.15-1.47) | |||||||||||||
| rs3775292 (intron 3, C > G) | |||||||||||||||||
| Cancer type | |||||||||||||||||
| Colorectal cancer | 2 | 0.93 | 0.609a | 82.2 | 0.89 | 0.657a | 68 | 0.93 | 0.464a | 63.8 | 0.92 | 0.797a | 78.2 | 2 | 0.94 | 0.233 | 0 |
| (0.69-1.25) | (0.54-1.48) | (0.76-1.13) | (0.50-1.69) | (0.86-1.04) | |||||||||||||
| Breast Cancer | 1 | 1.06 | 0.601 | 1.14 | 0.607 | 1.07 | 0.535 | 1.12 | 0.656 | 1 | 1.06 | 0.499 | |||||
| (0.86-1.30) | (0.69-1.90) | (0.87-1.30) | (0.68-1.86) | (0.90-1.26) | |||||||||||||
| Skin malignant melanoma | 1 | 0.91 | 0.411 | 1.09 | 0.753 | 0.93 | 0.503 | 1.12 | 0.659 | 1 | 0.96 | 0.683 | |||||
| (0.73-1.14) | (0.64-1.84) | (0.75-1.15) | (0.67-1.89) | (0.80-1.16) | |||||||||||||
| rs5743312 (C9948T, intron 3, C > T) | |||||||||||||||||
| Ethnicity | |||||||||||||||||
| Asian | NA | NA | NA | NA | 2 | 1.17 | 0.017 | 25.3 | |||||||||
| (1.03-1.33) | |||||||||||||||||
| Caucasian | NA | NA | NA | NA | 2 | 0.99 | 0.995 | 0 | |||||||||
| (0.83-1.21) | |||||||||||||||||
| Sample size | |||||||||||||||||
| Large | 2 | 1.02 | 0.82 | 0 | 1.89 | <0.001 | 4 | 1.08 | 0.268 | 37.2 | 1.87 | 0.001 | 0 | 2 | 1.11 | 0.305a | 61.7 |
| (0.88-1.17) | (1.32-2.70) | (0.94-1.24) | (1.31-2.65) | (0.99-1.24) | |||||||||||||
| Small | 1 | 0.99 | 0.97 | 1.70 | 0.569 | 1.04 | 0.9 | 1.70 | 0.566 | 2 | 1.02 | 0.894 | 0 | ||||
| (0.51-1.93) | (0.28-10.45) | (0.55-1.98) | (0.28-10.40) | (0.78-1.32) | |||||||||||||
NA, not available. Other footnotes as in Table 4. The bold means the significant results.
Heterogeneity
The heterogeneities that originated within the collection of selected studies and within each subgroup of studies are shown in Tables 4 and 5, respectively. Slight heterogeneities were found when comparing different studies. To explore the influence of individual studies on the pooled results, we analyzed the sensitivity of our methodology by removing one study at a time from the pooled analyses. No significant heterogeneity was found for any genetic model, which suggested that our results were relatively reliable.
Publication bias
The Begg’s rank correlation and Egger’s linear regression tests were conducted to evaluate publication bias. According to the results of these tests, a slight publication bias for rs3775290 in M2 was indicated (Table 6).
Table 6.
The results of Begg’s and Egger’s test for the publication bias
| Comparison model | Begg’s test | Egger’s test | ||
|---|---|---|---|---|
| Z value | P value | t value | P value | |
| rs3775290 (C13766T, C > T) | ||||
| M1 | −0.98 | 0.327 | −0.88 | 0.443 |
| M2 | 0.98 | 0.327 | 3.08 | 0.054 |
| M3 | −0.98 | 0.327 | −0.78 | 0.493 |
| M4 | 1.47 | 0.142 | 1.7 | 0.188 |
| M5 | −0.19 | 0.851 | 0.87 | 0.432 |
| rs3775291 (G13909A, L412F, G > A) | ||||
| M1 | 0.49 | 0.621 | 0.62 | 0.559 |
| M2 | 0.99 | 0.322 | 1.06 | 0.329 |
| M3 | 0.49 | 0.621 | 1.02 | 0.346 |
| M4 | 0.99 | 0.322 | 1.02 | 0.348 |
| M5 | 1.25 | 0.211 | 1.48 | 0.183 |
| rs3775292 (C/G, intron 3, C > G) | ||||
| M1 | −1.36 | 0.174 | −1.14 | 0.371 |
| M2 | 0.00 | 1.000 | 3.88 | 0.061 |
| M3 | 0.00 | 1.000 | −0.68 | 0.567 |
| M4 | 0.00 | 1.000 | 3.11 | 0.090 |
| M5 | 0.68 | 0.497 | 0.14 | 0.901 |
| rs5743312 (C9948T, intron 3, C > T) | ||||
| M1 | −0.52 | 0.602 | −0.39 | 0.764 |
| M2 | −0.52 | 0.602 | −0.76 | 0.587 |
| M3 | −0.52 | 0.602 | 0.75 | 0.590 |
| M4 | −0.52 | 0.602 | −0.75 | 0.589 |
| M5 | 0.00 | 1.000 | −0.86 | 0.479 |
NA, not available. Other footnotes as in Table 4.
Discussion
In this meta-analysis, we found that TLR3 rs5743312 was associated with an increased overall risk of developing cancer. This was also true for the large sample size subgroup. TLR3 rs3775290 and rs3775291 polymorphisms were found to be associated with increased cancer risks in the stratified analysis, whereas no association was found between TLR3 rs3775292 and cancer risk.
The TLR3 rs5743312 polymorphism is located in intron 3, and no previous studies have shown any association between this polymorphism and cancer risk. However, after data integration, we concluded that this SNP is the only significant site in the TLR3 gene with respect to cancer risk. According to our meta-analysis, the remaining 3 SNPs exhibited no association with cancer risks. The subgroup analyses for the rs5743312 SNP showed the same tendency as the whole group analysis. Previous studies have suggested that intronic SNPs may exhibit specific functions, such as directing alternative splicing [23, 24]. As intronic polymorphisms have been shown to exhibit critical functions, it would be prudent to include intronic SNPs, such as rs5743312, in future studies.
rs3775290 is located in exon 4 of the TLR3 gene and is also a hotspot TLR3 SNP. In our stratification analyses, TLR3 rs3775290 was found to be associated with cancer risk in Asian populations. This may be due to the variability in genetic background between Asians and Caucasians (Fig. 2). In the HapMap database, rs3775290 showed a ratio of 0.217:0.783 for the wild-type A:variant G allele in the HapMap-CEU population and of 0.408:0.592 for the A:G allele in the HapMap-CHB and HapMap-JPT populations (http://www.ncbi.nlm.nih.gov/SNP/snp_ref.cgi?rs=3775290). Variations in ethnic backgrounds play an important role in genetic susceptibility, and genetic differences between Asians and Caucasians may be the reason that these different ethnic populations follow different life styles and are thus exposed to different environmental factors [25]. Population groups that are carrying different genotypes or allele frequencies of the rs3775290 polymorphism may show differences in cancer susceptibility [26]. The C to T variant of rs3775290 results in a silent mutation in phenylalanine residue 459. When a SNP leads to a silent mutation, it does not necessarily indicate that the SNP has no impact on protein dynamics. For example, a silent mutation located in an exon might affect interactions between genetic elements and additional molecules, such as metal ions or transcription factors [27].
Fig. 2.
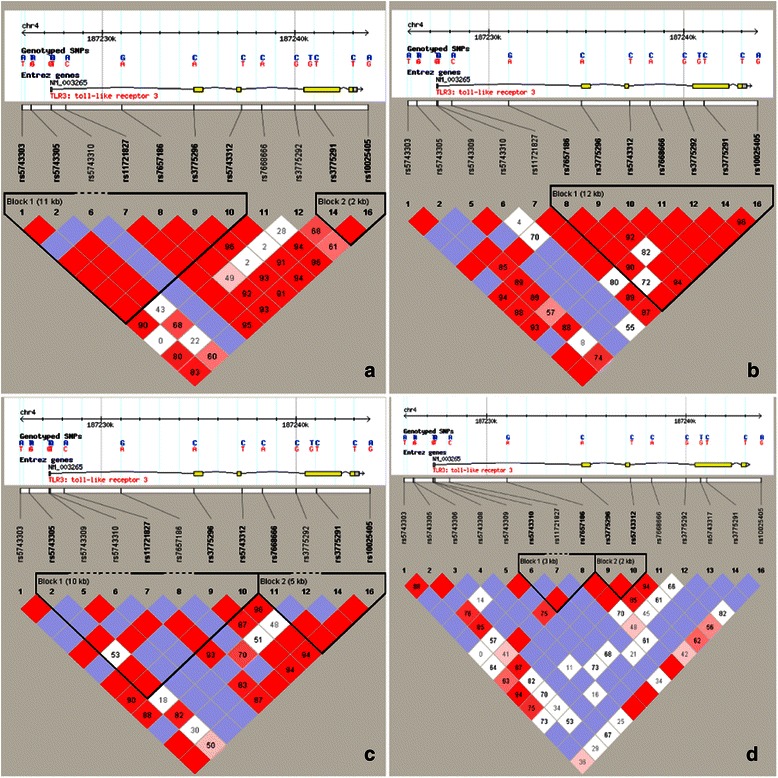
The linkage disequilibrium plot of TLR3 gene polymorphisms found in individuals of different ethnicities, expanding 20 kb from 5′ to 3′ each (downloaded from the HapMap database). a, European; b, Chinese Han; c, Japanese; d, African. Different SNP distributions arise in response to differences in ethnicity. Therefore, differences in ethnicity should be considered when designing the study. Furthermore, some TLR3 SNPs might be in linkage disequilibrium, suggesting that a haplotype study may be more effective for predicting or screening susceptible populations
To date, the rs3775291 polymorphism has been the most investigated SNP in TLR3. In the stratified analysis, TLR3 rs3775291 was also found to be associated with cancer risk in Asians in a heterozygote model (Table 5). Furthermore, when examining our findings on a subgroup-by-subgroup basis, we found that the small sample size subgroup showed that rs3775291 was associated with a significantly increased cancer risk in the M2 and M4 models, whereas the hospital-based subgroup showed this association in the M1, M3, and M5 models. These findings may have arisen because the 3 corresponding studies each obtained significant results. It is possible that in the Asian subgroup, the rs3775291 SNP was associated with cancer risks due to differences in genetic and environmental backgrounds between Asians and Caucasians. In the subgroup analyses, we found a significant association in the small sample size subgroup but not in the large sample size subgroup. In the large sample size subgroup, the ORs in the 5 included studies were all approximately 1.0, whereas the ORs in the studies conducted by Li et al. [9], Zeljic et al. [19], and Moumad et al. [21] indicated a significant association in the small sample size subgroup. The same was true for the hospital-based analysis. Reliable results could be obtained in these cases based on the high quality of the studies designed to explore the real associations of TLR3 SNPs with cancer risk. Differences in patients’ genetic or environmental backgrounds might certainly be a common mechanism behind the conclusions of our stratification analysis. Differences in how studies were controlled or documented might also provide an explanation for these conclusions. For example, the studies conducted by Li et al. [9] and Moumad et al. [21] were well controlled, which might explain why positive results were obtained following the analysis of the small sample size subgroup. Furthermore, rs3775291 is located in exon 4, and the G to A variant results in the change of a leucine to phenylalanine at residue 412, which might provide a mechanistic explanation for the effects of this SNP.
Similar to rs5743312, rs3775292 is an intronic polymorphism that has also been investigated in detail, as its location in intron 3 is near rs3775290 and rs3775291. Thus, this polymorphism might be associated with variability in alternative splicing and further associated with the linkage disequilibrium between rs3775290 and rs3775291 [24]. Two of the included studies showed no association of these SNPs with cancer risk, whereas one showed an association. However, our integrated meta-analysis results did not find any association between the rs3775292 SNP and cancer risk. Additional studies will be required to confirm these results.
Our meta-analysis had several limitations. First, only studies that were published in English or Chinese were included in our analysis, thereby creating potential publication bias. Second, the pooled sample size was relatively limited and therefore could support only preliminary evaluations of the association between various TLR3 polymorphisms and the incidence of various types of cancer. Additionally, we were not always able to obtain original data from the published literature, such as the age and sex of the patients, or the environmental factors that might have affected the hosts. Thus, we used unadjusted information, whereas a more precise analysis could be conducted if detailed information on the original data were available. Therefore, additional studies are required to improve the reliability of these results.
Conclusion
In summary, this meta-analysis indicated that the variant allele of TLR3 rs5743312 is potentially associated with increased cancer risks both in the whole collection of studies and in the large sample size subgroup. In the stratified analysis, the variant genotype of the TLR3 rs3775290 polymorphism was associated with an increased cancer risk in the Asian subgroup. TLR3 rs3775291 was also associated with an increased cancer risk in the Asian, hospital-based source of controls, and small sample size subgroups. No association was found between TLR3 rs3775292 and cancer risk. Additional well-designed, large-scale, and functional studies on TLR3 SNPs are required to confirm our findings.
Acknowledgements
We thank Martha L. Slattery from Department of Internal Medicine, University of Utah Health Sciences Center, Salt Lake City, Utah, USA for generously providing original data about the genotype of TLR3 rs3775291 and rs3775292 in colorectal cancer and control groups.
Footnotes
Ben-Gang Wang and De-Hui Yi contributed equally to this work.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
YFL designed the study and revised the manuscript; BGW and DHY extracted the data and wrote the manuscript. All authors read and approved the final manuscript.
Contributor Information
Ben-Gang Wang, Email: wangbengang2008@163.com.
De-Hui Yi, Email: ydh1900@sina.com.
Yong-Feng Liu, Email: liuyongfengsubmit@163.com.
References
- 1.Zhang SY, Herman M, Ciancanelli MJ, Perez de Diego R, Sancho-Shimizu V, Abel L, et al. TLR3 immunity to infection in mice and humans. Curr Opin Immunol. 2013;25:19–33. doi: 10.1016/j.coi.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Frazao JB, Errante PR, Condino-Neto A. Toll-like receptors’ pathway disturbances are associated with increased susceptibility to infections in humans. Arch Immunol Ther Exp (Warsz) 2013;61:427–43. doi: 10.1007/s00005-013-0243-0. [DOI] [PubMed] [Google Scholar]
- 3.Netea MG, Wijmenga C, O’Neill LA. Genetic variation in Toll-like receptors and disease susceptibility. Nat Immunol. 2012;13:535–42. doi: 10.1038/ni.2284. [DOI] [PubMed] [Google Scholar]
- 4.He JF, Jia WH, Fan Q, Zhou XX, Qin HD, Shugart YY, et al. Genetic polymorphisms of TLR3 are associated with nasopharyngeal carcinoma risk in Cantonese population. BMC Cancer. 2007;7:194. doi: 10.1186/1471-2407-7-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Singh V, Srivastava N, Kapoor R, Mittal RD. Single-nucleotide polymorphisms in genes encoding Toll-like receptor-2, -3, -4, and -9 in a case-control study with bladder cancer susceptibility in a North Indian population. Arch Med Res. 2013;44:54–61. doi: 10.1016/j.arcmed.2012.10.008. [DOI] [PubMed] [Google Scholar]
- 6.Mandal RK, George GP, Mittal RD. Association of Toll-like receptor (TLR) 2, 3 and 9 genes polymorphism with prostate cancer risk in North Indian population. Mol Biol Rep. 2012;39:7263–9. doi: 10.1007/s11033-012-1556-5. [DOI] [PubMed] [Google Scholar]
- 7.Pandey S, Mittal B, Srivastava M, Singh S, Srivastava K, Lal P, et al. Evaluation of Toll-like receptors 3 (c.1377C/T) and 9 (G2848A) gene polymorphisms in cervical cancer susceptibility. Mol Biol Rep. 2011;38:4715–21. doi: 10.1007/s11033-010-0607-z. [DOI] [PubMed] [Google Scholar]
- 8.Etokebe GE, Knezevic J, Petricevic B, Pavelic J, Vrbanec D, Dembic Z. Single-nucleotide polymorphisms in genes encoding toll-like receptor-2, -3, -4, and -9 in case-control study with breast cancer. Genet Test Mol Biomarkers. 2009;13:729–34. doi: 10.1089/gtmb.2009.0045. [DOI] [PubMed] [Google Scholar]
- 9.Li G, Zheng Z. Toll-like receptor 3 genetic variants and susceptibility to hepatocellular carcinoma and HBV-related hepatocellular carcinoma. Tumour Biol. 2013;34:1589–94. doi: 10.1007/s13277-013-0689-z. [DOI] [PubMed] [Google Scholar]
- 10.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22:719–48. [PubMed] [Google Scholar]
- 11.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–88. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 12.Thakkinstian A, McElduff P, D’Este C, Duffy D, Attia J. A method for meta-analysis of molecular association studies. Stat Med. 2005;24:1291–306. doi: 10.1002/sim.2010. [DOI] [PubMed] [Google Scholar]
- 13.Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50:1088–101. doi: 10.2307/2533446. [DOI] [PubMed] [Google Scholar]
- 14.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–34. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lei F. The genetic contribution of TLR3-mediated signaling genes susceptibility and progress of breast cancer. Shanghai: Fudan University; 2010. [Google Scholar]
- 16.Gast A, Bermejo JL, Claus R, Brandt A, Weires M, Weber A, et al. Association of inherited variation in Toll-like receptor genes with malignant melanoma susceptibility and survival. PLoS One. 2011;6:e24370. doi: 10.1371/journal.pone.0024370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Slattery ML, Herrick JS, Bondurant KL, Wolff RK. Toll-like receptor genes and their association with colon and rectal cancer development and prognosis. Int J Cancer. 2012;130:2974–80. doi: 10.1002/ijc.26314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Resler AJ, Malone KE, Johnson LG, Malkki M, Petersdorf EW, McKnight B, et al. Genetic variation in TLR or NFkappaB pathways and the risk of breast cancer: a case-control study. BMC Cancer. 2013;13:219. doi: 10.1186/1471-2407-13-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zeljic K, Supic G, Jovic N, Kozomara R, Brankovic-Magic M, Obrenovic M, et al. Association of TLR2, TLR3, TLR4 and CD14 genes polymorphisms with oral cancer risk and survival. Oral Dis. 2014;20:416–24. doi: 10.1111/odi.12144. [DOI] [PubMed] [Google Scholar]
- 20.Yeyeodu ST, Kidd LR, Oprea-Ilies GM, Burns BG, Vancleave TT, Shim JY, et al. IRAK4 and TLR3 sequence variants may alter breast cancer risk among African-American women. Front Immunol. 2013;4:338. doi: 10.3389/fimmu.2013.00338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moumad K, Lascorz J, Bevier M, Khyatti M, Ennaji MM, Benider A, et al. Genetic polymorphisms in host innate immune sensor genes and the risk of nasopharyngeal carcinoma in North Africa. Genes, Genomes and Genetics. 2013;3:971-7. [DOI] [PMC free article] [PubMed]
- 22.Zidi S, Verdi H, Yilmaz-Yalcin Y, Yazici AC, Gazouani E, Mezlini A, et al. Impact of Toll-like receptors 2/3/4/9, IL-1-alpha/beta and TNF-alpha polymorphisms in cervical cancer susceptibility in Tunisia: genetic polymorphisms implicated in the occurence of cervical cancer. Pathol Oncol Res. 2014 May 17, [Epub ahead of print].
- 23.De Brasi CD, Bowen DJ. Molecular characteristics of the intron 22 homologs of the coagulation factor VIII gene: an update. J Thromb Haemost. 2008;6:1822–4. doi: 10.1111/j.1538-7836.2008.03094.x. [DOI] [PubMed] [Google Scholar]
- 24.Biamonti G, Catillo M, Pignataro D, Montecucco A, Ghigna C. The alternative splicing side of cancer. Semin Cell Dev Biol. 2014;32:30–6. doi: 10.1016/j.semcdb.2014.03.016. [DOI] [PubMed] [Google Scholar]
- 25.Dick DM. Gene-environment interaction in psychological traits and disorders. Annu Rev Clin Psychol. 2011;7:383–409. doi: 10.1146/annurev-clinpsy-032210-104518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao LB, Pan XM, Sun H, Wang X, Rao L, Li LJ, et al. The association between ATM D1853N polymorphism and breast cancer susceptibility: a meta-analysis. J Exp Clin Cancer Res. 2010;29:117. doi: 10.1186/1756-9966-29-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schattner P, Diekhans M. Regions of extreme synonymous codon selection in mammalian genes. Nucleic Acids Res. 2006;34:1700–10. doi: 10.1093/nar/gkl095. [DOI] [PMC free article] [PubMed] [Google Scholar]


