Abstract
To clarify the epidemiology of influenza A viruses in coordinated swine production systems to which no animals from outside the system are introduced, we conducted virologic surveillance during September 2012–September 2013. Animal age, geographic location, and farm type were found to affect the prevalence of these viruses.
Keywords: influenza, viruses, swine, surveillance, H1N1, H3N2, H3N2, zoonoses, United States, North America
Influenza A viruses (IAVs) are the etiologic agents of acute respiratory disease in many mammalian species. Although originating in wild aquatic birds, IAVs have been successful in crossing the species barrier, and specific subtypes have become endemic among humans and domestic swine populations (1). In the United States, influenza was first described in swine herds during the 1918 pandemic and has circulated among domestic pigs for nearly a century (2). The ability of swine IAVs to infect humans and cause pandemics such as that of the influenza A(H1N1) virus observed during 2009 (3,4) and the sporadic transmission of various swine influenza viruses, including H1N1 (5), H3N2 (6), and variant H3N2 (7), are public health concerns and highlight the need for increased vigilance and understanding of IAV epidemiology among swine.
Here we report the results from 13-months of active surveillance of IAV in coordinated swine production systems in the United States. The objectives of this study were 1) to determine the prevalence of IAV within farms in a closed production system and 2) to determine which sampled population is most affected by IAV.
The Study
Multisite coordinated production systems are the common method of swine production in the United States. These systems consist of multiple farms operating in tandem, with each farm responsible for 1 stage of the production process (Figure 1). These systems are closed, meaning there are no introductions of animals from outside the system. Each farm in the system has a specific purpose: 1) to breed, gestate, farrow, and raise to the point of weaning replacement breeding stock of a specified genotype (multiplier farms [MF]); 2) to raise replacement female pigs, commonly called gilts, to 5–6 months of age for breeding (gilt development farms or units [GDU]); and 3) to breed, gestate, deliver, and raise to the point of weaning piglets specifically for meat production (breed-to-wean farms [BTW]). Gilts from GDUs are moved to MF or BTW farms for breeding. All farms house multiple age cohorts, although in different rooms or buildings. During suckling of piglets on MF and BTW farms, as well as in GDU farms, 1 cohort of piglets differing in age by <1 week are housed in a single room/building, and 1 cohort is removed before the entry of the next.
Figure 1.
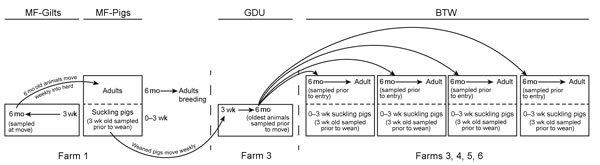
Flow of animals through a closed swine production system. Coordinated swine production systems maximize desired animal traits and weight gain. High-quality breeding sows grown and bred in multiplier farms (MF-Gilts) were sampled transfer to commercial gilt development farms (GDU), where they were sampled again at exit (6 months). At ≈3 weeks of age, piglets were sampled before weaning (MF-Pigs). Mature gilts were transported from the GDU to 1 of 4 commercial breed-to-wean (BTW) farms, where samples were collected from gilts before entry and piglets before weaning.
Four coordinated, multisite production systems, each consisting of 1 MF farm that includes both gilts and pigs to produce replacement female stock, 1 GDU farm to raise replacement female gilts from 3 to ≈26 weeks of age, and 4 BTW farms to raise pigs for meat, were selected to monitor the dynamics of IAV transmission in swine breeding herds. Systems are located across the United States. System 1’s MF and GDU sites are located in Illinois and its BTW sites in Georgia. Systems 2, 3, and 4 are located entirely in Illinois, Oklahoma, and Nebraska, respectively.
Nasal swab samples were collected from pigs monthly during September 2012–September 2013 from each farm in all 4 systems. The animal type and time of sampling differed on the different farm types. On the MF and BTW farms, samples were collected from 30 piglets at ≈3 weeks of age and from 30 gilts. On the MFs, 30 gilts were sampled before entry to the farm. On the GDU farms, 30 animals at ≈26 weeks of age were sampled before movement to a BTW. In the BTWs, gilts were sampled before breeding (4–8 weeks after arrival) and piglets were sampled immediately before weaning (Figure 1). This strategy enabled assessment of IAV status before the movement of animals to the next stage of production. Laboratory methods are summarized in the Technical Appendix.
During the 13-month period spanning September 2012–September 2013, a total of 14,954 swab samples were collected and tested for the presence of the IAV matrix gene by real-time reverse transcription PCR. Of the samples collected, 741 (5.0%) tested positive, which is consistent with previous surveillance studies (8).
Bivariate analysis found statistically significant correlations between infection and location in Illinois, GDU farm type, and system 2 (p<0.001 for all), but not for age (Table 1). We then constructed a logistic regression model that assessed the effect of age, system, location, and farm type on having a positive influenza result. Age was statistically significant by this model (p = 0.004); the odds ratio for piglets at weaning whose samples tested positive for IAV was 1.3 (95% CI 1.1–1.6) compared to that for gilts (Table 2), which is consistent with previous studies (9). However, this finding could be related to interaction between age and state/system. System 2 again had higher odds of positive results (OR 1.7, 95% CI 1.3–2.3) compared with system 1. MF pigs were found to have a lower risk for infection (OR 0.7, 95% CI 0.5–0.9) and GDU pigs to have a higher risk for infection (OR 1.6, 95% CI 1.2–2.1) when compared with BTW pigs. Finally, Illinois had higher odds for IAV infection (OR 3.2, 95% CI 2.6–4.0) compared with the other 3 states.
Table 1. Epidemiologic data for influenza A virus among swine in coordinated swine production systems, United States, September 2012–September 2013*.
| Variable | No. samples | No. (%) positive samples | p value |
|---|---|---|---|
| State* | |||
| Georgia | 2,520 | 73 (2.9) | |
| Illinois | 4,490 | 408 (9.1) | <0.001 |
| Nebraska | 3,894 | 126 (3.2) | |
| Oklahoma |
4,050 |
134 (3.3) |
|
| Age group | |||
| Gilt | 8,028 | 375 (4.7) | 0.85 |
| Piglet |
6,926 |
366 (5.3) |
|
| Farm type | |||
| MF, gilts† | 1,526 | 67 (4.4) | |
| MF, piglets | 1,559 | 72 (4.6) | |
| GDU | 1,455 | 115 (7.9) | <0.001 |
| BTW |
10,414 |
487 (4.7) |
|
| System | |||
| 1 | 3,673 | 142 (3.9) | |
| 2 | 3,337 | 339 (10.2) | <0.001 |
| 3 | 4,050 | 134 (3.3) | |
| 4 | 3,894 | 126 (3.2) | |
*Piglets were sampled before weaning; gilts were sampled at entry to MFs and before moving from GDUs to BTWs. MF, multiplier farm; GDU, gilt development unit; BTW, breed-to-wean.
Table 2. Correlation between age, state, farm type or production system, and influenza A virus status in swine in coordinated production systems, United States, September 2012–September 2013*.
| Factor | aOR (95% CI) | p value |
|---|---|---|
| Age group |
|
|
| Piglet vs. gilt |
1.3 (1.1–1.6) |
0.004 |
| State |
|
|
| Illinois vs. Oklahoma |
1.9 (1.4–2.6) |
<0.001 |
| Farm type | ||
| MF piglets vs. BTW | 0.7 (0.5–0.9) | 0.011 |
| GDU vs. BTW |
1.6 (1.2–2.1) |
<0.001 |
| System number |
|
|
| 2 vs. 1 | 1.7 (1.3–2.3) | <0.001 |
*Only statistically significant factors are shown; full analysis is provided in the Technical Appendix. aOR, adjusted odds ratio; CI, confidence interval; MF, multiplier farm; GDU, gilt development unit; BTW, breed-to-wean.
IAV subtypes were determined for 25.2% of the IAV positive samples (Figure 2). All 3 common porcine influenza subtypes (H1N1, H1N2, and H3N2) were detected during the 13-month surveillance period. Increased prevalence of IAV was detected in piglets (MF pigs, GDU, BTW pigs) in all 4 systems, particularly in systems 1, 2, and 4, from winter to early summer (Figure 2, panels A, B, D), which is consistent with other studies (8). Multiple subtype detection occurred only on BTW farms in system 2 (Figure 2, panel B).
Figure 2.
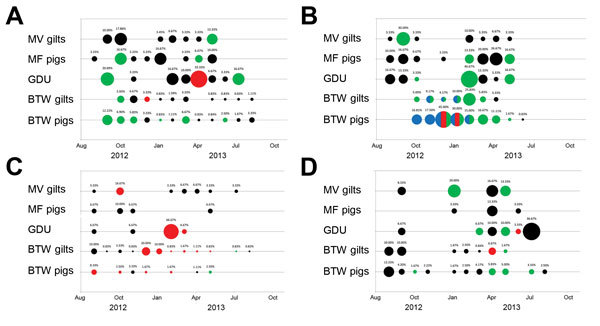
Prevalence of influenza A virus by system, farm type, and month, United States, September 2012–September 2013, positive RNA nasal swab samples were categorized by age range (gilt, female pig 5–6 months of age or piglet, ≈3 weeks of age) and production point farm (multiplier farms [MF] Gilt, MF Pig, GDU, BTW Gilt, BTW Pig). Each circle depicts a month during which positive influenza samples were collected; percentage of positive swabs is listed above each circle. Influenza A subtypes are indicated by circle color: green, H1N1; blue, H1N2; red, H3N2; black, untyped. Multicolored circles indicate the detection of >1 subtype.
Conclusions
We found that IAV infection was present at all stages of swine production within coordinated production systems. Animal age, geographic location, and type of farm affected risk for infection. We also found continuing virus circulation in all populations year round, although prevalence was higher from winter through early summer. To fully elucidate the factors that contribute to persistent IAV infection in swine farms and therefore develop evidence-based control strategies, further research is needed.
Logistic regression model of influenza A virus status among swine in coordinated production systems, USA, September 2012-September 2013.
Acknowledgments
We thank the swine producers and veterinary staff for their participation and cooperation in this study. We thank Adam Rubrum, Jerry Parker, Rich Elia, Mark Weilnau, Scott Krauss, and Pamela McKenzie for their excellent technical assistance, data management, and administrative support. Additionally, we thank Julie Groff for assistance with graphic design.
This study was supported by the National Institute of Allergy and Infectious Diseases, the National Institutes of Health Department of Health and Human Services (contract no. HHSN266200700005C), and the American Lebanese Syrian Associated Charities.
Biography
Dr. Kaplan is a postdoctoral research associate at St. Jude Children’s Research Hospital in Memphis. His research interests include virus ecology, virus evolution, and the molecular determinants of virus host range.
Footnotes
Suggested citation for this article: Kaplan BS, DeBeauchamp J, Stigger-Rosser E, Franks J, Crumpton JC, Turner J. Influenza virus surveillance in coordinated swine production systems, United States. Emerg Infect Dis. 2015 Oct [date cited]. http://dx.doi.org/10.3201/eid2110.140633
References
- 1.Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y. Evolution and ecology of influenza A viruses. Microbiol Rev. 1992;56:152–79 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown IH. The epidemiology and evolution of influenza viruses in pigs. Vet Microbiol. 2000;74:29–46 . 10.1016/S0378-1135(00)00164-4 [DOI] [PubMed] [Google Scholar]
- 3.Garten RJ, Davis CT, Russell CA, Shu B, Lindstrom S, Balish A, et al. Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science. 2009;325:197–201. 10.1126/science.1176225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dawood FS, Jain S, Finelli L, Shaw MW, Lindstrom S, Garten RJ, et al. Novel Swine-Origin Influenza A (H1N1) Virus Investigation Team, Emergence of a novel swine-origin influenza A (H1N1) virus in humans. N Engl J Med. 2009;360:2605–15. 10.1056/NEJMoa0903810 [DOI] [PubMed] [Google Scholar]
- 5.Epperson S, Jhung M, Richards S, Quinlisk P, Ball L, Moll M, et al. Human infections with influenza A(H3N2) variant virus in the United States, 2011–2012. Clin Infect Dis. 2013;57(Suppl 1):S4–11. 10.1093/cid/cit272 [DOI] [PubMed] [Google Scholar]
- 6.Wong KK, Gambhir M, Finelli L, Swerdlow DL, Ostroff S, Reed C. Transmissibility of variant influenza from swine to humans: a modeling approach. Clin Infect Dis. 2013;57(Suppl 1):S16–22. 10.1093/cid/cit303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Freidl GS, Meijer A, de Bruin E, de Nardi M, Munoz O, Capua I, et al. Influenza at the animal-human interface: a review of the literature for virological evidence of human infection with swine or avian influenza viruses other than A(H5N1). Euro Surveill. 2014;19:20793. [DOI] [PubMed] [Google Scholar]
- 8.Corzo CA, Culhane M, Juleen K, Stigger-Rosser E, Ducatez MF, Webby RJ, et al. Active surveillance for influenza A virus among swine, midwestern United States, 2009–2011. Emerg Infect Dis. 2013;19:954–60. 10.3201/eid1906.121637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Allerson MW, Davies PR, Gramer MR, Torremorell M. Infection dynamics of pandemic 2009 H1N1 influenza virus in a two-site swine herd. Transbound Emerg Dis. 2014;61:490–9. 10.1111/tbed.12053 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Logistic regression model of influenza A virus status among swine in coordinated production systems, USA, September 2012-September 2013.


