Abstract
The mammalian outer, middle and inner ears have different embryonic origins and evolved at different times in the vertebrate lineage. The outer ear is derived from first and second branchial arch ectoderm and mesoderm, the middle ear ossicles are derived from neural crest mesenchymal cells that invade the first and second branchial arches, whereas the inner ear and its associated vestibule-acoustic (VIIIth) ganglion are derived from the otic placode. In this review, we discuss recent findings in the development of these structures and describe the contributions of members of a Forkhead transcription factor family, the Foxi family to their formation. Foxi transcription factors are critical for formation of the otic placode, survival of the branchial arch neural crest, and developmental remodeling of the branchial arch ectoderm.
1: THE ANATOMY AND EMBRYONIC ORIGINS OF THE INNER, MIDDLE AND OUTER EARS
1.1: Components of the mammalian ear
The mammalian auditory apparatus comprises three compartments, termed the inner, middle, and outer ears. Each compartment arises from a separate embryonic origin (Figure 1), but the final structures work in concert to detect sounds and translate them into electrical signals transmitted by sensory neurons to the brain.
Figure 1. Simple schematic diagram of the auditory apparatus.
The diagram shows the external ear and canal (red), the middle ear cavity (blue) and ear ossicles (yellow) and the inner ear (green), with the embryonic origins of each component.
The Inner Ear
The mature inner ear is found inside a highly calcified bony labyrinth. The soft tissue of the inner ear, termed the epithelial or membranous labyrinth, is composed of three parts containing six sensory patches to detect sound and motion. First, the three semi-circular canals, each with a sensory crista housed in an ampulla, serve to detect angular acceleration. Second, the utricle and the saccule, each containing a sensory macula, detect linear acceleration and gravity. Finally, the cochlea serves to detect sound with its sensory patch, the organ of Corti, running the length of the cochlear duct (Groves and Fekete, 2012). The sensory patches consist of highly polarized, mechanically sensitive sensory epithelial cells, called hair cells. Hair cells are surrounded by supporting cells that provide physical and trophic support and help to maintain potassium balance in the endolymph bathing each sensory organ.
The middle ear
In aquatic vertebrates, sound vibrations from the surrounding water pass directly into the inner ear where they are translated into neural signals. Land tetrapods, on the other hand, require additional structures to allow impedance matching between air pressure sound waves and fluid vibrations within the epithelial labyrinth. This process is achieved by vibrations of one or more small middle ear bones, or ossicles, against the wall of the cochlea. The ossicles, the air filled space they occupy, and a vibrating tympanic membrane comprise the vertebrate middle ear. While birds and reptiles possess a single ossicle, the columella, mammals have three, the malleus, the incus, and the stapes. When sound waves strike the tympanic membrane it vibrates, creating pulses that move the malleus. Movements of the malleus cause the incus to pivot, which presses the stapes in and out against the oval window of the inner ear with the same frequency as the original air-borne sound, causing vibrations in the fluid inside the cochlea. The middle ear ossicles occupy an air-filled space that is connected to the external environment by the Eustachian tube, with one end in the middle ear and the other in the throat. The Eustachian tube permits adjustments in air pressure in response to external pressure changes and drainage of any fluid that might accumulate in the middle ear space. The middle ear itself is lined with a ciliated epithelium that covers the entire middle ear cavity in birds, but only the ventral two thirds in mammals, where the remainder is non-ciliated epithelium derived from cranial neural crest cells (Thompson and Tucker, 2013). It is now accepted that the tympanic middle ear evolved independently several times in different vertebrate lineages (Manley, 2010).
The outer ear
The tympanic membrane separates the middle ear from the outer ear, consisting in land mammals of an ear canal and an external pinna. Birds, most reptiles, aquatic mammals, and monotremes also possess ear canals but lack an obvious pinna structure. The pinna assists in selecting directionality of sound input by diffraction of incoming sound waves, and in many mammals can be rotated to detect sounds from a specific source. The relationship between the diffractive capacity of an object and sound wavelength suggests that pinnae arose in small mammals with the ability to hear moderately high frequency sounds (Clack and Allin, 2004).
1.2: Development of the inner ear primordium – from non-neural ectoderm to the otic placode
Complex sequential signaling leads to the formation of the pre-placodal region from non-neural ectoderm
The entire inner ear, together with the vestibulo-acoustic ganglion that will connect its sensory regions to the hindbrain, develops from a thickened region of embryonic ectoderm next to the hindbrain termed the otic placode (Groves, 2005; Streit, 2001). As discussed in other chapters in this volume, the otic placode is one of several cranial placodes that form the olfactory and lens epithelium, the epibranchial and ophthalmic trigeminal sensory ganglia, the inner ear and in some vertebrate taxa, the lateral line system (Baker and Bronner-Fraser, 2001; Saint-Jeannet and Moody, 2014; Schlosser, 2006; Schlosser, 2010; Streit, 2004; Streit, 2007). The transition from early embryonic ectoderm to a definitive placode proceeds through a series of sequential developmental decisions. These can be divided conveniently into three stages: the choice between neural and non-neural ectoderm, the division of non-neural ectoderm into neural crest and early “pre-placodal” progenitors in the medio-lateral axis, and the singling out of individual placodes from pre-placodal progenitors along rostro-caudal axis of the head. These developmental decisions have been well-summarized in a number of recent reviews (Grocott et al., 2012; Groves and LaBonne, 2014; Patthey and Gunhaga, 2011; Patthey and Gunhaga, 2013), but we will briefly describe some of these steps below.
An emerging consensus is that early embryonic ectoderm shows evidence of division into presumptive neural and non-neural ectoderm even before the start of gastrulation in vertebrates. The action of FGF signals from tissues underlying early ectoderm (such as hypoblast, future definitive endoderm, or anterior visceral endoderm), together with antagonists of the Wnt and BMP signaling pathways leads to the expression of pre-neural genes, such as ERNI, Otx2, Geminin and Sox3 (Bally-Cuif et al., 1995; Groves and LaBonne, 2014; Kroll et al., 1998; Papanayotou et al., 2008; Rex et al., 1997; Streit et al., 2000). Coincident with this early induction, BMP and Wnt signaling in more ventral ectoderm populations specifies non-neural ectoderm, defined by the expression of Dlx, Gata2/3 and Foxi gene families (Brown et al., 2005; Hans and Westerfield, 2007; Hoffman et al., 2007; Knight et al., 2003; Li and Cornell, 2007; Luo et al., 2001; Matsuo-Takasaki et al., 2005; McLarren et al., 2003; Ohyama and Groves, 2004a; Papalopulu and Kintner, 1993; Pera and Kessel, 1999; Phillips et al., 2006; Pieper et al., 2012; Sheng and Stern, 1999; Woda et al., 2003).
The exposure of pre-neural ectoderm to neural inducing factors leads to expression of markers of the definitive neural plate, such as Sox2 (Rex et al., 1997; Streit et al., 1997; Uchikawa et al., 2003), whereas non-neural ectoderm genes such as Dlx5/6 and Gata3 begin to be restricted to the border between neural and non-neural ectoderm (Feledy et al., 1999; Khudyakov and Bronner-Fraser, 2009; Kwon et al., 2010; Pieper et al., 2012; Streit, 2002; Woda et al., 2003). Evidence from a number of vertebrate species suggests this border is sharpened by cross-repressive interactions between neural and non-neural transcription factors and by positive feedback within each domain (Feledy et al., 1999; Kwon et al., 2010; Linker et al., 2009; Luo et al., 2001; Matsuo-Takasaki et al., 2005; McLarren et al., 2003; Pieper et al., 2012; Tribulo et al., 2003; Woda et al., 2003). As ectoderm further differentiates, a thin strip at the neural plate border receives both FGF and Wnt signaling; however, it is also subject to continuing BMP inhibition from secreted antagonists (Ahrens and Schlosser, 2005; Brugmann et al., 2004; Litsiou et al., 2005). This border region divides into two molecularly distinct populations around the time of head formation: the pre-placodal region (PPR) confined to the anterior region of the embryo, and premigratory neural crest that is generated from all levels of the embryo other than the forebrain (Betancur et al., 2010; Grocott et al., 2012; Groves and LaBonne, 2014; McCabe and Bronner-Fraser, 2009; Milet and Monsoro-Burq, 2012; Prasad et al., 2012; Sauka-Spengler and Bronner-Fraser, 2008; Schlosser, 2006; Schlosser, 2010; Steventon et al., 2014; Stuhlmiller and Garcia-Castro, 2012). The pre-placodal region is defined by expression of members of the Six (sine oculis homeobox) transcription factors and their Eya (eyes absent) co-factor partners (Bailey and Streit, 2006; Saint-Jeannet and Moody, 2014; Streit, 2007). This domain lies lateral to the neural crest progenitor domain at the boundary of the neural plate.
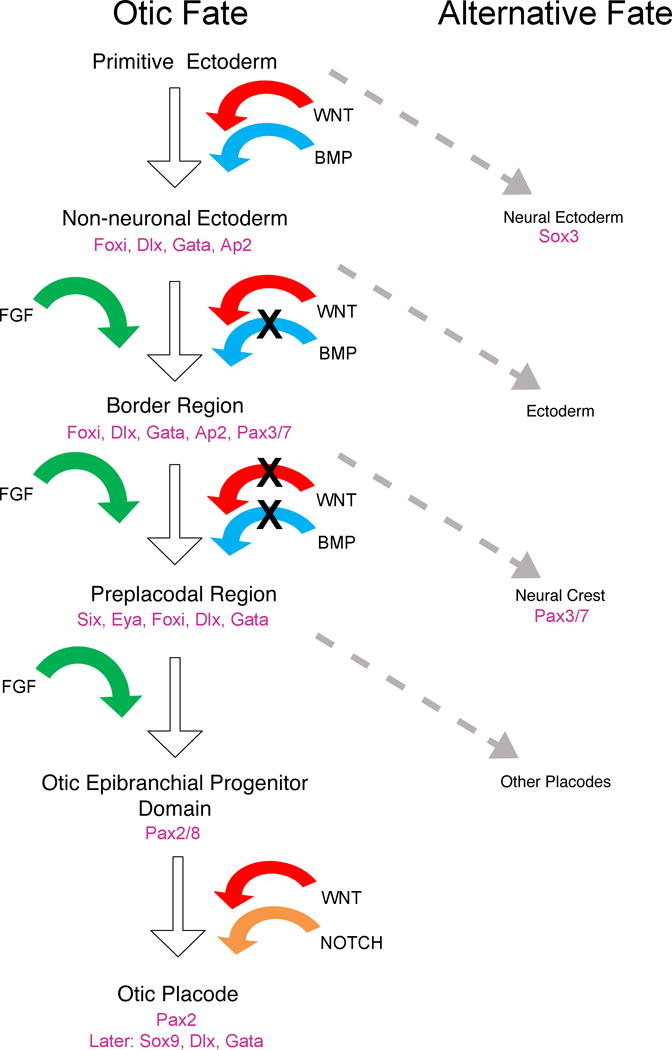
To date, no genetic fate mapping studies have been performed to determine the derivatives of Six- or Eya-expressing progenitors, although dye labeling studies suggest this region will contribute to all craniofacial placodes including the otic placode (Bhattacharyya et al., 2004; Kozlowski et al., 1997; Streit, 2002; Xu et al., 2008). In addition to these definitive pre-placodal markers, non-neural ectoderm genes such as foxi1 (in fish), Foxi3 (in amniotes) and Dlx5/6 and Gata3 become restricted to the pre-placodal region (Grocott et al., 2012; Groves and LaBonne, 2014). The expression and maintenance of pre-placodal markers is dependent on FGF signaling and the inhibition of Wnt and Bmp signaling (Ahrens and Schlosser, 2005; Brugmann et al., 2004; Litsiou et al., 2005), while cells closer to the neural plate respond to FGF, BMP and Wnt signaling and express early neural crest markers such as Msx1 and Pax3/7 (Groves and LaBonne, 2014; Milet and Monsoro-Burq, 2012; Prasad et al., 2012). Figure 2 summarizes the series of signals involved in specification of the pre-placodal region from non-neural ectoderm before and after gastrulation.
Figure 2. Requirements for otic induction from primitive ectoderm.
The figure depicts a consensus scheme of otic induction for amniotes. For each step, the necessary signaling factors for the otic pathway are shown. The alternative pathway is chosen in the absence of the specific combination of signaling and/or with additional signals. Gene expression defining tissue identity for individual steps are indicated. X represents the requirement for an inhibitor of a signaling pathway, such as BMP or Wnt inhibitors.
Induction of the otic placode by FGFs and its refinement by Wnt and Notch signaling
As discussed above, the pre-placodal ectoderm gives rise to all craniofacial sensory placodes of the vertebrate head (Grocott et al., 2012; Litsiou et al., 2005; Sato et al., 2010; Streit, 2007), and the progenitors for each of the placodes are indistinguishable during early establishment of the pre-placodal region at the neural plate boundary. The differentiation of distinct placodal fates requires regional signals, and in the case of the otic placode, data from all major vertebrate groups suggests that FGF signaling plays an early and critical role (Ohyama et al., 2007; Riley and Phillips, 2003; Solomon et al., 2004). Interestingly, both the location and identity of FGFs involved in otic placode induction vary between species; for example, FGF3 from the hindbrain and FGF10 from cranial mesoderm are necessary for otic placode induction in mice (Urness et al., 2010; Wright and Mansour, 2003), while in zebrafish, this role is served by Fgf3 and Fgf8 that are both expressed in the hindbrain (Leger and Brand, 2002; Maroon et al., 2002; Phillips et al., 2001). The absolute necessity for FGF signaling in otic placode induction has been shown in multiple organisms. (Alvarez et al., 2003; Ladher et al., 2005; Leger and Brand, 2002; Yang et al., 2013; Zelarayan et al., 2007). The earliest otic markers expressed by pre-placodal ectoderm in response to FGF signals are members of the Pax2/5/8 family (Groves and Bronner-Fraser, 2000; Heller and Brandli, 1999; Ohyama and Groves, 2004b; Pfeffer et al., 1998). Fate-mapping studies in mice and chick suggest that the Pax2-expressing domain contains all the progenitors of the inner ear but also gives rise to the epibranchial placodes and some parts of the epidermis (Ohyama and Groves, 2004b; Ohyama et al., 2007; Streit, 2001). This region of Pax2-expressing cells has been referred to as the otic-epibranchial progenitor domain, or OEPD (Freter et al., 2008; Ladher et al., 2010).
Studies in amniotes suggest that one mechanism that refines the broad domain of Pax2-expressing cells into the otic placode and epibranchial placodes and ectoderm is the strength and duration of FGF signaling (Freter et al., 2008; Ladher et al., 2010). Some of the earliest markers of the otic placode are negative regulators of the FGF pathway, such as Sprouty genes and dual-specificity phosphatases such as Dusp6 and Dusp9 (Chambers and Mason, 2000; Urness et al., 2008). The importance of attenuating FGF signaling in otic placode induction was demonstrated by a failure of induction following prolonged ectopic activation of FGF signaling (Freter et al., 2008); extended FGF signaling is instead required to promote epibranchial placode fate (Ladher et al., 2010). Two other mechanisms that demarcate otic from non-otic derivatives within the Pax2 domain are Wnt and Notch signaling (Groves and Fekete, 2012). A gradient of Wnt signals from the midline and border of the neural plate directs Pax2-expressing cells towards an otic fate rather than epidermal derivatives (Ohyama et al., 2007). Disruption of canonical Wnt signaling (for example, by conditional deletion of β-catenin in Pax2-expressing cells) greatly reduces the size of the otic placode, while activation of Wnt signals by expression of constitutively active forms of β-catenin drive the entire Pax2 domain towards an otic fate (Freter et al., 2008; Ohyama et al., 2006). The final step in the refinement of otic and non-otic fates is up-regulation of ligands of the Notch signaling pathway such as Jagged1 in presumptive otic ectoderm in response to high levels of Wnt signaling (Jayasena et al., 2008). Jagged1-Notch signaling in this region positively feeds back to further increase the strength of Wnt signaling in regions of Pax2-expressing cells close to the neural plate, but not in more lateral regions where Wnt levels are too low to induce Jag1 expression and activate the Notch pathway (Jayasena et al., 2008). In this way, a continuous gradient of Wnt becomes discontinuous, fixing otic and non-otic fates (Figure 2).
Although Pax2/5/8 gene family members are the earliest known markers of the otic placode, these genes do not appear to be necessary for otic placode induction. However, they are necessary for later events in inner ear development (Bouchard et al., 2010; Burton et al., 2004). To date, no combination of Six and Eya mouse knockouts, knock-down, or dominant-negative mutants in other vertebrates prevents the induction of the otic placode (Bricaud and Collazo, 2006; Zhang et al., 2004; Zheng et al., 2003; Zou et al., 2006), though these genes are also individually necessary for later aspects of ear development and some double mutants of these genes can have very small otic vesicles (Chen et al., 2009; Grifone et al., 2005; Zou et al., 2004). Mutation or knock-down of Dlx gene family members can greatly reduce the size of the inner ear (Esterberg and Fritz, 2009; Liu et al., 2003; Solomon and Fritz, 2002; Solomon et al., 2004), but Dlx genes again do not appear to be necessary for otic placode induction (Robledo and Lufkin, 2006; Robledo et al., 2002). To date, the only genes individually necessary for otic placode induction appear to be foxi1 in fish (Hans et al., 2007; Hans et al., 2004; Nissen et al., 2003; Solomon et al., 2003a) and Foxi3 in chick and mouse (Edlund et al., 2014; Khatri et al., 2014). We will return to the role of Foxi genes in inner ear induction later in the chapter. First, we will discuss the development of the first and second pharyngeal arches that generate much of the middle and external ears, together with the jaw.
1.3: The middle and outer ears develop from the first two branchial arches
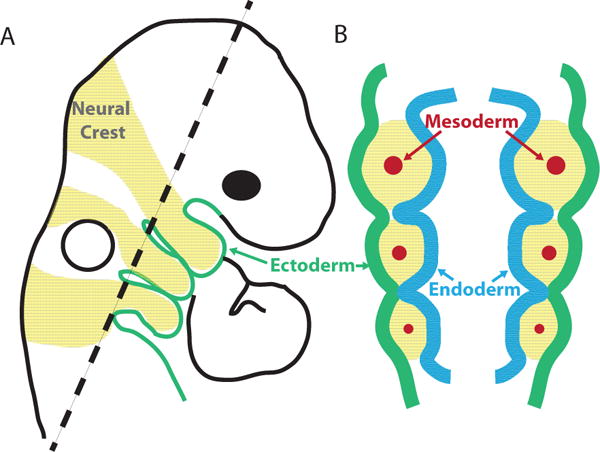
During vertebrate embryogenesis, structures in the face and neck develop from transient branchial arches, also called pharyngeal arches. Following establishment of the embryonic body plan and the development of a basic neural tube, five to six pairs of arches expand from the ventral side of the embryo at the level of the hindbrain and eventually fuse medially (Figure 3A). The arches are arranged in a simple serial pattern and contain elements of all three embryonic germ layers. The surface of each branchial arch (BA) is composed externally of ectoderm, and each arch is lined internally with endoderm. Between the epithelial layers lies a mesodermal core surrounded by immigrating neural crest cells from the midbrain and hindbrain. The boundaries between arches are defined by points of direct contact between ectoderm and endoderm, termed pharyngeal pouches (Figure 3B). The endoderm of the pouches is critical for branchial arch development (Grevellec and Tucker, 2010) and the detailed morphology of pharyngeal tissue types has been comprehensively reviewed by Graham, Liu and colleagues (Graham et al., 2004; Graham and Richardson, 2012; Graham and Smith, 2001; Szabo-Rogers et al., 2010)
Figure 3. Arrangement of germ layers and neural crest cells in the branchial arches.
(A) sagittal view of an embryonic day 9.5 (E9.5) mouse embryo showing branchial arches (BA) 1–3. The external surface of each arch is ectoderm. Neural crest cells (NC) in three distinct streams populate the arches. The most anterior stream contains NC from the midbrain and rhombomeres 1 and 2 from the hindbrain. Rhombomeres 3 and 5, the white regions between streams of yellow neural crest produce very few neural crest cells. (B) Schematic of a coronal section through the arches of the embryo in (A). Branchial arches consist of external ectoderm, an endodermal lining, a core of mesoderm, and neural crest cells from the midbrain and hindbrain. The boundaries of the arches are defined by points of contact between endoderm and ectoderm: the pharyngeal pouches.
Branchial arch ectoderm, mesoderm, and endoderm contribute to middle and outer ear structures
As branchial arch development progresses, the dorsal (proximal) territory of the first arch bifurcates into two processes, a posterior mandibular process and an anterior maxillary process. A portion of the outer ear derives from posterior ectoderm and mesoderm of the mandibular process and anterior ectoderm and mesoderm of BA2 (Alasti and Van Camp, 2009). Three auricular hillocks arise on the surface of each arch, then expand and remodel, while the ectoderm of the two arches fuses to form the external pinna that is the visible portion of the mammalian ear. Ectoderm from BA1 invaginates and develops into the outer ear canal that leads to the tympanic membrane (Minoux et al., 2013).
Meanwhile, ear structures also develop from pharyngeal pouch endoderm. The Eustachian tube and a portion of the middle ear cavity lining develop from the first pouch. The middle ear consists of an air filled cavity containing one ossicle in birds and reptiles (the columella) or three ossicles in mammals (the malleus, incus, and stapes) that transmit vibrations from the ear drum to the cochlea. In birds and reptiles the epithelial lining of this cavity is completely derived from first pouch endoderm. In land tetrapods, however, the three ossicles condense adjacent to the endodermal epithelium of the first pouch (Mallo, 1998; Mallo, 2001). The endoderm must therefore rupture to line the middle ear cavity, where it comprises only the ventral portion of the cavity epithelium. The dorsal portion is composed of epithelialized neural crest cells, as described below (Thompson and Tucker, 2013). An excellent recent review of middle ear evolution is given by Manley (Manley, 2010).
Cranial neural crest cells contribute to middle ear structures
One of the developmental innovations unique to vertebrates is the neural crest, a specialized population of cells that exit the neuroepithelium and populate the periphery of the body. Neural crest cells have the potential to differentiate into a variety of cell types and structures. Cranial crest cells derived from the midbrain and hindbrain are particularly plastic, generating bone and cartilage that forms much of the face, jaw, and anterior skull, as well as melanocytes and peripheral neural structures. The process by which cranial neural crest populations adopt their final fate is complicated, involving interactions between all three embryonic primary germ layers and finely tuned signaling, much of which takes place in the branchial arches (Creuzet et al., 2005; Graham and Smith, 2001; Medeiros and Crump, 2012; Minoux and Rijli, 2010).
By the time arch-derived structures begin to take shape, the bulk of mesenchymal cells are cranial neural crest cells that delaminated from the posterior midbrain and the hindbrain. Migration occurs in separate, distinct, and stereotypical streams of cells, with cells delaminating from the posterior midbrain and rhombomeres 1 and 2 of the hindbrain populating BA1, and cells from rhombomere 4 populating BA2. As these cells migrate, they often express Hox genes characteristic of their respective rhombomeres of origin, contributing to the establishment of a distinct identity for each branchial arch.
The neural crest in the mandibular process gives rise to Meckel’s cartilage that develops in mammals into two of the three ossicles of the middle ear, the incus and the malleus, as well as supporting the mandible during ossification (Chai et al., 2000). Neural crest cells in the maxillary process differentiate into the upper jaw, or maxilla, associated connective tissue, and the squamosal bone. The second branchial arch (BA2) contains neural crest cells that develop into Reichert’s cartilage from which the third ear ossicle, the stapes, is derived (Sienknecht, 2013). Additionally, in mammals the three neural-crest derived middle ear ossicles are surrounded by mesenchymal neural crest cells at the site of the future middle ear cavity. After the incus and malleus separate from Meckel’s cartilage and the stapes separates from Reichert’s cartilage, the surrounding crest cells retract to create an air filled space and undergo a mesenchymal to epithelial transition to form the dorsal portion of the middle ear cavity lining (Thompson and Tucker, 2013).
1.4: Signals and transcriptional regulators involved in the development of the first and second branchial arches
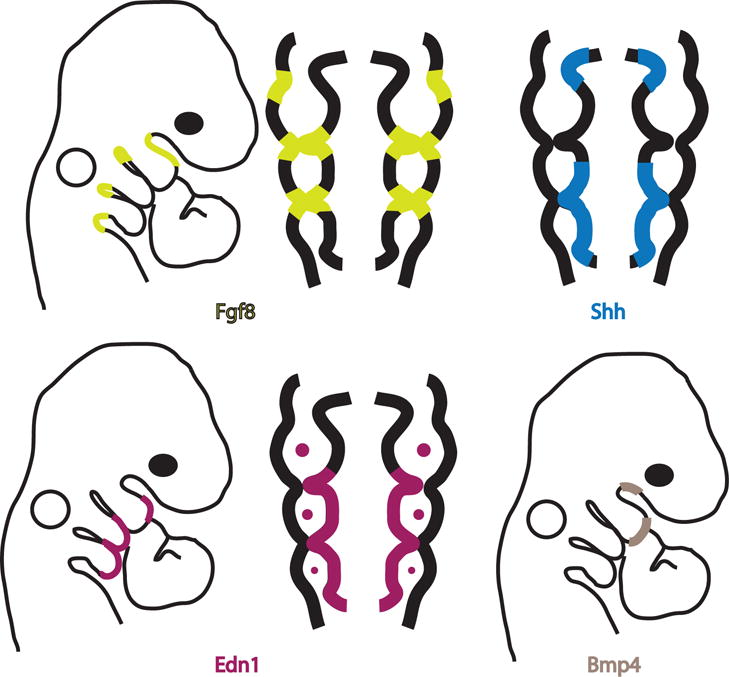
The ectoderm and endoderm surrounding the arches produce multiple signaling factors, including FGFs, BMPs, Endothelins, and Sonic Hedgehog, that communicate to the underlying mesenchyme – a mixture of mesoderm and neural crest cells (Figure 4). Fgfs are expressed in the epithelial tissues of the branchial arches and are particularly enriched in the pouch endoderm and overlying ectoderm in the clefts between the arches (Crossley and Martin, 1995; Wall and Hogan, 1995). In the developmental context of the branchial arches, FGFs function as survival factors for neural crest entering the arches, and as signals that induce expression of arch patterning transcription factors in mesenchyme and ectoderm.
Figure 4. Signaling factors from the pharyngeal ectoderm and endoderm.
At embryonic day 9.5 (E9.5), four major signaling factors are secreted from the ectoderm and endoderm. Fgf8 is expressed in the ectoderm between the arches and between the mandibular and maxillary processes of BA1 and in the endoderm at the tips of the pharyngeal pouches. Shh is expressed in endoderm underlying BA2 and BA3. Edn1 is expressed in ventral arch ectoderm, endoderm underlying the mandibular process, BA2, and BA3, and in the mesoderm of each arch. Bmp4 is expressed in the ectoderm overlying the ventral domain and the maxillary process of branchial arch 1 (BA1).
One important function of signaling factors from the arch ectoderm is defining the dorsal-ventral signals that pattern post-migratory neural crest cells in the arches (reviewed in (Medeiros and Crump, 2012)). In order to establish the dorsal-ventral axis in BA1, dorsally expressed Fgf in the ectoderm is restricted by Bone Morphogenetic Protein 4 (BMP4), expressed in the ventral (distal) portion of the BA1 ectoderm. Ectopic Bmp4 expression or implantation of a BMP4 soaked bead reduces Fgf8 expression in the branchial arches of chick embryos (Shigetani et al., 2000). Interestingly, in the arches BMP4 induces expression of its own antagonists, Noggin and Chordin (Stottmann et al., 2001). In addition to maintaining the boundaries of ectodermal Fgf8 expression, BMP4 also plays important roles in dorsal-ventral patterning of BA1. A signaling network in which BMP4 and Endothelin 1 (EDN1) overlap to establish the dorsal-ventral axis is conserved from fish to mammals (Alexander et al., 2011; Ozeki et al., 2004; Ruest et al., 2004). BMP4 promotes expression of ventral territory transcription factors in the arch mesenchyme and represses expression of dorsal transcription factors (Liu et al., 2005). Conditional deletion of Bmp4 from both mouse arch ectoderm and endoderm results in almost complete absence of the mandible and shift toward the midline of the tympanic ring and Meckel’s cartilage (Liu et al., 2005).
There is evidence in zebrafish that Bmp4 induces ectodermal expression of Edn1 (Alexander et al., 2011) and as the arches develop, factors induced by Bmp4 come to define the most ventral region of BA1, whereas Edn1 targets occupy a more intermediate domain. One key role of Edn1 signaling is to establish the nested expression pattern of Dlx transcription factors that delineate the dorsal-ventral axis of BA1 (Ozeki et al., 2004). Zebrafish and mouse Edn1-null mutants transform mandibular arch structures into structures normally derived from the maxillary process, with a mirror duplication observable in skeletal staining (Miller et al., 2000; Ozeki et al., 2004; Ruest et al., 2004).
A fourth signaling pathway, the Hedgehog pathway is also of critical importance for patterning the pharyngeal region. Sonic hedgehog (Shh) is expressed in the foregut endoderm prior to arch outgrowth (Wall and Hogan, 1995). Ablation of Shh-expressing endoderm in chick embryos leads to absence of BA1 derived structures, defects that can be rescued by implantation of Shh soaked beads in the approximate location of the missing endoderm (Brito et al., 2006). These ablation and bead rescue experiments demonstrate a role for Shh in establishing much of the ectodermal patterning required for arch development, including Fgf8 and Bmp4 expression (Haworth et al., 2007).
2: FORKHEAD PROTEINS AS TRANSCRIPTION FACTORS AND PIONEER FACTORS
In 1989, a new transcription factor containing a previously uncharacterized protein motif was identified in Drosophila melanogaster. Named Forkhead (FKH), the protein was first shown to function in gut development (Weigel et al., 1989). Shortly thereafter a homolog, HNF-3, was identified in rat and the motif conserved between the two proteins was determined to be a DNA binding domain, now called a Forkhead box (Lai et al., 1990; Weigel and Jackle, 1990). In the ensuing years, an extraordinary number of additional factors containing the Forkhead box motif have been found across diverse animal species. In an effort to systematize nomenclature of these related genes, a consensus naming strategy was established for chordates in which each gene containing the motif was labeled “Fox” for forkhead box, sorted into a class lettered A-S based on closest homology, and assigned a unique number within that class (Jackson et al., 2010; Kaestner et al., 2000). For example, Hnf-3 is renamed Foxa2 to indicate that it is a Forkhead-box protein in the a class distinct from Foxa1.
Forkhead proteins are archetypal pioneer factors
While the classical description of transcription factors views these proteins as binding their target sites in DNA and influencing transcription of target genes within a short time after binding, a subset of transcription factors can influence transcriptional activity in a delayed fashion. Often termed pioneer factors, such proteins can act through more than one mechanism, both passive and active. Pioneer factors that operate in a passive role are necessary but not sufficient for transcriptional activation (Bossard and Zaret, 1998; Zaret and Carroll, 2011). Rather, they bind target sequences that control gene expression via multiple transcription factors acting in concert, and binding of the pioneer factor merely reduces the number of steps required to initiate gene expression at the appropriate timepoint. On the other hand, many pioneer factors can actively influence gene regulation through manipulation of chromatin structure. They are able to bind the outer face of DNA in regions of tightly compacted chromatin and then open the chromatin to improve accessibility to other transcription factors (Cirillo et al., 2002; Cirillo and Zaret, 1999; Cuesta et al., 2007; Perlmann, 1992). Interestingly, some pioneer factors demonstrate the ability to remain bound to chromatin during mitosis (Yan et al., 2006; Zaret and Carroll, 2011). Although chromatin remains transcriptionally inactive during this time, the presence of pioneer factors at target sequences upon completion of cell division allows rapid reactivation of target gene transcription (Blobel et al., 2009; Dey et al., 2009; Kadauke et al., 2012). This mechanism has been described as “bookmarking” the DNA in a specific cell lineage, as a form of epigenetic mark, to aid in preparing lineage-specific genes for subsequent rapid transcription.
The shared DNA-binding domain in FOX proteins is composed of three α helices and two loops resembling insect wings, and was thus called a winged-helix domain (Lai et al., 1993). Interestingly, the winged helix domain bears a distinct resemblance and possibly an evolutionary relationship to the linker histones H5 and H1 (Zaret and Carroll, 2011). The similarity of the FOX winged helix domain to linker histones suggests that FOX proteins might have access to DNA in regions of compacted chromatin. Indeed, this appears to be the case for more than one member of the Forkhead family; three FOXA factors, FOXD3, FOXE1, and FOXO have been shown to be pioneer factors that can actively open chromatin to allow for binding of additional transcription factors (Cuesta et al., 2007; Hatta and Cirillo, 2007; Xu et al., 2009; Zaret and Carroll, 2011). Additionally, these FOX factors and other pioneer factors commonly bind in an unusually tight manner to chromatin (Zaret and Carroll, 2011).
Perhaps the best-characterized Forkhead pioneer factors are the FOXA proteins, originally identified as early arrivals to mouse albumin regulatory sites during endoderm differentiation (Gualdi et al., 1996). The presence of redundantly expressed FOXA1 or FOXA2 at this albumin regulatory site is required for early liver development (Bossard and Zaret, 1998; Lee et al., 2005). The FOXA factors, in the cellular context of liver progenitors, bind their target sites in compacted chromatin and open it for further transcription factor binding (Cirillo et al., 2002). FOXA1 protein remains bound to chromatin during mitosis, both at specific target sites and non-specific sites throughout the compacted chromosomes (Caravaca et al., 2013). This suggests a bookmarking role for FOXA1 at its specific target sites, and non-specific binding as a mechanism for retaining FOXA1 close at hand to jump-start activity at nearby target sites upon the conclusion of telophase. Of note, a classical transcription factor role has been proposed for FOXA2 in inducing expression of Tbx1 in the endoderm (Yamagishi et al., 2003). Overexpression of Foxa2 alone in HeLa cells was sufficient to activate a luciferase reporter fused to the FOXA binding site of the Tbx1 enhancer. While this may be an artifact of the in vitro system, it is important not to discount the possibility that a transcription factor may behave as either a pioneer factor or classical transcription factor depending on the target gene and cellular context. Whether acting as classical transcription factors or pioneer factors, with their abundance in chordate genomes, their presence in almost every tissue throughout development, and their ability as pioneer factors to prime genomic regions for rapid activation, FOX transcription factors are critical and universal developmental regulators.
3: THE ROLE OF FOXI FAMILY MEMBERS IN INNER EAR DEVELOPMENT
3.1: The role of Foxi1 in mammalian ear development
The first Foxi family member to be identified in mammals was Fkh10, now re-named Foxi1 (Hulander et al., 1998). Foxi1 mutant mice display hyperactivity, circling behavior, and deafness (Hulander et al., 1998) and although the inner ear initially develops normally, much of the epithelial labyrinth becomes swollen and expanded by late embryonic stages, consistent with a failure in endolymphatic fluid regulation in the ear (Hulander et al., 2003). Foxi1 is initially expressed in the primordium of the endolymphatic duct in the developing otocyst (Ohyama and Groves, 2004a), and is later restricted to the epithelium of the endolymphatic duct and sac (Hulander et al., 2003; Raft et al., 2014; Vidarsson et al., 2009). FOXI1 is known to regulate a number of genes involved in ion transport, such as Slc26A4 which encodes the pendrin anion exchanger implicated in Pendred syndrome (Everett et al., 1997), and human FOXI1 mutations can also cause Pendred syndrome (Yang et al., 2007). FOXI1 also regulates subunits of other ion transporters such as the vacuolar H+-ATPases (Vidarsson et al., 2009), and mice with mutations in some of these subunits also have enlarged endolymphatic compartments (Lorente-Canovas et al., 2013). Interestingly, the apparent functional homolog of Foxi1 in zebrafish is foxi3, which also appears to regulate the development and function of transport epithelial cells, the ionocytes of the fish epidermis (Cruz et al., 2013; Hsiao et al., 2007; Janicke et al., 2007; Janicke et al., 2010; Solomon et al., 2003b; Thermes et al., 2010).
3.2: Expression, regulation and function of Foxi1/3 factors in inner ear development – from non-neural ectoderm to otic placode via the pre-placodal domain
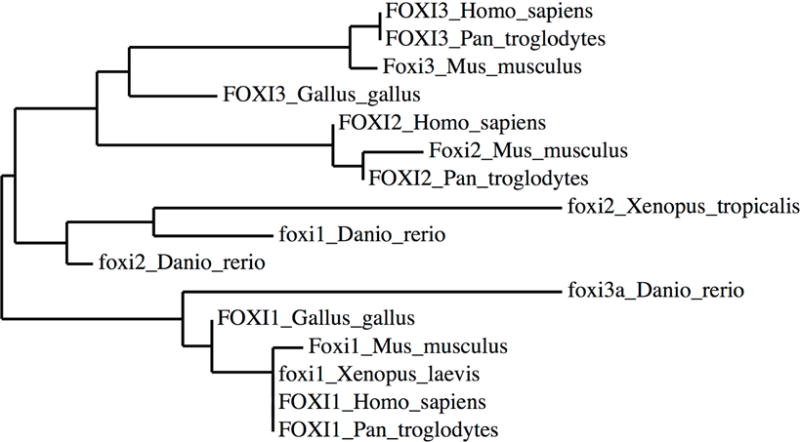
Both DNA sequence data and phenotypic analysis of mutants suggest that zebrafish foxi1 and mouse and chicken Foxi3 should be considered to be homologs (Figure 5). For convenience, we will refer to the genes as Foxi1/3 in the following sections to refer to results that have been confirmed in both fish and mouse and/or chicken. Foxi1/3 transcripts are initially expressed in non-neural ectoderm surrounding the future neural plate (Khatri et al., 2014; Khatri and Groves, 2013; Kwon et al., 2010; Lee et al., 2003; Nissen et al., 2003; Ohyama and Groves, 2004a; Solomon et al., 2003a; Solomon et al., 2003b), overlapping with other transcription factor markers of non-neural ectoderm such as Gata3, Dlx5/6, and Ap2α. In common with these other non-neural ectoderm genes, Foxi1/3 becomes restricted to the pre-placodal domain with definitive pre-placodal region markers such as Six and Eya gene family members(Grocott et al., 2012; Groves and LaBonne, 2014; Kwon et al., 2010; Streit, 2007). After formation of the otic placode, Foxi1/3 is down-regulated in the otic region but continues to be expressed in epibranchial placode ectoderm and ectoderm of the future pharyngeal arches (Edlund et al., 2014; Khatri et al., 2014; Khatri and Groves, 2013; Ohyama and Groves, 2004a; Solomon et al., 2003a; Solomon et al., 2003b). Later in development, Foxi3 is also expressed in pharyngeal clefts and pouches (Edlund et al., 2014; Ohyama and Groves, 2004a) and in placodes associated with epidermal appendages such as tooth primordia and hair follicles (Drogemuller et al., 2008; Shirokova et al., 2013).
Pre-placodal Foxi1/3 expression is regulated by the same signals that initiate expression of other non-neural ectoderm genes. Zebrafish foxi1 is dependent on BMP signaling before, but not after, gastrulation (Bhat et al., 2013; Hans et al., 2004; Kwon et al., 2010; Solomon et al., 2003a). A number of studies have reached different conclusions regarding a possible role for Fgf signaling in regulating foxi1 expression in zebrafish (Hans et al., 2007; Nechiporuk et al., 2007; Padanad et al., 2012; Phillips et al., 2004), although the precise time and manner in which Fgf signaling was manipulated may explain some of the differences in these experiments. Furthermore, another zebrafish study suggests that, in addition to Bmps and Fgfs, foxi1 is also regulated by retinoic acid (Hans and Westerfield, 2007). Expression of Pax8, induced by foxi1, as well as expression of foxi1 itself, expand ectopically in the presence of retinoic acid, whereas pax8 expression decreases in response to RA inhibitors. This action of retinoic acid may be direct (rather than indirectly affecting anterior-posterior patterning) as dlx3b expression in the preplacodal domain is not affected by manipulating retinoic acid levels (Hans and Westerfield, 2007). Finally, in chick, ectodermal Foxi3 expression in gastrulating embryos is regulated by signals released from the hypoblast, as has also been shown for Dlx5/6 genes (Khatri et al., 2014; Pera et al., 1999). These hypoblast signals appear to be functionally distinct from later signals in cranial mesendoderm, which regulate Six and Eya family genes in the pre-placodal region (Khatri et al., 2014; Litsiou et al., 2005).
In both zebrafish and chick, Foxi1/3 genes are part of a transcriptional regulatory network that gives the tissue its competence to adopt a pre-placodal identity and to respond to FGF signals. In zebrafish Foxi1, Gata3 and Tfap2a form a complex, cross-regulatory feedback network that is at least partially dependent on Bmp signaling (Bhat et al., 2013; Kwon et al., 2010). Although the same experiments have yet to be performed in chick, recent evidence suggest that amniote Foxi3, Dlx5, and Six/Eya genes can cross-regulate each other. Ectopic expression of Dlx5 or Six1 and Eya2 leads to ectopic Foxi3 expression in non-neural ectoderm (Khatri et al., 2014). Furthermore, Foxi3 overexpression at the same developmental stage leads to activation of these three genes in non-neural ectoderm. Together, these fish and chick data support the idea that Foxi1/3 is part of a transcriptional regulatory network in non-neural ectoderm that is essential for the transition of pre-placodal ectoderm into the otic placode.
The only other amniote Foxi homolog, Foxi2, does not appear to be expressed in the otic placode, although it is notably expressed in the future epidermis surrounding the otic placode, in progenitors for delaminating epibranchial sensory neurons, and in branchial arch ectoderm (Freter et al., 2008; Khatri and Groves, 2013; Ohyama and Groves, 2004a). Zebrafish foxi2 is expressed in chordamesoderm during somitogenesis and is later found in the retina and the branchial arches (Solomon et al., 2003b). Genetic manipulations that reduce the size of the otic placode in amniotes, such as conditional deletion of β-catenin in Pax2-expressing progenitors, lead to an expansion of the Foxi2 ectodermal domain, while manipulations that expand the otic placode lead to a corresponding reduction in Foxi2 expression (Freter et al., 2008; Jayasena et al., 2008; Ohyama et al., 2006; Urness et al., 2010). Homozygous Foxi2 mutant mice have been generated in our laboratory, but appear to have no obvious defect (Ohyama, Edlund, Birol, and Groves, unpublished observations).
3.3: Functional role of Foxi1/3 in otic placode induction
foxi1 was first identified in two different zebrafish mutagenesis screens as the hearsay and foo mutants, both of which have significant jaw defects and either very small or completely absent otocysts (Nissen et al., 2003; Solomon et al., 2003a). The consensus from a number of studies suggests that foxi1 acts to provide ectoderm with competence to respond to FGF signaling during otic placode induction (Bhat et al., 2013; Hans et al., 2007; Hans et al., 2013; Hans and Westerfield, 2007; Kwon et al., 2010; Padanad et al., 2012; Padanad and Riley, 2011). Zebrafish foxi1 is also required for epibranchial placode induction (Lee et al., 2003; Nechiporuk et al., 2007; Nechiporuk et al., 2005), consistent with the derivation of both the otic placode and epibranchial placodes from a common Pax2-expressing domain. However, an understanding of the precise role of foxi1 in zebrafish leading from the establishment of the pre-placodal domain to the induction of the otic placode in response to FGF signals is complicated by several observations. Analysis of otic placode markers shows that expression of pax2, one of the first otic markers, is decreased, but still present in foxi1 mutants (Hans et al., 2007; Hans et al., 2004; Nissen et al., 2003; Solomon et al., 2003a; Solomon et al., 2004). Abolishing otic induction completely in zebrafish requires knocking down foxi1 together with either three other non-neural ectoderm genes (tfap2a and 2c and gata3 (Bhat et al., 2013; Kwon et al., 2010)) or with dlx3b and dlx4b (Hans et al., 2007; Solomon et al., 2004). In these embryos pax2a expression is completely abolished and the ear does not form even in the presence of fgf8 overexpression. (Hans et al., 2007; Hans et al., 2004; Solomon et al., 2004). Moreover, in zebrafish unlike mammals, the early otic placode markers pax2a and pax8 appear to be expressed over slightly different time courses (Hans et al., 2004; Mackereth et al., 2005; Ohyama and Groves, 2004b), and appear to be regulated by different signals. Pax8 can be regulated independently by either foxi1 or FGF (Hans et al., 2004; Phillips et al., 2004; Solomon et al., 2004), whereas pax2a is regulated by both dlx3b/4b and FGF signals (Hans et al., 2007; Hans et al., 2004; Solomon et al., 2004). Further complicating matters, knockdown of pax2a, pax2b and pax8 leads to the disappearance of the inner ear in zebrafish, but not in mice (Bouchard et al., 2010; Mackereth et al., 2005). Finally, dlx3b and dlx4b do not appear to have counterparts in amniotes that share expression pattern and function. Dlx5 and Dlx6 are expressed much earlier and more broadly in amniotes than dlx3b/4b in fish (McLarren et al., 2003; Pera et al., 1999; Streit, 2002; Streit, 2007), and are not necessary for otic placode induction (Robledo and Lufkin, 2006; Robledo et al., 2002). Therefore the relationship of these genes to foxi1 is difficult to compare with the interactions between Foxi3 and Dlx5/6 in amniotes.
The function of Foxi3 in otic placode induction in amniotes has received much less attention, and it is not clear at present whether the pathways regulated by Foxi3 are mechanistically more simple, or simply less well explored. Knockdown of Foxi3 in chick embryos at pre-placodal stages can effectively abolish Pax2 expression, as well as greatly attenuate the ability of pre-placodal ectoderm to express Pax2 when cultured in the presence of FGFs (Khatri et al., 2014). Foxi3 mutant mice appear to entirely lack an inner ear (Edlund et al., 2014), although it is not clear whether this represents a complete loss of otic tissue, or whether a small amount of Pax2-expressing otic tissue remains as in zebrafish (Hans et al., 2007; Hans et al., 2004; Nissen et al., 2003; Solomon et al., 2003a; Solomon et al., 2004). We are currently characterizing the otic placode phenotype of Foxi3 mutant mice in more detail; our preliminary results suggest that, like foxi1 mutant zebrafish, Foxi3 mutant mice also have defects in their epibranchial ganglia in addition to lacking the inner ear (Birol, Ohyama, Edlund and Groves, unpublished observations).
4: THE ROLE OF FOXI FAMILY MEMBERS IN MIDDLE EAR, OUTER EAR AND JAW DEVELOPMENT
4.1: The role of Foxi1/3 in Jaw, Middle Ear, and Outer Ear Development
In zebrafish, Foxi1 is necessary not only for otic placode induction, but also for jaw development(Nissen et al., 2003; Solomon et al., 2003a). foxi1 is expressed in pharyngeal endoderm and ectoderm of zebrafish, but not in neural crest cells. Yet, the jaw phenotype in these fish is at least partially attributable to neural crest apoptosis that occurs in the first and second arches, concurrent with decreased fgf3 expression in the endoderm (Edlund et al., 2014; Nissen et al., 2003). In zebrafish Foxi1 remains bound to chromatin over multiple cycles of mitosis in cell cultures and may be a chromatin-remodeling pioneer factor that does not itself actively induce expression of many genes, but rather poises genes required for specific developmental processes for rapid expression upon receipt of the appropriate signals (Yan et al., 2006).
Foxi3 is expressed in the pharyngeal region of mouse embryos in a segmented pattern between the branchial arches (Edlund et al., 2014; Ohyama and Groves, 2004a)). Foxi3 is expressed in the endoderm of each pharyngeal pouch as well as the overlying cleft ectoderm. It is important to note that Foxi3 is expressed neither in branchial arch mesoderm nor in the cranial neural crest cells that populate arch mesenchyme (Edlund et al., 2014; Ohyama and Groves, 2004a). Further supporting homology between the amniote Foxi3 gene and the zebrafish foxi1 gene is the catastrophic suite of branchial arch defects present in Foxi3 mutant mice highly reminiscent of Hearsay mutant zebrafish. Foxi3 mutants are postnatal lethal but born with completely absent middle and outer ears and severely truncated and deformed jaws (Edlund et al., 2014). The mutant pups are readily identifiable by their lack of a mouth, with a continuous ectodermal covering over the lower half of the face, and by the absence of outer ear pinnae. Skeletal preps of the heads of E18.5 Foxi3 mutant embryos reveal a number of severe defects in bone and cartilage development. In Foxi3 mutants, only a small anterior portion of the mandible develops which is fused to the distal end of maxilla. The maxilla itself is also malformed (Edlund et al., 2014). In zebrafish foxi1 mutants, the size of jaw cartilages is significantly reduced and jaw morphology is substantially disrupted (Nissen et al., 2003; Solomon et al., 2003a).
The middle ear ossicles are evolutionary derivatives of the two anterior arch cartilages: Meckel’s cartilage in BA1 and Reichert’s cartilage in BA2 (Sienknecht, 2013). Both cartilages are neural crest derived, thus the severe defects in crest-derived tissue in Foxi3 mutant mice also affects middle ear structures. There is no sign of the tympanic ring or incus, malleus, or stapes of the middle ear, and the entire temporal bone and inner ear are absent. The absence of the outer ear may be attributable to aberrant ectodermal morphology in Foxi3 mutant mice, which show a failure to demarcate distinct arches (Edlund et al., 2014). In Foxi3 mutants, cranial neural crest cells undergo apoptosis as they populate the branchial arches. Since neural crest cells do not express Foxi3, this suggests that FOXI3 may regulate the expression of trophic or survival factors in arch ectoderm or endoderm.
Of the four signaling pathways regulating branchial arch development described in Section 1.4, FGF signaling has the clearest role in neural crest cell survival. The presence of strong ectodermal Fgf expression promotes post-migratory neural crest survival in the branchial arches. Increased levels of apoptosis in the arch mesenchyme are observed in fgf3 knockdown zebrafish, in chicks treated with Fgf8 RNAi, and in mouse Fgf8 hypomorphs and arch ectoderm-conditional Fgf8 knockouts (Abu-Issa et al., 2002; Creuzet et al., 2004; Frank et al., 2002; Nissen et al., 2003; Trumpp et al., 1999). Experiments with mice carrying hypomorphic alleles of Fgf8 and with mice in which Fgf8 is locally knocked out of the arch ectoderm reveal its importance in promoting normal development of the neural crest-derived skeleton. In two studies on compound heterozygote mice, each carrying one null allele and one hypomorphic allele of Fgf8, neural crest development is affected in all branchial arches (Abu-Issa et al., 2002; Frank et al., 2002; Inman et al., 2013; Macatee et al., 2003; Trumpp et al., 1999). In BA1, the maxilla, mandible, and middle ear ossicles, among other skeletal elements either fail to form or are hypoplastic. This is likely attributable to the substantial increase in cell death in the BA1 mesenchyme.
As apoptosis can be reduced by artificial introduction of FGF when arch morphology is already defective, it is probable that FGF signaling directly promotes survival of migrating neural crest cells (Edlund et al., 2014). FGFs from the ectoderm also play a role in patterning the underlying mesenchyme. Expression of transcription factors, including Dlx2, Gsc, Barx1, Pax9, and Lhx6, which contribute to defining the dorsal territory of BA1, requires ectodermal signaling (Ferguson et al., 2000; Neubuser et al., 1997; Tucker et al., 1999). foxi1 and Foxi3 expression are required for normal pharyngeal pouch morphology in zebrafish and mouse respectively, FOXI factors establish signaling centers in the developing branchial arches necessary for crest survival, and the craniofacial phenotype seen in Foxi3 mouse mutants is due to reduced FGF8 signaling in the pharyngeal region. The activity of FOXI3 in pharyngeal epithelia is required for early expression of Fgf8 in arch ectoderm; this pathway is conserved in zebrafish where fgf3 is expressed in branchial arch ectoderm and requires the expression of foxi1. In zebrafish foxi1 morphants, ectopic expression of fgf3 in pharyngeal ectoderm can reduce neural crest cell death (Edlund et al., 2014).
4.2: Foxi3 has a conserved role in mammalian pharyngeal development
The pharyngeal development function of FOXI3 is conserved in mammals as evidenced by phenotypes seen in three breeds of hairless dogs, all of which possess identical mutations in the Foxi3 gene (Drogemuller et al., 2008). The mutation, a seven-base pair duplication introduces a premature stop codon prior to the DNA binding domain and is presumed to create a null allele. In addition to a striking lack of hair, possibly related to a role for FOXI3 in regulating the mammalian ectodysplasin pathway (Shirokova et al., 2013), heterozygous dogs have malformed and missing teeth and occasional malformations in the outer ear (Drogemuller et al., 2008; Tassano, 2014). A human patient with a deletion in one copy of chromosome 2 encompassing seven genes including Foxi3 has also been identified. The patient was found to have mandibular asymmetry and unilateral malformation of the outer ear (Tassano, 2014).
It is clear from studies in zebrafish, chick, mouse, dog, and from one case study of a human patient, that FOXI1/3 homologs are highly conserved developmental regulators critical for proper formation of all portions of the ear. Because of the structural and phenotypic similarities between mammalian FOXI3 and zebrafish Foxi1, it is likely that if Foxi1 is a pioneer factor, so is FOXI3. Their conserved role in mammalian branchial arch development suggests that examination of genes and signaling pathways regulated by FOXI3, perhaps through epigenetic changes and chromatin remodeling, may provide insight into the developmental processes involved with shaping jaws and ears from simple branchial arches.
5: CONCLUSIONS
5.1: Ear development is a complex process that requires Foxi1/3 in multiple steps
In land vertebrates the ear consists of three compartments derived from multiple embryonic tissues, while in fish only the inner ear is present. The inner ear arises first during development via sequential refinement of ectodermal identity. Intercellular signaling and subsequent changes in transcriptional profiles of increasingly smaller patches of ectoderm confer otic identity on a small portion of pre-placodal ectoderm. This ectoderm forms the otic placode from which the inner ear develops. Mice and fish lacking Foxi3 or foxi1, respectively, fail to form complete otic placodes, a process normally mediated by FGF signaling, and lack all inner ear structures upon completion of embryonic development. In addition, the middle and outer ears are absent in Foxi3 mutant mice due to failure to form distinct and properly patterned branchial arches. In the presumptive branchial arches of Foxi3 mutant mice, post-migratory neural crest cells undergo apoptosis. This function of Foxi is conserved in morphant zebrafish, which also have severe branchial arch derived skeletal defects, although middle and outer ears are not present in fish. In Foxi3 mutant mice, Fgf8 expression in the branchial ectoderm is delayed. Artificial introduction of FGF can rescue neural crest cell apoptosis in zebrafish Foxi1 morphants, indicating that the proximal cause of cell death in Foxi mutants is likely to be reduced FGF signaling.
5.2: Pioneer factors may play a role in morphological diversity
From an evolutionary perspective, it is interesting to speculate about the ways in which minor perturbations of cranial neural crest can lead to the broad spectrum of facial structures present in vertebrates. While apoptosis of neural crest cells in the arches, at least in laboratory genetic experiments, frequently results in severe facial skeletal defects, it is intriguing that induction of apoptosis or even ablation of sizeable populations of trunk neural crest cell precursors often presents no appreciable obstacle to animal development (Vaglia and Hall, 1999). Trunk neural crest is not usually skeletogenic, with the notable exception of turtle shells. Interestingly, these skeletogenic trunk crest cells migrate in a distinct second wave and express cranial neural crest markers (Cebra-Thomas et al., 2013). Aside from the turtle shell, structures derived from trunk crest: the peripheral nervous system and melanocytes, remain relatively unchanged between classes of vertebrates. Is it possible that the sensitivity of cranial neural crest to changes in total cell number represents a rich substrate for natural selection?
Recent evidence suggests a pivotal role for non-coding DNA in subtle variations of facial shape. While small changes to coding sequences of genes involved with craniofacial development can give rise to disabling phenotypes, sequence changes and even deletions of elements that regulate expression of these same genes can instead generate normal variations in face shape (Attanasio et al., 2013). The authors of the study found that single enhancer deletions resulted in subtle but quantifiable changes to the shape of the mouse skull. It seems then, that enhancers may be underappreciated in craniofacial developmental studies. It is possible that some changes in enhancer function are related to chromatin compaction, that is, alterations in the timing or extent of enhancer-containing chromatin in open conformation may permit transcription factor binding in new temporal and spatial patterns. One mechanism potentially at play is changes in pioneer factor activity. Pioneer factors can function to actively open and expose chromatin, priming regions of DNA for transcription factor binding and maintaining cellular lineage throughout multiple rounds of mitosis (Zaret and Carroll, 2011). FOXI1/3 is included among the forkhead transcription factors proposed as pioneer factors. Zebrafish Foxi1, the homolog of mammalian FOXI3, has been shown to remain bound to DNA during mitosis in cultured cells (Yan et al., 2006). Yan and colleagues examined chromatin fractions and found Foxi1 both in active chromatin and in chromatin tightly bound to the nuclear matrix, suggesting a dual role for Foxi1 in chromatin organization. Interestingly, the study showed very small changes in gene expression in response to ectopic Foxi1 induction. Although transcription factor overexpression in cultured cells may not produce results representative of in vivo activity, the evidence suggests that zebrafish Foxi1 acts not to immediately promote target gene expression, but rather as a pioneer factor that alters the set of genes accessible to other transcription factors necessary for developmental lineage specification. It is tempting to speculate that Foxi1 and FOXI3 could be pioneer factors that influence timing and expression of a suite of pharyngeal patterning factors and might be key in evolution of diverse jaw and ear shapes and sizes. Regardless, when subtle temporal or spatial changes spontaneously occur to branchial arch gene expression, new and potentially adaptive facial structures can arise. Thus, on an individual level the sensitivity of cranial neural crest to genetic perturbations may occasionally result in deformities incompatible with survival, but from a population perspective, accumulation of subtle facial shape changes may advantageous enough that the very same cranial neural crest sensitivity is a powerful force of evolution in vertebrates.
References
- Abu-Issa R, Smyth G, Smoak I, Yamamura K, Meyers EN. Fgf8 is required for pharyngeal arch and cardiovascular development in the mouse. Development. 2002;129:4613–25. doi: 10.1242/dev.129.19.4613. [DOI] [PubMed] [Google Scholar]
- Ahrens K, Schlosser G. Tissues and signals involved in the induction of placodal Six1 expression in Xenopus laevis. Dev Biol. 2005;288:40–59. doi: 10.1016/j.ydbio.2005.07.022. [DOI] [PubMed] [Google Scholar]
- Alasti F, Van Camp G. Genetics of microtia and associated syndromes. J Med Genet. 2009;46:361–9. doi: 10.1136/jmg.2008.062158. [DOI] [PubMed] [Google Scholar]
- Alexander C, Zuniga E, Blitz IL, Wada N, Le Pabic P, Javidan Y, Zhang T, Cho KW, Crump JG, Schilling TF. Combinatorial roles for BMPs and Endothelin 1 in patterning the dorsal-ventral axis of the craniofacial skeleton. Development. 2011;138:5135–46. doi: 10.1242/dev.067801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez Y, Alonso MT, Vendrell V, Zelarayan LC, Chamero P, Theil T, Bosl MR, Kato S, Maconochie M, Riethmacher D, Schimmang T. Requirements for FGF3 and FGF10 during inner ear formation. Development. 2003;130:6329–38. doi: 10.1242/dev.00881. [DOI] [PubMed] [Google Scholar]
- Attanasio C, Nord AS, Zhu Y, Blow MJ, Li Z, Liberton DK, Morrison H, Plajzer-Frick I, Holt A, Hosseini R, Phouanenavong S, Akiyama JA, Shoukry M, Afzal V, Rubin EM, FitzPatrick DR, Ren B, Hallgrimsson B, Pennacchio LA, Visel A. Fine tuning of craniofacial morphology by distant-acting enhancers. Science. 2013;342:1241006. doi: 10.1126/science.1241006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey AP, Streit A. Sensory organs: making and breaking the pre-placodal region. Current topics in developmental biology. 2006;72:167–204. doi: 10.1016/S0070-2153(05)72003-2. [DOI] [PubMed] [Google Scholar]
- Baker CV, Bronner-Fraser M. Vertebrate cranial placodes I. Embryonic induction. Dev Biol. 2001;232:1–61. doi: 10.1006/dbio.2001.0156. [DOI] [PubMed] [Google Scholar]
- Bally-Cuif L, Gulisano M, Broccoli V, Boncinelli E. c-otx2 is expressed in two different phases of gastrulation and is sensitive to retinoic acid treatment in chick embryo. Mechanisms of development. 1995;49:49–63. doi: 10.1016/0925-4773(94)00301-3. [DOI] [PubMed] [Google Scholar]
- Betancur P, Bronner-Fraser M, Sauka-Spengler T. Assembling neural crest regulatory circuits into a gene regulatory network. Annu Rev Cell Dev Biol. 2010;26:581–603. doi: 10.1146/annurev.cellbio.042308.113245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat N, Kwon HJ, Riley BB. A gene network that coordinates preplacodal competence and neural crest specification in zebrafish. Dev Biol. 2013;373:107–17. doi: 10.1016/j.ydbio.2012.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya S, Bailey AP, Bronner-Fraser M, Streit A. Segregation of lens and olfactory precursors from a common territory: cell sorting and reciprocity of Dlx5 and Pax6 expression. Dev Biol. 2004;271:403–14. doi: 10.1016/j.ydbio.2004.04.010. [DOI] [PubMed] [Google Scholar]
- Blobel GA, Kadauke S, Wang E, Lau AW, Zuber J, Chou MM, Vakoc CR. A reconfigured pattern of MLL occupancy within mitotic chromatin promotes rapid transcriptional reactivation following mitotic exit. Mol Cell. 2009;36:970–83. doi: 10.1016/j.molcel.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossard P, Zaret KS. GATA transcription factors as potentiators of gut endoderm differentiation. Development. 1998;125:4909–17. doi: 10.1242/dev.125.24.4909. [DOI] [PubMed] [Google Scholar]
- Bouchard M, de Caprona D, Busslinger M, Xu P, Fritzsch B. Pax2 and Pax8 cooperate in mouse inner ear morphogenesis and innervation. BMC Dev Biol. 2010;10:89–89. doi: 10.1186/1471-213X-10-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bricaud O, Collazo A. The transcription factor six1 inhibits neuronal and promotes hair cell fate in the developing zebrafish (Danio rerio) inner ear. J Neurosci. 2006;26:10438–10451. doi: 10.1523/JNEUROSCI.1025-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brito JM, Teillet MA, Le Douarin NM. An early role for sonic hedgehog from foregut endoderm in jaw development: ensuring neural crest cell survival. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:11607–12. doi: 10.1073/pnas.0604751103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown ST, Wang J, Groves AK. Dlx gene expression during chick inner ear development. J Comp Neurol. 2005;483:48–65. doi: 10.1002/cne.20418. [DOI] [PubMed] [Google Scholar]
- Brugmann SA, Pandur PD, Kenyon KL, Pignoni F, Moody SA. Six1 promotes a placodal fate within the lateral neurogenic ectoderm by functioning as both a transcriptional activator and repressor. Development. 2004;131:5871–81. doi: 10.1242/dev.01516. [DOI] [PubMed] [Google Scholar]
- Burton Q, Cole LK, Mulheisen M, Chang W, Wu DK. The role of Pax2 in mouse inner ear development. Dev Biol. 2004;272:161–175. doi: 10.1016/j.ydbio.2004.04.024. [DOI] [PubMed] [Google Scholar]
- Caravaca JM, Donahue G, Becker JS, He X, Vinson C, Zaret KS. Bookmarking by specific and nonspecific binding of FoxA1 pioneer factor to mitotic chromosomes. Genes Dev. 2013;27:251–60. doi: 10.1101/gad.206458.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cebra-Thomas JA, Terrell A, Branyan K, Shah S, Rice R, Gyi L, Yin M, Hu Y, Mangat G, Simonet J, Betters E, Gilbert SF. Late-emigrating trunk neural crest cells in turtle embryos generate an osteogenic ectomesenchyme in the plastron. Dev Dyn. 2013;242:1223–35. doi: 10.1002/dvdy.24018. [DOI] [PubMed] [Google Scholar]
- Chai Y, Jiang X, Ito Y, Bringas P, Jr, Han J, Rowitch DH, Soriano P, McMahon AP, Sucov HM. Fate of the mammalian cranial neural crest during tooth and mandibular morphogenesis. Development. 2000;127:1671–9. doi: 10.1242/dev.127.8.1671. [DOI] [PubMed] [Google Scholar]
- Chambers D, Mason I. Expression of sprouty2 during early development of the chick embryo is coincident with known sites of FGF signalling. Mechanisms of development. 2000;91:361–4. doi: 10.1016/s0925-4773(99)00288-9. [DOI] [PubMed] [Google Scholar]
- Chen B, Kim EH, Xu PX. Initiation of olfactory placode development and neurogenesis is blocked in mice lacking both Six1 and Six4. Dev Biol. 2009;326:75–85. doi: 10.1016/j.ydbio.2008.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirillo LA, Lin FR, Cuesta I, Friedman D, Jarnik M, Zaret KS. Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol Cell. 2002;9:279–89. doi: 10.1016/s1097-2765(02)00459-8. [DOI] [PubMed] [Google Scholar]
- Cirillo LA, Zaret KS. An early developmental transcription factor complex that is more stable on nucleosome core particles than on free DNA. Mol Cell. 1999;4:961–9. doi: 10.1016/s1097-2765(00)80225-7. [DOI] [PubMed] [Google Scholar]
- Clack JA, Allin E. The evolution of single- and multiple-ossicle ears in fish and tetrapods. In: Manley GA, Popper AN, Fay RR, editors. Evolution of the Vertebrate Auditory System. Springer Verlag; New York: 2004. pp. 128–163. [Google Scholar]
- Creuzet S, Couly G, Le Douarin NM. Patterning the neural crest derivatives during development of the vertebrate head: insights from avian studies. J Anat. 2005;207:447–59. doi: 10.1111/j.1469-7580.2005.00485.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creuzet S, Schuler B, Couly G, Le Douarin NM. Reciprocal relationships between Fgf8 and neural crest cells in facial and forebrain development. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:4843–7. doi: 10.1073/pnas.0400869101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crossley PH, Martin GR. The mouse Fgf8 gene encodes a family of polypeptides and is expressed in regions that direct outgrowth and patterning in the developing embryo. Development. 1995;121:439–51. doi: 10.1242/dev.121.2.439. [DOI] [PubMed] [Google Scholar]
- Cruz SA, Chao PL, Hwang PP. Cortisol promotes differentiation of epidermal ionocytes through Foxi3 transcription factors in zebrafish (Danio rerio) Comparative biochemistry and physiology Part A, Molecular & integrative physiology. 2013;164:249–57. doi: 10.1016/j.cbpa.2012.09.011. [DOI] [PubMed] [Google Scholar]
- Cuesta I, Zaret KS, Santisteban P. The forkhead factor FoxE1 binds to the thyroperoxidase promoter during thyroid cell differentiation and modifies compacted chromatin structure. Mol Cell Biol. 2007;27:7302–14. doi: 10.1128/MCB.00758-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey A, Nishiyama A, Karpova T, McNally J, Ozato K. Brd4 marks select genes on mitotic chromatin and directs postmitotic transcription. Molecular biology of the cell. 2009;20:4899–909. doi: 10.1091/mbc.E09-05-0380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drogemuller C, Karlsson EK, Hytonen MK, Perloski M, Dolf G, Sainio K, Lohi H, Lindblad-Toh K, Leeb T. A mutation in hairless dogs implicates FOXI3 in ectodermal development. Science. 2008;321:1462. doi: 10.1126/science.1162525. [DOI] [PubMed] [Google Scholar]
- Edlund RK, Ohyama T, Kantarci H, Riley BB, Groves AK. Foxi transcription factors promote pharyngeal arch development by regulating formation of FGF signaling centers. Dev Biol. 2014;390:1–13. doi: 10.1016/j.ydbio.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esterberg R, Fritz A. dlx3b/4b are required for the formation of the preplacodal region and otic placode through local modulation of BMP activity. Dev Biol. 2009;325:189–99. doi: 10.1016/j.ydbio.2008.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett LA, Glaser B, Beck JC, Idol JR, Buchs A, Heyman M, Adawi F, Hazani E, Nassir E, Baxevanis AD, Sheffield VC, Green ED. Pendred syndrome is caused by mutations in a putative sulphate transporter gene (PDS) Nat Genet. 1997;17:411–22. doi: 10.1038/ng1297-411. [DOI] [PubMed] [Google Scholar]
- Feledy JA, Beanan MJ, Sandoval JJ, Goodrich JS, Lim JH, Matsuo-Takasaki M, Sato SM, Sargent TD. Inhibitory patterning of the anterior neural plate in Xenopus by homeodomain factors Dlx3 and Msx1. Dev Biol. 1999;212:455–464. doi: 10.1006/dbio.1999.9374. [DOI] [PubMed] [Google Scholar]
- Ferguson CA, Tucker AS, Sharpe PT. Temporospatial cell interactions regulating mandibular and maxillary arch patterning. Development. 2000;127:403–12. doi: 10.1242/dev.127.2.403. [DOI] [PubMed] [Google Scholar]
- Frank DU, Fotheringham LK, Brewer JA, Muglia LJ, Tristani-Firouzi M, Capecchi MR, Moon AM. An Fgf8 mouse mutant phenocopies human 22q11 deletion syndrome. Development. 2002;129:4591–603. doi: 10.1242/dev.129.19.4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freter S, Muta Y, Mak SS, Rinkwitz S, Ladher RK. Progressive restriction of otic fate: the role of FGF and Wnt in resolving inner ear potential. Development. 2008;135:3415–24. doi: 10.1242/dev.026674. [DOI] [PubMed] [Google Scholar]
- Graham A, Begbie J, McGonnell I. Significance of the cranial neural crest. Dev Dyn. 2004;229:5–13. doi: 10.1002/dvdy.10442. [DOI] [PubMed] [Google Scholar]
- Graham A, Richardson J. Developmental and evolutionary origins of the pharyngeal apparatus. EvoDevo. 2012;3:24. doi: 10.1186/2041-9139-3-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham A, Smith A. Patterning the pharyngeal arches. Bioessays. 2001;23:54–61. doi: 10.1002/1521-1878(200101)23:1<54::AID-BIES1007>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Grevellec A, Tucker AS. The pharyngeal pouches and clefts: Development, evolution, structure and derivatives. Semin Cell Dev Biol. 2010;21:325–32. doi: 10.1016/j.semcdb.2010.01.022. [DOI] [PubMed] [Google Scholar]
- Grifone R, Demignon J, Houbron C, Souil E, Niro C, Seller MJ, Hamard G, Maire P. Six1 and Six4 homeoproteins are required for Pax3 and Mrf expression during myogenesis in the mouse embryo. Development. 2005;132:2235–49. doi: 10.1242/dev.01773. [DOI] [PubMed] [Google Scholar]
- Grocott T, Tambalo M, Streit A. The peripheral sensory nervous system in the vertebrate head: a gene regulatory perspective. Dev Biol. 2012;370:3–23. doi: 10.1016/j.ydbio.2012.06.028. [DOI] [PubMed] [Google Scholar]
- Groves AK. The Induction of the Otic Placode. In: Popper AN, Kelley MW, Wu DK, editors. Development of the Inner Ear. Springer Verlag; New York: 2005. pp. 10–42. [Google Scholar]
- Groves AK, Bronner-Fraser M. Competence, specification and commitment in otic placode induction. Development. 2000;127:3489–99. doi: 10.1242/dev.127.16.3489. [DOI] [PubMed] [Google Scholar]
- Groves AK, Fekete DM. Shaping sound in space: the regulation of inner ear patterning. Development. 2012;139:245–57. doi: 10.1242/dev.067074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groves AK, LaBonne C. Setting appropriate boundaries: fate, patterning and competence at the neural plate border. Dev Biol. 2014;389:2–12. doi: 10.1016/j.ydbio.2013.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gualdi R, Bossard P, Zheng M, Hamada Y, Coleman JR, Zaret KS. Hepatic specification of the gut endoderm in vitro: cell signaling and transcriptional control. Genes Dev. 1996;10:1670–1682. doi: 10.1101/gad.10.13.1670. [DOI] [PubMed] [Google Scholar]
- Hans S, Christison J, Liu D, Westerfield M. Fgf-dependent otic induction requires competence provided by Foxi1 and Dlx3b. BMC Dev Biol. 2007;7:5. doi: 10.1186/1471-213X-7-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hans S, Irmscher A, Brand M. Zebrafish Foxi1 provides a neuronal ground state during inner ear induction preceding the Dlx3b/4b-regulated sensory lineage. Development. 2013;140:1936–45. doi: 10.1242/dev.087718. [DOI] [PubMed] [Google Scholar]
- Hans S, Liu D, Westerfield M. Pax8 and Pax2a function synergistically in otic specification, downstream of the Foxi1 and Dlx3b transcription factors. Development. 2004;131:5091–102. doi: 10.1242/dev.01346. [DOI] [PubMed] [Google Scholar]
- Hans S, Westerfield M. Changes in retinoic acid signaling alter otic patterning. Development. 2007;134:2449–58. doi: 10.1242/dev.000448. [DOI] [PubMed] [Google Scholar]
- Hatta M, Cirillo LA. Chromatin opening and stable perturbation of core histone:DNA contacts by FoxO1. J Biol Chem. 2007;282:35583–93. doi: 10.1074/jbc.M704735200. [DOI] [PubMed] [Google Scholar]
- Haworth KE, Wilson JM, Grevellec A, Cobourne MT, Healy C, Helms JA, Sharpe PT, Tucker AS. Sonic hedgehog in the pharyngeal endoderm controls arch pattern via regulation of Fgf8 in head ectoderm. Dev Biol. 2007;303:244–58. doi: 10.1016/j.ydbio.2006.11.009. [DOI] [PubMed] [Google Scholar]
- Heller N, Brandli AW. Xenopus Pax-2/5/8 orthologues: novel insights into Pax gene evolution and identification of Pax-8 as the earliest marker for otic and pronephric cell lineages. Dev Genet. 1999;24:208–19. doi: 10.1002/(SICI)1520-6408(1999)24:3/4<208::AID-DVG4>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Hoffman TL, Javier AL, Campeau SA, Knight RD, Schilling TF. Tfap2 transcription factors in zebrafish neural crest development and ectodermal evolution. J Exp Zool B Mol Dev Evol. 2007;308:679–91. doi: 10.1002/jez.b.21189. [DOI] [PubMed] [Google Scholar]
- Hsiao CD, You MS, Guh YJ, Ma M, Jiang YJ, Hwang PP. A positive regulatory loop between foxi3a and foxi3b is essential for specification and differentiation of zebrafish epidermal ionocytes. PloS one. 2007;2:e302. doi: 10.1371/journal.pone.0000302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulander M, Kiernan AE, Blomqvist SR, Carlsson P, Samuelsson EJ, Johansson BR, Steel KP, Enerback S. Lack of pendrin expression leads to deafness and expansion of the endolymphatic compartment in inner ears of Foxi1 null mutant mice. Development. 2003;130:2013–25. doi: 10.1242/dev.00376. [DOI] [PubMed] [Google Scholar]
- Hulander M, Wurst W, Carlsson P, Enerback S. The winged helix transcription factor Fkh10 is required for normal development of the inner ear. Nat Genet. 1998;20:374–6. doi: 10.1038/3850. [DOI] [PubMed] [Google Scholar]
- Inman KE, Purcell P, Kume T, Trainor PA. Interaction between Foxc1 and Fgf8 during mammalian jaw patterning and in the pathogenesis of syngnathia. PLoS Genet. 2013;9:e1003949. doi: 10.1371/journal.pgen.1003949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson BC, Carpenter C, Nebert DW, Vasiliou V. Update of human and mouse forkhead box (FOX) gene families. Human genomics. 2010;4:345–52. doi: 10.1186/1479-7364-4-5-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janicke M, Carney TJ, Hammerschmidt M. Foxi3 transcription factors and Notch signaling control the formation of skin ionocytes from epidermal precursors of the zebrafish embryo. Dev Biol. 2007;307:258–71. doi: 10.1016/j.ydbio.2007.04.044. [DOI] [PubMed] [Google Scholar]
- Janicke M, Renisch B, Hammerschmidt M. Zebrafish grainyhead-like1 is a common marker of different non-keratinocyte epidermal cell lineages, which segregate from each other in a Foxi3-dependent manner. Int J Dev Biol. 2010;54:837–50. doi: 10.1387/ijdb.092877mj. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayasena CS, Ohyama T, Segil N, Groves AK. Notch signaling augments the canonical Wnt pathway to specify the size of the otic placode. Development. 2008;135:2251–61. doi: 10.1242/dev.017905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadauke S, Udugama MI, Pawlicki JM, Achtman JC, Jain DP, Cheng Y, Hardison RC, Blobel GA. Tissue-specific mitotic bookmarking by hematopoietic transcription factor GATA1. Cell. 2012;150:725–37. doi: 10.1016/j.cell.2012.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaestner KH, Knochel W, Martinez DE. Unified nomenclature for the winged helix/forkhead transcription factors. Genes Dev. 2000;14:142–6. [PubMed] [Google Scholar]
- Khatri SB, Edlund RK, Groves AK. Foxi3 Is necessary for the induction of the chick otic placode in response to FGF signaling. Dev Biol. 2014 doi: 10.1016/j.ydbio.2014.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatri SB, Groves AK. Expression of the Foxi2 and Foxi3 transcription factors during development of chicken sensory placodes and pharyngeal arches. Gene expression patterns: GEP. 2013;13:38–42. doi: 10.1016/j.gep.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khudyakov J, Bronner-Fraser M. Comprehensive spatiotemporal analysis of early chick neural crest network genes. Dev Dyn. 2009;238:716–23. doi: 10.1002/dvdy.21881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight RD, Nair S, Nelson SS, Afshar A, Javidan Y, Geisler R, Rauch GJ, Schilling TF. lockjaw encodes a zebrafish tfap2a required for early neural crest development. Development. 2003;130:5755–5768. doi: 10.1242/dev.00575. [DOI] [PubMed] [Google Scholar]
- Kozlowski DJ, Murakami T, Ho RK, Weinberg ES. Regional cell movement and tissue patterning in the zebrafish embryo revealed by fate mapping with caged fluorescein. Biochem Cell Biol. 1997;75:551–62. [PubMed] [Google Scholar]
- Kroll KL, Salic AN, Evans LM, Kirschner MW. Geminin, a neuralizing molecule that demarcates the future neural plate at the onset of gastrulation. Development. 1998;125:3247–3258. doi: 10.1242/dev.125.16.3247. [DOI] [PubMed] [Google Scholar]
- Kwon HJ, Bhat N, Sweet EM, Cornell RA, Riley BB. Identification of early requirements for preplacodal ectoderm and sensory organ development. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1001133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladher RK, O’Neill P, Begbie J. From shared lineage to distinct functions: the development of the inner ear and epibranchial placodes. Development. 2010;137:1777–1785. doi: 10.1242/dev.040055. [DOI] [PubMed] [Google Scholar]
- Ladher RK, Wright TJ, Moon AM, Mansour SL, Schoenwolf GC. FGF8 initiates inner ear induction in chick and mouse. Genes Dev. 2005;19:603–13. doi: 10.1101/gad.1273605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai E, Clark KL, Burley SK, Darnell JE., Jr Hepatocyte nuclear factor 3/fork head or “winged helix” proteins: a family of transcription factors of diverse biologic function. Proceedings of the National Academy of Sciences of the United States of America. 1993;90:10421–3. doi: 10.1073/pnas.90.22.10421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai E, Prezioso VR, Smith E, Litvin O, Costa RH, Darnell JE., Jr HNF-3A, a hepatocyte-enriched transcription factor of novel structure is regulated transcriptionally. Genes Dev. 1990;4:1427–36. doi: 10.1101/gad.4.8.1427. [DOI] [PubMed] [Google Scholar]
- Lee CS, Friedman JR, Fulmer JT, Kaestner KH. The initiation of liver development is dependent on Foxa transcription factors. Nature. 2005;435:944–7. doi: 10.1038/nature03649. [DOI] [PubMed] [Google Scholar]
- Lee SA, Shen EL, Fiser A, Sali A, Guo S. The zebrafish forkhead transcription factor Foxi1 specifies epibranchial placode-derived sensory neurons. Development. 2003;130:2669–79. doi: 10.1242/dev.00502. [DOI] [PubMed] [Google Scholar]
- Leger S, Brand M. Fgf8 and Fgf3 are required for zebrafish ear placode induction, maintenance and inner ear patterning. Mechanisms of development. 2002;119:91–108. doi: 10.1016/s0925-4773(02)00343-x. [DOI] [PubMed] [Google Scholar]
- Li W, Cornell RA. Redundant activities of Tfap2a and Tfap2c are required for neural crest induction and development of other non-neural ectoderm derivatives in zebrafish embryos. Dev Biol. 2007;304:338–354. doi: 10.1016/j.ydbio.2006.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linker C, De Almeida I, Papanayotou C, Stower M, Sabado V, Ghorani E, Streit A, Mayor R, Stern CD. Cell communication with the neural plate is required for induction of neural markers by BMP inhibition: evidence for homeogenetic induction and implications for Xenopus animal cap and chick explant assays. Dev Biol. 2009;327:478–486. doi: 10.1016/j.ydbio.2008.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litsiou A, Hanson S, Streit A. A balance of FGF, BMP and WNT signalling positions the future placode territory in the head. Development. 2005;132:4051–62. doi: 10.1242/dev.01964. [DOI] [PubMed] [Google Scholar]
- Liu D, Chu H, Maves L, Yan YL, Morcos PA, Postlethwait JH, Westerfield M. Fgf3 and Fgf8 dependent and independent transcription factors are required for otic placode specification. Development. 2003;130:2213–2224. doi: 10.1242/dev.00445. [DOI] [PubMed] [Google Scholar]
- Liu W, Selever J, Murali D, Sun X, Brugger SM, Ma L, Schwartz RJ, Maxson R, Furuta Y, Martin JF. Threshold-specific requirements for Bmp4 in mandibular development. Dev Biol. 2005;283:282–93. doi: 10.1016/j.ydbio.2005.04.019. [DOI] [PubMed] [Google Scholar]
- Lorente-Canovas B, Ingham N, Norgett EE, Golder ZJ, Karet Frankl FE, Steel KP. Mice deficient in H+-ATPase a4 subunit have severe hearing impairment associated with enlarged endolymphatic compartments within the inner ear. Disease models & mechanisms. 2013;6:434–42. doi: 10.1242/dmm.010645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo T, Matsuo-Takasaki M, Sargent TD. Distinct roles for Distal-less genes Dlx3 and Dlx5 in regulating ectodermal development in Xenopus. Mol Reprod Dev. 2001;60:331–337. doi: 10.1002/mrd.1095. [DOI] [PubMed] [Google Scholar]
- Macatee TL, Hammond BP, Arenkiel BR, Francis L, Frank DU, Moon AM. Ablation of specific expression domains reveals discrete functions of ectoderm- and endoderm-derived FGF8 during cardiovascular and pharyngeal development. Development. 2003;130:6361–74. doi: 10.1242/dev.00850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackereth MD, Kwak SJ, Fritz A, Riley BB. Zebrafish pax8 is required for otic placode induction and plays a redundant role with Pax2 genes in the maintenance of the otic placode. Development. 2005;132:371–82. doi: 10.1242/dev.01587. [DOI] [PubMed] [Google Scholar]
- Mallo M. Embryological and genetic aspects of middle ear development. Int J Dev Biol. 1998;42:11–22. [PubMed] [Google Scholar]
- Mallo M. Formation of the middle ear: recent progress on the developmental and molecular mechanisms. Dev Biol. 2001;231:410–9. doi: 10.1006/dbio.2001.0154. [DOI] [PubMed] [Google Scholar]
- Manley GA. An evolutionary perspective on middle ears. Hear Res. 2010;263:3–8. doi: 10.1016/j.heares.2009.09.004. [DOI] [PubMed] [Google Scholar]
- Maroon H, Walshe J, Mahmood R, Kiefer P, Dickson C, Mason I. Fgf3 and Fgf8 are required together for formation of the otic placode and vesicle. Development. 2002;129:2099–108. doi: 10.1242/dev.129.9.2099. [DOI] [PubMed] [Google Scholar]
- Matsuo-Takasaki M, Matsumura M, Sasai Y. An essential role of Xenopus Foxi1a for ventral specification of the cephalic ectoderm during gastrulation. Development. 2005;132:3885–3894. doi: 10.1242/dev.01959. [DOI] [PubMed] [Google Scholar]
- McCabe KL, Bronner-Fraser M. Molecular and tissue interactions governing induction of cranial ectodermal placodes. Dev Biol. 2009;332:189–195. doi: 10.1016/j.ydbio.2009.05.572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLarren KW, Litsiou A, Streit A. DLX5 positions the neural crest and preplacode region at the border of the neural plate. Dev Biol. 2003;259:34–47. doi: 10.1016/s0012-1606(03)00177-5. [DOI] [PubMed] [Google Scholar]
- Medeiros DM, Crump JG. New perspectives on pharyngeal dorsoventral patterning in development and evolution of the vertebrate jaw. Dev Biol. 2012;371:121–35. doi: 10.1016/j.ydbio.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milet C, Monsoro-Burq AH. Neural crest induction at the neural plate border in vertebrates. Dev Biol. 2012;366:22–33. doi: 10.1016/j.ydbio.2012.01.013. [DOI] [PubMed] [Google Scholar]
- Miller CT, Schilling TF, Lee K, Parker J, Kimmel CB. sucker encodes a zebrafish Endothelin-1 required for ventral pharyngeal arch development. Development. 2000;127:3815–28. doi: 10.1242/dev.127.17.3815. [DOI] [PubMed] [Google Scholar]
- Minoux M, Kratochwil CF, Ducret S, Amin S, Kitazawa T, Kurihara H, Bobola N, Vilain N, Rijli FM. Mouse Hoxa2 mutations provide a model for microtia and auricle duplication. Development. 2013;140:4386–97. doi: 10.1242/dev.098046. [DOI] [PubMed] [Google Scholar]
- Minoux M, Rijli FM. Molecular mechanisms of cranial neural crest cell migration and patterning in craniofacial development. Development. 2010;137:2605–21. doi: 10.1242/dev.040048. [DOI] [PubMed] [Google Scholar]
- Nechiporuk A, Linbo T, Poss KD, Raible DW. Specification of epibranchial placodes in zebrafish. Development. 2007;134:611–23. doi: 10.1242/dev.02749. [DOI] [PubMed] [Google Scholar]
- Nechiporuk A, Linbo T, Raible DW. Endoderm-derived Fgf3 is necessary and sufficient for inducing neurogenesis in the epibranchial placodes in zebrafish. Development. 2005;132:3717–30. doi: 10.1242/dev.01876. [DOI] [PubMed] [Google Scholar]
- Neubuser A, Peters H, Balling R, Martin GR. Antagonistic interactions between FGF and BMP signaling pathways: a mechanism for positioning the sites of tooth formation. Cell. 1997;90:247–55. doi: 10.1016/s0092-8674(00)80333-5. [DOI] [PubMed] [Google Scholar]
- Nissen RM, Yan J, Amsterdam A, Hopkins N, Burgess SM. Zebrafish foxi one modulates cellular responses to Fgf signaling required for the integrity of ear and jaw patterning. Development. 2003;130:2543–54. doi: 10.1242/dev.00455. [DOI] [PubMed] [Google Scholar]
- Ohyama T, Groves AK. Expression of mouse Foxi class genes in early craniofacial development. Dev Dyn. 2004a;231:640–6. doi: 10.1002/dvdy.20160. [DOI] [PubMed] [Google Scholar]
- Ohyama T, Groves AK. Generation of Pax2-Cre mice by modification of a Pax2 bacterial artificial chromosome. Genesis. 2004b;38:195–9. doi: 10.1002/gene.20017. [DOI] [PubMed] [Google Scholar]
- Ohyama T, Groves AK, Martin K. The first steps towards hearing: mechanisms of otic placode induction. Int J Dev Biol. 2007;51:463–72. doi: 10.1387/ijdb.072320to. [DOI] [PubMed] [Google Scholar]
- Ohyama T, Mohamed OA, Taketo MM, Dufort D, Groves AK. Wnt signals mediate a fate decision between otic placode and epidermis. Development. 2006;133:865–75. doi: 10.1242/dev.02271. [DOI] [PubMed] [Google Scholar]
- Ozeki H, Kurihara Y, Tonami K, Watatani S, Kurihara H. Endothelin-1 regulates the dorsoventral branchial arch patterning in mice. Mechanisms of development. 2004;121:387–95. doi: 10.1016/j.mod.2004.02.002. [DOI] [PubMed] [Google Scholar]
- Padanad MS, Bhat N, Guo B, Riley BB. Conditions that influence the response to Fgf during otic placode induction. Dev Biol. 2012;364:1–10. doi: 10.1016/j.ydbio.2012.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padanad MS, Riley BB. Pax2/8 proteins coordinate sequential induction of otic and epibranchial placodes through differential regulation of foxi1, sox3 and fgf24. Dev Biol. 2011;351:90–8. doi: 10.1016/j.ydbio.2010.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papalopulu N, Kintner C. Xenopus Distal-less related homeobox genes are expressed in the developing forebrain and are induced by planar signals. Development. 1993;117:961–975. doi: 10.1242/dev.117.3.961. [DOI] [PubMed] [Google Scholar]
- Papanayotou C, Mey A, Birot AM, Saka Y, Boast S, Smith JC, Samarut J, Stern CD. A mechanism regulating the onset of Sox2 expression in the embryonic neural plate. PLoS Biol. 2008;6 doi: 10.1371/journal.pbio.0060002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patthey C, Gunhaga L. Specification and regionalisation of the neural plate border. Eur J Neurosci. 2011;34:1516–28. doi: 10.1111/j.1460-9568.2011.07871.x. [DOI] [PubMed] [Google Scholar]
- Patthey C, Gunhaga L. Signaling pathways regulating ectodermal cell fate choices. Exp Cell Res. 2013 doi: 10.1016/j.yexcr.2013.08.002. [DOI] [PubMed] [Google Scholar]
- Pera E, Kessel M. Expression of DLX3 in chick embryos. Mechanisms of development. 1999;89:189–193. doi: 10.1016/s0925-4773(99)00207-5. [DOI] [PubMed] [Google Scholar]
- Pera E, Stein S, Kessel M. Ectodermal patterning in the avian embryo: epidermis versus neural plate. Development. 1999;126:63–73. doi: 10.1242/dev.126.1.63. [DOI] [PubMed] [Google Scholar]
- Perlmann T. Glucocorticoid receptor DNA-binding specificity is increased by the organization of DNA in nucleosomes. Proceedings of the National Academy of Sciences of the United States of America. 1992;89:3884–8. doi: 10.1073/pnas.89.9.3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer PL, Gerster T, Lun K, Brand M, Busslinger M. Characterization of three novel members of the zebrafish Pax2/5/8 family: dependency of Pax5 and Pax8 expression on the Pax2.1 (noi) function. Development. 1998;125:3063–74. doi: 10.1242/dev.125.16.3063. [DOI] [PubMed] [Google Scholar]
- Phillips BT, Bolding K, Riley BB. Zebrafish fgf3 and fgf8 encode redundant functions required for otic placode induction. Dev Biol. 2001;235:351–65. doi: 10.1006/dbio.2001.0297. [DOI] [PubMed] [Google Scholar]
- Phillips BT, Kwon HJ, Melton C, Houghtaling P, Fritz A, Riley BB. Zebrafish msxB, msxC and msxE function together to refine the neural-nonneural border and regulate cranial placodes and neural crest development. Dev Biol. 2006;294:376–390. doi: 10.1016/j.ydbio.2006.03.001. [DOI] [PubMed] [Google Scholar]
- Phillips BT, Storch EM, Lekven AC, Riley BB. A direct role for Fgf but not Wnt in otic placode induction. Development. 2004;131:923–31. doi: 10.1242/dev.00978. [DOI] [PubMed] [Google Scholar]
- Pieper M, Ahrens K, Rink E, Peter A, Schlosser G. Differential distribution of competence for panplacodal and neural crest induction to non-neural and neural ectoderm. Development. 2012;139:1175–87. doi: 10.1242/dev.074468. [DOI] [PubMed] [Google Scholar]
- Prasad MS, Sauka-Spengler T, LaBonne C. Induction of the neural crest state: control of stem cell attributes by gene regulatory, post-transcriptional and epigenetic interactions. Dev Biol. 2012;366:10–21. doi: 10.1016/j.ydbio.2012.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raft S, Andrade LR, Shao D, Akiyama H, Henkemeyer M, Wu DK. Ephrin-B2 governs morphogenesis of endolymphatic sac and duct epithelia in the mouse inner ear. Dev Biol. 2014;390:51–67. doi: 10.1016/j.ydbio.2014.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rex M, Orme A, Uwanogho D, Tointon K, Wigmore PM, Sharpe PT, Scotting PJ. Dynamic expression of chicken Sox2 and Sox3 genes in ectoderm induced to form neural tissue. Dev Dyn. 1997;209:323–332. doi: 10.1002/(SICI)1097-0177(199707)209:3<323::AID-AJA7>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Riley BB, Phillips BT. Ringing in the new ear: resolution of cell interactions in otic development. Dev Biol. 2003;261:289–312. doi: 10.1016/s0012-1606(03)00245-8. [DOI] [PubMed] [Google Scholar]
- Robledo RF, Lufkin T. Dlx5 and Dlx6 homeobox genes are required for specification of the mammalian vestibular apparatus. Genesis. 2006;44:425–37. doi: 10.1002/dvg.20233. [DOI] [PubMed] [Google Scholar]
- Robledo RF, Rajan L, Li X, Lufkin T. The Dlx5 and Dlx6 homeobox genes are essential for craniofacial, axial, and appendicular skeletal development. Genes Dev. 2002;16:1089–101. doi: 10.1101/gad.988402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruest LB, Xiang X, Lim KC, Levi G, Clouthier DE. Endothelin-A receptor-dependent and -independent signaling pathways in establishing mandibular identity. Development. 2004;131:4413–23. doi: 10.1242/dev.01291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saint-Jeannet JP, Moody SA. Establishing the pre-placodal region and breaking it into placodes with distinct identities. Dev Biol. 2014 doi: 10.1016/j.ydbio.2014.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato S, Ikeda K, Shioi G, Ochi H, Ogino H, Yajima H, Kawakami K. Conserved expression of mouse Six1 in the pre-placodal region (PPR) and identification of an enhancer for the rostral PPR. Dev Biol. 2010;344:158–71. doi: 10.1016/j.ydbio.2010.04.029. [DOI] [PubMed] [Google Scholar]
- Sauka-Spengler T, Bronner-Fraser M. A gene regulatory network orchestrates neural crest formation. Nat Rev Mol Cell Biol. 2008;9:557–68. doi: 10.1038/nrm2428. [DOI] [PubMed] [Google Scholar]
- Schlosser G. Induction and specification of cranial placodes. Dev Biol. 2006;294:303–51. doi: 10.1016/j.ydbio.2006.03.009. [DOI] [PubMed] [Google Scholar]
- Schlosser G. Making senses development of vertebrate cranial placodes. Int Rev Cell Mol Biol. 2010;283:129–234. doi: 10.1016/S1937-6448(10)83004-7. [DOI] [PubMed] [Google Scholar]
- Sheng G, Stern CD. Gata2 and Gata3: novel markers for early embryonic polarity and for non-neural ectoderm in the chick embryo. Mechanisms of development. 1999;87:213–216. doi: 10.1016/s0925-4773(99)00150-1. [DOI] [PubMed] [Google Scholar]
- Shigetani Y, Nobusada Y, Kuratani S. Ectodermally derived FGF8 defines the maxillomandibular region in the early chick embryo: epithelial-mesenchymal interactions in the specification of the craniofacial ectomesenchyme. Dev Biol. 2000;228:73–85. doi: 10.1006/dbio.2000.9932. [DOI] [PubMed] [Google Scholar]
- Shirokova V, Jussila M, Hytonen MK, Perala N, Drogemuller C, Leeb T, Lohi H, Sainio K, Thesleff I, Mikkola ML. Expression of Foxi3 is regulated by ectodysplasin in skin appendage placodes. Dev Dyn. 2013;242:593–603. doi: 10.1002/dvdy.23952. [DOI] [PubMed] [Google Scholar]
- Sienknecht UJ. Developmental origin and fate of middle ear structures. Hear Res. 2013;301:19–26. doi: 10.1016/j.heares.2013.01.019. [DOI] [PubMed] [Google Scholar]
- Solomon KS, Fritz A. Concerted action of two dlx paralogs in sensory placode formation. Development. 2002;129:3127–3136. doi: 10.1242/dev.129.13.3127. [DOI] [PubMed] [Google Scholar]
- Solomon KS, Kudoh T, Dawid IB, Fritz A. Zebrafish foxi1 mediates otic placode formation and jaw development. Development. 2003a;130:929–940. doi: 10.1242/dev.00308. [DOI] [PubMed] [Google Scholar]
- Solomon KS, Kwak SJ, Fritz A. Genetic interactions underlying otic placode induction and formation. Dev Dyn. 2004;230:419–433. doi: 10.1002/dvdy.20067. [DOI] [PubMed] [Google Scholar]
- Solomon KS, Logsdon JM, Fritz A. Expression and phylogenetic analyses of three zebrafish FoxI class genes. Dev Dyn. 2003b;228:301–307. doi: 10.1002/dvdy.10373. [DOI] [PubMed] [Google Scholar]
- Steventon B, Mayor R, Streit A. Neural crest and placode interaction during the development of the cranial sensory system. Dev Biol. 2014;389:28–38. doi: 10.1016/j.ydbio.2014.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stottmann RW, Anderson RM, Klingensmith J. The BMP antagonists Chordin and Noggin have essential but redundant roles in mouse mandibular outgrowth. Dev Biol. 2001;240:457–73. doi: 10.1006/dbio.2001.0479. [DOI] [PubMed] [Google Scholar]
- Streit A. Origin of the vertebrate inner ear: evolution and induction of the otic placode. J Anat. 2001;199:99–103. doi: 10.1046/j.1469-7580.2001.19910099.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streit A. Extensive cell movements accompany formation of the otic placode. Dev Biol. 2002;249:237–254. doi: 10.1006/dbio.2002.0739. [DOI] [PubMed] [Google Scholar]
- Streit A. Early development of the cranial sensory nervous system: from a common field to individual placodes. Dev Biol. 2004;276:1–15. doi: 10.1016/j.ydbio.2004.08.037. [DOI] [PubMed] [Google Scholar]
- Streit A. The preplacodal region: an ectodermal domain with multipotential progenitors that contribute to sense organs and cranial sensory ganglia. Int J Dev Biol. 2007;51:447–61. doi: 10.1387/ijdb.072327as. [DOI] [PubMed] [Google Scholar]
- Streit A, Berliner AJ, Papanayotou C, Sirulnik A, Stern CD. Initiation of neural induction by FGF signalling before gastrulation. Nature. 2000;406:74–8. doi: 10.1038/35017617. [DOI] [PubMed] [Google Scholar]
- Streit A, Sockanathan S, Perez L, Rex M, Scotting PJ, Sharpe PT, Lovell-Badge R, Stern CD. Preventing the loss of competence for neural induction: HGF/SF, L5 and Sox-2. Development. 1997;124:1191–1202. doi: 10.1242/dev.124.6.1191. [DOI] [PubMed] [Google Scholar]
- Stuhlmiller TJ, Garcia-Castro MI. Current perspectives of the signaling pathways directing neural crest induction. Cell Mol Life Sci. 2012;69:3715–37. doi: 10.1007/s00018-012-0991-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo-Rogers HL, Smithers LE, Yakob W, Liu KJ. New directions in craniofacial morphogenesis. Dev Biol. 2010;341:84–94. doi: 10.1016/j.ydbio.2009.11.021. [DOI] [PubMed] [Google Scholar]
- Tassano E, Jagannathan V, Drögemüller C, Leoni M, Hytönen MK, Severino M, Gimelli S, Cuoco C, Di Rocco M, Sanio K, Groves AK, Leeb T, Gimelli G. Congenital aural atresia associated with agenesis of internal carotid artery in a girl with FOXI3 deletion. Journal of Molecular Medicine. 2014 doi: 10.1002/ajmg.a.36895. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thermes V, Lin CC, Hwang PP. Expression of Ol-foxi3 and Na(+)/K(+)-ATPase in ionocytes during the development of euryhaline medaka (Oryzias latipes) embryos. Gene expression patterns: GEP. 2010;10:185–92. doi: 10.1016/j.gep.2010.04.001. [DOI] [PubMed] [Google Scholar]
- Thompson H, Tucker AS. Dual origin of the epithelium of the mammalian middle ear. Science. 2013;339:1453–6. doi: 10.1126/science.1232862. [DOI] [PubMed] [Google Scholar]
- Tribulo C, Aybar MJ, Nguyen VH, Mullins MC, Mayor R. Regulation of Msx genes by a Bmp gradient is essential for neural crest specification. Development. 2003;130:6441–6452. doi: 10.1242/dev.00878. [DOI] [PubMed] [Google Scholar]
- Trumpp A, Depew MJ, Rubenstein JL, Bishop JM, Martin GR. Cre-mediated gene inactivation demonstrates that FGF8 is required for cell survival and patterning of the first branchial arch. Genes Dev. 1999;13:3136–48. doi: 10.1101/gad.13.23.3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker AS, Yamada G, Grigoriou M, Pachnis V, Sharpe PT. Fgf-8 determines rostral-caudal polarity in the first branchial arch. Development. 1999;126:51–61. doi: 10.1242/dev.126.1.51. [DOI] [PubMed] [Google Scholar]
- Uchikawa M, Ishida Y, Takemoto T, Kamachi Y, Kondoh H. Functional analysis of chicken Sox2 enhancers highlights an array of diverse regulatory elements that are conserved in mammals. Dev Cell. 2003;4:509–519. doi: 10.1016/s1534-5807(03)00088-1. [DOI] [PubMed] [Google Scholar]
- Urness LD, Li C, Wang X, Mansour SL. Expression of ERK signaling inhibitors Dusp6, Dusp7, and Dusp9 during mouse ear development. Dev Dyn. 2008;237:163–9. doi: 10.1002/dvdy.21380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urness LD, Paxton CN, Wang X, Schoenwolf GC, Mansour SL. FGF signaling regulates otic placode induction and refinement by controlling both ectodermal target genes and hindbrain Wnt8a. Dev Biol. 2010;340:595–604. doi: 10.1016/j.ydbio.2010.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaglia JL, Hall BK. Regulation of neural crest cell populations: occurrence, distribution and underlying mechanisms. Int J Dev Biol. 1999;43:95–110. [PubMed] [Google Scholar]
- Vidarsson H, Westergren R, Heglind M, Blomqvist SR, Breton S, Enerback S. The forkhead transcription factor Foxi1 is a master regulator of vacuolar H-ATPase proton pump subunits in the inner ear, kidney and epididymis. PloS one. 2009;4:e4471. doi: 10.1371/journal.pone.0004471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall NA, Hogan BL. Expression of bone morphogenetic protein-4 (BMP-4), bone morphogenetic protein-7 (BMP-7), fibroblast growth factor-8 (FGF-8) and sonic hedgehog (SHH) during branchial arch development in the chick. Mechanisms of development. 1995;53:383–92. doi: 10.1016/0925-4773(95)00453-x. [DOI] [PubMed] [Google Scholar]
- Weigel D, Jackle H. The fork head domain: a novel DNA binding motif of eukaryotic transcription factors? Cell. 1990;63:455–6. doi: 10.1016/0092-8674(90)90439-l. [DOI] [PubMed] [Google Scholar]
- Weigel D, Jurgens G, Kuttner F, Seifert E, Jackle H. The homeotic gene fork head encodes a nuclear protein and is expressed in the terminal regions of the Drosophila embryo. Cell. 1989;57:645–58. doi: 10.1016/0092-8674(89)90133-5. [DOI] [PubMed] [Google Scholar]
- Woda JM, Pastagia J, Mercola M, Artinger KB. Dlx proteins position the neural plate border and determine adjacent cell fates. Development. 2003;130:331–342. doi: 10.1242/dev.00212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright TJ, Mansour SL. Fgf3 and Fgf10 are required for mouse otic placode induction. Development. 2003;130:3379–90. doi: 10.1242/dev.00555. [DOI] [PubMed] [Google Scholar]
- Xu H, Dude CM, Baker CVH. Fine-grained fate maps for the ophthalmic and maxillomandibular trigeminal placodes in the chick embryo. Dev Biol. 2008;317:174–186. doi: 10.1016/j.ydbio.2008.02.012. [DOI] [PubMed] [Google Scholar]
- Xu J, Watts JA, Pope SD, Gadue P, Kamps M, Plath K, Zaret KS, Smale ST. Transcriptional competence and the active marking of tissue-specific enhancers by defined transcription factors in embryonic and induced pluripotent stem cells. Genes Dev. 2009;23:2824–38. doi: 10.1101/gad.1861209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi H, Maeda J, Hu T, McAnally J, Conway SJ, Kume T, Meyers EN, Yamagishi C, Srivastava D. Tbx1 is regulated by tissue-specific forkhead proteins through a common Sonic hedgehog-responsive enhancer. Genes Dev. 2003;17:269–81. doi: 10.1101/gad.1048903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Xu L, Crawford G, Wang Z, Burgess SM. The forkhead transcription factor FoxI1 remains bound to condensed mitotic chromosomes and stably remodels chromatin structure. Mol Cell Biol. 2006;26:155–68. doi: 10.1128/MCB.26.1.155-168.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, O’Neill P, Martin K, Maass JC, Vassilev V, Ladher R, Groves AK. Analysis of FGF-dependent and FGF-independent pathways in otic placode induction. PloS one. 2013;8:e55011. doi: 10.1371/journal.pone.0055011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T, Vidarsson H, Rodrigo-Blomqvist S, Rosengren SS, Enerback S, Smith RJ. Transcriptional control of SLC26A4 is involved in Pendred syndrome and nonsyndromic enlargement of vestibular aqueduct (DFNB4) Am J Hum Genet. 2007;80:1055–63. doi: 10.1086/518314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret KS, Carroll JS. Pioneer transcription factors: establishing competence for gene expression. Genes Dev. 2011;25:2227–41. doi: 10.1101/gad.176826.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelarayan LC, Vendrell V, Alvarez Y, Dominguez-Frutos E, Theil T, Alonso MT, Maconochie M, Schimmang T. Differential requirements for FGF3, FGF8 and FGF10 during inner ear development. Dev Biol. 2007;308:379–91. doi: 10.1016/j.ydbio.2007.05.033. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Knosp BM, Maconochie M, Friedman RA, Smith RJH. A comparative study of Eya1 and Eya4 protein function and its implication in branchio-oto-renal syndrome and DFNA10. J Assoc Res Otolaryngol. 2004;5:295–304. doi: 10.1007/s10162-004-4044-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W, Huang L, Wei ZB, Silvius D, Tang B, Xu PX. The role of Six1 in mammalian auditory system development. Development. 2003;130:3989–4000. doi: 10.1242/dev.00628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou D, Silvius D, Fritzsch B, Xu PX. Eya1 and Six1 are essential for early steps of sensory neurogenesis in mammalian cranial placodes. Development. 2004;131:5561–72. doi: 10.1242/dev.01437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou D, Silvius D, Rodrigo-Blomqvist S, Enerback S, Xu PX. Eya1 regulates the growth of otic epithelium and interacts with Pax2 during the development of all sensory areas in the inner ear. Dev Biol. 2006;298:430–41. doi: 10.1016/j.ydbio.2006.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]


