Abstract
Background
Respiratory illness was reported among humans and swine at an agricultural fair in 2011; 3 human infections with an influenza A(H3N2) variant (H3N2v) virus were confirmed. Using epidemiologic investigation data, we sought to estimate H3N2v transmissibility from swine to humans.
Methods
We developed a model of H3N2v transmission among swine and humans and fit it to data from a cohort of 100 agricultural club members reporting swine contact to estimate transmissibility. A sensitivity analysis was performed varying H3N2v prevalence in the club cohort. Using the best-fit transmission probability, we simulated the number of swine-acquired infections among all fair attendees.
Results
We estimated the best-fit probability of swine-to-human H3N2v transmission per minute of swine contact. Applying this probability to 14 910 people with swine contact at the fair, we estimate that there were 80 (95% confidence interval [CI], 40–133) H3N2v infections among persons aged <20 years and 58 (95% CI, 29–96) H3N2v infections among person aged ≥20 years.
Conclusions
Using early data from investigation of a new virus with unclear transmission properties, we estimated the transmissibility of H3N2v from swine to humans and the burden of H3N2v among fair attendees. Although the risk of H3N2v virus infection is small for fair attendees with minimal swine contact, large populations attend agricultural events each year, and human cases will likely occur when infected swine are present.
Keywords: influenza virus, swine, variant influenza, transmission
From 2005 through 2010, 21 variant influenza virus infections (ie, infections in humans with influenza viruses that normally infect swine) were reported to the Centers for Disease Control and Prevention (CDC) at a frequency of between 1 and 6 cases per year. In July 2011, a new influenza A(H3N2) variant virus emerged that was a reassortant virus derived from a swine virus and 2009 H1N1 pandemic (pH1N1) virus; this virus is hereafter referred to as H3N2v when it infects humans and H3N2pM when it infects swine. From July to November 2011, 12 cases of H3N2v were confirmed. This new virus had acquired the pH1N1 matrix (M) gene, which may increase transmissibility in animal models [1, 2]. H3N2v had not been detected before 2011, and although cases appeared to be occurring more frequently, the transmissibility of this variant virus from swine to humans had not been studied.
In dynamic models of disease transmission, the rate at which people become infected depends on the pathogen's transmissibility, the balance of susceptible and infectious individuals over time, and patterns of contact between susceptible and infectious individuals. In a 2-population (swine and human) model where swine are transmitting infection to humans, the proportion of humans infected is driven by the amount of contact between swine and humans, the probability of transmission when contact occurs, and the changing proportion of infectious swine. Given the incidence of infection among swine, the incidence among humans, and a measure of swine-human contact, one can infer the transmissibility of the pathogen.
In August 2011, an outbreak of respiratory illness occurred among humans and swine at an agricultural fair (Fair A) during which 3 human infections with H3N2v virus were con-firmed. Using data from an epidemiologic investigation of the outbreak [3], we developed a mathematical model to estimate the transmissibility of H3N2v from swine to humans at the fair. Using this model, we conducted an outbreak simulation to estimate the total number of H3N2v infections among Fair A attendees acquired from swine at the fair.
METHODS
Epidemiologic Investigation
The model was based on a retrospective cohort study of agricultural club members who participated in Fair A [3]. In this study, swine contact was defined as touching swine or visiting the swine barn, swine show, or swine auction. Suspected H3N2v cases were defined as club members having ≥1 respiratory symptom (cough, sore throat, or runny nose) and ≥1 symptom from 1 of the following categories ≤7 days after attending the fair: fever (>38°C or subjective fever), gastrointestinal (diarrhea or vomiting), or constitutional (fatigue, muscle aches, joint pain). A total of 127 club members were interviewed about their symptoms and swine exposure at the fair using a standard questionnaire. Our model was limited to 100 (79%) members reporting swine contact at Fair A. Those reporting no swine contact were considered not to be at risk for swine-acquired H3N2v virus infection in the model. Members reporting swine contact had a median age of 13 years (range, 4–19 years), and 13 (13%) members met criteria for suspected H3N2v infection. For the initial model, we assumed all suspected cases were attributable to H3N2v infection, although none underwent diagnostic testing for influenza. The duration of swine contact per day was not collected during interviews; an average value was estimated for all members based on informal open-ended discussions with members. Some swine exhibitors reported respiratory illness among swine during or in the week after the fair [3], but none of the swine were tested for influenza. In the model, we assumed that illness among swine was attributable to H3N2pM.
Model Base Parameters
Swine–human contact (Csh) was represented by the amount of time that the 100 swine-exposed club members spent in the swine barn, which was estimated at an average of 60 minutes per person per day (Table 1). Immunity to H3N2v among club members was estimated at 10% based on prior studies of the prevalence of cross-reactive antibodies to H3N2v in children using similar variant influenza viruses [4–6]. The estimated fraction of humans with preexisting immunity to H3N2v was removed from the initial susceptible population. The incubation period of H3N2v in humans was estimated at 2 days based on the cohort study data. The duration of infectiousness was estimated at 5 days based on prior studies of seasonal influenza viruses [7].
Table 1.
Model Base Parameters
| Parameter | Definition | Value | Source |
|---|---|---|---|
| T h | Total number of humans | Club members: 127 | Fair A investigation [3] |
| General attendees: 70 000 | Fair A investigation | ||
| N h | Number of humans with swine contact | Club members: 100 (age <20 y) | Fair A investigation |
| General attendees: 14 910 (6468 age <20 y; 8442 age ≥20 y) | Fair B investigationa | ||
| N s | Number of swine | 208 | Fair A investigation |
| I h | Number of infected humans | Initial value: 0 | Assumption |
| I s | Number of infected swine | Initial value: 1 | Fair A investigationb |
| C sh | Mean duration of contact between humans and swine | Club members: 60 min/d | Fair A investigation |
| General attendees: 5 min/d | |||
| P sh | Transmission probability of H3N2v, swine to human | Estimated by model | |
| P sh75 | Transmission probability of H3N2v, swine to human, assuming 75% of illnesses among club members were attributable to H3N2v | Estimated by model | |
| R ss | Influenza A(H3N2) reproduction number, swine to swine | 2 | [8] |
| 1/κh | Incubation period, humans | Deterministic model: 2 d | Fair A investigation |
| Stochastic model: Gaussian distribution, mean = 2 d, variance = 0.5 | |||
| 1/σs | Duration of infectiousness, swine | 5 d | [8, 9] |
| 1/σh | Duration of infectiousness, humans | 5 d | [7] |
| Immh | Preexisting immunity to H3N2v among humans | Age <20 y: 10% | [4–6, 10] |
| Age ≥20 y: 50% | |||
| F total | Duration of fair | 9 d | Fair A investigation |
Overall attendance was estimated for Fair A; age distribution of attendees and prevalence of swine contact by age group estimated from Fair B investigation.
Based on the observation that 1 febrile swine was present at the start of Fair A.
There were 208 swine at the fair, and 1 swine was reported to have a febrile illness at the start of the fair. This swine was assumed to be the index swine case of H3N2pM virus infection, although it was not tested for influenza virus. The reproduction number of H3N2pM among swine (Rss) was estimated at 2 based on experimental studies of swine influenza transmission in swine [8]. The duration of infectiousness in swine was estimated at 5 days [8, 9]. The period of swine-to-human and swine-to-swine contact was limited to the duration of the fair (9 days).
Model of H3N2v Transmission
We developed a population dynamic (susceptible-exposed-infectious-recovered) model of swine-to-human H3N2v transmission (Figure 1). Swine could be susceptible to H3N2pM (Ss), infected with H3N2pM (Is), or recovered from H3N2pM (Rs). Ns is the total number of swine. Humans transitioned between 4 states: susceptible (Sh); exposed (Eh), which includes humans exposed to infected swine but not yet symptomatic; infected (Ih); and recovered (Rh). The exposed state was specified for humans to simulate an incubation period between the acquisition of infection from swine and the onset of illness in the human.
Figure 1.
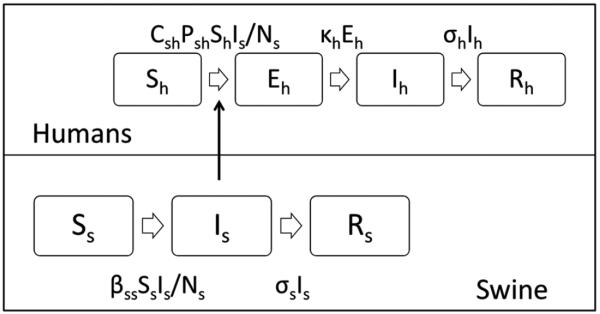
Model of influenza A(H3N2) variant (H3N2v) transmission from swine to humans. Abbreviations: Sh, susceptible humans; Eh, exposed humans; Ih, infected humans; Rh, recovered humans; Ss, susceptible swine; Is, infectious swine; Rs, recovered swine; Csh, minutes of swine barn contact per person per day; Psh, swine-to-human H3N2v transmission probability per minute of contact with infectious swine; κh, rate of progression from exposed to infected among humans (1/incubation period); σh, recovery rate among humans (1/duration of illness); βss, force of infection from swine to swine, σs, recovery rate among swine (1/duration of illness).
The model included 2 modes of H3N2v/H3N2pM transmission: swine-to-swine and swine-to-human. Swine were housed in a barn in close proximity to each other for the duration of the fair and were much more likely to be exposed for long periods of time to ill swine than to ill humans; therefore, new infections among swine were assumed to have been acquired from other swine and not from infected humans. The rate of infection in humans was set to be proportional to the prevalence of infection among swine in the model. Because contact with infected swine is a risk factor for H3N2v infection and sustained human-to-human transmission had not been described with this virus, new infections in humans were assumed to be acquired from infected swine and not from infected humans.
The model (Figure 1) was defined by the set of ordinary differential equations:
dSh/dt = −Csh × Psh × Sh × (Is/Ns)
dEh/dt = Csh × Psh × Sh × (Is/Ns) − κh × Eh
dIh/dt = κh × Eh − σh × Ih
dRh/dt = σh × Ih
dSs/dt = −βss × Ss × (Is/Ns)
dIs/dt = βss × Ss × (Is/Ns) − σs × Is
dRs/dt = σs × Is,
where Csh is the minutes of swine contact per person per day, Psh is the H3N2v swine-to-human transmission probability per minute of contact with the infectious swine population, κh is the rate of progression from exposed to infected among humans (1/incubation period), σh is the recovery rate among humans (1/duration of illness), βss is the force of infection from swine to swine, and σs is the recovery rate among swine (1/duration of illness). βss is Rss × σs, where Rss is the swine-to-swine H3N2pM reproduction number.
The model was fit to the cumulative incidence of suspected H3N2v infections among the agricultural club cohort by solving the set of differential equations for transmission probability by successive iterations using the Runge-Kutta 4 method of integration. A stochastic model using a Poisson distribution around the best-fit transmission probability and a Gaussian distribution around incubation period (μ = 2 days, σ2 = 0.5) was run for 100 simulations to illustrate the range of possible outcomes around the deterministic estimate. The deterministic and stochastic models were implemented using Berkeley Madonna software (version 8.3.18 for Windows, Berkeley, California).
The log likelihood of transmission probability was calculated by comparing simulated vs observed incident human H3N2v virus infections while varying the transmission probability. A 95% confidence interval (CI) was calculated for transmission probability based on the likelihood ratio test.
Simulation of H3N2v Outbreak Among All Fair A Attendees With Swine Contact
Based on the best-fit transmission probability of H3N2v from swine to members of the agricultural club cohort, we then simulated the number of swine-acquired H3N2v infections at Fair A, which was attended by approximately 70 000 people. Data on the age distribution and prevalence and duration of swine contact for Fair A attendees were not available; these parameter estimates for Fair A attendees were based on a survey conducted at Fair B, a fair in the same county occurring 3 weeks after Fair A. Thirty-three percent of the Fair B attendees were aged <20 years; the prevalence of swine exposure was 28% among Fair B attendees aged <20 years and 18% among attendees aged ≥20 years (Table 1). The mean swine-to-human contact rate among Fair B attendees reporting swine contact was estimated at 5 minutes per day. Based on prior serologic studies, preexisting immunity to H3N2v was estimated at 10% among persons aged <20 years and 50% among persons aged ≥20 years [4–6, 10].
Sensitivity Analysis
Our base analysis assumed that all suspected cases among the agricultural club cohort were due to H3N2v. We conducted a sensitivity analysis in which 75% of cases were assumed to be attributable to H3N2v. This assumption was based on limited serologic testing which found that 4 of 6 children <4 years of age who provided convalescent serum samples were seropositive to H3N2v [3]. The model was refit using the lower cumulative incidence to estimate the transmission probability (Psh75). Psh75 was used to simulate the expected number of H3N2v virus infections among Fair A attendees with swine contact.
RESULTS
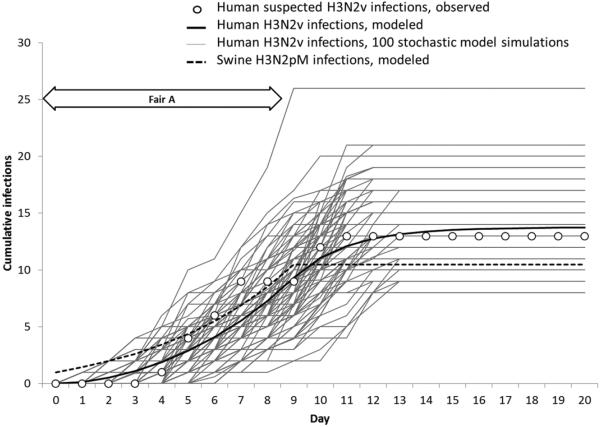
Assuming Rss of 2 for swine-to-swine transmission [8], we estimated that 10.5 of the 208 swine became infected with H3N2pM (Figure 2). The probability of H3N2v transmission to a susceptible human for each minute of contact with the infectious swine population (Psh) was estimated to be 0.024 (95% CI, .012–.040). The proportion of infectious swine in the swine barn (Is/Ns), ranged from 1 of 208 (0.005) at the start of the fair to 5.60 of 208 (0.027) on the last day of the fair. The 100 stochastic simulation runs, which assumed Psh = 0.024, produced a total number of H3N2v infections among the cohort ranging from 8 to 26, illustrating 100 possible outcomes of the model based on probability distributions around this estimate of Psh and the incubation period of H3N2v.
Figure 2.
Cumulative influenza A(H3N2) virus infections among agricultural club cohort members (H3N2v) and swine (H3N2pM), Fair A, August 2011.
Among the population of 70 000 Fair A attendees, 14 910 were estimated to have swine contact and thus were at risk for swine-acquired H3N2v. Among the 14 910, of whom 10% of persons <20 years of age and 50% of persons ≥20 years of age were assumed to have immunity to H3N2v, we applied a Psh of 0.024 to estimate the number of swine-to-human H3N2v transmission events that occurred at Fair A. The simulated number of swine-acquired H3N2v infections among fair attendees aged <20 years was 80 (95% CI, 40–133; Figure 3). The simulated number of swine-acquired H3N2v infections among fair attendees aged ≥20 years was 58 (95% CI, 29–96). Among fair attendees with swine contact, 1.2% (95% CI, .6%–2.1%) became infected with H3N2v in the <20-year-old age group and 0.7% (95% CI, .3%–1.1%) became infected in the ≥20-year-old age group.
Figure 3.
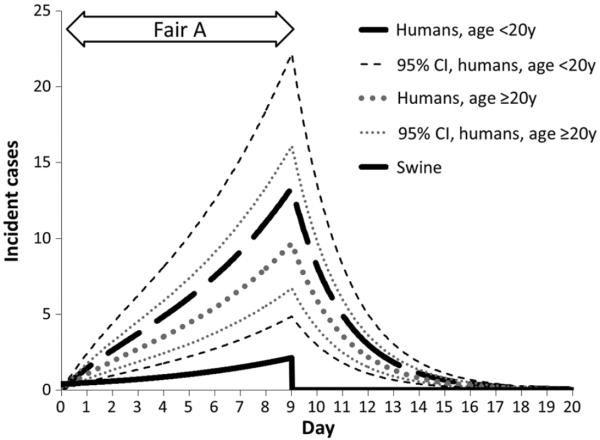
Simulated incident influenza A(H3N2) virus infections among humans (H3N2v) and swine (H3N2pM) by day and age group (for humans), Fair A attendees, August 2011. Abbreviation: CI, confidence interval.
For the sensitivity analysis in which 75% of the suspected cases among the club members were assumed to be attributable to H3N2v, the Psh75 was 0.017 (95% CI, .008–.029) per minute of contact with the infectious swine population. The simulated number of H3N2v infections among fair attendees with swine contact with Psh75 = 0.017 was 0.9% (95% CI, .4%–1.5%) among persons aged <20 years and 0.5% (95% CI, .2%–.9%) among persons aged ≥20 years.
DISCUSSION
Using early epidemiologic data from the first known fair-associated outbreak of H3N2pM/H3N2v, we developed a model to estimate the transmission potential of H3N2v from swine to humans. The transmission probability estimated from the investigation data was then applied to simulate the outbreak among a large population of fair attendees. Although only a third of fair attendees were <20 years old, more infections were expected to occur in this age group than among adults based on their levels of prior immunity to H3N2v and their patterns of swine contact.
This model estimates a probability of H3N2v virus infection of 0.024 for each minute a person spent in contact with the infectious swine population at Fair A. The risk to an individual fair attendee of H3N2v infection depends not only on the transmission probability, but also on that individual's contact with swine, his/her immunity to H3N2v, and whether the swine contacted by that individual were in the infectious stage of H3N2pM. The proportion of fair attendees infected in the simulation model should not be generalized to other fairs where the incidence of H3N2pM among swine or the patterns of swine-to-human contact may be different. The transmission probability estimated by this model is best applied to estimating the relative impact of H3N2v on fairs where the model components, including the number of swine, the prevalence of infection in swine, the number and age distribution of fair attendees, and the duration of swine-to-human contact, are known. The model can also be used to simulate the impact of interventions such as limiting duration of swine-to-human contact or dismissing swine from the fair.
Although H3N2v virus infections are expected to be rare among fair attendees who may have only brief contact with infectious swine, local and state fairs are often widely attended. Fairs present opportunities for many people to come into contact with a relatively small number of potentially infected swine. The International Association of Fairs and Expositions estimates that approximately 150 million people attend fairs in North America every year (International Association of Fairs and Expositions, written communication, 2 November 2012). The estimates produced by this model were compatible with outbreak investigation findings; at Fair A, 89 suspected, probable, and confirmed cases were identified among all fair attendees [3], and at one fair attended by approximately 20 000 people where an outbreak of H3N2pM was confirmed among swine, 73 confirmed H3N2v infections were identified (M. Jhung, written communication, 25 October 2012). Despite the low attack rates simulated by this model, several infections with variant influenza virus may be expected if infectious swine are present at a fair.
Efforts to prevent influenza among swine at fairs could decrease the risk of human infections. Screening swine for illness before admission to the swine barn and promptly removing or isolating any ill swine, in addition to canceling swine events at fairs where a swine influenza outbreak is occurring, might reduce contact between infected and susceptible swine and mitigate a swine influenza outbreak. These measures were implemented in 2012 at fairs by states with H3N2v outbreaks and likely led to a decrease in human infections [11–13]. Vaccination of swine, particularly with a homologous strain, reduces the likelihood of infections among swine and reduces viral shedding [8, 9, 14]. Decreasing viral shedding by vaccination may limit opportunities for swine-to-human transmission of influenza viruses. Reducing contact with infected swine, especially among children who are at greater risk for H3N2v infection due to low preexisting immunity [4], may prevent outbreaks of variant influenza. In August 2012, the CDC issued a recommendation that children <5 years of age and persons with underlying health conditions that place them at high risk for influenza complications [15] should avoid swine contact at agricultural fairs during the 2012 fair season [16].
This study is subject to certain limitations. The data used for fitting the model are limited to suspected, not confirmed, H3N2v (among humans) and H3N2pM (among swine) infections. This approach can be justified by the fact that the Fair A outbreak occurred before the typical influenza season and H3N2v infections were confirmed among attendees. However, noninfluenza respiratory viruses may have contributed to illnesses reported among agricultural club members. If only a proportion of the illnesses observed in the cohort were due to H3N2v, but the number of swine infected and the patterns of swine contact remained the same, the virus would be less transmissible from swine to humans, and fewer infections would be predicted among the entire fair population. In the sensitivity analysis, the proportion of attendees affected was lower than the proportion in base analysis, although this difference was not statistically significant. Minutes of swine exposure were not assessed for individual cohort members or other Fair A attendees; instead, the average duration of swine–human contact was estimated for both groups. In the model, the best-fit transmission probability from swine to human is inversely proportional to the minutes of swine exposure assumed for the cohort members. The values used for relative duration of swine contact among cohort members and other Fair A attendees may be imprecise. In addition, swine contact can be characterized by measures other than the number of minutes spent in the swine barn, such as whether the swine were handled directly or observed from a distance; these other measures of swine contact were not included in the model and may be different between cohort members and other fair attendees. The reproduction number of H3N2pM among swine was estimated from experimental studies of influenza A transmission; this estimate determined the number of swine in the model who were infectious. Although illness was observed among swine, no swine were tested for influenza at Fair A. The true number of infectious swine could have been higher if many asymptomatic infectious swine were present, or lower if some of the illness observed among swine was not due to influenza. The study was not designed to assess human-to-human transmission of H3N2v because the transmissibility of H3N2v among humans and patterns of contact among agricultural members and fair attendees were not known. Therefore, this model estimates the number of swine-acquired H3N2v infections, and not the total number of H3N2v infections, among fair attendees. If some infections among the cohort members were acquired from humans rather than swine, the transmission probability from swine to human would be lower. However, among hundreds of H3N2v cases identified in 2012, human-to-human transmission seems to be inefficient and limited [17].
Despite these limitations, developing this model based on early investigation data allowed us to estimate the transmission risk of an emerging zoonotic pathogen with unclear transmission properties. It also allowed simulation of the burden of H3N2v infections among all Fair A attendees. This method demonstrates how mathematical modeling can be used with epidemio-logic investigations of zoonotic outbreaks to better understand transmissibility. More than 300 fair-associated H3N2v cases have been identified in multiple states since July 2012, and most cases have occurred among children who attended agricultural fairs [17]. Future investigations of H3N2v outbreaks, especially where swine contact can be measured and infections in swine and humans can be confirmed with diagnostic testing, will further inform this model and improve estimates of transmission risk.
Acknowledgments
We thank the following individuals for their contributions to the field investigation: Maria Moll, MD, Erica Smith, MPH, Jeffrey Miller, MD, Virginia Dato, MD, Kumar Nalluswami, MD, Atmaram Nambiar, MD, and James Lute, PhD, at the Pennsylvania Department of Health; James Lando, MD, and Sharon Silvestri, RN, at the Allegheny County Health Department; Erin Moore, DVM, at the Pennsylvania Department of Agriculture; and Adena Greenbaum, MD, Rahul Ganatra, BS, Matthew Biggerstaff, MPH, Eugene Lam, MD, Aaron Storms, MD, Kathy Hancock, PhD, Alicia Branch, PhD, Susan Trock, DVM, Alexander Klimov, PhD, Bo Shu, MD, Lynnette Brammer, MPH, Scott Epperson, MPH, and Michael Jhung, MD, at the Centers for Disease Control and Prevention (CDC).
Financial support. This work was supported by the CDC and the Pennsylvania Department of Health.
Supplement sponsorship. This article was published as part of a supplement entitled “The Emergence of Influenza A (H3N2)v Virus: What We Learned From the First Wave, July 2011–April 2012,” sponsored by the Centers for Disease Control and Prevention.
Footnotes
Publisher's Disclaimer: The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the CDC.
Potential conflicts of interest. All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Chou YY, Albrecht RA, Pica N, et al. The M segment of the 2009 new pandemic H1N1 influenza virus is critical for its high transmission efficiency in the guinea pig model. J Virol. 2011;85:11235–41. doi: 10.1128/JVI.05794-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lakdawala SS, Lamirande EW, Suguitan AL, Jr, et al. Eurasian-origin gene segments contribute to the transmissibility, aerosol release, and morphology of the 2009 pandemic H1N1 influenza virus. PLoS Pathog. 2011;7:e1002443. doi: 10.1371/journal.ppat.1002443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wong KK, Greenbaum A, Moll ME, et al. Outbreak of influenza A (H3N2) variant virus infection among attendees of an agricultural fair, Pennsylvania, USA, 2011. Emerg Infect Dis. 2012;18:1937–44. doi: 10.3201/eid1812.121097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention Antibodies cross-reactive to influenza A (H3N2) variant virus and impact of 2010–11 seasonal influenza vaccine on cross-reactive antibodies—United States. MMWR Morb Mortal Wkly Rep. 2012;61:237–41. [PubMed] [Google Scholar]
- 5.Skowronski DM, De Serres G, Janjua NZ, et al. Cross-reactive antibody to swine influenza A(H3N2) subtype virus in children and adults before and after immunisation with 2010/11 trivalent inactivated influenza vaccine in Canada, August to November 2010. Euro Surveill. 2012;17:20066. doi: 10.2807/ese.17.04.20066-en. [DOI] [PubMed] [Google Scholar]
- 6.Skowronski DM, Janjua NZ, De Serres G, et al. Cross-reactive and vaccine-induced antibody to emerging swine influenza A(H3N2)v. J Infect Dis. 2012;206:1852–61. doi: 10.1093/infdis/jis500. [DOI] [PubMed] [Google Scholar]
- 7.Carrat F, Vergu E, Ferguson NM, et al. Time lines of infection and disease in human influenza: a review of volunteer challenge studies. Am J Epidemiol. 2008;167:775–85. doi: 10.1093/aje/kwm375. [DOI] [PubMed] [Google Scholar]
- 8.Romagosa A, Allerson M, Gramer M, et al. Vaccination of influenza A virus decreases transmission rates in pigs. Vet Res. 2011;42:120. doi: 10.1186/1297-9716-42-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Romagosa A, Gramer M, Joo HS, Torremorell M. Sensitivity of oral fluids for detecting influenza A virus in populations of vaccinated and non-vaccinated pigs. Influenza Other Respi Viruses. 2012;6:110–8. doi: 10.1111/j.1750-2659.2011.00276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Waalen K, Kilander A, Dudman SG, Ramos-Ocao R, Hungnes O. Age-dependent prevalence of antibodies cross-reactive to the influenza A (H3N2) variant virus in sera collected in Norway in 2011. Euro Surveill. 2012;17:20170. [PubMed] [Google Scholar]
- 11.Disis J. Indiana State Fair: Pig barn closed with reports of sick animals. Indianapolis Star; [Accessed 21 November 2012]. Aug 6, 2012. http://www.indystar.com/article/20120806/LOCAL/120806029/-b-Indiana-State-Fair-b-Pig-barn-closed-reports-sick-animals. [Google Scholar]
- 12.Snowbeck C. Minnesota State Fair: Veterinarians will monitor pigs for new flu. St Paul Pioneer Press; [Accessed 21 November 2012]. Aug 14, 2012. http://www.twincities.com/statefair/ci_21312786/minnesota-state-fair-officials-look-out-new-strain. [Google Scholar]
- 13.Woods J. 3 pigs at state fair have swine flu. The Columbus Dispatch; [Accessed 21 November 2012]. Aug 3, 2012. http://www.dispatch.com/content/stories/local/2012/08/03/2-pigs-at-state-fair-have-swine-flu.html. [Google Scholar]
- 14.Vincent AL, Ciacci-Zanella JR, Lorusso A, et al. Efficacy of inactivated swine influenza virus vaccines against the 2009 A/H1N1 influenza virus in pigs. Vaccine. 2010;28:2782–7. doi: 10.1016/j.vaccine.2010.01.049. [DOI] [PubMed] [Google Scholar]
- 15.Centers for Disease Control and Prevention Prevention and control of influenza with vaccines: recommendations of the Advisory Committee on Immunization Practices (ACIP), 2010. MMWR Recomm Rep. 2010;59:1–62. [PubMed] [Google Scholar]
- 16.Centers for Disease Control and Prevention [Accessed 2 September 2012];Take action to prevent the spread of flu between people and pigs at fairs. Available at: http://www.cdc.gov/flu/swineflu/h3n2v-fairs-factsheet.htm.
- 17.Centers for Disease Control and Prevention Evaluation of rapid influenza diagnostic tests for influenza A (H3N2)v virus and updated case count—United States, 2012. MMWR Morb Mortal Wkly Rep. 2012;61:619–21. [PubMed] [Google Scholar]