Abstract
Advances in the treatment of HIV infection have dramatically reduced the death rate from AIDS and improved the quality of life of many HIV-infected individuals. However, the possible long-term toxicity associated with antiretroviral therapy (ART), stigma and cost, all contribute to the necessity of finding a cure for HIV infection. In infected individuals taking ART, HIV persists in a small number of cells that can survive for the lifetime of the infected person. These persistently infected cells, usually referred as the ‘reservoirs for HIV infection’, are the main barriers to a cure. The diversity of the tissues and cellular types in which HIV persists, as well as the multiplicity of the molecular mechanisms contributing to HIV persistence, complicate the efforts to develop a safe, effective, and globally accessible cure for HIV. In this review, we summarise recent data that contribute to our understanding of HIV persistence during ART by addressing three questions pertaining to the HIV reservoir: (1) when is the reservoir established; (2) where is the reservoir maintained; and (3) how does the reservoir persist?
Keywords: HIV reservoir, HIV latency, HIV cure, acute infection, CD4+ T cells, tissues, molecular mechanisms
Introduction
Despite its unquestionable success at reducing HIV replication and improving the quality of life of many people living with HIV/AIDS, combination antiretroviral therapy (ART) does not eradicate the virus [1,2], due to the early establishment of a long-lived viral reservoir [3–5]. The best clinical evidence for the existence of a reservoir for HIV is provided by the rapid viral rebound observed in the vast majority of individuals who interrupt ART. HIV persistence during ART is the reason why current therapies are not curative and has been the subject of intense research during the past 15 years since ART was implemented.
There are multiple reasons underlying HIV persistence during ART, which include the following:
-
►
residual levels of viral replication that may not be fully suppressed in drug-privileged anatomical compartments;
-
►
the persistence of a small pool of cells carrying silent integrated genomes that can be reactivated and reignite infection;
-
►
persistent immune dysfunctions that fail at controlling residual replication and reactivation from latently infected cells.
The use of the word ‘reservoir’ to define the pool of cells in which replication-competent HIV persists during ART is still a matter of debate, as there is no consensus on the main mechanism by which HIV persists during ART. None the less, viral reservoirs can be defined as cell types or anatomical sites in association with which replication-competent forms of the virus persist with more stable kinetic properties than the main pool of actively replicating virus [6,7]. Identifying these cells and tissues and characterising the mechanisms by which HIV persists in these sites is a prerequisite to the design of therapeutic strategies aimed at eradicating HIV.
When is the reservoir established?
Soon after the implementation of ART in 1996, the possibility that ART initiated very early in infection could prevent the establishment of the long-lived HIV reservoir and shorten the duration of HIV persistence after prolonged therapy was proposed [8]. The rationale for this intervention originates from the fact that the latent reservoir is not created but rather revealed by ART, as latently infected CD4+ T cells are generated during untreated HIV infection (Figure 1). Therefore, a reasonable hypothesis is that by reducing the duration of exposure to the virus through early ART initiation, one would limit the overall number of infected cells, thereby reducing the possibility for some of them to revert to a resting state or to directly establish latency.
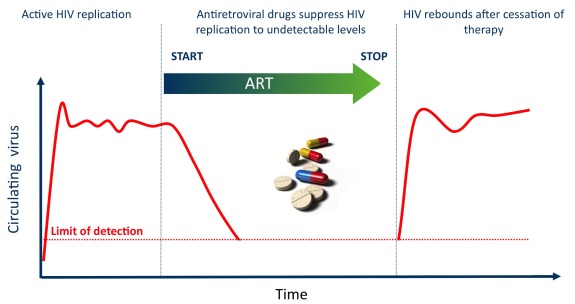
Figure 1.
Clinical definition of the HIV reservoir. Untreated HIV infection is characterised by high levels of viral replication that can be measured in the plasma of HIV-infected individuals. ART reduces viral replication to undetectable levels by standard viral load measurements. When ART is interrupted, HIV replication resumes, revealing that HIV persisted in cellular and anatomical ‘reservoirs’ during ART and that these reservoirs can re-ignite infection.
By 1998, Chun et al. demonstrated that initiation of ART in infected individuals as early as 10 days after the onset of symptoms of primary infection does not prevent the generation of latently infected CD4+ T cells carrying infectious virus [8]. This is in line with recent data generated in the non-human primate model of SIV infection, in which institution of ART as early as 3 days post infection could not prevent the establishment of a viral reservoir, evidenced by viral rebound after ART interruption [9]. Importantly, the time to viral rebound correlates with total viraemia during acute infection and with proviral DNA at the time of ART discontinuation [10], suggesting that the size of the reservoir is a critical parameter that can predict a clinical readout such as the time to viral rebound. Although these two studies suggest that even very early ART intervention may not be able to prevent the establishment of a reservoir for HIV, the capacity of early ART to reduce the size of this persistent reservoir has been demonstrated in several independent studies, using a variety of virological readouts [11–15]. The precise timing at which the reservoir is established is difficult to determine, as latency is likely to occur primarily in tissues that are difficult to access in recently infected individuals. The ‘when’ question may be easier to address if considered together with the ‘how’ question: the well-accepted model of the generation of latently infected cells proposes that they originate from activated cells, most likely specific for HIV antigens [16,17], that are infected and differentiate into long-lived resting memory cells [18]. As a consequence, the latent reservoir may not be established before the generation of memory CD4+ T cells. In contrast, if HIV latency can be directly established in resting CD4+ T cells without the need for these cells to go through an activation state [19,20], the reservoir may be seeded in the first days following infection. Identifying the precise timing during which the latent reservoir is established is technically challenging as at the early stages the bulk of infected cells are likely to be productively infected, which complicates the effort to identify the minute fraction of latently infected CD4+ T cells.
In addition to the quantitative restriction by early ART, the reservoir is less genetically diverse in subjects who start ART early in infection [21]. More importantly, it may be more easily targeted by autologous HIV-specific CD8+ T cell responses, as viruses archived at a later stage of the disease are more likely to present escape mutations [22]. Therefore, the benefits of early ART intervention in the context of curative strategies are both qualitative and quantitative.
Of note, early ART is the only currently available intervention that limits the size of the latent HIV reservoir and leads to clinical benefits in HIV-infected individuals. A prime example is the case of the ‘Mississippi child’ who started ART by 2 days of life for 18 months and was able to remain virally suppressed for 27 months in the absence of ART [23]. Similarly, ‘post-treatment controllers’ from the VISCONTI cohort of adults started ART within the first 2 months of infection and were able to control viraemia without ART for more than 5 years [24,25]. The mechanisms by which early ART can lead to natural viral control in a subset of individuals are still under investigation. Although the mechanisms are likely to differ between individuals who are able to control replication-competent virus for different periods of time (‘Boston patients’ [26], Mississippi child [23], post-treatment controllers [25], elite controllers [27]), a reduced frequency of infected cells is common to all. This reinforces the clinical relevance of early ART interventions, which greatly limit the size of the reservoir, in curative strategies.
Where is the reservoir maintained?
Cellular reservoirs
Although alternative reservoirs may contribute to HIV persistence (detailed below), CD4+ T cells represent the best-characterised reservoir for HIV in virally suppressed subjects on long-term ART [3,4,28,29]. CD4+ T cells can be further subdivided into subsets according to their memory status or their effector functions upon stimulation (Figure 2).
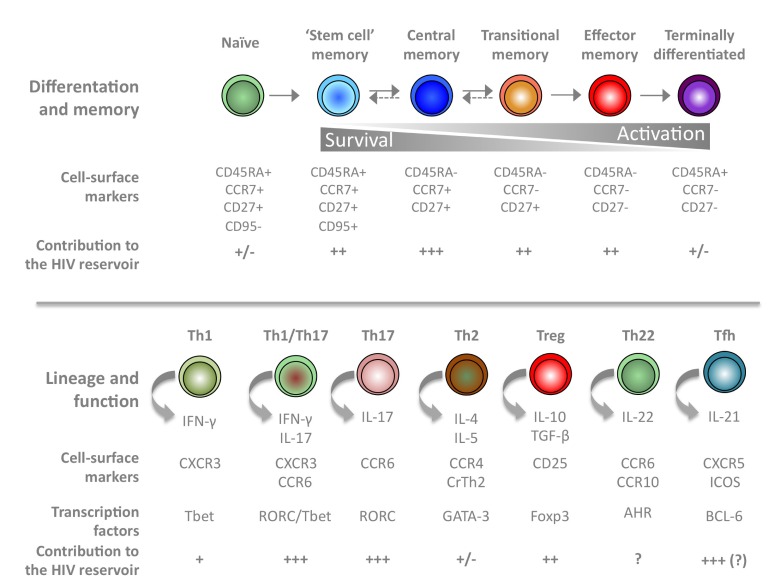
Figure 2.
Contribution of CD4+ T cell subsets to the HIV reservoir during ART. CD4+ T cell subsets can be classified according to their differentiation and memory status (top) or to their effector functions (bottom). Cell-surface markers and the production of specific cytokines can be used to identify each individual subset. The relative contribution of each subset to the HIV reservoir is indicated.
(Adapted from Geginat et al. Semin Immunol 2013; 25: 252–262; Geginat et al. Front Immunol 2014; 16 Dec 2014.)
Within the CD4 compartment, naïve cells and recent thymic emigrants can carry HIV DNA and replication-competent HIV [30–32], but their frequency of infection is usually much lower than that of memory CD4+ T cells [21,29,33,34]. Within the memory compartment, central (TCM), transitional (TTM) and effector memory (TEM) cells are the three major reservoirs for HIV in individuals on suppressive therapy and harbour replication-competent virus [29], although their contribution to the functional reservoir may vary [35]. In addition, two groups have recently shown that the recently identified CD4+ T memory stem cells (TSCM) harbour high per-cell levels of HIV-1 DNA and make increasing contributions to the total reservoir measured by HIV DNA over time [36,37].
As an alternative to the CD4+ T cell subsets that distinguish different stages of T cell differentiation (TCM, TTM and TEM), the CD4 compartment can be divided into subsets that demarcate different functional programs and homing capacities including Th1, Th2, Th17, regulatory T cells (Tregs) and follicular T helper cells (Tfh). It is well established that the effector function of CD4+ T cells, which is based on the expression of particular transcription factors, chemokine receptors and cytokine secretion upon stimulation, influence the capacity of this particular cell to serve as a long-term reservoir for HIV during ART [38,39]. In particular, regulatory Tregs [40] and Tfh cells [41] may represent preferential cellular reservoirs for HIV in virally suppressed individuals. CD4+ T cells expressing CCR6, a marker of Th17 cells with homing capacity to the gut, are highly sensitive to HIV infection [42–44]. This could be attributed to the intrinsic nature of Th17 cells rather than to their homing potential, since CD4+ T cells expressing the integrin beta 7, another marker for homing potential, do not display this increased sensitivity to HIV infection [38]. Indeed, the majority of HIV type 1 DNA in circulating CD4+ T lymphocytes is present in non-gut-homing resting memory CD4+ T cells [45].
In addition to CD4+ T cells, non-conventional cellular HIV reservoirs have been described. They include CD8+ T cells [46,47], the double negative CD3+CD4-CD8-subset [48,49] and cells from the myeloid lineage including circulating monocytes [50–53]. In the NHP model of SIV infection, myeloid cells containing viral DNA show evidence of T cell phagocytosis in vivo, suggesting that their viral DNA may be attributed to phagocytosis of infected T cells [54], questioning the role of myeloid cells as a major source of SIV in vivo. The contribution of these potential alternative reservoirs to HIV persistence in humans is difficult to assess, as the majority of these cells may reside in tissues that are not easily accessible. Furthermore, obtaining sorted myeloid cells with sufficient yield and purity from these tissues is very challenging. Nevertheless, in HIV-infected humans, a few lines of evidence suggest that cells from the myeloid lineage could serve as a long-term reservoir for the virus in mucosal tissues such as the lung [55,56] and duodenum [57]. Of note, these studies usually only demonstrate the presence of viral nucleic acids or viral proteins in myeloid cells isolated from blood and tissues. Whether replication-competent virus can persist in these cells after prolonged ART is largely unknown.
Anatomical reservoirs
As mentioned above, HIV primarily infects CD4+ T cells and cells from the myeloid lineage. Although these cells are found in the circulating blood, the bulk of the cellular targets for HIV primarily reside in lymphoid tissues [58]. Most of the studies aimed at measuring viral persistence (both residual viral replication and latency) have been conducted in the peripheral blood, although it is clear that tissue reservoirs such as the gut and lymph nodes are important sites for HIV persistence [59,60]. Studies in mice have revealed the existence of tissue-resident memory T cells that play an important role in protective immunity to site-specific pathogens and mucosal sites, such as lung and intestine, contain tissue-retained memory populations that do not recirculate [61,62]. In humans, a recent study conducted in blood, lymphoid and mucosal tissues obtained from organ donors revealed that the subset composition and phenotype of peripheral blood T cells does not reflect that of spleen, lymph nodes or mucosal tissues, suggesting that blood is a distinct compartment [63]. This reinforces the importance of conducting studies aimed at understanding the mechanism of HIV persistence and quantifying persistent HIV directly in tissue samples because these reservoirs may not be reflected in the circulating blood. Most of the studies conducted in tissue reservoirs for HIV have used PCR-based assays to assess the levels of HIV persistence, as recovering replication-competent HIV from these sites is technically challenging.
As mentioned above, T cell homeostasis driven by cytokine or T cell receptor-mediated signals in Tcell subsets varies with their differentiation stage and their tissue localisation, and cannot be inferred from blood [64]. TEM CD4+ Tcells producing IL-2 predominate in mucosal tissues and accumulate as TCM cells in lymphoid tissue [63]. In the gut, high frequencies of activated cells [63] as well as persistent elevated levels of inflammation may favour residual viral replication even after prolonged ART. Conversely, the high frequency of the relatively quiescent TCM cells in lymph nodes [63,65], which are in constant interactions with stromal cells and antigen-presenting cells to ensure the homeostasis of the CD4 compartment, may promote the survival of latently infected cells in this site. Most HIV DNA and RNA in the blood is found in TCM cells, whereas in ileum and rectum, most HIV DNA and RNA was found in TEM cells [66]. The characterisation of the mechanisms of HIV persistence in these anatomical reservoirs will require the use of assays that can distinguish residual viral replication from latency and sorting procedures that will ensure high levels of purity of the cell population examined in different anatomical sites.
The importance of the gastrointestinal tract as a privileged site for HIV persistence after prolonged ART is still debated. In the blood, the majority of HIV DNA is present in non-gut-homing resting memory CD4+ T cells [45]. Several studies have directly examined the frequency of infection in biopsies obtained from different regions of the gastrointestinal tract. In some studies, infection frequencies measured in the gastrointestinal tract are higher than in the matched blood samples [59,60], whereas other investigators found no significant differences in the frequency of infection of blood and rectal CD4+ T cells [67]. Similar to peripheral blood mononuclear cells (PBMCs), the decay in the frequency of infected cells in the gastrointestinal tract appears to be minimal [68,69] and the frequencies found in the two compartments are well correlated [29,70]. This questions the existence of a compartmentalisation between the blood and gut reservoirs. Compartmentalisation is suggested by the existence of gut-residing CD4+ T cells [63] and the fact that different HIV-1 quasispecies populate different parts of the gut [71]. In addition, several studies suggest that HIV DNA resides in cells other than CD4+ T cells, including myeloid cells [72] and more specifically tissue macrophages [57] in the gastrointestinal tract, although this may partially be attributed to contamination by Tcells [21], at least in some of the samples tested. The contributions of the gut reservoir to viral rebound upon ART interruption is still unclear [73,74]. Of note, the relative contribution of viral latency and residual replication to viral persistence during suppressive ART in the gut are largely unknown, for the technical reasons mentioned above.
Replication-competent virus has been recovered in cells isolated from lymph nodes in virally suppressed individuals [28,41] and non-human primates [75]. The recent identification of follicular T helper cells, which reside in the germinal centres of lymph nodes, as a major compartment for HIV production, and perhaps persistence [41], will most probably refocus attention around these lymphoid organs. Interestingly, a recent study indicates that B cell follicles constitute ‘sanctuaries’ for persistent SIV replication in the presence of potent antiviral CD8 responses, as a result of the relative exclusion of cytotoxic T lymphocytes (CTL) from this site [76]. Moreover, concentrations of antiretroviral drugs are much lower in lymph nodes than in peripheral blood, which correlates with continued virus replication measured by detection of viral RNA in productively infected cells [77]. Therefore, lymph nodes may play a major role in HIV persistence through residual levels of viral replication in Tfh cells. In addition, TCM cells, which represent a preferential cellular subset for latent HIV, are enriched in lymph nodes. Therefore, lymph nodes could contribute to HIV persistence both through residual levels of viral replication and latency.
In addition to the gut and lymph nodes, several other tissue compartments could serve as a long-term reservoir for HIV. While it is still unclear if the brain could serve as a long-lived reservoir for replication-competent HIV during suppressive ART [78], it clearly contains cells with an integrated provirus in untreated HIV-infected individuals [79,80]. The male [81] and female [82,83] genital tracts could also serve as long-lived reservoirs for HIV during ART. Other previously neglected potential reservoirs such as the kidney [84] and the liver, which may play an important role in viral persistence, at least in the humanised mouse model [85], warrant additional studies. In all cases, it would be important to demonstrate that these potential reservoirs fulfil the criteria of a long-lived viral reservoir during ART, including the replication competency of the persistent viral genomes.
How is the reservoir maintained?
Residual viral replication during ART
ART achieves prolonged suppression of viral replication; however, through the use of highly sensitive PCR assays that are able to detect single copies of HIV RNA, residual plasma viraemia has been shown to exist even in patients who have been virally suppressed for prolonged periods of time [18,86,87]. This residual viraemia could originate from reactivation of virus in latently infected memory CD4+ T cells that are undergoing antigenic stimulation. Interestingly, activated CD4+ T cells in the peripheral blood have been shown to spontaneously release viral particles even in the absence of stimuli [88]. Viraemia could also come from productively infected CD4+ T cells sequestered in lymphoid tissues. Low levels of viral replication in lymphoid organs can play a role in HIV persistence via the spread of virus through cell-to-cell contact in the virological synapse [89]. Significantly, residual plasma viraemia in virally suppressed individuals was shown to correlate with the size of the CD4+ T cell reservoir, but not with markers of immune activation, suggesting that immune system activation alone is not responsible for the observed low-level viraemia [90].
Maintenance of latently infected cells
The persistence of HIV in memory CD4+ T cell subsets is ensured by T cell survival and homeostatic proliferation in response to interleukin-7 (IL-7) signalling [29]. IL-7 mediates the proliferation of latently infected CD4+ T cells without disrupting latency [91], and when administered to virally suppressed subjects, induces a modest but significant expansion of the reservoir [92]. In addition to homeostatic proliferation, latently infected CD4+ T cells undergo proliferation in response to antigenic stimulation, as suggested by phylogenetic analyses showing high numbers of identical sequence expansions in virally suppressed individuals [93,94]. Recently, several groups sequenced the integration sites of the HIV genome within the host DNA and found that specific HIV-integration sites are linked to clonal expansion [95–97]. These studies also indicate that integration of the viral DNA into cancer genes contributes to persistent infection [96]. While the majority of these expanded integrated viral genomes is likely to be incompetent for HIV replication [97,98], it is possible that these defective integrants retain the capacity to generate viral RNA and perhaps viral proteins that could contribute to abnormally elevated levels of immune activation in virally suppressed individuals.
Molecular mechanisms of HIV latency
In maintaining the HIV reservoir, latently infected resting memory CD4+ T cells potentially have the greatest clinical significance. Their long life-span ensures that the virus can be maintained in quiescent cells for years. Resting memory CD4+ T cells support a favourable environment for the maintenance of HIV latency, and multiple molecular mechanisms have been proposed to mediate its induction, including the site of viral integration, transcriptional interference, chromatin remodelling, restriction of transcription host factors and requirements for the HIV accessory protein Tat (transactivator of transcription).
In a study of viral integration sites in resting CD4+ T cells from virally suppressed individuals, a surprising 93% of proviruses were found in actively transcribed genes [99]. This site preference is most likely due to interaction of the pre-integration complex with cellular host factors associated with gene transcription. Integration into actively transcribed genes can lead to transcriptional interference caused by the elongating RNA polymerase II complex transcribing through the 5′ LTR. This leads to interference with pre-initiation complex formation and silencing of HIV transcription [100] that could only be partially reversed through cellular activation by TNF-α[101]. Studies in a primary cell-based model of HIV latency have shown that latent proviruses have an orientation bias when compared to productively infected cells within the same model system [102]. These data suggest that mechanistically transcriptional interference is a significant factor in silencing HIV transcription.
Another contribution to HIV latency is establishment of a repressive chromatin environment. Epigenetic modifications alter the physical structure of chromatin and affect transcription levels including CpG methylation and histone methylation. A Jurkat model demonstrated that initiation of HIV latency was associated with CCAAT-box binding transcription factor 1 (CBF-1)-dependent histone deacetylase (HDAC)-1 recruitment to the 5′LTR and histone H3 lysine 9 (H3K9) trimethylation [103,104]. H3K9/27 trimethylation was also shown to be involved in the establishment of latency in primary cells [105]. DNA methylation at CpG islands is a repressive epigenetic modification that can inhibit transcription factor binding and can recruit HDAC-2 for histone deacetylation. The role of DNA methylation in HIV latency, however, is controversial. In studies using Jurkat cell lines or an in vitro primary CD4+ T cell model of latency, densely methylated proviral DNA was associated with a reduced capacity to reverse latency [106]. Subsequent studies using resting CD4+ T cells isolated from a cohort of virally suppressed HIV-infected individuals demonstrated very low levels of CpG dinucleotides within the 5′LTR, suggesting DNA methylation may not have a significant role in the maintenance of HIV latency [107]. In addition, the role of histone modification in HIV latency has also been of interest, and has led to the recent clinical trials using HDAC inhibitors as an eradication strategy [108–110]. Histone deacetylases are a family of enzymes functioning to remove the acetyl groups from lysine residues, one of the signals required for binding of activating transcription factors [111,112]. HDAC activity has been associated with the repression of HIV transcription [113,114]. Among the different classes of HDACs, the class I HDACs, HDAC-1, -2 and -3, are recruited to the HIV-1 LTR in cell-line models of HIV-1 latency [111,113–115]. In support of the role of HDACs in HIV latency, class I HDAC inhibitors have been shown to induce HIV expression in both in vitro cell models of latency and in resting CD4+ T cells from HIV-infected patients [116–120]. Although promising conceptually as an eradication strategy, therapeutically, the results have been mixed. The HDAC inhibitors vorinostat and panobinostat induced a significant increase in cell-associated HIV RNA in the clinical setting [108–110]. However, these drugs had no effect on the size the of the latent HIV reservoir. One of the assumptions made in HIV eradication strategies is that latent provirus reactivation will induce either cell death in the now productively infected cell, or allow it to be recognised by HIV-specific CTL. Of note, a recent study showed that HDAC inhibitors have a negative impact on CTL activities by impairing IFN-© production and their ability to recognise and eliminate HIV-infected target cells in vitro [121]. These data highlight the importance of considering how the anti-latency compounds will influence multiple arms of the immune response to maximise clinical effectiveness.
HIV latency is also affected by the availability of host transcription factors. The specific recruitment of factors such as NF-κB, Sp1 and NFAT are required for HIV transcription. NF-κB binds to the HIV 5′ LTR and initiates pre-initiation complex formation and transcription initiation [113]. Phosphorylation of RNA polymerase II on the serine 5 position in the heptapeptide repeats of its C-terminal domain (CTD) promotes transcription initiation but not transcript elongation. In order for efficient elongation to occur, sustained activation of NF-κB expression is required to lead to the synthesis of the HIV Tat protein. Tat functions to recruit P-TEFb, which results in the phosphorylation of serine 2 of the RNA polymerase II CTD and transcription elongation.
Sequestering of NF-κB and other factors required for transcription initiation, such as NFAT, in the cytoplasm of quiescent CD4+ T cells also contributes to establishment of HIV latency [122,123]. P-TEFb becomes incorporated into an inactive complex with HEXIM and 7SK RNA, which restricts availability of P-TEFb for efficient transcription elongation [124,125]. The sequestering of P-TEFb plays a significant role in limiting Tat synthesis, a critical factor in HIV transcription.
The HIV Tat protein functions in transcriptional elongation. In the absence of Tat, the majority of the RNA polymerase II complexes prematurely terminate transcription near the promoter [126]. Tat binds to the transactivation response element (TAR), a sequence in the 5′-non-coding region of HIV mRNA that forms a stable stem-loop structure. Efficient elongation of HIV transcripts requires the recruitment of a complex of proteins comprising CycT1 and CDK9/P-TEFb [127]. CycT1 induces CDK9/P-TEFb kinase activity to facilitate phosphorylation of a number of proteins within the elongating transcription complex, including the RNA polymerase II CTD [128]. In addition to recruiting factors for transcriptional elongation, Tat also functions to counteract the activities of negative elongation factors such as the RNA-binding NELF complex associated with RNAPII pausing [129,130]. Without efficient elongation, host cell factors such as Setx, Srn2 59–39 exonuclease and microprocessor complexes function to prematurely terminate HIV transcripts [131], setting up a negative feedback loop that further downmodulates HIV transcription and induces latency [132]. Interestingly, two recent studies by the Weinberger group suggested that rather than a limiting factor that unnecessarily restricts viral replication, Tat activity functions as a regulation mechanism that mediates viral latency for HIV survival [133,134]. Using a computer modelling approach, they demonstrated that the state of HIV expression could be separated from the state of activation of the host cell, and that the two processes were in fact independent [133]. They then used a synthetic biology approach to alter either Tat expression or cellular activation and found that Tat expression was the controlling factor of virus expression irrespective of the host cell state. Mathematical modelling was further applied to explain that the function of Tat to control viral expression may have evolved to promote cell survival and, consequently, enhanced virus propagation [134]. After initial entry of the virus at mucosal surfaces, productive infection of all target cells would induce rapid cell death and therefore limit the capability of the virus to efficiently establish infection. Induction of viral latency would promote cell survival and allow the virus to be spread to other sites in the body when those cells migrated to other tissues. Their mathematical model predicted that establishment of latency benefited the virus over continued productive infection, and resulted in higher infection rates. Interestingly, these data would suggest that latency is one of the earliest steps in HIV infection, in order to limit the death of target cells. Although the implication for intervention strategies is that there may not be a clinical intervention early enough that could block the establishment of a latent reservoir, the aim of limiting the size of the reservoir to preserve immune system function and reduce inflammation is well documented.
Conclusion
The interdependency of residual replication, proviral latency and immune dysfunction complicates our understanding of the mechanisms by which HIV persists in virally suppressed individuals. Indeed, the pool of latently infected CD4+ T cells can be replenished by ongoing replication, a phenomenon that is not fully controlled by the host immune response. Recent data also suggest that viral latency (as defined by the lack of virion production) and transcriptional latency (no transcriptional activity of the LTR) may not always overlap. This concept of ‘leaky latency’ is attracting lots of interest, as it does not only question the widely accepted model of latency, but also suggests that the so-called ‘latent reservoir’ may produce low amounts of viral products (RNA and perhaps proteins) that could contribute to the persistent immune dysfunctions seen in virally suppressed individuals.
While the virological mechanisms of HIV persistence have been extensively investigated in many cohorts of HIV-infected individuals receiving suppressive ART, only a few studies have evaluated the antiviral immune response (both innate and adaptive) during ART and how they influence HIV persistence. This neglected area is now getting much more attention, particularly in the context of ‘shock and kill’ strategies to reduce the size of the reservoir, which will most probably require an immune component to be successful.
Acknowledgements
This work was supported by the Delaney AIDS Research Enterprise (DARE) to find a cure (NIH grant number 1U19AI096109) and by the Foundation for AIDS Research (amfAR Research Consortium on HIV Eradication 108928-56-RGRL).
References
- 1. Richman DD, Margolis DM, Delaney M et al. The challenge of finding a cure for HIV infection. Science 2009; 323: 1304– 1307. [DOI] [PubMed] [Google Scholar]
- 2. International AIDS Society Scientific Working Group on HIV Cure, Deeks SG, Autran B et al. Towards an HIV cure: a global scientific strategy. Nat Rev Immunol 2012; 12: 607– 614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wong JK, Hezareh M, Gunthard HF et al. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science 1997; 278: 1291– 1295. [DOI] [PubMed] [Google Scholar]
- 4. Finzi D, Hermankova M, Pierson T et al. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science 1997; 278: 1295– 1300. [DOI] [PubMed] [Google Scholar]
- 5. Chun TW, Stuyver L, Mizell SB et al. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc Natl Acad Sci U S A 1997; 94: 13193– 13197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Eisele E, Siliciano RF.. Redefining the viral reservoirs that prevent HIV-1 eradication. Immunity 2012; 37: 377– 388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Blankson JN, Persaud D, Siliciano RF.. The challenge of viral reservoirs in HIV-1 infection. Ann Rev Med 2002; 53: 557– 593. [DOI] [PubMed] [Google Scholar]
- 8. Chun TW, Engel D, Berrey MM et al. Early establishment of a pool of latently infected, resting CD4(+) T cells during primary HIV-1 infection. Proc Natl Acad Sci U S A 1998; 95: 8869– 8873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Whitney JB, Hill AL, Sanisetty S et al. Rapid seeding of the viral reservoir prior to SIV viraemia in rhesus monkeys. Nature 2014; 512: 74– 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Williams JP, Hurst J, Stohr W et al. HIV-1 DNA predicts disease progression and post-treatment virological control. Elife 2014; 3: e03821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ananworanich J, Schuetz A, Vandergeeten C et al. Impact of multi-targeted antiretroviral treatment on gut T cell depletion and HIV reservoir seeding during acute HIV infection. PloS One 2012; 7: e33948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ananworanich J, Dube K, Chomont N.. How does the timing of antiretroviral therapy initiation in acute infection affect HIV reservoirs? Curr Opin HIV AIDS 2015; 10: 18– 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Buzon MJ, Martin-Gayo E, Pereyra F et al. Long-term antiretroviral treatment initiated in primary HIV-1 infection affects the size, composition and decay kinetics of the reservoir of HIV-1 infected CD4 T cells. J Virol 2014: 10056– 10065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Archin NM, Vaidya NK, Kuruc JD et al. Immediate antiviral therapy appears to restrict resting CD4+ cell HIV-1 infection without accelerating the decay of latent infection. Proc Natl Acad Sci U S A 2012; 109: 9523– 9528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Strain MC, Little SJ, Daar ES et al. Effect of treatment, during primary infection, on establishment and clearance of cellular reservoirs of HIV-1. J Infect Dis 2005; 191: 1410– 1418. [DOI] [PubMed] [Google Scholar]
- 16. Douek DC, Brenchley JM, Betts MR et al. HIV preferentially infects HIV-specific CD4+ T cells. Nature 2002; 417: 95– 98. [DOI] [PubMed] [Google Scholar]
- 17. Demoustier A, Gubler B, Lambotte O et al. In patients on prolonged HAART, a significant pool of HIV infected CD4 T cells are HIV-specific. AIDS 2002; 16: 1749– 1754. [DOI] [PubMed] [Google Scholar]
- 18. Maldarelli F, Palmer S, King MS et al. ART suppresses plasma HIV-1 RNA to a stable set point predicted by pretherapy viremia. PLoS Pathog 2007; 3: e46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Swiggard WJ, Baytop C, Yu JJ et al. Human immunodeficiency virus type 1 can establish latent infection in resting CD4+ T cells in the absence of activating stimuli. J Virol 2005; 79: 14179– 14188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Saleh S, Solomon A, Wightman F et al. CCR7 ligands CCL19 and CCL21 increase permissiveness of resting memory CD4+ T cells to HIV-1 infection: a novel model of HIV-1 latency. Blood 2007; 110: 4161– 4164. [DOI] [PubMed] [Google Scholar]
- 21. Josefsson L, Stockenstrom S, Faria NR et al. The HIV-1 reservoir in eight patients on long-term suppressive antiretroviral therapy is stable with few genetic changes over time. Proc Natl Acad Sci U S A 2013; 110: E4987– 4996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Deng K, Pertea M, Rongvaux A et al. Broad CTL response is required to clear latent HIV-1 due to dominance of escape mutations. Nature 2015; 517: 381– 385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Persaud D, Gay H, Ziemniak C et al. Absence of detectable HIV-1 viremia after treatment cessation in an infant. N Engl J Med 2013; 369: 1828– 1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hocqueloux L, Prazuck T, Avettand-Fenoel V et al. Long-term immunovirologic control following antiretroviral therapy interruption in patients treated at the time of primary HIV-1 infection. AIDS 2010; 24: 1598– 1601. [DOI] [PubMed] [Google Scholar]
- 25. Saez-Cirion A, Bacchus C, Hocqueloux L et al. Post-Treatment HIV-1 controllers with a long-term virological remission after the interruption of early initiated antiretroviral therapy ANRS VISCONTI Study. PLoS Pathog 2013; 9: e1003211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Henrich TJ, Hu Z, Li JZ et al. Long-term reduction in peripheral blood HIV type 1 reservoirs following reduced-intensity conditioning allogeneic stem cell transplantation. J Infect Dis 2013; 207: 1694– 1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Graf EH, Mexas AM, Yu JJ et al. Elite suppressors harbor low levels of integrated HIV DNA and high levels of 2-LTR circular HIV DNA compared to HIV+ patients on and off HAART. PLoS Pathog 2011; 7: e1001300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chun TW, Carruth L, Finzi D et al. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature 1997; 387: 183– 188. [DOI] [PubMed] [Google Scholar]
- 29. Chomont N, El-Far M, Ancuta P et al. HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nat Med 2009; 15: 893– 900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wightman F, Solomon A, Khoury G et al. Both CD31(+) and CD31 naive CD4(+) T cells are persistent HIV type 1-infected reservoirs in individuals receiving antiretroviral therapy. J Infect Dis 2010; 202: 1738– 1748. [DOI] [PubMed] [Google Scholar]
- 31. Fabre-Mersseman V, Dutrieux J, Louise A et al. CD4(+) recent thymic emigrants are infected by HIV invivo, implication for pathogenesis. AIDS 2011; 25: 1153– 1162. [DOI] [PubMed] [Google Scholar]
- 32. Bacchus C, Cheret A, Avettand-Fenoel V et al. A single HIV-1 cluster and a skewed immune homeostasis drive the early spread of HIV among resting CD4+ cell subsets within one month post-infection. PloS One 2013; 8: e64219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Brenchley JM, Hill BJ, Ambrozak DR et al. T-cell subsets that harbor human immunodeficiency virus (HIV) in vivo: implications for HIV pathogenesis. J Virol 2004; 78: 1160– 1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ostrowski MA, Chun TW, Justement SJ et al. Both memory and CD45RA+/CD62L+ naive CD4(+) T cells are infected in human immunodeficiency virus type 1-infected individuals. J Virol 1999; 73: 6430– 6435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Soriano-Sarabia N, Bateson RE, Dahl NP et al. Quantitation of replication-competent HIV-1 in populations of resting CD4+ T cells. J Virol 2014; 88: 14070– 14077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Buzon MJ, Sun H, Li C et al. HIV-1 persistence in CD4(+) T cells with stem cell-like properties. Nat Med 2014; 20: 139– 142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jaafoura S, de Goer de Herve MG, Hernandez-Vargas EA et al. Progressive contraction of the latent HIV reservoir around a core of less-differentiated CD4(+) memory T Cells. Nat Commun 2014; 5: 5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Monteiro P, Gosselin A, Wacleche VS et al. Memory CCR6+CD4+ T cells are preferential targets for productive HIV type 1 infection regardless of their expression of integrin beta7. J Immunol 2011; 186: 4618– 4630. [DOI] [PubMed] [Google Scholar]
- 39. Bernier A, Cleret-Buhot A, Zhang Y et al. Transcriptional profiling reveals molecular signatures associated with HIV permissiveness in Th1Th17 cells and identifies peroxisome proliferator-activated receptor gamma as an intrinsic negative regulator of viral replication. Retrovirology 2013; 10: 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tran TA, de Goer de Herve MG, Hendel-Chavez H et al. Resting regulatory CD4 T cells: a site of HIV persistence in patients on long-term effective antiretroviral therapy. PloS One 2008; 3: e3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Perreau M, Savoye AL, De Crignis E et al. Follicular helper T cells serve as the major CD4 T cell compartment for HIV-1 infection, replication, and production. J Exp Med 2013; 210: 143– 156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. El Hed A, Khaitan A, Kozhaya L et al. Susceptibility of human Th17 cells to human immunodeficiency virus and their perturbation during infection. J Infect Dis 2010; 201: 843– 854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gosselin A, Monteiro P, Chomont N et al. Peripheral blood CCR4+CCR6+ and CXCR3+CCR6+CD4+ T cells are highly permissive to HIV-1 infection. J Immunol 2010; 184: 1604– 1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Alvarez Y, Tuen M, Shen G et al. Preferential HIV infection of CCR6+ Th17 cells is associated with higher levels of virus receptor expression and lack of CCR5 ligands. J Virol 2013; 87: 10843– 10854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. McBride K, Xu Y, Bailey M et al. The majority of HIV type 1 DNA in circulating CD4+ T lymphocytes is present in non-gut-homing resting memory CD4+ T cells. AIDS Res Hum Retroviruses 2013; 29: 1330– 1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Livingstone WJ, Moore M, Innes D et al. Frequent infection of peripheral blood CD8-positive T-lymphocytes with HIV-1. Edinburgh Heterosexual Transmission Study Group. Lancet 1996; 348: 649– 654. [DOI] [PubMed] [Google Scholar]
- 47. De Maria A, Pantaleo G, Schnittman SM et al. Infection of CD8+ T lymphocytes with HIV. Requirement for interaction with infected CD4+ cells and induction of infectious virus from chronically infected CD8+ cells. J Immunol 1991; 146: 2220– 2226. [PubMed] [Google Scholar]
- 48. Cheney KM, Kumar R, Purins A et al. HIV type 1 persistence in CD4- /CD8- double negative T cells from patients on antiretroviral therapy. AIDS Res Hum Retroviruses 2006; 22: 66– 75. [DOI] [PubMed] [Google Scholar]
- 49. Kaiser P, Joos B, Niederost B et al. Productive human immunodeficiency virus type 1 infection in peripheral blood predominantly takes place in CD4/CD8 double-negative T lymphocytes. J Virol 2007; 81: 9693– 9706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Delobel P, Sandres-Saune K, Cazabat M et al. Persistence of distinct HIV-1 populations in blood monocytes and naive and memory CD4 T cells during prolonged suppressive HAART. AIDS 2005; 19: 1739– 1750. [DOI] [PubMed] [Google Scholar]
- 51. Gibellini D, Borderi M, De Crignis E et al. HIV-1 DNA load analysis in peripheral blood lymphocytes and monocytes from naive and HAART-treated individuals. J Infect 2008; 56: 219– 225. [DOI] [PubMed] [Google Scholar]
- 52. Zhu T, Muthui D, Holte S et al. Evidence for human immunodeficiency virus type 1 replication in vivo in CD14(+) monocytes and its potential role as a source of virus in patients on highly active antiretroviral therapy. J Virol 2002; 76: 707– 716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lambotte O, Taoufik Y, Goer MG et al. Detection of infectious HIV in circulating monocytes from patients on prolonged highly active antiretroviral therapy. J Acquir Immune Defic Syndr 2000; 23: 114– 119. [DOI] [PubMed] [Google Scholar]
- 54. Calantone N, Wu F, Klase Z et al. Tissue myeloid cells in SIV-infected primates acquire viral DNA through phagocytosis of infected T cells. Immunity 2014; 41: 493– 502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Cribbs SK, Lennox J, Caliendo AM et al. Healthy HIV-1-infected individuals on highly active antiretroviral therapy harbor HIV-1 in their alveolar macrophages. AIDS Res Hum Retroviruses 2015; 31: 64– 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Jambo KC, Banda DH, Kankwatira AM et al. Small alveolar macrophages are infected preferentially by HIV and exhibit impaired phagocytic function. Mucosal Immunol 2014; 7: 1116– 1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zalar A, Figueroa MI, Ruibal-Ares B et al. Macrophage HIV-1 infection in duodenal tissue of patients on long term HAART. Antiviral Res 2010; 87: 269– 271. [DOI] [PubMed] [Google Scholar]
- 58. Ganusov VV, De Boer RJ.. Do most lymphocytes in humans really reside in the gut? Trends Immunol 2007; 28: 514– 518. [DOI] [PubMed] [Google Scholar]
- 59. Chun TW, Nickle DC, Justement JS et al. Persistence of HIV in gut-associated lymphoid tissue despite long-term antiretroviral therapy. J Infect Dis 2008; 197: 714– 720. [DOI] [PubMed] [Google Scholar]
- 60. Yukl SA, Gianella S, Sinclair E et al. Differences in HIV burden and immune activation within the gut of HIV-positive patients receiving suppressive antiretroviral therapy. J Infect Dis 2010; 202: 1553– 1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Klonowski KD, Williams KJ, Marzo AL et al. Dynamics of blood-borne CD8 memory T cell migration in vivo. Immunity 2004; 20: 551– 562. [DOI] [PubMed] [Google Scholar]
- 62. Teijaro JR, Turner D, Pham Q et al. Cutting edge: Tissue-retentive lung memory CD4 T cells mediate optimal protection to respiratory virus infection. J Immunol 2011; 187: 5510– 5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Sathaliyawala T, Kubota M, Yudanin N et al. Distribution and compartmentalization of human circulating and tissue-resident memory T cell subsets. Immunity 2013; 38: 187– 197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Thome JJ, Yudanin N, Ohmura Y et al. Spatial map of human T cell compartmentalization and maintenance over decades of life. Cell 2014; 159: 814– 828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Sallusto F, Lenig D, Forster R et al. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature 1999; 401: 708– 712. [DOI] [PubMed] [Google Scholar]
- 66. Yukl SA, Shergill AK, Ho T et al. The distribution of HIV DNA and RNA in cell subsets differs in gut and blood of HIV-positive patients on ART: implications for viral persistence. J Infect Dis 2013; 208: 1212– 1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Descours B, Lambert-Niclot S, Mory B et al. Direct quantification of cell associated HIV-DNA in isolated rectal and blood memory CD4 T cells revealed their similar and low infection levels in long-term treated patients. J Acquir Immune Defic Syndr 2013; 62: 255– 259. [DOI] [PubMed] [Google Scholar]
- 68. Poles MA, Boscardin WJ, Elliott J et al. Lack of decay of HIV-1 in gut-associated lymphoid tissue reservoirs in maximally suppressed individuals. J Acquir Immune Defic Syndr 2006; 43: 65– 68. [DOI] [PubMed] [Google Scholar]
- 69. Belmonte L, Olmos M, Fanin A et al. The intestinal mucosa as a reservoir of HIV-1 infection after successful HAART. AIDS 2007; 21: 2106– 2108. [DOI] [PubMed] [Google Scholar]
- 70. Avettand-Fenoel V, Prazuck T, Hocqueloux L et al. HIV-DNA in rectal cells is well correlated with HIV-DNA in blood in different groups of patients, including long-term non-progressors. AIDS 2008; 22: 1880– 1882. [DOI] [PubMed] [Google Scholar]
- 71. Marle G, Gill MJ, Kolodka D et al. Compartmentalization of the gut viral reservoir in HIV-1 infected patients. Retrovirology 2007; 4: 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Yukl SA, Sinclair E, Somsouk M et al. A comparison of methods for measuring rectal HIV levels suggests that HIV DNA resides in cells other than CD4+ T cells, including myeloid cells. AIDS 2014; 28: 439– 442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Lerner P, Guadalupe M, Donovan R et al. The gut mucosal viral reservoir in HIV-infected patients is not the major source of rebound plasma viremia following interruption of highly active antiretroviral therapy. J Virol 2011; 85: 4772– 4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Rothenberger MK, Keele BF, Wietgrefe SW et al. Large number of rebounding/founder HIV variants emerge from multifocal infection in lymphatic tissues after treatment interruption. Proc Natl Acad Sci U S A 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. North TW, Higgins J, Deere JD et al. Viral sanctuaries during highly active antiretroviral therapy in a nonhuman primate model for AIDS. J Virol 2010; 84: 2913– 2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Fukazawa Y, Lum R, Okoye AA et al. B cell follicle sanctuary permits persistent productive simian immunodeficiency virus infection in elite controllers. Nat Med 2015; 21: 132– 139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Fletcher CV, Staskus K, Wietgrefe SW et al. Persistent HIV-1 replication is associated with lower antiretroviral drug concentrations in lymphatic tissues. Proc Natl Acad Sci U S A 2014; 111: 2307– 2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Gray LR, Roche M, Flynn JK et al. Is the central nervous system a reservoir of HIV-1? Curr Opin HIV AIDS 2014; 9: 552– 558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Churchill MJ, Gorry PR, Cowley D et al. Use of laser capture microdissection to detect integrated HIV-1 DNA in macrophages and astrocytes from autopsy brain tissues. J Neurovirol 2006; 12: 146– 152. [DOI] [PubMed] [Google Scholar]
- 80. Thompson KA, Cherry CL, Bell JE, McLean CA.. Brain cell reservoirs of latent virus in presymptomatic HIV-infected individuals. Am J Pathol 2011; 179: 1623– 1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Deleage C, Moreau M, Rioux-Leclercq N et al. Human immunodeficiency virus infects human seminal vesicles in vitro and in vivo. Am J Pathol 2011; 179: 2397– 2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Launay O, Tod M, Tschope I et al. Residual HIV-1 RNA and HIV-1 DNA production in the genital tract reservoir of women treated with HAART: the prospective ANRS EP24 GYNODYN study. Antivir Ther 2011; 16: 843– 852. [DOI] [PubMed] [Google Scholar]
- 83. Fiscus SA, Cu-Uvin S, Eshete AT et al. Changes in HIV-1 subtypes B and C genital tract RNA in women and men after initiation of antiretroviral therapy. Clin Infect Dis 2013; 57: 290– 297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Canaud G, Dejucq-Rainsford N, Avettand-Fenoel V et al. The kidney as a reservoir for HIV-1 after renal transplantation. J Am Soc Nephrol 2014; 25: 407– 419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Marsden MD, Kovochich M, Suree N et al. HIV latency in the humanized BLT mouse. J Virol 2012; 86: 339– 347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Palmer S, Wiegand AP, Maldarelli F et al. New real-time reverse transcriptase-initiated PCR assay with single-copy sensitivity for human immunodeficiency virus type 1 RNA in plasma. J Clin Microbiol 2003; 41: 4531– 4536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Palmer S, Maldarelli F, Wiegand A et al. Low-level viremia persists for at least 7 years in patients on suppressive antiretroviral therapy. Proc Natl Acad Sci U S A 2008; 105: 3879– 3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Chun TW, Nickle DC, Justement JS et al. HIV-infected individuals receiving effective antiviral therapy for extended periods of time continually replenish their viral reservoir. J Clin Invest 2005; 115: 3250– 3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Kulpa DA, Brehm JH, Fromentin R et al. The immunological synapse: the gateway to the HIV reservoir. Immunol Rev 2013; 254: 305– 325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Chun TW, Murray D, Justement JS et al. Relationship between residual plasma viremia and the size of HIV proviral DNA reservoirs in infected individuals receiving effective antiretroviral therapy. J Infect Dis 2011; 204: 135– 138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Bosque A, Famiglietti M, Weyrich AS et al. Homeostatic proliferation fails to efficiently reactivate HIV-1 latently infected central memory CD4+ T cells. PLoS Pathog 2011; 7: e1002288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Vandergeeten C, Fromentin R, DaFonseca S et al. Interleukin-7 promotes HIV persistence during antiretroviral therapy. Blood 2013; 121: 4321– 4329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Stockenstrom S, Odevall L, Lee E et al. Longitudinal genetic characterization reveals that cell proliferation maintains persistent HIV-1 during effective HIV therapy. J Infect Dis 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Wagner TA, McKernan JL, Tobin NH et al. An increasing proportion of monotypic HIV-1 DNA sequences during antiretroviral treatment suggests proliferation of HIV-infected cells. J Virol 2013; 87: 1770– 1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Maldarelli F, Wu X, Su L et al. HIV latency. Specific HIV integration sites are linked to clonal expansion and persistence of infected cells. Science 2014; 345: 179– 183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Wagner TA, McLaughlin S, Garg K et al. HIV latency. Proliferation of cells with HIV integrated into cancer genes contributes to persistent infection. Science 2014; 345: 570– 573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Cohn LB, Silva IT, Oliveira TY et al. HIV-1 integration landscape during latent and active infection. Cell 2015; 160: 420– 432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Imamichi H, Natarajan V, Adelsberger JW et al. Lifespan of effector memory CD4+ T cells determined by replication-incompetent integrated HIV-1 provirus. AIDS 2014; 28: 1091– 1099. [DOI] [PubMed] [Google Scholar]
- 99. Han Y, Lassen K, Monie D et al. Resting CD4+ T cells from human immunodeficiency virus type 1 (HIV-1)-infected individuals carry integrated HIV-1 genomes within actively transcribed host genes. J Virol 2004; 78: 6122– 6133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Lenasi T, Contreras X, Peterlin BM.. Transcriptional interference antagonizes proviral gene expression to promote HIV latency. Cell Host Microbe 2008; 4: 123– 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Han Y, Lin YB, An W et al. Orientation-dependent regulation of integrated HIV-1 expression by host gene transcriptional readthrough. Cell Host Microbe 2008; 4: 134– 146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Shan L, Yang HC, Rabi SA et al. Influence of host gene transcription level and orientation on HIV-1 latency in a primary-cell model. J Virol 2011; 85: 5384– 5393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Tyagi M, Karn J.. CBF-1 promotes transcriptional silencing during the establishment of HIV-1 latency. EMBO J 2007; 26: 4985– 4995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Pearson R, Kim YK, Hokello J et al. Epigenetic silencing of human immunodeficiency virus (HIV) transcription by formation of restrictive chromatin structures at the viral long terminal repeat drives the progressive entry of HIV into latency. J Virol 2008; 82: 12291– 12303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Tyagi M, Pearson RJ, Karn J.. Establishment of HIV latency in primary CD4+ cells is due to epigenetic transcriptional silencing and P-TEFb restriction. J Virol 2010; 84: 6425– 6437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Blazkova J, Trejbalova K, Gondois-Rey F et al. CpG methylation controls reactivation of HIV from latency. PLoS Pathog 2009; 5: e1000554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Blazkova J, Murray D, Justement JS et al. Paucity of HIV DNA methylation in latently infected, resting CD4+ T cells from infected individuals receiving antiretroviral therapy. J Virol 2012; 86: 5390– 5392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Archin NM, Liberty AL, Kashuba AD et al. Administration of vorinostat disrupts HIV-1 latency in patients on antiretroviral therapy. Nature 2012; 487: 482– 485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Elliott JH, Wightman F, Solomon A et al. Activation of HIV transcription with short-course vorinostat in HIV-infected patients on suppressive antiretroviral therapy. PLoS Pathog 2014; 10: e1004473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Rasmussen TA, Tolstrup M, Brinkman CR et al. Panobinostat, a histone deacetylase inhibitor, for latent- virus reactivation in HIV-infected patients on suppressive antiretroviral therapy: a phase 1/2, single group, clinical trial. Lancet HIV 2014; 1: 13– 21. [DOI] [PubMed] [Google Scholar]
- 111. Marban C, Suzanne S, Dequiedt F et al. Recruitment of chromatin-modifying enzymes by CTIP2 promotes HIV-1 transcriptional silencing. EMBO J 2007; 26: 412– 423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Coull JJ, Romerio F, Sun JM et al. The human factors YY1 and LSF repress the human immunodeficiency virus type 1 long terminal repeat via recruitment of histone deacetylase 1. J Virol 2000; 74: 6790– 6799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Williams SA, Chen LF, Kwon H et al. NF-kappaB p50 promotes HIV latency through HDAC recruitment and repression of transcriptional initiation. EMBO J 2006; 25: 139– 149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Jiang G, Espeseth A, Hazuda DJ, Margolis DM.. c-Myc and Sp1 contribute to proviral latency by recruiting histone deacetylase 1 to the human immunodeficiency virus type 1 promoter. J Virol 2007; 81: 10914– 10923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Archin NM, Keedy KS, Espeseth A et al. Expression of latent human immunodeficiency type 1 is induced by novel and selective histone deacetylase inhibitors. AIDS 2009; 23: 1799– 1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Ylisastigui L, Archin NM, Lehrman G et al. Coaxing HIV-1 from resting CD4 T cells: histone deacetylase inhibition allows latent viral expression. AIDS 2004; 18: 1101– 1108. [DOI] [PubMed] [Google Scholar]
- 117. Archin NM, Espeseth A, Parker D et al. Expression of latent HIV induced by the potent HDAC inhibitor suberoylanilide hydroxamic acid. AIDS Res Hum Retroviruses 2009; 25: 207– 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Edelstein LC, Micheva-Viteva S, Phelan BD, Dougherty JP.. Short communication: activation of latent HIV type 1 gene expression by suberoylanilide hydroxamic acid (SAHA), an HDAC inhibitor approved for use to treat cutaneous T cell lymphoma. AIDS Res Hum Retroviruses 2009; 25: 883– 887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Savarino A, Mai A, Norelli S et al. ‘ Shock and kill’ effects of class I-selective histone deacetylase inhibitors in combination with the glutathione synthesis inhibitor buthionine sulfoximine in cell line models for HIV-1 quiescence. Retrovirology 2009; 6: 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Reuse S, Calao M, Kabeya K et al. Synergistic activation of HIV-1 expression by deacetylase inhibitors and prostratin: implications for treatment of latent infection. PloS One 2009; 4: e6093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Jones RB, O’Connor R, Mueller S et al. Histone deacetylase inhibitors impair the elimination of HIV-infected cells by cytotoxic T-lymphocytes. PLoS Pathog 2014; 10: e1004287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Kinoshita S, Su L, Amano M et al. The T cell activation factor NF-ATc positively regulates HIV-1 replication and gene expression in T cells. Immunity 1997; 6: 235– 244. [DOI] [PubMed] [Google Scholar]
- 123. Nabel G, Baltimore D.. An inducible transcription factor activates expression of human immunodeficiency virus in T cells. Nature 1987; 326: 711– 713. [DOI] [PubMed] [Google Scholar]
- 124. Contreras X, Barboric M, Lenasi T, Peterlin BM.. HMBA releases P-TEFb from HEXIM1 and 7SK snRNA via PI3K/Akt and activates HIV transcription. PLoS Pathog 2007; 3: 1459– 1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Zhou Q, Yik JH.. The Yin and Yang of P-TEFb regulation: implications for human immunodeficiency virus gene expression and global control of cell growth and differentiation. Microbiol Mol Biol Rev 2006; 70: 646– 659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Kao SY, Calman AF, Luciw PA, Peterlin BM.. Anti-termination of transcription within the long terminal repeat of HIV-1 by tat gene product. Nature 1987; 330: 489– 493. [DOI] [PubMed] [Google Scholar]
- 127. Zhou Q, Li T, Price DH.. RNA polymerase II elongation control. Ann Rev Biochem 2012; 81: 119– 143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Kim YK, Bourgeois CF, Isel C et al. Phosphorylation of the RNA polymerase II carboxyl-terminal domain by CDK9 is directly responsible for human immunodeficiency virus type 1 Tat-activated transcriptional elongation. Mol Cell Biol 2002; 22: 4622– 4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Jadlowsky JK, Wong JY, Graham AC et al. Negative elongation factor is required for the maintenance of proviral latency but does not induce promoter-proximal pausing of RNA polymerase II on the HIV long terminal repeat. Mol Cell Biol 2014; 34: 1911– 1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Kwak H, Lis JT.. Control of transcriptional elongation. Ann Rev Genet 2013; 47: 483– 508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Contreras X, Benkirane M, Kiernan R.. Premature termination of transcription by RNAP II: the beginning of the end. Transcription 2013; 4: 72– 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Wagschal A, Rousset E, Basavarajaiah P et al. Microprocessor, Setx, Xrn2, and Rrp6 co-operate to induce premature termination of transcription by RNAPII. Cell 2012; 150: 1147– 1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Razooky BS, Pai A, Aull K et al. A Hardwired HIV Latency Program. Cell 2015; 160: 990– 1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Rouzine IM, Weinberger AD, Weinberger LS.. An evolutionary role for HIV latency in enhancing viral transmission. Cell 2015; 160: 1002– 1012. [DOI] [PMC free article] [PubMed] [Google Scholar]