Abstract
Aminoglycosides, amikacin (AK) and kanamycin (KM) are second line anti-tuberculosis drugs used to treat tuberculosis (TB) and resistance to them affects the treatment. Membrane and membrane associated proteins have an anticipated role in biological processes and pathogenesis and are potential targets for the development of new diagnostics/vaccine/therapeutics. In this study we compared membrane and membrane associated proteins of AK and KM resistant and susceptible Mycobacterium tuberculosis isolates by 2DE coupled with MALDI-TOF/TOF-MS and bioinformatic tools. Twelve proteins were found to have increased intensities (PDQuest Advanced Software) in resistant isolates and were identified as ATP synthase subunit alpha (Rv1308), Trigger factor (Rv2462c), Dihydrolipoyl dehydrogenase (Rv0462), Elongation factor Tu (Rv0685), Transcriptional regulator MoxR1(Rv1479), Universal stress protein (Rv2005c), 35kDa hypothetical protein (Rv2744c), Proteasome subunit alpha (Rv2109c), Putative short-chain type dehydrogenase/reductase (Rv0148), Bacterioferritin (Rv1876), Ferritin (Rv3841) and Alpha-crystallin/HspX (Rv2031c). Among these Rv2005c, Rv2744c and Rv0148 are proteins with unknown functions. Docking showed that both drugs bind to the conserved domain (Usp, PspA and SDR domain) of these hypothetical proteins and GPS-PUP predicted potential pupylation sites within them. Increased intensities of these proteins and proteasome subunit alpha might not only be neutralized/modulated the drug molecules but also involved in protein turnover to overcome the AK and KM resistance. Besides that Rv1876, Rv3841 and Rv0685 were found to be associated with iron regulation signifying the role of iron in resistance. Further research is needed to explore how these potential protein targets contribute to resistance of AK and KM.
Introduction
Mycobacterium tuberculosis is the etiological factor of tuberculosis (TB), causes significant morbidity and mortality worldwide. In 2013, WHO reported 8.6 million people developed TB and 1.3 million died from the disease [1]. Increasing spreads of multidrug-resistant tuberculosis (MDR-TB) has worsened the situation and treatment of MDR-TB leads to the use of second line drugs. Emergence of extensively drug resistant tuberculosis (XDR-TB) indicates not only search for new diagnostic markers, drugs, amendment in second line treatment regimens but also to explore the unknown mechanisms of resistance in M. tuberculosis for developing novel drug targets. Aminoglycosides, AK and KM are important anti-mycobacterial drugs for category-II TB patients. Category II TB patients include those who had failed previous TB treatment, relapsed after treatment, or defaulted during previous treatment. Cumulative mechanisms associated with resistance to aminoglycosides include majorly mutation in ribosomal protein/16S rRNA [2], cell wall impermeability [3], enzymatic inactivation of drugs [4], trapping of drug [5], decreased inner membrane transport and active efflux pumps [6]. Two-third of M. tuberculosis isolates showed KM and AK resistance due to rrs mutation, however remaining 1/3rd do not have these mutations suggesting the involvement of some other mechanism(s) for resistance. Developments in molecular and cellular biology have imposed doubts on the ability of genetic analysis alone to predict any complex phenotypes. As primarily proteins manifest most of the biological processes, information about the actual state of cell can be obtained by analyzing the protein patterns. 2-DE coupled with MALDI-TOF-MS and bioinformatic tools have now been accepted as major analytical tools for detection, identification and characterization of protein species [7–8]. Most of the published proteomic studies concentrate mainly on soluble proteins and there are few comprehensive reports [9–14] on membrane proteins. The identification and characterization of membrane or membrane associated proteins of M. tuberculosis is important due to their anticipated role in virulence and bacterial-host interactions. Membranes and membrane associated proteins are likely to function as enzymes, receptors, transporters or signal transducers that could be of vital importance to the microbe and hence could qualify as drug targets [15–18]. Comparative proteomic studies addressing whole cell proteins with second line aminoglycosides drug resistance isolates have been reported [8]. However, membrane and membrane associated proteome of aminoglycosides resistant M. tuberculosis isolates have not been addressed. To address this, we analyzed the membranes and membrane associated proteins of AM and KM resistant M. tuberculosis by proteomic and bioinformatic approach. Such information could be helpful for the development of newer diagnostics and therapeutic agents for better treatment particularly drug resistance TB.
Materials and Methods
M. tuberculosis isolates and drug susceptibility testing
Five total suseptible (rifampicin, isoniazid, ethambutol, pyrazinamide, streptomycin, kanamycin and amikacin) and five AK & KM resistant (sensitive to first line drugs) M. tuberculosis isolates were obtained from Mycobacterial Repository Centre of National JALMA Institute for Leprosy and Other Mycobacterial Diseases, Agra, India. Drug susceptibility testing (DST) for all the drugs were performed by LJ proportion [19] and REMA method [20–21]. REMA method uses the oxidation–reduction of colorimetric indicator resazurin for determination of drug resistance and minimal inhibitory concentration (MICs) of antimicrobial agents against M. tuberculosis. Resazurin, which is blue in its oxidized state, turns pink when reduced by viable cells.
Membrane and membrane associated protein fraction preparation
Mycobacterial cell lysate was prepared as described by [8 & 22] with slight modifications. Briefly, cells were suspended in sonication buffer with 1% v/v Triton X–100 and then broken by intermittent sonication at 4°C for 20 min. Homogenate was centrifuged at 12,000 g for 20 min at 4°C. Resulting supernatants were ultracentrifuged at 150,000 x g for 90 min. and the pellet (cell membrane) was collected, washed and dissolved in 2D rehydration buffer. Protein concentrations were estimated by Bradford method [23] using BSA as standard. Protein extractions were performed for three times in biological and technical replicas.
2DE, In gel digestion & MS
IEF & SDS-PAGE were carried out using the published protocol of “in gel rehydration” with slight modifications [8 & 24]. Gel images were analyzed using PDQuest Advanced software version 8.0.0 (BIORAD, Hercules, CA, USA). Protein spots which showed increased intensities with more than 1.5 fold were selected for identification. Equal amount of proteins were loaded in all gels and experiments were repeated in biological and technical replicates at least three times. In-gel digestion of proteins and MALDI-TOF/MS was carried out using published protocol [8 & 25]. Mass spectra of digested proteins were acquired using Autoflex II TOF/TOF 50 (Bruker Daltonik GmbH, Leipzig, Germany).
Validation by MS/MS analysis
Matched precursor peptide ions of identified proteins were selected for subsequent fragmentation using PSD for MS/ MS. Lift_ATT.lift method was open in flex control software; parent peak mass spectrum was acquired by hitting laser for 400–550 shots followed by acquisition of fragments of selected precursor ion for the same no. of shots. Both parent and fragment spectrums were pooled to generate MS/MS spectrum of a particular peptide. MS/MS spectrum was submitted to database using MASCOT wizard described in MS protocol [8]. The same parameters were used for MS/MS search in addition with fragment mass tolerance from 0.2 to 1.0 Da.
Bioinformatic analysis
Protein sequences of selected proteins were retrieved from Tuberculist server http://tuberculist.epfl.ch/ and their probable functions were predicted using published protocol of BLASTp, InterProScan, KEGG, docking and GPS-PUP [26–31].
Results
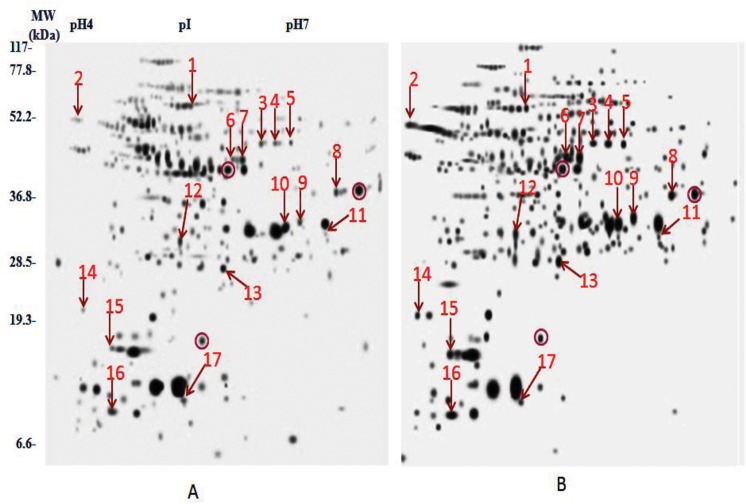
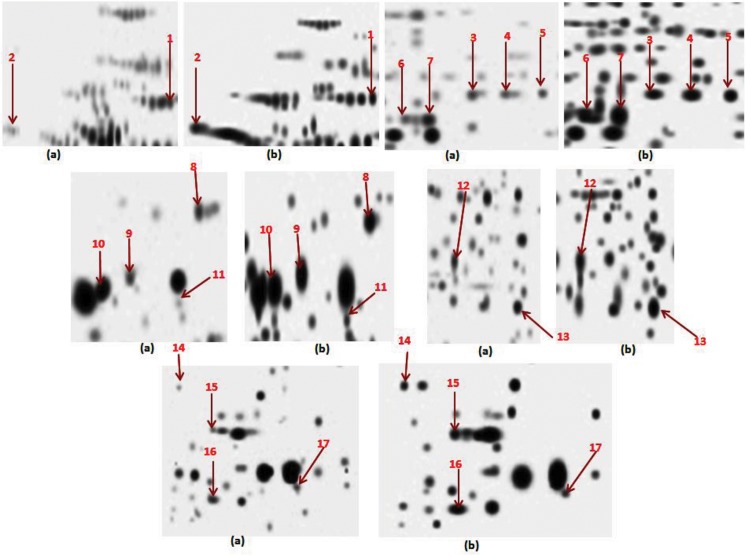
The main aim of the study was to compare the membranes and membrane associated proteins profiles of AM and KM resistant (lacks rrs mutation) with total sensitive isolates. rrs gene encoding 16S rRNA, have been associated with amikacin and kanamycin resistance. Results of DST by REMA methods are represented in Table 1. 2DE profile run in triplicates for all isolates was employed to compare the protein profiles and composite images are shown in Fig 1. Comparison of 2D gels by PDQuest Advanced software revealed seventeen protein spots (identified as twelve protein with its species) with consistently increased intensities in resistant as compared to sensitive isolates (cut limit ≥ 1.5 fold change in spot intensity). Student t-test was used for the statistical analysis by PDQuest Advanced software. The system picks up the spots with differential intensity of significant levels built in the system. To rule out the chance of any artifact, proteins showing equal intensity were considered as internal control (encircled in Fig 1). Protein spots encircled in Fig 1 were taken as internal controls to monitor the equal loading on the gels. Magnified regions of these protein spots are shown in Fig 2. Proteins spots of increased intensities were identified by MALDI-TOF-MS (Table 2) and their identity were further revalidated by MS/MS (Table 3) taking at least three peptides to be matched. Detailed information of MS and MS/MS of all the proteins were shown in supporting files (S1 Text and S2 Text). The identified proteins were ATP synthase subunit alpha (Rv1308), Trigger factor (Rv2462c), Dihydrolipoyl dehydrogenase (Rv0462), Elongation factor Tu (Rv0685), Transcriptional regulator MoxR1(Rv1479), Universal stress protein (Rv2005c), 35kDa hypothetical protein (Rv2744c), Proteasome subunit alpha (Rv2109c), Putative short-chain type dehydrogenase/reductase (Rv0148), Bacterioferritin (Rv1876), Ferritin (Rv3841) and Alpha-crystallin/HspX (Rv2031c). Out of twelve, Rv1308, Rv0462, Rv2109c, Rv0148, Rv1876 and Rv3841 belonged to intermediary metabolism and respiration, Rv2005c and Rv2031c to virulence/detoxification/adaptation, Rv2462c to cell wall and cell processes, Rv2744c to conserved hypothetical, Rv1479 to regulatory proteins and Rv0685 to information pathways categories. The level of difference in protein spot intensity has been represented as densitometric ratio in Table 2. These proteins were also reported in membrane fraction of M. tuberculosis complex by various authors [9–14].
Table 1. Drug susceptibility profile of M. tuberculosis isolates included in this study.
| S. No. | Isolates Code | Drug susceptibility profile by Proportion method | MIC by REMA method | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| SM | RIF | INH | EMB | PZA | KM | SM μg/ml | AK μg/ml | KM μg/ml | ||
| 1 | H37Rv | S | S | S | S | S | S | ≤0.2 | ≤0.025 | 0.05 |
| 2 | S 1 | S | S | S | S | S | S | 2.0 | 0.1 | 0.1 |
| 3 | S 2 | S | S | S | S | S | S | 0.5 | 0.2 | 0.2 |
| 4 | S 3 | S | S | S | S | S | S | ≤0.2 | ≤0.025 | 0.05 |
| 5 | S 4 | S | S | S | S | S | S | 2.0 | 0.2 | 0.1 |
| 6 | S 5 | S | S | S | S | S | S | 1.0 | 0.1 | 0.2 |
| 7 | R 1 | S | S | S | S | S | R | 0.5 | 12 | 16 |
| 8 | R 2 | S | S | S | S | S | R | ≤0.2 | 16 | 32 |
| 9 | R 3 | S | S | S | S | S | R | 1.0 | 16 | 16 |
| 10 | R 4 | S | S | S | S | S | R | 1.0 | 32 | 12 |
| 11 | R 5 | S | S | S | S | S | R | ≤0.2 | 12 | 32 |
S: sensitive; R: resistant; Rifampicin (RIF), Isoniazid (INH), Ethambutol (EMB), Streptomycin (SM), Pyrazinamide (PZA), Amikacin (AK), Kanamycin (KM)
Fig 1. Composite images of 2DE profile M. tuberculosis isolates (a) Total susceptible (b) AM and KM resistant (Encircled spots are taken as internal control).
Fig 2. Magnified regions of 2D gels showing proteins of increased intensity (a) Sensitive (b) Resistant.
Table 2. Details of proteins identified by Mass Spectrometry.
| Spot No. | Accession Number | Protein identified | MASCOT Score | Nominal Mass (Da) | pI | Sequence Coverage % | ORF No. | Densitometric ratio of protein intensity between sensitive and resistant isolates | Functional category * |
|---|---|---|---|---|---|---|---|---|---|
| D1 | P63673 (ATPA_MYCTU) | ATP synthase subunit alpha | 172 | 59252 | 5.03 | 35% | Rv1308 | 1: 1.54 | 1 |
| D 2 | O53189 (TIG_MYCTU) | Trigger factor | 55 | 50586 | 4.43 | 21% | Rv2462c | 1: 1.80 | 2 |
| D 3 | P66004 (DLDH_MYCTU) | Dihydrolipoyl dehydrogenase | 125 | 49208 | 5.53 | 35% | Rv0462 | 1: 1.62 | 1 |
| D 4 | P66004 (DLDH_MYCTU) | Dihydrolipoyl dehydrogenase | 116 | 49208 | 5.53 | 35% | Rv0462 | 1: 1.53 | 1 |
| D 5 | P66004 (DLDH_MYCTU) | Dihydrolipoyl dehydrogenase | 52 | 49208 | 5.53 | 13% | Rv0462 | 1: 1.60 | 1 |
| D 6 | P0A558 (EFTU_MYCTU) | Elongation factor Tu | 193 | 43566 | 5.28 | 60% | Rv0685 | 1: 1.58 | 3 |
| D 7 | P0A558 (EFTU_MYCTU) | Elongation factor Tu | 143 | 43566 | 5.28 | 50% | Rv0685 | 1: 1.73 | 3 |
| D 8 | Q79FN7 (Q79FN7_MYCTU) | Transcriptional regulator MoxR1 | 116 | 40738 | 5.96 | 32% | Rv1479 | 1: 1.52 | 4 |
| D 9 | P64921 (Y2005_MYCTU) | Universal stress protein | 61 | 30966 | 5.53 | 23% | Rv2005c | 1: 1.99 | 5 |
| D 10 | P0C5C4 (35KD_MYCTU) | 35kDa protein | 124 | 29240 | 5.71 | 33% | Rv2744c | 1: 2.00 | 6,2 |
| D11 | P0C5C4 (35KD_MYCTU) | 35kDa protein | 79 | 29240 | 5.71 | 61% | Rv2744c | 1: 1.69 | 6,2 |
| D 12 | O33244 (PSA_MYCTU) | Proteasome subunit alpha | 69 | 26865 | 5.41 | 27% | Rv2109c | 1: 2.09 | 1 |
| D 13 | P96825 (Y0148_MYCTU) | Putative short-chain type dehydrogenase/reductase | 181 | 29760 | 5.26 | 59% | Rv0148 | 1: 1.91 | 1 |
| D14 | P63697 (BFR_MYCTU) | Bacterioferritin | 54 | 18239 | 4.50 | 27% | Rv1876 | 1: 2.70 | 1 |
| D 15 | P96237 (BFRB_MYCTU) | Ferritin | 114 | 20429 | 4.73 | 38% | Rv3841 | 1: 1.83 | 1 |
| D 16 | P0A5B7 (ACR_MYCTU) | Alpha-crystallin | 148 | 16217 | 5.00 | 78% | Rv2031c | 1: 1.64 | 5 |
| D 17 | P0A5B7 (ACR_MYCTU) | Alpha-crystallin | 93 | 16217 | 5.00 | 54% | Rv2031c | 1: 1.98 | 5 |
*Note: 1- intermediary metabolism and respiration, 2- cell wall and cell processes, 3- information pathways, 4- regulatory proteins, 5- virulence, detoxification, adaptation, 6- conserved hypothetical’s
Table 3. MS/MS analysis of identified proteins.
| Spot No. | Peak Mass (Da) | Protein Identified | Nominal Mass | Mascot Score | pI | Sequence of peptides | ORF No. |
|---|---|---|---|---|---|---|---|
| D1 | 894.4477 | ATP synthase subunit alpha | 59252 | 25 | 5.03 | LDLSQYR | Rv1308 |
| 1264.7051 | ATP synthase subunit alpha | 59252 | 18 | 5.03 | VVNPLGQPIDGR | Rv1308 | |
| 1289.6714 | ATP synthase subunit alpha | 59252 | 21 | 5.03 | ASEEEILTEIR | Rv1308 | |
| 1297.7342 | ATP synthase subunit alpha | 59252 | 19 | 5.03 | QGVKEPLQTGIK | Rv1308 | |
| 1313.7349 | ATP synthase subunit alpha | 59252 | 60 | 5.03 | HVLIIFDDLTK | Rv1308 | |
| 1319.7631 | ATP synthase subunit alpha | 59252 | 30 | 5.03 | ALELQAPSVVHR | Rv1308 | |
| 1553.7943 | ATP synthase subunit alpha | 59252 | 48 | 5.03 | EAYPGDVFYLHSR | Rv1308 | |
| 1602.9144 | ATP synthase subunit alpha | 59252 | 65 | 5.03 | TGEVLSVPVGDGFLGR | Rv1308 | |
| 1747.9579 | ATP synthase subunit alpha | 59252 | 52 | 5.03 | ASEEEILTEIRDSQK | Rv1308 | |
| 1886.0888 | ATP synthase subunit alpha | 59252 | 98 | 5.03 | LSDDLGGGSLTGLPIIETK | Rv1308 | |
| 2612.4725 | ATP synthase subunit alpha | 59252 | 51 | 5.03 | GFAATGGGSVVPDEHVEALDEDKLAK | Rv1308 | |
| D2 | 1291.7448 | Trigger factor | 50586 | 25 | 4.43 | NQLPTMFADVR | Rv2462c |
| 1378.8073 | Trigger factor | 50586 | 32 | 4.43 | FNELLVEQGSSR | Rv2462c | |
| 1671.9877 | Trigger factor | 50586 | 81 | 4.43 | EAMLDQIVNDALPSR | Rv2462c | |
| 1801.1078 | Trigger factor | 50586 | 94 | 4.43 | LIAGLDDAVVGLSADESR | Rv2462c | |
| 1903.1486 | Trigger factor | 50586 | 109 | 4.43 | INVEVPFAELEPDFQR | Rv2462c | |
| 2158.3327 | Trigger factor | 50586 | 29 | 4.43 | VRINVEVPFAELEPDFQR | Rv2462c | |
| D3 | 1621.8785 | Dihydrolipoyl dehydrogenase | 49208 | 118 | 5.53 | NYGVDVTIVEFLPR | Rv0462 |
| 1890.9713 | Dihydrolipoyl dehydrogenase | 49208 | 63 | 5.53 | VLQAIGFAPNVEGYGLDK | Rv0462 | |
| 1909.9795 | Dihydrolipoyl dehydrogenase | 49208 | 48 | 5.53 | SIIIAGAGAIGMEFGYVLK | Rv0462 | |
| 1980.9351 | Dihydrolipoyl dehydrogenase | 49208 | 50 | 5.53 | AFGISGEVTFDYGIAYDR | Rv0462 | |
| 2015.0726 | Dihydrolipoyl dehydrogenase | 49208 | 13 | 5.53 | THYDVVVLGAGPGGYVAAIR | Rv0462 | |
| 2276.1980 | Dihydrolipoyl dehydrogenase | 49208 | 100 | 5.53 | LVPGTSLSANVVTYEEQILSR | Rv0462 | |
| 2688.4487 | Dihydrolipoyl dehydrogenase | 49208 | 25 | 5.53 | LGVTILTATKVESIADGGSQVTVTVTK.D | Rv0462 | |
| 2774.4226 | Dihydrolipoyl dehydrogenase | 49208 | 111 | 5.53 | HGELLGGHLVGHDVAELLPELTLAQR | Rv0462 | |
| D4 | 1161.6462 | Dihydrolipoyl dehydrogenase | 49208 | 21 | 5.53 | WDLTASELAR | Rv0462 |
| 1171.6699 | Dihydrolipoyl dehydrogenase | 49208 | 39 | 5.53 | NAELVHIFTK | Rv0462 | |
| 1621.9626 | Dihydrolipoyl dehydrogenase | 49208 | 51 | 5.53 | NYGVDVTIVEFLPR | Rv0462 | |
| 1891.1342 | Dihydrolipoyl dehydrogenase | 49208 | 85 | 5.53 | VLQAIGFAPNVEGYGLDK | Rv0462 | |
| 1981.1184 | Dihydrolipoyl dehydrogenase | 49208 | 14 | 5.53 | AFGISGEVTFDYGIAYDR | Rv0462 | |
| 2015.2435 | Dihydrolipoyl dehydrogenase | 49208 | 15 | 5.53 | THYDVVVLGAGPGGYVAAIR | Rv0462 | |
| D5 | 895.5120 | Dihydrolipoyl dehydrogenase | 49208 | 19 | 5.53 | FPFTANAK | Rv0462 |
| 1161.7045 | Dihydrolipoyl dehydrogenase | 49208 | 28 | 5.53 | WDLTASELAR | Rv0462 | |
| 1170.6847 | Dihydrolipoyl dehydrogenase | 49208 | 20 | 5.53 | AHGVGDPSGFVK | Rv0462 | |
| 1171.7299 | Dihydrolipoyl dehydrogenase | 49208 | 33 | 5.53 | NAELVHIFTK | Rv0462 | |
| 1397.9056 | Dihydrolipoyl dehydrogenase | 49208 | 19 | 5.53 | AAQLGLSTAIVEPK | Rv0462 | |
| D6 | 1404.5958 | Elongation factor Tu | 43566 | 24 | 5.28 | AFDQIDNAPEER | Rv0685 |
| 1413.7572 | Elongation factor Tu | 43566 | 67 | 5.28 | QVGVPYILVALNK | Rv0685 | |
| 1555.7710 | Elongation factor Tu | 43566 | 34 | 5.28 | VLHDKFPDLNETK | Rv0685 | |
| 1701.8430 | Elongation factor Tu | 43566 | 28 | 5.28 | GITINIAHVEYQTDK | Rv0685 | |
| 1801.8729 | Elongation factor Tu | 43566 | 76 | 5.28 | .ELLAAQEFDEDAPVVR | Rv0685 | |
| 2074.9777 | Elongation factor Tu | 43566 | 53 | 5.28 | ADAVDDEELLELVEMEVR | Rv0685 | |
| 2195.1034 | Elongation factor Tu | 43566 | 31 | 5.28 | ETDKPFLMPVEDVFTITGR | Rv0685 | |
| 2356.1634 | Elongation factor Tu | 43566 | 34 | 5.28 | WVASVEELMNAVDESIPDPVR | Rv0685 | |
| D7 | 1404.6006 | Elongation factor Tu | 43566 | 28 | 5.28 | AFDQIDNAPEER | Rv0685 |
| 1413.7786 | Elongation factor Tu | 43566 | 70 | 5.28 | QVGVPYILVALNK | Rv0685 | |
| 1681.8413 | Elongation factor Tu | 43566 | 110 | 5.28 | LLDQGQAGDNVGLLLR | Rv0685 | |
| 1801.8014 | Elongation factor Tu | 43566 | 57 | 5.28 | ELLAAQEFDEDAPVVR | Rv0685 | |
| 2033.8394 | Elongation factor Tu | 43566 | 15 | 5.28 | HTPFFNNYRPQFYFR | Rv0685 | |
| 2074.8391 | Elongation factor Tu | 43566 | 48 | 5.28 | ADAVDDEELLELVEMEVR | Rv0685 | |
| 2194.9570 | Elongation factor Tu | 43566 | 52 | 5.28 | ETDKPFLMPVEDVFTITGR | Rv0685 | |
| 2355.9810 | Elongation factor Tu | 43566 | 40 | 5.28 | WVASVEELMNAVDESIPDPVR | Rv0685 | |
| D8 | 1785.8129 | Transcriptional regulator MoxR1 | 40738 | 27 | 5.96 | IQFTPDLVPTDIIGTR | Rv1479 |
| 1982.8948 | Transcriptional regulator MoxR1 | 40738 | 29 | 5.96 | DYVIPQDVIEVIPDVLR | Rv1479 | |
| 2196.0884 | Transcriptional regulator MoxR1 | 40738 | 43 | 5.96 | GRDYVIPQDVIEVIPDVLR | Rv1479 | |
| 2244.1285 | Transcriptional regulator MoxR1 | 40738 | 94 | 5.96 | LVLTYDALADEISPEIVINR | Rv1479 | |
| 2308.0928 | Transcriptional regulator MoxR1 | 40738 | 37 | 5.96 | LQEIAANNFVHHALVDYVVR | Rv1479 | |
| 2491.0784 | Transcriptional regulator MoxR1 | 40738 | 21 | 5.96 | EEFDTELGPVVANFLLADEINR | Rv1479 | |
| 2832.2880 | Transcriptional regulator MoxR1 | 40738 | 65 | 5.96 | QGREEFDTELGPVVANFLLADEINR | Rv1479 | |
| D9 | 859.4570 | Universal stress protein | 30966 | 21 | 5.53 | LAGWQER | Rv2005c |
| 932.4974 | Universal stress protein | 30966 | 56 | 5.53 | YPDVPVSR | Rv2005c | |
| 1330.8178 | Universal stress protein | 30966 | 11 | 5.53 | GLLGSVSSSLVRR | Rv2005c | |
| 1367.8154 | Universal stress protein | 30966 | 44 | 5.53 | SASAQLVVVGSHGR | Rv2005c | |
| 1737.0408 | Universal stress protein | 30966 | 53 | 5.53 | LAGWQERYPDVPVSR | Rv2005c | |
| 1924.1982 | Universal stress protein | 30966 | 44 | 5.53 | GGLTGMLLGSVSNAVLHAAR | Rv2005c | |
| D10 | 1029.5576 | 35 kDa protein | 29240 | 20 | 5.71 | LLSQLEQAK | Rv2744c |
| 1412.7607 | 35 kDa protein | 29240 | 72 | 5.71 | VQIQQAIEEAQR | Rv2744c | |
| 1424.7377 | 35 kDa protein | 29240 | 13 | 5.71 | TLHDQALSAAAQAK | Rv2744c | |
| 1525.8869 | 35 kDa protein | 29240 | 56 | 5.71 | QLADIEKLQVNVR | Rv2744c | |
| 1615.8335 | 35 kDa protein | 29240 | 10 | 5.71 | QALTLADQATAAGDAAK | Rv2744c | |
| 1822.9247 | 35 kDa protein | 29240 | 132 | 5.71 | YANAIGSAELAESSVQGR | Rv2744c | |
| 2783.3882 | 35 kDa protein | 29240 | 84 | 5.71 | ATEYNNAAEAFAAQLVTAEQSVEDLK | Rv2744c | |
| D11 | 1412.8493 | 35 kDa protein | 29240 | 50 | 5.71 | VQIQQAIEEAQR | Rv2744c |
| 1525.9723 | 35 kDa protein | 29240 | 49 | 5.71 | QLADIEKLQVNVR | Rv2744c | |
| 1823.0114 | 35 kDa protein | 29240 | 21 | 5.71 | YANAIGSAELAESSVQGR | Rv2744c | |
| 1864.0864 | 35 kDa protein | 29240 | 136 | 5.71 | THQALTQQAAQVIGNQR | Rv2744c | |
| D12 | 1054.5026 | Proteasome subunit alpha | 26865 | 25 | 5.41 | FNEFDNLR | Rv2109c |
| 2020.1235 | Proteasome subunit alpha | 26865 | 74 | 5.41 | SVVALAYAGGVLFVAENPSR | Rv2109c | |
| 2219.2512 | Proteasome subunit alpha | 26865 | 24 | 5.41 | AKSVVALAYAGGVLFVAENPSR | Rv2109c | |
| 2924.3203 | Proteasome subunit alpha | 26865 | 29 | 5.41 | AGSADTSGGDQPTLGVASLEVAVLDANRPR | Rv2109c | |
| D13 | 884.4423 | Putative short-chain type dehydrogenase/reductase | 29760 | 13 | 5.26 | AAWPHFR | Rv0148 |
| 1190.5693 | Putative short-chain type dehydrogenase/reductase | 29760 | 27 | 5.26 | WAEITDLSGAK | Rv0148 | |
| 1313.6913 | Putative short-chain type dehydrogenase/reductase | 29760 | 23 | 5.26 | VHLYGGYHVLR | Rv0148 | |
| 1339.6028 | Putative short-chain type dehydrogenase/reductase | 29760 | 53 | 5.26 | MSFENWDAVLK | Rv0148 | |
| 1581.9283 | Putative short-chain type dehydrogenase/reductase | 29760 | 90 | 5.26 | LGLVGLINTLALEGAK | Rv0148 | |
| 1748.8280 | Putative short-chain type dehydrogenase/reductase | 29760 | 14 | 5.26 | DGTGAGSAMADEVVAEIR | Rv0148 | |
| 1921.9303 | Putative short-chain type dehydrogenase/reductase | 29760 | 36 | 5.26 | AVANYDSVATEDGAANIIK | Rv0148 | |
| 2162.1125 | Putative short-chain type dehydrogenase/reductase | 29760 | 102 | 5.26 | EYALTLAGEGASVVVNDLGGAR | Rv0148 | |
| 2388.1836 | Putative short-chain type dehydrogenase/reductase | 29760 | 58 | 5.26 | VALFGNDGANFDKPPSVQDVAAR | Rv0148 | |
| D14 | 1414.6818 | Bacterioferritin | 18239 | 79 | 4.50 | ILLLDGLPNYQR | Rv1876 |
| 1776.6893 | Bacterioferritin | 18239 | 55 | 4.50 | MQDNWGFTELAAHTR | Rv1876 | |
| 1924.8015 | Bacterioferritin | 18239 | 45 | 4.50 | EQFEADLAIEYDVLNR | Rv1876 | |
| D15 | 932.4086 | Ferritin | 20429 | 31 | 4.73 | NQFDRPR | Rv3841 |
| 1084.5530 | Ferritin | 20429 | 54 | 4.73 | VEIPGVDTVR | Rv3841 | |
| 1228.6306 | Ferritin | 20429 | 27 | 4.73 | EALALALDQER | Rv3841 | |
| 1265.5885 | Ferritin | 20429 | 19 | 4.73 | HFYSQAVEER | Rv3841 | |
| 1550.8525 | Ferritin | 20429 | 105 | 4.73 | AGANLFELENFVAR | Rv3841 | |
| 1632.8621 | Ferritin | 20429 | 15 | 4.73 | EVDVAPAASGAPHAAGGR | Rv3841 | |
| D16 | 1095.5822 | Alpha-crystallin | 16217 | 36 | 5.00 | TEQKDFDGR | Rv2031c |
| 1162.6453 | Alpha-crystallin | 16217 | 66 | 5.00 | SEFAYGSFVR | Rv2031c | |
| 1715.0573 | Alpha-crystallin | 16217 | 42 | 5.00 | GILTVSVAVSEGKPTEK | Rv2031c | |
| 1752.8950 | Alpha-crystallin | 16217 | 96 | 5.00 | DFDGRSEFAYGSFVR | Rv2031c | |
| 1869.0448 | Alpha-crystallin | 16217 | 19 | 5.00 | AELPGVDPDKDVDIMVR | Rv2031c | |
| 2037.1053 | Alpha-crystallin | 16217 | 29 | 5.00 | TVSLPVGADEDDIKATYDK | Rv2031c | |
| 2950.6204 | Alpha-crystallin | 16217 | 13 | 5.00 | SLFPEFSELFAAFPSFAGLRPTFDTR | Rv2031c | |
| D17 | 1162.4795 | Alpha-crystallin | 16217 | 65 | 5.00 | SEFAYGSFVR | Rv2031c |
| 1458.6522 | Alpha-crystallin | 16217 | 17 | 5.00 | TVSLPVGADEDDIK | Rv2031c | |
| 1714.8305 | Alpha-crystallin | 16217 | 43 | 5.00 | GILTVSVAVSEGKPTEK | Rv2031c | |
| 1752.6882 | Alpha-crystallin | 16217 | 32 | 5.00 | DFDGRSEFAYGSFVR | Rv2031c | |
| 1868.8202 | Alpha-crystallin | 16217 | 53 | 5.00 | AELPGVDPDKDVDIMVR | Rv2031c | |
| 2036.8814 | Alpha-crystallin | 16217 | 56 | 5.00 | TVSLPVGADEDDIKATYDK | Rv2031c | |
| 2293.0996 | Alpha-crystallin | 16217 | 17 | 5.00 | ATYDKGILTVSVAVSEGKPTEK | Rv2031c |
BLAST and InterProScan analysis
BLASTP analysis was performed for proteins of unknown function. Rv0148 was found to be highly conserved in mycobacterial and bacterial species as putative short chain dehydrogenase/reductase protein. InterProScan analysis of Rv0148 showed motifs (PF00106) from residues 8–183 which provides a signature for short chain dehydrogenase. Rv2005c was found to be highly conserved in mycobacterial and bacterial species as universal stress protein, in some mycobacterial species it appeared as hypothetical protein with unknown function. InterProScan analysis of Rv2005c showed the presence of two signature motifs of Usp domain with amino acid residues from10-148 and 162–293 (PF00582) and three signatures motifs of universal stress protein with amino acid residues from 159–177, 253–265 and 271–293 (PRINTS: PR01438). Rv2744c was found to be conserved alanine rich hypothetical protein, exhibited significant homology with hypothetical and phase shock protein A (pspA) of all M. tuberculosis complex and NTMs. InterProScan analysis of Rv2744c showed the presence of PspA domain with amino acid residues from 3–242 (PF04012).
Multiple Sequence Alignment
Multiple sequence alignment of mtu (M. tuberculosis) proteins was performed for the set of five organism’s mbo (M. bovis), maf (M. africanum), mav (M. avium), mle (M. leprae) and hsa (Homo sapiens) {Table 4}. Results showed that hypothetical proteins (Rv0148, Rv2005c and Rv2744c) exhibited 100% homology (except Rv2744c- 99.60% and 99.30% homology) to their corresponding proteins in M. bovis and M. africanum which are members of tuberculosis complex. In M. avium, > 87% homology has been seen, except Rv2005c (64.40%). Less than 48% of homology has been seen with M. leprae and Homo sapiens to their corresponding proteins as well as with other proteins.
Table 4. Multiple sequence alignment of the hypothetical proteins with defined set of organisms.
| ORF Number | Mbo (M. bovis) | Maf (M. africanum) | Mav (M. avium) | Mle (M. leprae) | Has (Homo. sapiens) |
|---|---|---|---|---|---|
| Rv0148 | 100% | 100% | 87.30% | 23.42% | 47.50% |
| Rv2005c | 100% | 100% | 64.40% | 28.10% | 21.70% |
| Rv2744c | 99.6% | 99.30% | 88.00% | 26.60% | 24.60% |
3D modeling and docking
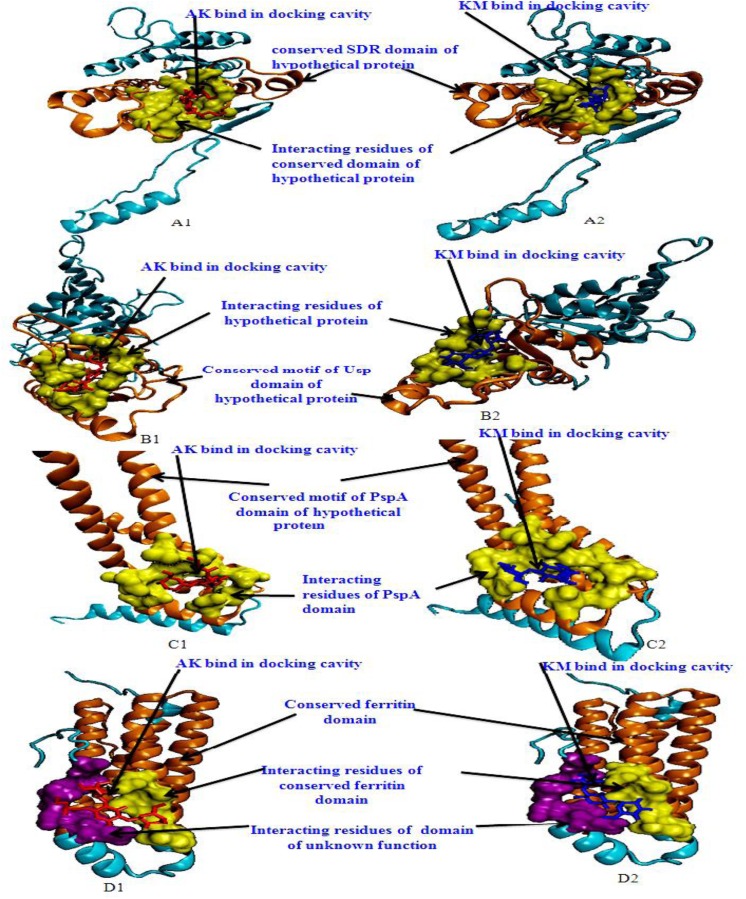
Molecular docking analysis of selected 3D models (showing less than 2% discrepancy from Ramachandran plot) of hypothetical proteins was performed to detect their binding with AK and KM. Parameters used for selection of 3D models and molecular docking are represented in Table 5. Docking of Rv0148 and Rv2005c (Fig 3) showed the interaction of both drugs into the central cavity of conserved motif of SDR domain and Usp domain of hypothetical proteins respectively. Interacting residues were almost common for both drugs, which suggests similar binding site for both. Docking with Rv2744c show that both drugs interact at the similar interacting residue of conserved PspA domain of hypothetical protein. With Rv3841 both drugs interacted with amino acids of conserved ferritin domain as well as domain of unknown function.
Table 5. 3D modeling and docking parameters used for bioinformatic analysis.
| ORF No. | TM-score | RMSD value (Å) | Drug | Global Energy | Attractive Vander wall forces | Repulsive Vander wall forces | ACE | Interacting amino acids | Remarks |
|---|---|---|---|---|---|---|---|---|---|
| Rv0148 | 0.81±0.09 | 4.6±3.0Å | AK | -29.30 | -20.55 | 19.86 | -10.46 | 18,99,123,149,150,152,153,158,161,164,168,194–196,198,200 & 201 | AK binds properly within the central cavity of conserved SDR domain |
| Rv0148 | 0.81±0.09 | 4.6±3.0Å | KM | -45.45 | -22.03 | 10.90 | -14.08 | 99,103,123,149,150–152,164,168,194–196,198,200 & 201 | KM also binds within the central cavity of conserved SDR domain |
| Rv2005c | 0.95±0.05 | 2.8±2.0Å | AK | -32.66 | -20.26 | 8.27 | -06.23 | 15,16,42,43,64,67,71,100,101,120,121 & 130–132 | AK binds in central cavity of conserved motif of Usp domain |
| Rv2005c | 0.95±0.05 | 2.8±2.0Å | KM | -31.64 | -18.15 | 3.76 | -05.29 | 14,15,16,42,43,44,64,67,71,100,120,121 & 122 | KM also binds in central cavity of conserved motif of Usp domain of hypothetical protein |
| Rv2744c | 0.30±0.10 | 15.3±3.4Å | AK | -22.56 | -22.44 | 7.00 | 00.59 | 19,20,22,25,29,36,38,39,40,41 & 44 | AK interact to conserved motif of PspA domain |
| Rv2744c | 0.30±0.10 | 15.3±3.4Å | KM | -25.82 | -16.56 | 2.90 | -03.51 | 19,20,22,25,29,36,38,39,40,41,44 & 230 | KM also interact to conserved motif of PspA domain of hypothetical protein |
| Rv3841 | 0.92±0.06 | 2.3±1.7Å | AK | -26.34 | -16.62 | 4.97 | -7.23 | 134,137,138,141,144,163, 164,165,166,167,168,169& 170 | AK binds to close vicinity of conserved ferritin domain & domain of unknown function |
| Rv3841 | 0.92±0.06 | 2.3±1.7Å | KM | -34.66 | -19.57 | 7.05 | -9.35 | 134,135,137,138,141,144,163,164,165,166,167,168,169 &170 | KM also binds to close vicinity of conserved ferritin domain & domain of unknown function |
Fig 3. 3D model of hypothetical proteins & ferritin showing docking with AK & KM: A1 and A2 shows molecular docking of Rv0148 with AM (red) & KM (blue) respectively, orange color shows SDR domain, yellow color shows interacting residues of SDR domain.
B1 and B2 shows molecular docking of Rv2005c with AM (red) & KM (blue) respectively, orange color shows Usp domain, yellow color shows interacting residues of Usp domain. C1 and C2 shows docking of Rv2744c with AM (red) & KM (blue) respectively, orange color shows PspA domain of hypothetical protein, yellow color shows interacting residues of PspA domain. D1 and D2 shows docking of Rv3841 with AM (red) & KM (blue) respectively, orange color shows conserved ferritin domain of protein, yellow color shows interacting residues of conserved ferritin domain, purple color shows interacting residues of unknown domain.
Prediction of pupylation sites
By utilizing the default threshold (medium), GPS-PUP predicted six pupylation sites at position K7, K71, K94, K120, K134, and K135 in Rv2744c. Rv2005c and Rv0148 showed two pupylation sites at position K80, K248 and K280, K285 respectively (Table 6).
Table 6. Predicted / identified pupylation sites within identified proteins.
| ORF No. | Position of lysine residue undergoes pupylation | Peptide | Score | Cut-off |
|---|---|---|---|---|
| Rv2005c | 80 | ANAVKLAKEAVGADR | 3.488 | 2.452 |
| 248 | VCDRPARKLVQKSAS | 3.858 | 2.452 | |
| Rv0148 | 280 | ITDLSGAKIAGFKL* | 3.11 | 2.452 |
| 285 | GAKIAGFKL****** | 2.748 | 2.452 | |
| Rv2744c | 7 | *MANPFVKAWKYLMA | 3.15 | 2.452 |
| 71 | RQLADIEKLQVNVRQ | 3.315 | 2.452 | |
| 94 | TAAGDAAKATEYNNA | 2.787 | 2.452 | |
| 120 | EQSVEDLKTLHDQAL | 3.063 | 2.452 | |
| 134 | LSAAAQAKKAVERNA | 2.835 | 2.452 | |
| 135 | SAAAQAKKAVERNAM | 2.496 | 2.452 |
Discussion
In this study we used a proteomic approach to compare membrane and membrane associated proteins of AK and KM resistant and susceptible isolates by 2DE, MALDI-TOF/MS and bioinformatic tools. Resistant isolates were also sequenced and analyzed for known rrs mutations. These isolates did not exhibit mutations at the reported sites. Proteins with increased intensities in the resistant isolates were identified, which might be used as diagnostic markers or drug targets for therapeutics. 2DE/MS has an advantage over the traditional methods (SDS-PAGE, chromatography and sequencing) as not only the identification of a large number of unknown proteins but also protein species separation. Several reports for identification of diagnostics and drug targets employing proteomic approaches exist [32–33]. However, to the best of our knowledge, no such membrane proteome analysis with AK & KM resistant M. tuberculosis isolates has been reported.
Our study revealed twelve proteins with increased intensity in AM and KM resistant as compared to total susceptible isolates. Five spots matched with already identified protein species and therefore total seventeen protein spots were found to be upregulated. Out of twelve, Rv1308, Rv0462, Rv2109c, Rv0148, Rv1876 and Rv3841 belonged to intermediary metabolism and respiration, Rv2005c and Rv2031c to virulence/detoxification/adaptation, Rv2462c to cell wall and cell processes, Rv2744c to conserved hypothetical, Rv1479 to regulatory proteins and Rv0685 to information pathways categories. These proteins were membrane associated [9–14] but were not purely membrane proteins having transmembrane helix. This might be due to the selection of consistently increased intensities of spots in resistant as compared to sensitive isolates (cut limit ≥ 1.5 fold changes in spot intensity).This suggests that membranes with trans membrane helix do not show consistently increased intensities up to 1.5 fold.
Rv1308 (ATP synthase subunit alpha) is a regulatory subunit that produces ATP in the presence of proton gradient across the membrane. Mycobacteria reside in specialized niches and may require adaptations in the energy metabolism. It has been reported that it not only stipulates in replicating mycobacteria, but also in the dormant state [34–36]. Rv0462 (Dihydrolipoyl dehydrogenase/Lpd) is involved in energy metabolism and antioxidant defense. It is the third enzyme of M. tuberculosis’s pyruvate dehydrogenase complex and first enzyme of peroxynitrite reductase/peroxidase, which helps M. tuberculosis to resist host reactive nitrogen intermediates. Without Lpd, M. tuberculosis cannot metabolize branched-chain amino acids and potentially toxic branched-chain intermediates accumulate [37]. Heo et al [38] reported that this protein induces dendritic cells maturation, Th1-mediated responses and may contribute to vaccine development against M. tuberculosis infection. Rv2109c (Proteasome subunit alpha) is involved in protein degradation and required for the virulence of M. tuberculosis. It not only degrades proteins that are toxic due to oxidative or nitrosative damage but also allows other proteins to participate in NO detoxification or repair of macromolecules [39]. Fortune et al [40] reported its involvement in regulating the synthesis of secreted or surface proteins that alter host immunity and thus favor persistence. Discovery of pupylome in M. tuberculosis [41] and other conserved proteasomal components like proteasome b/a subunits, recognition ATPase, pupylase, and depupylome revealed the protein regulation and turnover through proteasome. Identity of these proteins began to explain why defects in protein degradation attenuate virulence in vivo [42–43]. Rv0148 has been identified as probable short-chain dehydrogenases/reductases and possess two binding domains- NAD and substrate. M. tuberculosis acquires Rv0148 gene via horizontal gene transfer from eukaryotics. As the gene has been retained in the genome through selective advantage, it might play a key role in pathogenesis and immunomodulation [44].
Rv1876 (bacterioferritin) and Rv3841 (ferritin), unique for iron homeostasis and are increased intensities under iron-rich and decreased under iron deprived conditions [45]. Very little is known about the protein-protein interactions that carry iron for storage or promote the mobilization of stored iron from ferritin like molecules. Iron assimilation and utilization in M. tuberculosis plays a crucial role in growth, virulence and latency. Function of these may not be just limited to iron uptake; they may be contributing to other metabolic activities, the mechanism of which is still unclear. Pandey and Rodriguez [46] suggested that ferritin (bfrB) is mandatory to maintain iron homeostasis in M. tuberculosis and ferritin lacking bacilli are more susceptible to killing by antibiotics. Our results showed increased intensities of both ferritin and bacterioferritin in membrane fraction. Earlier we reported that only bacterioferritin intensity increased in whole cell lysate and suggested its role in imparting resistance to AK and KM [8]. It is assumed that the heme group present might seize its site of action by providing abnormal site for binding or modulating the protein to block its binding site which needs to be further explored. Although the pathways and enzymes involved in iron metabolism in M. tuberculosis are well established, still our information on iron dependent post-transcriptional, translational regulations, outer membrane iron transporters, trafficking and partitioning of siderophores/Fe-loaded apoproteins in mycobacteria is inadequate. Consequently, it can be a promising antimycobacterial target.
Rv2031c (Alpha-crystallin/HspX), a heat shock protein and Rv2005c (universal stress protein) are not only involved in cell protection to diverse stimuli like stress, dormancy, heat, drug and hypoxia by preventing protein aggregation but have established roles in resistance/stress/virulence/dormancy [47]. Rv2031c is regulated by the two-component regulatory system (DosR/DevR regulon) and is a latency stage disease marker [48–49]. Heat shock proteins assist in M. tuberculosis survival and also provide signal to the immune response [50]. Rv2031c and Rv2005c were predicted as a strong vaccine candidate by Zvi A et al [51].
Rv2462c (trigger factor), involved in protein export, acts as chaperone by maintaining the newly synthesized protein in an open conformation. It was reported to have increased intensity during nutrient deprivation in M. tuberculosis [52]. Rifat et al [53] reported that its intensity is regulated by inorganic phosphate limitation and suggested to play an important role in the survival of M. tuberculosis during chronic infection.
Rv2744c (35-kDa antigen), a hypothetical protein, is homologous to phage shock protein A (PspA) of E. coli and a predominant binding partner and substrate of PepD for proteolysis [54]. It was found to exhibit increased intensity upon exposure to vancomycin and cell wall damaging antibiotics suggesting their role in resistance to cell envelope stress [55]. PepD proteolytically regulates Rv2744c levels to maintain cell wall/cell envelope homeostasis in M. tuberculosis. It is also speculated that cleavage of Rv2744c by PepD may represent a mechanism for terminating the membrane stress response following cessation of the inducing stimulus. Future studies are aimed at delineating the specific mechanism by which it participates in cell wall homeostasis, and defining the other factors that participate in this stress response pathway.
Rv1479 (MoxR1) is a probable transcriptional regulator involved in regulatory function. Jungblut et al [56] found that four MoxR protein species were with different mobility. Hu et al [57] reported that MoxR1 m-RNA expression was more than four-fold in persisters compared to stationary phase of mycobacteria. Recently Rv1479 has been reported in M. tuberculosis pellicles [58]. Our study assumes that increased intensity of this protein overcomes the burden of the transcriptional regulation.
Rv0685 (Elongation factor–Tu) is a conserved protein involved in the elongation phase of translation and post-translational modifications. It also has RNA chaperone activities, ensuring that tmRNA adopts an optimal conformation during aminoacylation [59]. Ef-Tu phosphorylation is implicated in acclimation to the stress conditions encountered during the course of infection. M. tuberculosis phosphoproteome revealed Ef-Tu to be phosphorylated, Sajid et al [60] reported that phosphorylation of Ef-Tu by Protein Kinase B reduced its interaction with GTP, suggesting reduction in protein synthesis. In our study intensity of Rv0685 might be increased due to interruption of translational steps (primary target sites of aminoglycosides) and accumulation of Rv0685 might occur.
In the present study we observed that on performing docking analysis, both drugs interacted with conserved residues of Usp, SDR, PspA and ferritin domain of Rv2005c, Rv0148, Rv2744c and Rv3841 respectively which might alter their functions. It is predicted that these proteins might be exhibiting increased intensities to compensate the effect of drugs. Further we also found pupylation sites in Rv0148, Rv2005c and Rv2744c. Pupylation is a PTM through which small disordered protein Pup is conjugated to lysine residues of proteins marking them for proteasomal degradation. As modification with pup is reversible, pupylation is also likely to have a regulatory role [42]. Pup-proteasome system controlled by pupylation contributes to the virulence/survival strategy of M. tuberculosis in the host and makes the bacteria more resistant to various stresses [61]. Therefore it is assumed that pupylation of modulated protein (drug-protein complex) might be undergo turnover by proteasome machinery to overcome stress through protein-protein interaction. We assume that increased intensities of proteins might be contributing in imparting resistance against AK and KM. Further, detailed study in this direction may help in searching for new targets for drug development.
Conclusion
In a nutshell, this is the first report on the membrane and membrane associated proteins of AK and KM resistance in M. tuberculosis using proteomic coupled with bioinformatic approaches. Among the twelve proteins which were found to have increased intensities only nine were with defined roles and three with unknown functions. Molecular docking showed proper interaction of both drugs with hypothetical proteins (Rv2005c, Rv2744c and Rv0148) as well as ferritin. GPS-PUP analysis suggested presence of pupylation sites within these proteins. It is depicted that increased intensities of these proteins and proteasome sub unit alpha might not only be neutralizing/modulating the drug molecules but are also involved in protein turnover to overcome AK and KM resistance. Apart from that we found three proteins–ferritin, bacterioferritin and elongation factor-Tu, involved in iron storage, homeostasis, detoxification, and regulation/metabolism. We assume that iron regulation/metabolism might be playing some crucial role in contributing resistance to AK and KM. Increased elongation factor-Tu (Rv0685) might be due to interruption of translational steps by these drugs. These findings need further exploitation for the development of newer therapeutic agents or molecular markers which can directly be targeted to a gene/protein responsible for resistance so that an extreme condition like XDR-TB can be prevented, which could ultimately lead to broaden the narrow gauge of new or existing therapeutics.
Supporting Information
(RAR)
(RAR)
Acknowledgments
The authors are grateful to Director, NJIL & OMD for the support. DS is SRF (ICMR, New Delhi). We thank Mr. Jaypal for assistance.
Abbreviations
- TB
Tuberculosis
- AK
Amikacin
- KM
Kanamycin
- 2DE
Two Dimensional Gel Electrophoresis
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by ICMR New Delhi.
References
- 1.WHO Report 2013. Global tuberculosis control 2013. Available from: http://apps.who.int/iris/bitstream/10665/91355/1/9789241564656_eng.pdf.
- 2. Beauclerk AAD, and Cundliffe E. Site of action of two ribosomal RNA methylases responsible for resistance to aminoglycoside. J Mol Biol. 1987; 193: 661–671. [DOI] [PubMed] [Google Scholar]
- 3. Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev. 2003; 67: 593–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Welch KT, Virga KG, Whittemore NA, Ozen C, Wright E, Brown CL, et al. Discovery of non-carbohydrate inhibitors of aminoglycoside-modifying enzymes. Bioorg Med Chem. 2005; 13: 6252–6363. [DOI] [PubMed] [Google Scholar]
- 5. Magnet S, Courvalin P, Lambert T. Resistance modulation cell division type efflux pump involved in aminoglycoside resistance in Acinetobacter baumannii BM4454. Antimicrob Agents Chemother. 2001; 45: 3375–3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Magnet S, Smith TA, Zheng R, Nordmann P, Blanchard JS. Aminoglycosides resistance resulting from tight drug binding to an altered aminoglycosides acetyl transferase. Antomicrob Agents Chemother. 2003; 47: 1577–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Singhal N, Sharma P, Kumar M, Joshi B, Bisht D. Analysis of intracellular expressed proteins of Mycobacterium tuberculosis clinical isolates. Proteome Sci. 2006; 10:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kumar B, Sharma D, Sharma P, Katoch VM, Venkatesan K, Bisht D. Proteomic analysis of Mycobacterium tuberculosis isolates resistant to kanamycin and amikacin. J proteomics 2013; 94: 68–77. 10.1016/j.jprot.2013.08.025 [DOI] [PubMed] [Google Scholar]
- 9. Gu S, Chen J, Dobos KM, Bradbury EM, Belisle JT, Chen X. Comprehensive proteomic profiling of the membrane constituents of a Mycobacterium tuberculosis strain. Mol Cell Proteomics 2003; 2:1284–96. [DOI] [PubMed] [Google Scholar]
- 10. Sinha S, Arora S, Kosalai K, Namane A, Pym AS, Cole ST. Proteome analysis of the plasma membrane of Mycobacterium tuberculosis .Comp Funct Genomics 2002; 3:470–83. 10.1002/cfg.211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Xiong Y, Chalmers MJ, Gao FP, Cross TA, Marshall AG. Identification of Mycobacterium tuberculosis H37Rv integral membrane proteins by one-dimensional gel electrophoresis and liquid chromatography electrospray ionization tandem mass spectrometry. J Proteome Res 2005; 4: 855–861. [DOI] [PubMed] [Google Scholar]
- 12. Mattow J, Siejak F, Hagens K, Schmidt F, Koehler C, Treumann A, et al. An improved strategy for selective and efficient enrichment of integral plasma membrane proteins of mycobacteria. Proteomics 2007; 7:1687–1701. [DOI] [PubMed] [Google Scholar]
- 13. Malen H, De Souza GA, Pathak S, Softeland T, Wiker HG. Comparison of membrane proteins of Mycobacterium tuberculosis H37Rv and H37Ra strains. BMC Microbiology 2011; 11: 18 10.1186/1471-2180-11-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. de Souza GA, Arntzen MO, Fortuin S, Schurch AC, Malen H, McEvoy CR, et al. Proteogenomic analysis of polymorphisms and gene annotation divergences in prokaryotes using a clustered mass spectrometry-friendly database. Mol Cell Proteomics 2011; 10(1):M110.002527 10.1074/mcp.M110.002527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sigler K, and Hofer M. Biotechnological aspects of membrane function. Crit. Rev. Biotechnol. 1997; 17: 69–86. [DOI] [PubMed] [Google Scholar]
- 16. Hoffmann C, Leis A, Niederweis M, Plitzko JM, Engelhardt H. Disclosure of the mycobacterial outer membrane: cryo-electron tomography and vitreous sections reveal the lipid bilayer structure. Proc. Natl. Acad. Sci. 2008; 105: 3963–3967. 10.1073/pnas.0709530105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zuber B, Chami M, Houssin C, Dubochet J, Griffiths G, Daffe M. Direct visualization of the outer membrane of mycobacteria and corynebacteria in their native state. J Bacteriol 2008; 190: 5672–5680. 10.1128/JB.01919-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Velayati AA, Farnia P, Ibrahim TA, Haroun RZ, Kuan HO, Ghanavi J, et al. Differences in cell wall thickness between resistant and nonresistant strains of Mycobacterium tuberculosis: Using transmission electron microscopy. Chemother 2009; 55: 303–307. [DOI] [PubMed] [Google Scholar]
- 19. Canetti G, Fox W, Khomenko A, Mahler HT, Menon NK, Mitchison DA, et al. Advances in techniques of testing mycobacterial drug sensitivity, and the use of sensitivity tests in tuberculosis control programmes. Bull World Health Organ 1969; 41: 21–43. [PMC free article] [PubMed] [Google Scholar]
- 20. Palomino JC, Martin A, Camacho M, Guerra H, Swings J, Portaels P. Resazurin microtiter assay plate: simple and inexpensive method for detection of drug resistance in Mycobacterium tuberculosis . Antimicrob Agents Chemother 2002; 44: 2720–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jadaun GP, Agarwal C, Sharma H, Ahmed Z, Upadhyay P, Faujdar J, et al. Determination of ethambutol MICs for Mycobacterium tuberculosis and Mycobacterium avium isolates by resazurin microtitre assay. J. Antimicrob. Chemother. 2007; 60: 152–155. [DOI] [PubMed] [Google Scholar]
- 22. Brodie AF, Kalra VK, Lee SH, Cohen NS. Properties of energy-transducing systems in different types of membrane preparations from Mycobacterium phlei-preparation, resolution, and reconstitution. Methods Enzymol. 1979; 55: 175–200. [DOI] [PubMed] [Google Scholar]
- 23. Bradford MM. A rapid and sensitive method for quantitation of microgram quantities of protein utilizing the principle of protein-dye-binding. Anal. Biochem. 1976; 72: 248–254. [DOI] [PubMed] [Google Scholar]
- 24. Gorg A, Obermaier C, Boguth G, Harder A, Scheibe B, Wildgruber R, et al. The current state of two-dimensional electrophoresis with immobilized pH gradients. Electrophoresis 2000; 21: 1037–1053. [DOI] [PubMed] [Google Scholar]
- 25. Samanovic MI, Li H, Darwin KH. The Pup-Proteasome System of Mycobacterium tuberculosis . Subcell Biochem. 2013; 66: 267–95. 10.1007/978-94-007-5940-4_10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990; 215: 403–410. [DOI] [PubMed] [Google Scholar]
- 27. Pearson WR and Lipman DJ. Improved tools for biological sequence comparison. Proc Natl Acad Sci USA 1988; 85: 2444–2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Schneidman-Duhovny D, Inbar Y, Nussinov R, Wolfson HJ. PatchDock and SymmDock: servers for rigid and symmetric docking. Nucleic Acids Res 2005; 33 Web Server: W363–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Andrusier N, Nussinov R, Wolfson HJ. FireDock: Fast interaction refinement in molecular docking. Proteins 2007; 69: 139–159. [DOI] [PubMed] [Google Scholar]
- 30. Mashiach E, Schneidman-Duhovny D, Andrusier N, Nussinov R, Wolfson HJ. FireDock: A Web server for fast interaction refinement in molecular docking. Nucleic Acids Res 2008; 36 Web Server: W229–232. 10.1093/nar/gkn186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Liu Z, Ma Q, Cao J, Gao X, Ren J, Xue Y. GPS-PUP: computational prediction of pupylation sites in prokaryotic proteins. Mol Biosyst. 2011; 7:2737–2740. 10.1039/c1mb05217a [DOI] [PubMed] [Google Scholar]
- 32. Zhang J, Wang K, Zhang J, Liu SS, Dai L, Zhang JY. Using proteomic approach to identify tumor-associated proteins as biomarkers in human esophageal squamous cell carcinoma. J. Proteome Res. 2011; 10: 2863–72. 10.1021/pr200141c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhang L, Wang Q, Wang W, Liu Y, Wang J, Yue J, et al. Identification of putative biomarkers for the serodiagnosis of drug-resistant Mycobacterium tuberculosis . Proteome Sci. 2012; 10: 12 10.1186/1477-5956-10-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Koul A, Vranckx L, Dendouga N, Balemans W, Van den Wyngaert I, Vergauwen K, et al. Diarylquinolines are bactericidal for dormant mycobacteria as a result of disturbed ATP homeostasis. J. Biol. Chem. 2008; 283: 25273–25280. 10.1074/jbc.M803899200 [DOI] [PubMed] [Google Scholar]
- 35. Koul A, Arnoult E, Lounis N, Guillemont J, Andries K. The challenge of new drug discovery for tuberculosis. Nature 2011; 469: 483–490. 10.1038/nature09657 [DOI] [PubMed] [Google Scholar]
- 36. Rao SP, Alonso S, Rand L, Dick T, Pethe K. The proton motive force is required for maintaining ATP homeostasis and viability of hypoxic, non replicating Mycobacterium tuberculosis . Proc Natl Acad Sci USA 2008; 105: 11945–11950. 10.1073/pnas.0711697105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Venugopal A, Bryk R, Shi S, Rhee K, Rath P, Schnappinger D, et al. Virulence of Mycobacterium tuberculosis depends on lipoamide dehydrogenase, a member of three multienzyme complexes. Cell Host Microbe. 2011; 9: 21–31. 10.1016/j.chom.2010.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Heo DR, Shin SJ, Kim WS, Noh K, Park JW, Son KH, et al. Mycobacterium tuberculosis lpdC, Rv0462, induces dendritic cell maturation and Th1 polarization. Biochem Biophys Res Commun. 2011; 411: 642–7. 10.1016/j.bbrc.2011.07.013 [DOI] [PubMed] [Google Scholar]
- 39. Vandal OH, Roberts JA, Odaira T, Schnappinger D, Nathan CF, Ehrt S. Acid-susceptible mutants of Mycobacterium tuberculosis share hyper susceptibility to cell wall and oxidative stress and to the host environment. J Bacteriol 2009; 191(2): 625–31. 10.1128/JB.00932-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Fortune SM, Solache A, Jaeger A, Hill PJ, Belisle JT, Bloom BR, et al. Mycobacterium tuberculosis inhibits macrophage responses to IFN-gamma through myeloid differentiation factor 88-dependent and -independent mechanisms. J Immunol 2004; 172(10): 6272–6280. [DOI] [PubMed] [Google Scholar]
- 41. Pearce MJ, Mintseris J, Ferreyra J, Gygi SP, Darwin KH. Ubiquitin-like protein involved in the proteasome pathway of Mycobacterium tuberculosis . Science 2008; 322:1104–1107. 10.1126/science.1163885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Burns KE, Cerda-Maira FA, Wang T, Li H, Bishai WR, Darwin KH. Depupylation of prokaryotic ubiquitin-like protein from mycobacterial proteasome substrates. Mol. Cell. 2010; 39: 821–827. 10.1016/j.molcel.2010.07.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Samanovic MI, Li H, Darwin KH. The Pup-Proteasome System of Mycobacterium tuberculosis . Subcell Biochem 2013; 66: 267–95. 10.1007/978-94-007-5940-4_10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kinsella RJ, and McInerney JO. Eukaryotic genes in Mycobacterium tuberculosis? Possible alternative explanations. Trends Genet 2003; 12: 687–689. [DOI] [PubMed] [Google Scholar]
- 45. Hwang SA, and Actor JK. Lactoferrin modulation of BCG-infected dendritic cell functions. Int. Immunol. 2009; 21: 1185–1197. 10.1093/intimm/dxp084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pandey R, and Rodriguez GM. A ferritin mutant of Mycobacterium tuberculosis is highly susceptible to killing by antibiotics and is unable to establish a chronic infection in mice. Infect. Immun. 2012; 80: 3650–3659. 10.1128/IAI.00229-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sherman DR, Voskuil M, Schnappinger D, Liao R, Harrell MI, Schoolnik GK. Regulation of the Mycobacterium tuberculosis hypoxic response gene encoding alpha-crystallin. Proc Natl Acad Sci USA 2001; 98: 7534–7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Geluk A, Lin MY, van Meijgaarden KE, Leyten EM, Franken KL, Ottenhoff TH, et al. T-cell recognition of the HspX protein of Mycobacterium tuberculosis correlates with latent M. tuberculosis infection but not with M. bovis BCG vaccination. Infect Immun 2007; 75: 2914–2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gautam US, Sikri K, Tyagi JS. The residue threonine 82 of DevR (DosR) is essential for DevR activation and function in Mycobacterium tuberculosis despite its atypical location. J Bacteriol. 2011;193:4849–58. 10.1128/JB.05051-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Roupie V, Romano M, Zhang L, Korf H, Lin MY, Franken KL, et al. Immunogenicity of eight dormancy regulon-encoded proteins of Mycobacterium tuberculosis in DNA-vaccinated and tuberculosis-infected mice. Infect. Immun. 2007; 75: 941–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zvi A, Ariel N, Fulkerson J, Sadoff JC, Shafferman A. Whole genome identification of Mycobacterium tuberculosis vaccine candidates by comprehensive data mining and bioinformatic analyses. BMC Med. Genomics 2008; 1: 18 10.1186/1755-8794-1-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hampshire T, Soneji S, Bacon J, James BW, Hinds J, Laing K, et al. Stationary phase gene expression of Mycobacterium tuberculosis following a progressive nutrient depletion: a model for persistent organisms? Tuberculosis 2004; 84: 228–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Rifat D, Bishai WR, Karakousis PC. Phosphate depletion: a novel trigger for Mycobacterium tuberculosis persistence. J. Infect. Dis. 2009; 200: 1126–1135. 10.1086/605700 [DOI] [PubMed] [Google Scholar]
- 54. White MJ, Savaryn JP, Bretl DJ, He H, Penoske RM, Terhune et al. The HtrA-like serine protease PepD interacts with and modulates the Mycobacterium tuberculosis 35-kDa antigen outer envelope protein. PLoS One 2011; 6(3): e18175 10.1371/journal.pone.0018175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Provvedi R, Boldrin F, Falciani F, Palù G, Manganelli Ril. Global transcriptional response to vancomycin in Mycobacterium tuberculosis . Microbiology 2009; 155: 1093–102. 10.1099/mic.0.024802-0 [DOI] [PubMed] [Google Scholar]
- 56. Jungblut PR, Schaible UE, Mollenkopf HJ, Zimny-Arndt U, Raupach B, Mattow J, et al. Comparative proteome analysis of Mycobacterium tuberculosis and Mycobacterium bovis BCG strains: towards functional genomics of microbial pathogens. Mol. Microbiol. 1999; 33: 1103–1117. [DOI] [PubMed] [Google Scholar]
- 57. Hu Y, and Coates AR. Increased levels of sigJ mRNA in late stationary phase cultures of Mycobacterium tuberculosis detected by DNA array hybridization. FEMS Microbiol Lett. 2001; 202: 59–65. [DOI] [PubMed] [Google Scholar]
- 58. Kerns PW, Ackhart DF, Basaraba RJ, Leid JG, Shirtliff ME. Mycobacterium tuberculosis pellicles express unique proteins recognized by the host humoral response. Pathog Dis 2014; 70:347–358. 10.1111/2049-632X.12142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Corvaisier S, Bordeau V, Felden B. Inhibition of transfer messenger RNA aminoacylation and trans-translation by aminoglycoside antibiotics. J. Biol. Chem. 2003; 278: 14788–14797. [DOI] [PubMed] [Google Scholar]
- 60. Sajid A, Arora G, Gupta M, Singhal A, Chakraborty K, Nandicoori VK, et al. Interaction of Mycobacterium tuberculosis elongation factor Tu with GTP is regulated by phosphorylation J Bacteriol 2011; 193(19): 5347–5358. 10.1128/JB.05469-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Darwin KH, Ehrt S, Gutierrez-Ramos JC, Weich N, Nathan CF. The proteasome of Mycobacterium tuberculosis is required for resistance to nitric oxide. Science 2003; 302: 1963–1966. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(RAR)
(RAR)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.