i. Summary
Biomedical and life sciences literature is unique because of its exponentially increasing volume and interdisciplinary nature. Biomedical literature access is essential for several types of users including biomedical researchers, clinicians, database curators, and bibliometricians. In the past few decades, several online search tools and literature archives, generic as well as biomedicine-specific, have been developed. We present this chapter in the light of three consecutive steps of literature access: searching for citations, retrieving full-text, and viewing the article. The first section presents the current state of practice of biomedical literature access, including an analysis of the search tools most frequently used by the users, including PubMed, Google Scholar, Web of Science, Scopus, and Embase, and a study on biomedical literature archives such as PubMed Central. The next section describes current research and the state-of-the-art systems motivated by the challenges a user faces during query formulation and interpretation of search results. The research solutions are classified into five key areas related to text and data mining, text similarity search, semantic search, query support, relevance ranking, and clustering results. Finally, the last section describes some predicted future trends for improving biomedical literature access, such as searching and reading articles on portable devices, and adoption of the open access policy.
Keywords: Biomedical Literature Search, Text Mining, Information Retrieval, Bioinformatics, Open Access, Relevance Ranking, Semantic Search, Text Similarity Search
1. Introduction
Literature search is the task of finding relevant information from the literature, e.g. finding the most influential articles on a topic, finding the answer to a specific question, or finding other (bibliographic or non-bibliographic) information on citations. Literature search is a fundamental step for every biomedical researcher in their scientific discovery process. Its roles range from reviewing past works at the beginning of a scientific study to the final step of results interpretation and discussion. Literature search is also important for clinicians seeking established and new findings for making important clinical decisions. Furthermore, since current biomedical research is heavily dependent on access to various kinds of online biological databases, literature search is also a key component of transforming knowledge encoded in nature language data, such as journal publications, into structured database records by dedicated database curators. In addition, literature search has other uses such as biomedical citation analysis for academic needs, and data collection for biomedical text mining research.
To meet the diverse needs of literature access by the scientific community worldwide, a number of Web-based search tools, e.g. PubMed [1] and Google Scholar [2], and online bibliographic archives, e.g. PubMed Central [3], have been developed over the last decades. As a result, the literature access process typically includes the following consecutive steps, searching for citations on a search tool, retrieving full-text on a bibliographic archive, and reading the article. Despite advances in information technologies, the ease of searching the biomedical literature has not kept pace for two main reasons. First, the size of the biomedical literature is large (dozens of millions) and it continues to grow rapidly (over a million per year), thus making the selection of proper search keywords and reviewing results a daunting task [4,5]. Second, biomedical research is becoming increasingly multi-disciplinary. As a result, the information most relevant to an individual researcher may appear in journals that are not usually considered relevant to his or her own research. For example, a 2006 study [6] found that half of the renal information is published in non-renal journals.
In response to the aforementioned challenges, there has been a recent surge in improving the literature access through the use of advanced information technologies in information retrieval (IR), data mining, and natural language processing (NLP). For instance, recent IR research includes relevance-ranking algorithms aimed at improving user retrieval effectiveness. Data mining algorithms can group similar results into clusters, thus providing users with a quick overview of the search results before focusing on individual papers. Text mining and NLP techniques can be used to automatically recognize named entities (e.g. genes) and their relations (e.g. protein-protein interaction) in the biomedical text, thus enabling novel entity-specific semantic searches as opposed to the traditional keyword based searches.
A number of literature search assistants using aforementioned information techniques have been developed over the years, some of which have been shown to be effective in real-world uses. For instance, by comparing words from the title and abstract of each citation, and the indexed MeSH terms using a weighted IR algorithm, related papers can be grouped together into clusters [7]. When used in most search tools, such a technique is known as “related articles” where users can easily find all papers relevant to a search result through a simple mouse click. The “related article” application has been frequently used [8] since its appearance in PubMed. Because of its success in PubMed, this feature has been adopted by many journal websites as well as commercial search tools.
Retrieving full-text of bibliographic archives poses another challenge for literature access. While most article abstracts are freely accessible, their full texts are still locked by the publishers: in order to read the full text, one would need either an institutional subscription or pay-per-view. Such an access model is inconvenient to the researchers and the global scholarly community [9]. In recognition of such a problem, a number of initiatives began to promote open access to the scientific literature. For instance, the Budapest Open Access Initiative reaffirmed in its 10th anniversary in 2012 that its goal is to make open access the default method for distributing new peer-reviewed research in every field and country. Agreed with such initiatives, a number of publishers and journals are adopting the open access paradigm for publishing articles. For instance, two major open access publishers include the BioMed Central (BMC) and Public Library of Science (PLoS). To accelerate open access, the U.S. National Library of Medicine started PubMed Central (PMC), a free digital repository of full-text articles in biomedical and life sciences in early 2000. With a little over 10 years development, PMC currently contains approximately three million items, and continues to grow at least 7% per year [10] despite some criticisms from professional societies and commercial publishers [11].
This chapter describes all of the above-mentioned issues in more depth. It first introduces some existing literature search tools, and bibliographic archives, that are commonly used to access the biomedical literature, in three consecutive steps: searching, retrieving, and reading articles. Next, it presents a selection of five key categories of text mining and IR applications that address challenges in searching literature. Finally, there is a discussion on the future trends of biomedical literature access, with a focus on the open access activities in the biomedical domain and recent transition to reading articles on portable devices.
2. Current Access to Biomedical Literature
Open access availability of biomedical literature has led an increasing number of users to resort to online methods of literature access. Journals and online databases are currently the most frequently accessed resources among biomedical information seekers, followed by books, proceedings, newsletters, technical reports, author web pages, etc. [12,13]. Given the rising quality, volume, and diversity of biomedical literature [14], the information seeking trend has advanced to multiple layers of information access. Current framework comprises a search tool that provides unified access to multiple literature archives; these archives store the full text of articles and offer multiple viewing media to read those articles. Current practice begins with the user crafting a keyword or faceted (structured) query, and submitting on the search tool. In response, the tool presents a ranked list of citations relevant to the user query. The user has the option to go to a specific citation, access the full-text on the linked literature archive, and view the article using a particular medium. Figure 1 demonstrates the three-step process of literature access.
Figure 1.
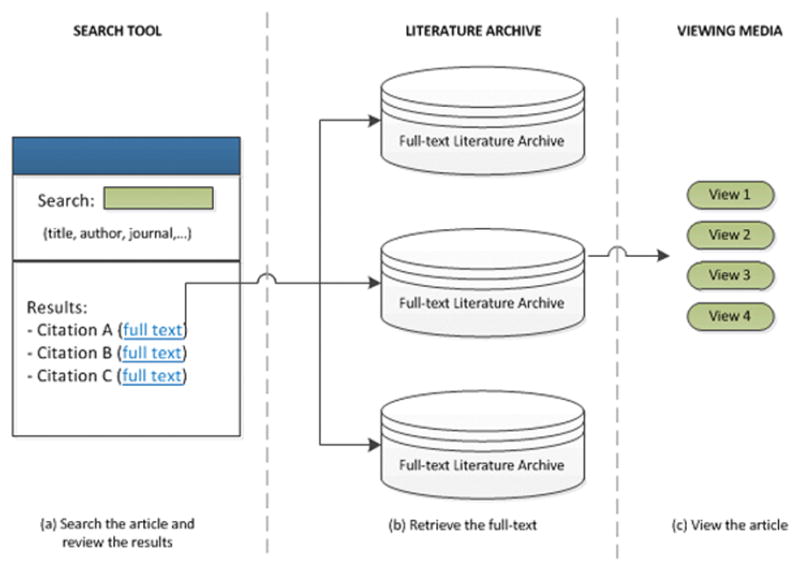
The Three Steps of Biomedical Literature Access (a) Searching the literature and reviewing results using a search tool (e.g. PubMed), (b) Retrieving the full-text on a literature archive (e.g., PubMed Central), (c) Consuming the article on a viewing media (e.g. PubTator)
2.1 Literature Search Tools
A search tool provides a single access point to multiple literature archives. At the core, the tool contains a citation database developed by indexing articles (abstract or full-text) from different sources. The tool interface serves two purposes: (i) provides search functionality supporting queries ranging from the standard keyword search to the comprehensive faceted search (e.g. search by author, journal, title, etc.), (ii) presents ranked list of citations relevant to the query, with several options to filter and re-sort the results; in addition to bibliographic information, each citation contains a link to retrieve the full-text of the article on a literature archive.
PubMed [1] is the most widely used search tool dedicated to biomedical and life sciences literature. Launched in 1996, PubMed is a publicly available citation database developed and maintained by the U.S. National Library of Medicine. To date, PubMed contains more than 22.9 million citations for biomedical literature belonging to MEDLINE indexed journals, manuscripts deposited in PubMed Central, and the NCBI Bookshelf. PubMed articles are indexed by the controlled vocabulary thesaurus, Medical Subject Headings (MeSH®). The search algorithm is based on PubMed’s automatic term mapping algorithm [15]. The PubMed citation database is updated daily. PubMed citations date back to the early 1950s, and approximately half a million also date back to 1809. The PubMed interface offers the keyword search, and allows the advanced queries by various fields such as author name, publication date, PubMed entering date, editor, grant number, status of MeSH indexing for MEDLINE citations, etc. A noteworthy feature of PubMed is the related citations algorithm [8] based on document similarity.
Embase [16] is a subscription-based biomedical citation database developed by Elsevier in 2000. This search service was developed primarily for biomedical and clinical practice with particular focus on drug discovery and development, drug safety, and pharmacovigilance research. Embase contains 25 million indexed records and indexes full-text articles from 8,306 journals, out of which 7,203 publish English language articles. Embase is often compared with MEDLINE, and contains 5 million records and covers 2,000 journals not included by MEDLINE. The Embase database is updated every day and nearly one million records are added per year. Embase has digitally scanned the articles from 1947 to 1973. While the official reported temporal coverage of Embase dates backs to 1947, some articles also date back to 1880s. The records are indexed by Emtree thesaurus for drug and chemical information. This allows for deep indexing of articles and flexible keyword searching using term mapping [17]. The search capability is enhanced using auto complete and synonym suggestion features. The results can be filtered by drug and disease mentions in the article.
While several other state of the art biomedical-specific search tools [14,18–20] have been designed since the inception of PubMed, these are not widely used as yet. Instead, other than PubMed, biomedical information seekers prefer rather generic tools that index articles from several disciplines in addition to biomedical and life sciences. Based on the popularity and discussion in previous studies [21–23], we describe one publicly available (Google Scholar [2]) and two subscription-based (Web of Science and Scopus [24,25]) tools, and describe their unique features.
Google Scholar [2], launched in 2004, is a Web search engine owned by Google Inc. Google Scholar indexes full-text articles from multiple disciplines from most peer-reviewed online journals of European and American publishers, scholarly books, and other non-peer reviewed journals. The size and coverage of biomedical articles in Google Scholar are not revealed; theoretically it consists of all biomedical articles available electronically. In addition to the keyword search, the tool offers searching by various fields such as author, publication date, journal, words occurring in title and body, with different methods of term matching. The results are sorted by relevance as determined by full text of each article, author, journal, and number of citations received.
Web of Science [24], developed by Thompson Reuters in 2004, is a citation database that covers over 12,000 top-tier international and regional journals, as per their selection process[26], in every area of the natural sciences, social sciences, and arts and humanities. The science citation database of Web of Science, which is likely to contain biomedical specific articles, covers more than 8,500 notable journals from 150 disciplines and is updated weekly. The temporal coverage is dated back to 1900 to the present day. The total number of biomedical citations cannot be approximated. The citations can be searched by various bibliographic fields, and the results display the total number of citations, comprehensive backward and forward citation maps, and additional keyword suggestions to improve the query. The result is ranked based on the overlap between the search terms and the terms in the articles. Also, the results can be filtered by Web of Science subject areas that are pre-assigned to journals.
Scopus [25], launched in 2004 by Elsevier, is a citation database for peer-reviewed literature from life sciences, health sciences, physical sciences, social sciences and humanities. Scopus, as of November 2012, includes citations from 19,500 peer-reviewed journals, 400 trade publications, 360 book series, and is updated one-to-two times weekly. Temporally, citations date back to 1823. Scopus contains more than 18,300 citations from the life, health, and physical science subject areas. The faceted search is comprehensive and includes fields such as publication date, document type, subject area, author, title, keywords, affiliation, etc. For each result citation, the number of incoming citations, Emtree drug terms, and Emtree medical terms are displayed. For a given citation, Scopus also displays the related articles computed based on shared references. The relevance rank of results is calculated based on relative frequency and location of the search terms in the article.
Table 1 summarizes various search tools based on some key features. The biomedical coverage and size of the generic search tools, Google Scholar, Web of Science, and Scopus, could not be accurately computed, as they do not provide a breakdown for biomedical and life science specific journals or articles. To get some insight on the coverage, we conducted a small experiment and submitted the query “type 2 diabetes mellitus” on various search tools. The results are shown in Table 2. Google Scholar returns the highest number of results. This is expected given the crawling nature of the search engine and the liberal inclusion criteria. Embase returns more citations than PubMed. While Web of Science returns the least number of results, it is discussed in higher number (10,356) of PubMed articles as compared to Scopus and Google Scholar which are discussed in 3,231 and 1,621 articles, respectively. The most recent and the oldest articles differ for each tool. With PubMed as reference point, Google Scholar shows the most up-to-date result. Also, the number of incoming citations for a 2001 article [27] is 7092, 3655, and 4722, on Google Scholar, Web of Science, and Scopus (and Embase), respectively. This highlights the differences in coverage of various tools.
Table 1.
Summary of various popular biomedical literature search tools
| PubMed | Google Scholar | Web of Science | Scopus | Embase | |
|---|---|---|---|---|---|
| Developer | U.S. National Library of Medicine | Google Inc. | Thompson Reuters | Elsevier | Elsevier |
| Launch Year | 1996 | 2004 | 2004 | 2004 | 2000 |
| Fee-based | No | No | Yes | Yes | Yes |
| Temporal Coverage | 1809 – present | Unknown | 1900 - present | 1823 - present | 1880-presemt |
| Total Biomedical Citations (approx.) | 22.9 million | Unknown | Unknown | 18,300 | 25 million |
| Covered Journals | MEDLINE indexed journals. Manuscripts from PubMed Central, NCBI Bookshelf | Peer-reviewed journals of America and Europe, scholarly books, Non-peer reviewed journals | 8,500 strictly selected science journals | Peer-reviewed journals, trade publications, book series | 7,600 biomedical and pharmacological journals from 90 countries, and 2500+ conferences |
| Update Frequency | Daily | Unknown | Weekly | 1–2 times Weekly | Daily |
| Relevance Ranking Features | Not applicable | Full-text, author, journal, number of incoming citations | Overlap of search terms with the terms in the article | Frequency and location of search terms in the article | Unknown |
| Non-bibliographic information | MeSH keywords, Related citations | Incoming citations | Keyword recommendations for query refinement, Incoming citations, Backward and forward citation maps | Emtree drug and medical terms, Related citations, Incoming Citations | Emtree drug, disease and other terms, PubMed link, incoming citations (linked to Scopus) |
Table 2.
Comparison of search results for “type 2 diabetes mellitus” on July 15 2013
| PubMed | Google Scholar | Web of Science | Scopus | EMBASE | |
|---|---|---|---|---|---|
| Number of results | 83,025 | 1,380,000 (approx.) | 52,351 | 117,875 | 207,444 |
| Publication year of the oldest article | 1967 | 1853 | 1951 | 1947 | 1909 |
| PubMed ID for the most recent article | 23847327 | 23846835A | 23504683A | 22968324 | 23668792 |
There exist other more recent articles not found in PubMed
Out of the above-mentioned tools, PubMed and Embase stand out in that they are the foremost developments, biomedical-specific, and the most frequently updated search tools. In addition, their inbuilt search algorithms utilize controlled vocabularies. The other three generic tools differ from PubMed and Embase in that they perform citation analysis and provide indications of scholarly impact of articles; Google scholar and Scopus provide the number of incoming citations for each article, and Web of Science offer thorough analysis including visual summaries of citation distributions. Also, all tools but PubMed employ a ranking algorithm that computes the relevance score of a given article with respect to search terms, incoming citations, journal, etc.
PubMed, Embase, and Scopus are similar in terms of their use of controlled vocabularies such as MeSH and Emtree in curating the articles. PubMed and Scopus are similar in their employment of the related citations algorithm, though internally quite different from each other. Web of Science is unique in that it has the keyword recommendation feature, and a strict criterion for journal selection. The selling point of Embase is that it covers significant number of biomedical articles and journals that are not covered by PubMed. Google Scholar is unique in the comprehensiveness of its ranking algorithm. Another advantage of Google Scholar is that it links to free full-text articles more than the other search tools that might point to a locked journal [22]. Google Scholar, however, unlike others, does not support bibliography management, such as integration with bibtex, RefWorks, EndNote, EndNote Web, etc. In sum, currently, there is no one-stop shop available for biomedical literature search as each tool has its own strengths and weaknesses. The choice of tool depends would thus depend on the subject matter, publication year, and the usage context, and a wise search strategy would use multiple tools instead of relying on one [21].
2.2 Full-text Literature Archives and Viewing Media
A literature search tool is integrated with multiple literature archives where full-text articles can be retrieved for further consumption. As of June 18 2013, out of the 22.9 million citations in PubMed, 4 million citations are linked to their free full text archives. Out of the citations linked to free full text archives, 2.3 million are archived in the PubMed Central [3] literature archive, and the remaining either contain direct links to journal’s website (e.g., Journal of Cell Biology, Oncotarget, Anticancer Research, BMJ Journals), or to comprehensive literature archives developed by major publishing companies.
PubMed Central (PMC) [3], launched in 2000, is a free federal digital archive of full-text biomedical and life sciences articles maintained by the U.S. National Library of Medicine. Currently, the PMC archives approximately 2.7 million articles provided by about 3,700 journals including full participation, NIH portfolio, and selective deposit journals. PMC also contains supplemental items optionally accompanying each article. Another domain-specific archive, EBSCO’s Cumulative Index to Nursing & Allied Health Literature (CINAHL) Plus with full text [28] is a subscription-based full-text literature archive designed for nurses, allied health professionals, researchers, nurse educators and students. The content is dated back to 1937 and includes full-text from 768 journals and 275 books from nursing and allied health disciplines. CINAHL is also a widely used search tool among nursing professionals.
Springer’s SpringerLink [29] was launched in 1996 and archives full-text content available from 1996. SpringerLink covers approximately 7.7 million full-text articles from electronic books and journals from all disciplines, out of which 6.4 million could be classified under the categories of biomedical, chemical, life, public health, and medical sciences. Supplementary material is also archived with each article.
ScienceDirect [30] is a subscription-based literature archive launched by Elsevier in 2000. ScienceDirect contains more than 11 million peer reviewed journal articles and book chapters from more than 2,500 peer-reviewed journals and more than 11,000 books, including 8,077 journals and book chapters from life and health sciences. ScienceDirect’s coverage goes back to 1823. Elsevier, which is also the host of Scopus search tool, has digitalized most of the pre-1996 content. Some additional content such as audio, video, datasets, and supplemental items are also archived. Since its launch, more than 700 million articles have been downloaded from the ScienceDirect website [31]. Recently, Elsevier has integrated its search tool, Scopus, and the literature archive, ScienceDirect into a new platform, SciVerse.
Wiley online library is a subscription-based full-text archive, developed in 2010 by Wiley-Blackwell publishing company. Wiley online library contains multidisciplinary collection of four million full-text articles from 1,500 journals, over 13,000 online books, and hundreds of reference works. The subject areas include chemistry, life sciences, medicine, nursing, dentistry and healthcare, veterinary medicine, physical sciences, and non-biomedical subjects [32]. The coverage of biomedical subject areas is not known.
Table 3 summarizes the above-discussed literature archives by their provider, launch year, temporal coverage, content coverage, and supported viewing media. Similar to the generic search tools, the total number of biomedical articles in the generic archives such as SpringerLink and ScienceDirect, could not be precisely computed. SpringerLink does provide a subject-wise breakdown, and archives the most number of full-text articles from biomedical and related areas. In PubMed, CINAHL is discussed in the highest number of articles (8595), followed by ScienceDirect (298), SpringerLink (116), and Wiley Online Library (38). These articles are related to information seeking and retrieval studies focused on biomedical articles. It should be noted that these numbers might not give a complete picture on the coverage of various archives, as there might be other studies published in journals not indexed by PubMed.
Table 3.
Comparison of biomedical full-text literature archives
| Literature Archive (Provider, Year) | Temporal Coverage | Full-text Biomedical Articles and archive coverage (approx.) | Viewing Media |
|---|---|---|---|
| PubMed Central (U.S. National Library of Medicine, 2000) | 1950 - present | 2.7 million from 3,700 journals, including full participation, NIH portfolio, selective deposit | Classic, PDF, EPUB, PubReader |
| CINAHL Plus with Full Text (EBSCO, 2010) | 1937-present | 768 journals and magazines, 275 books and monographs from nursing and allied health disciplines | |
| SpringerLink (Springer, 1996) | 1860 – present | 6.4 million from biomedical, chemical, life, public health, and medical sciences | Classic, PDF, EPUB |
| ScienceDirect (Elsevier, 2000) | 1823 - present | 8,077 life and health sciences journals and book chapters | |
| Wiley Online Library (Wiley-Blackwell, 2010) | Unknown | Journals, Online Books, and Reference Works (biomedical coverage unknown) | Classic, PDF |
Each literature archive offers one or more media or formats where the retrieved literature can be consumed (read) by the user. Currently, the aforementioned literature archives offer at least four types of viewing media. The first view is the classic view wherein the article can be viewed on the archive website itself. This view does not have any page breaks and needs to be read by scrolling vertically through a single long page. This is the default HTML format view offered by most literature sources for quick reference. The second viewing media is the PDF format (.pdf extension) wherein the article can be downloaded onto a device. All literature sources archive full-text in the PDF format that can be used to read on laptops, desktops, and Kindle, and can be printed into a hard copy. PDF format appears exactly as it would appear on a piece of paper; it allows paging, zooming, annotation, and commenting. The third viewing media is the open e-book standard EPUB (.epub extension) offered by PMC and ScienceDirect. This format offers a downloadable file that can be displayed on several devices and readers such as Calibre, iBooks, Google Books, Mobipocket, on various platforms such as Android, Windows, Mac OS X, iOS, Web, Google Chrome Extensions, etc. Finally, PMC offers a new view, PubReader [33], a user-friendly modification to the classic view that emulates the ease of reading the printed version of an article. PubReader was launched by the National Center for Biotechnology Information (NCBI) in 2012 and is coded in CSS and JavaScript. The PubReader display allows an article to be read on a Web browser through laptops, desktops, and tablet computers. PubReader offers ease of navigation and readability by organizing the article into columns and pages to fit into the target screen. In addition, ScienceDirect also offers mobile applications to be used on iPad, phones, and Tablet computers.
3. Text Mining Solutions to address Search Challenges
Given the exponential growth and increasing diversity of biomedical literature, the default querying mechanism (keyword or faceted search) would no longer be enough to meet the user needs. There is a need to provide alternative methods of writing queries and interactive support in query formulation [34–36]. Existing biomedical search tools have made a few efforts in this direction, such as keyword recommendation feature by Web of Science, and flexible keyword searching by Embase. Even when the user finds the right query to input, identifying the few most relevant articles among thousands of citations is not getting any easier. While most search tools employ a ranking algorithm to compute the relevance score of a given article, relevance remains an important topic in IR research [37,38]. Existing tools also provide filters to narrow down the results by different fields. Their ability to present the results in a summarized manner however remains largely unexplored.
In response to the shortcomings of the existing tools, the literature describes many alternative or experimental search interfaces. In this section, we discuss advanced NLP and IR techniques, primarily by discussing alternative interfaces implementing methods not yet available in the major literature search tools. We categorize these techniques into five sections: text similarity search, semantic search, query support, relevance ranking, and clustering; the former three primarily address the search challenges, and latter two address the result presentation issues.
3.1 Text Similarity Search
It can be difficult for users to make their exact information need explicit and then translate it into a query. Several alternative search interfaces have implemented another type of search where the query consists of one or more documents known to be relevant. The relevance of the documents to be retrieved is then calculated based on their similarity to the relevant documents.
eTBLAST is a tool for searching the literature for documents similar to a given passage of text, such as an abstract [39]. The tool extracts a set of keywords from the text and uses these to gather a subset of the literature. A final similarity score is computed for each document in the set by aligning the sentences in the input passage with the document retrieved. MedlineRanker allows the user to input a set of documents, and then finds the set of words most discriminative of the documents within the set [40]. These are then used as features in a classifier (Naïve Bayes), which is applied to unlabeled documents to return the most relevant results. While effective, this approach requires a sufficiently large training set, between 100 and 1000 abstracts.
Recent work by Ortuno et al. [41] partially alleviates the need for a large training set by allowing the user to enter a single abstract as query. The articles cited by the input article are then used to enrich the input set. This approach significantly improves the quality of the results over using only the input article and also typically returns significantly better results than pseudo relevance feedback. Tbahriti et al. [42] significantly improved the ability to determine whether two articles were related by classifying each sentence according to its purpose in the argumentative structure of the abstract (Purpose, Methods, Results, Conclusion). They found that the best results were obtained by increasing the weight of Purpose and Conclusion sentences relative to sentences classified as Methods or Results.
MScanner is similar to the other textual similarity tools in that it learns a classifier (Naïve Bayes) from a set of relevant documents input by the user [43]. In the case of MScanner, however, the only features are the set of MeSH terms associated with each article and the name of the journal where the article appears, resulting in a very high speed retrieval system. Other systems have experimented with using inputs other than text. Caipirini, for example, allows the user to specify a set of genes that are of interest and a set of genes that are not of interest [44]. The system locates abstracts mentioning the genes specified, and the system extracts keywords that appear more frequently than chance in these abstracts. These keywords are then used as features for a classifier (SVM), which provides a score representing the similarity of a text to the abstracts that mention the genes of interest versus the background set. This classifier is then applied to all of Medline and the top results are returned.
3.2 Semantic Search
A large part of the meaning of biomedical texts is captured by the entities they mention and the relationships discussed. This observation can be exploited to support semantic search in both the queries and in the way the results are displayed to the user. Systems supporting semantic search also differ in the types of entities and relationships extracted and the methodology employed.
MedEvi is a semantic search tool intended for finding evidence of specific relationships [45]. The tool recognizes 10 keywords representing entity types, such as “[gene]” and “[disease].” In addition, the tool orders results by preferring results containing the terms in the same order as they appear in the query and within close proximity. Kleio supports keyword searches of multiple prespecified fields [46], including both semantic types (including Protein, Metabolite, Disease, Organ, Acronyms, and Natural phenomenon) and article metadata (e.g. author). A specialized tool for specifically querying authors is Authority [47]. Authority uses a clustering approach over article metadata to determine whether ambiguous author names represent the same person or not. When the users queries for an author, the system displays the matching author clusters.
MEDIE allows queries to specify any combination of subject, verb and object [48]. For example, the query representing “What causes cancer?” would be verb=“cause” and object=“cancer.” This query returns a list of text fragments where the verb matches “cause” and its object is “cancer” or any of its hyponyms, such as “leukemia.” The results highlight genes and diseases in different colors. PubNet extracts entities and relationships from the articles returned by a standard PubMed query, then visualizes the results as a graph [49]. Entities supported include genes and proteins, MeSH terms and authors.
The EBIMed service uses keyword queries as input and provides a listing of the entities most common in documents matching the query [50]. EBIMed supports a fixed set of entity types (protein, cellular component, biological process, molecular function, drug and species) and locates both entities and relationships between two entities. All results are linked to the biological database that defines the entity. Quertle locates articles that describe relationships between the entities provided in a query, results are grouped by the relationship described [51,52]. Users may also switch to a keyword search with a single click. Quertle also supports a list of predefined query keywords that refer to entity types of varying granularity.
A Web-based text mining application named PubTator[53,54] was recently developed to support manual biocuration [55–58]. Because finding articles relevant to specific biological entities (such as gene/protein) is often the first step in biocuration, PubTator supports entity-specific semantic searches based on the use of several challenging-winning named entity recognition tools [59–64].
3.3 Query Support
An important aspect of improving the relevance of query results is to help the user translate their information need into a query. While text similarity, as discussed in Section 3.1, is a useful method for reducing this barrier, another method is to directly support the creation and revision of both keyword and faceted queries [34].
The iPubMed tool allows searching MEDLINE records to be more interactive through the search-as-you-type paradigm. Query results are dynamically updated after every keypress. iPubMed also supports approximate search, allowing users to dynamically correct spelling errors.
PubMed Assistant is a standalone system which includes a visual tool for creating Boolean queries and a query refinement tool that gathers useful keywords from results marked relevant by the user [65]. PubMed Assistant also supports integration with a citation manager.
Schardt et al. [66] demonstrated that search interfaces supporting the PICO system for focusing clinical queries improved the precision of the results. PICO is a framework for supporting evidence based medicine and is an acronym for Patient problem, Intervention, Comparison and Outcome [67,68]. SLIM is a tool emphasizing clinical queries which uses slider bars to quickly customize query results. Modifiable parameters include the age of the article, the journal subset, and both the age group and the study design of the clinical trial reported. askMEDLINE is a system which accepts clinical queries in the form of natural language questions. The system is particularly designed to support users who are not medical experts.
3.4 Relevance Ranking
Ranking results in order of their relevance to the query is a well-supported technique for reducing the workload of the user, and is supported in most existing tools for searching the literature except PubMed itself. While straightforward measurements such as TF-IDF are known to work well [37], there are still aspects that can be improved.
A common ranking technique in web search is to incorporate a measurement of the importance of the document into the score. The scientific record contains many types of bibliometric information that can be used to infer the quality or importance of an article. The PubFocus system, for example, ranks relevant documents according to an importance score that includes the impact factor of the journal and the volume of citations [69].
Bernstam et al. [70] demonstrated that algorithms that use citation data to determine document importance – including both simple citation counts and PageRank – significantly improve over algorithms that do not use citation data. Unfortunately, however, citation data suffers from “citation lag” – the period of time between when an article is published and when it is cited by another article. Tanaka et al. [71] partially overcome this limitation by using the data available at publication to learn which articles are likely to eventually be highly cited.
Lin [72] takes a different approach and instead uses the PubMed Related Articles tool to create a graph by linking similar document pairs. PageRank is then applied, producing a score for each document where higher scores imply the document contains more of the content from its neighbors in the graph. Scores are thus independent of any query but documents with higher scores will naturally be relevant for a wider range of queries.
Yeganova et al. [73] examined PubMed query logs and found that users frequently enter phrases such as “sudden death syndrome” without the quotes to indicate that the query contains a phrase. While PubMed interprets such queries as the conjunction of the individual terms, the authors demonstrate a qualitative difference between results that contain all terms and results that contain the terms as a phrase. They conclude that it would be beneficial to attempt to interpret such queries as containing a phrase, and in particular suggest that documents containing the terms in close proximity are more relevant than results that merely contain all terms.
The RefMed system employs relevance feedback to explicitly model the relevance of query results [74]. In relevance feedback, the system returns an initial set of results, allows the user to indicate whether each result is useful, and then uses the input as feedback to improve the next round of results. While relevance has traditionally been considered to be binary, RefMed uses a learning to rank algorithm (rankSVM) to allow the user to specify varying degrees of relevance along a scale.
The MiSearch tool uses an implicit form of relevance feedback to model the relevance of articles to the user [75]. The system automatically collects relevant documents by recording which documents are opened while browsing. This data is used to create a model of the likelihood that the user will open a document that can be used to rank the results of any query. Features for the model include authors, journal and PubMed indexing information.
3.5 Clustering Results
Clustering the results of the user query into topics helps in several ways. First, clustering the results helps to differentiate between the different meanings of ambiguous query terms. Second, in large sets of search results it can help the user focus on the subset of documents that interest them. Third, the clusters themselves can serve as an overview of the topic. This method has been considered in several PubMed derivatives that vary in their method of determining the clustering methodology. Popular variations include MeSH terms, other semantic content (such as UMLS concepts and GO terms), keywords, and document metadata (such as journal, authors, and date).
Anne O’Tate provides additional structure and a summary of the query results by clustering the content of the documents retrieved and also by extracting important words, the publication date, authors and their institutions [76]. Users are allowed to extend the query by any of the summarized information simply by clicking on it, and any query returning less than 50 results can be expanded to include the articles most closely related.
The McSyBi tool clusters query results both hierarchically and non-hierarchically [77]. Whereas the non-hierarchical clustering is primarily useful for focusing the query on particular subsets, the hierarchical clustering provides a brief summary of the query results. McSyBi also provides the ability for the user to adjust or reformulate the clustering by introducing a MeSH term, which is interpreted as a new binary feature for each document, depending on whether the document has been assigned the specified MeSH term. Users can also introduce a UMLS Semantic Type, which is considered present if the document is assigned at least one MeSH term with the specified type.
GoPubMed originally used the Gene Ontology (GO) [78] to organize the search results [79]. It currently groups search results according to categories “what” (biomedical concepts), “where” (affiliations and journals), “who” (author names), and “when” (date of publication). The “what” category is further subdivided into concepts from Gene Ontology, MeSH and UniProt. GO terms are located in the abstracts retrieved, even if they do not appear directly, and are highlighted when the abstract is displayed.
XplorMed is a tool for multifactored analysis of query results [80,81]. Results are displayed grouped both by coarse MeSH categories and important words that are shown both in summary and in context. Users may then explore the important words in more depth or display results ranked by inclusion of the important words.
Boyack et al. [7] sought to determine which clustering approach would produce the most coherent clusters over a large subset of MEDLINE. The analysis considered five analytical techniques: a vector space approach with TF-IDF vectors and cosine similarity, latent semantic analysis, topic modeling, the Poisson-based language model BM25 and PubMed Related Articles. The analysis also considered two data sources, MeSH subject headings and words from titles and abstracts. The article concluded that PubMed Related Articles created the most coherent clusters, closely followed by BM25, and also concluded that the clusters based on titles and abstracts are significantly better than those based only on MeSH headings.
SEACOIN (Search Explore Analyze COnnect INspire) is a system that merges important word analysis with clustering and a graphical visualization to achieve a simple interface suitable for novice users [82]. The SEACOIN visualization combines a word cloud that allows the user to add additional terms to the query, a multi-level tree-like graphic that allows users to see the relative number of documents containing different terms and term combinations, and a table listing the documents returned.
SimMed presents users with clusters of documents ranked by their degree of relevance to the query [83]. The interface emphasizes the clusters found to provide a summary of the query topic, thereby explicitly supporting exploratory searches. The clusters used are computed offline, allowing high retrieval performance.
4. Future Trends to Improve Biomedical Literature Access
In terms of the search tools, in addition to the research directions highlighted in Section 3, we can expect to see more reader-friendly and smart applications based on advanced IR and NLP techniques in order to help readers find and digest articles more effectively and efficiently. Furthermore, with the use of social media such as blogging and tweeting, new ways of sharing and recommending papers will gain more importance in the future, in addition to the traditional search-based mechanism. For instance, using social media makes it easier to make and share comments on papers, thus providing alternative views with respect to the impact of individual papers. In the future, biomedical literature search could also be personalized. That is, search results are tailored towards the interests of individual researchers based on their own work and/or past searches. In other words, the same query by two different users may return different search results. This is desirable in certain cases. For instance, a bench scientist and medical doctor are likely to search for different information (biological vs. clinical, respectively) even though they both search for the same drug and disease pair. In the general Web search domain, personalized search has been shown useful [84]. Therefore, such a feature could also be helpful to users when it comes to the biomedical literature search.
With regard to open access papers, we believe its size will continue to grow rapidly over the next 5 to 10 years. This is evidenced by the increasing number of publishers and journals interested in adopting the open access policy, as well as by the ever-growing interests from the scientific community. That is, more and more authors are considering open-access journals as their preferred choice for publishing their work. As this happens and together with Web and computer technology advances, we can imagine free access to most research articles anywhere, anytime on any devices.
Finally, with increased use of portable devices such as smartphones and computer tablets to access the Internet, there are growing needs and interests in searching and reading literature on those devices. Portable devices provide a great deal of benefits such as convenience, but also present new challenges. First, it is less likely to print out the papers with portable devices – a common way for reading papers. As a result, reading directly on those devices becomes necessary. But compared to desktop or laptop computers, the screen size of portable devices is usually much smaller. As such, readability becomes a real issue on those small-screen devices, especially when it comes to reading papers. This is because unlike reading emails or news articles, people do not generally read straight through an article. Instead, they often need to go back and forth when reading a journal article in order to understand and digest its content. Scrolling up and down on a modern computer screen is hard but still viable; this kind of operation becomes almost impossible on small-screen devices. As mentioned in Section 2.2, there has already been work on supporting convenient reading on small-screen devices from new reading apps to reader-friendly Web interfaces. Although these tools already provide better readability than the traditional Web browsers, further improvement is needed in order to make users to read and digest articles comfortably on those devices. We also expect advances in Web technology to help facilitate such a transition.
Acknowledgments
This research was supported by the Intramural Research Program at the National Institutes of Health, National Library of Medicine.
Contributor Information
Ritu Khare, Email: ritu.khare@nih.gov, National Center for Biotechnology Information, U.S. National Library of Medicine, NIH, Blg 38 A, Rm 1003B, 8600 Rockville Pike, Bethesda, MD 20894.
Robert Leaman, Email: robert.leaman@nih.gov, National Center for Biotechnology Information, U.S. National Library of Medicine, NIH, Blg 38 A, Rm 1003E, 8600 Rockville Pike, Bethesda, MD 20894.
Zhiyong Lu, Email: zhiyong.lu@nih.gov, National Center for Biotechnology Information, U.S. National Library of Medicine, NIH, Blg 38 A, Rm 1003A, 8600 Rockville Pike, Bethesda, MD 20894.
References
- 1.PubMed. US National Library of Medicine, National Institutes of Health; http://www.ncbi.nlm.nih.gov/pubmed. [Google Scholar]
- 2.Google Scholar. Google; http://scholar.google.com/ [Google Scholar]
- 3.PubMed Central. US National Library of Medicine, National Institutes of Health; http://www.ncbi.nlm.nih.gov/pmc/ [Google Scholar]
- 4.Hunter L, Cohen KB. Biomedical language processing: what’s beyond PubMed? Molecular cell. 2006;21 (5):589–594. doi: 10.1016/j.molcel.2006.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Islamaj Dogan R, Murray GC, Neveol A, et al. Understanding PubMed user search behavior through log analysis. Database: the journal of biological databases and curation. 2009;2009:bap018. doi: 10.1093/database/bap018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garg AX, Iansavichus AV, Kastner M, et al. Lost in publication: Half of all renal practice evidence is published in non-renal journals. Kidney international. 2006;70 (11):1995–2005. doi: 10.1038/sj.ki.5001896. [DOI] [PubMed] [Google Scholar]
- 7.Boyack KW, Newman D, Duhon RJ, et al. Clustering more than two million biomedical publications: comparing the accuracies of nine text-based similarity approaches. PloS one. 2011;6 (3):e18029. doi: 10.1371/journal.pone.0018029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin J, Wilbur WJ. PubMed related articles: a probabilistic topic-based model for content similarity. BMC bioinformatics. 2007;8:423. doi: 10.1186/1471-2105-8-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yiotis K. The Open Access Initiative: A New Paradigm for Scholarly Communications. Information Technology and Libraries. 2005;24(4) [Google Scholar]
- 10. [Accessed 13 July 2013];Wikipedia PubMed Central. http://en.wikipedia.org/wiki/PubMed_Central.
- 11.Davis PM. Public accessibility of biomedical articles from PubMed Central reduces journal readership--retrospective cohort analysis. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2013;27 (7):2536–2541. doi: 10.1096/fj.13-229922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grefsheim SF, Rankin JA. Information needs and information seeking in a biomedical research setting: a study of scientists and science administrators. Journal of the Medical Library Association: JMLA. 2007;95 (4):426–434. doi: 10.3163/1536-5050.95.4.426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hemminger BM, Lu D, Vaughan KTL, et al. Information Seeking Behavior of Academic Scientists. Journal of American Society for Information Science and Technology. 2007;58 (14):2205–2225. [Google Scholar]
- 14.Kim JJ, Rebholz-Schuhmann D. Categorization of services for seeking information in biomedical literature: a typology for improvement of practice. Briefings in bioinformatics. 2008;9 (6):452–465. doi: 10.1093/bib/bbn032. [DOI] [PubMed] [Google Scholar]
- 15.PubMed Tutorial, Automatic Term Mapping. US. National Library of Medicine, National Institutes of Health; http://www.nlm.nih.gov/bsd/disted/pubmedtutorial/020_040.html. [Google Scholar]
- 16.Embase: Biomedical Database. Elsevier; http://www.elsevier.com/online-tools/embase. [Google Scholar]
- 17. [Accessed 16 July 2013];Roche A-M Embase - Answers to your biomedical questions. http://www.slideshare.net/rocheam/embase-introduction.
- 18.Lu Z. PubMed and beyond: a survey of web tools for searching biomedical literature. Database: the journal of biological databases and curation. 2011;2011:baq036. doi: 10.1093/database/baq036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Falagas ME, Giannopoulou KP, Issaris EA, et al. World databases of summaries of articles in the biomedical fields. Archives of internal medicine. 2007;167 (11):1204–1206. doi: 10.1001/archinte.167.11.1204. [DOI] [PubMed] [Google Scholar]
- 20.Hoskins IC, Norris WE, Taylor R. Databases of biomedical literature: getting the whole picture. Archives of internal medicine. 2008;168 (1):113. doi: 10.1001/archinternmed.2007.26. author reply 113–114. [DOI] [PubMed] [Google Scholar]
- 21.Bakkalbasi N, Bauer K, Glover J, et al. Three options for citation tracking: Google Scholar, Scopus and Web of Science. Biomedical digital libraries. 2006;3:7. doi: 10.1186/1742-5581-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Falagas ME, Pitsouni EI, Malietzis GA, et al. Comparison of PubMed, Scopus, Web of Science, and Google Scholar: strengths and weaknesses. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2008;22 (2):338–342. doi: 10.1096/fj.07-9492LSF. [DOI] [PubMed] [Google Scholar]
- 23.Bar-Ilan J. Which h-index? — A comparison of WoS, Scopus and Google Scholar. Scientometrics. 2008;74 (2):257–271. [Google Scholar]
- 24.Web of Science. Thomson Reuters; http://thomsonreuters.com/web-of-science/ [PubMed] [Google Scholar]
- 25.Scopus - Document Search. Elsevier; http://www.scopus.com/home.url. [Google Scholar]
- 26.The Thomson Reuters Journal Selection Process. Thomson Reuters; http://wokinfo.com/essays/journal-selection-process/ [Google Scholar]
- 27.Tuomilehto J, Lindstrom J, Eriksson JG, et al. Prevention of type 2 diabetes mellitus by changes in lifestyle among subjects with impaired glucose tolerance. The New England journal of medicine. 2001;344 (18):1343–1350. doi: 10.1056/NEJM200105033441801. [DOI] [PubMed] [Google Scholar]
- 28.CINAHL Plus with Full Text. EBSCO; http://www.ebscohost.com/academic/cinahl-plus-with-full-text. [Google Scholar]
- 29.SpringerLink. Springer; http://link.springer.com/ [Google Scholar]
- 30.ScienceDirect.com. Search through over 11 million science, health, medical journal full text articles and books. Elsevier; http://www.sciencedirect.com/ [Google Scholar]
- 31.ScienceDirect Platform Brochure. Elsevier; http://www.info.sciverse.com/documents/files/content/pdf/SDPlatformBrochure_06.pdf. [Google Scholar]
- 32.Journals. Wiley Online Library; http://olabout.wiley.com/WileyCDA/Section/id-406089.html. [Google Scholar]
- 33.Lipman D. NLM Technical Bulletin. 2012. The PubReader View: A New Way to Read Articles in PMC. [Google Scholar]
- 34.Lu Z, Wilbur WJ, McEntyre JR, et al. Finding query suggestions for PubMed. AMIA Annual Symposium proceedings/AMIA Symposium AMIA Symposium. 2009;2009:396–400. [PMC free article] [PubMed] [Google Scholar]
- 35.Neveol A, Dogan RI, Lu Z. Author keywords in biomedical journal articles. AMIA Annual Symposium proceedings/AMIA Symposium AMIA Symposium. 2010;2010:537–541. [PMC free article] [PubMed] [Google Scholar]
- 36.Islamaj Dogan R, Lu Z. Click-words: learning to predict document keywords from a user perspective. Bioinformatics. 2010;26 (21):2767–2775. doi: 10.1093/bioinformatics/btq459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lu Z, Kim W, Wilbur WJ. Evaluating relevance ranking strategies for MEDLINE retrieval. AMIA Annual Symposium proceedings/AMIA Symposium AMIA Symposium; 2008. p. 439. [PMC free article] [PubMed] [Google Scholar]
- 38.Lu Z, Kim W, Wilbur WJ. Evaluating relevance ranking strategies for MEDLINE retrieval. Journal of the American Medical Informatics Association: JAMIA. 2009;16 (1):32–36. doi: 10.1197/jamia.M2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Errami M, Wren JD, Hicks JM, et al. eTBLAST: a web server to identify expert reviewers, appropriate journals and similar publications. Nucleic Acids Res. 2007;35 (Web Server issue):W12–15. doi: 10.1093/nar/gkm221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fontaine JF, Barbosa-Silva A, Schaefer M, et al. MedlineRanker: flexible ranking of biomedical literature. Nucleic Acids Res. 2009;37 (Web Server issue):W141–146. doi: 10.1093/nar/gkp353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ortuno FM, Rojas I, Andrade-Navarro MA, et al. Using cited references to improve the retrieval of related biomedical documents. BMC Bioinformatics. 2013;14:113. doi: 10.1186/1471-2105-14-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tbahriti I, Chichester C, Lisacek F, et al. Using argumentation to retrieve articles with similar citations: an inquiry into improving related articles search in the MEDLINE digital library. International journal of medical informatics. 2006;75 (6):488–495. doi: 10.1016/j.ijmedinf.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 43.Poulter GL, Rubin DL, Altman RB, et al. MScanner: a classifier for retrieving Medline citations. BMC Bioinformatics. 2008;9:108. doi: 10.1186/1471-2105-9-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Soldatos TG, O’Donoghue SI, Satagopam VP, et al. Caipirini: using gene sets to rank literature. BioData mining. 2012;5 (1):1. doi: 10.1186/1756-0381-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim JJ, Pezik P, Rebholz-Schuhmann D. MedEvi: retrieving textual evidence of relations between biomedical concepts from Medline. Bioinformatics. 2008;24 (11):1410–1412. doi: 10.1093/bioinformatics/btn117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nobata C, Cotter P, Okazaki N, et al. Kleio: a knowledge-enriched information retreival system for biology. Paper presented at the 31st annual international ACM SIGIR conference on research and development in information retreival.2008. [Google Scholar]
- 47.Torvik VI, Smalheiser NR. Author Name Disambiguation in MEDLINE. ACM transactions on knowledge discovery from data. 2009;3(3) doi: 10.1145/1552303.1552304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ohta T, Miyao Y, Ninomiya T, et al. An intelligent search engine and GUI-based efficient MEDLINE search tool based on deep syntactic parsing. Paper presented at the COLING/ACL Interactive presentation sessions; Sdney, Australia. 2006. [Google Scholar]
- 49.Douglas SM, Montelione GT, Gerstein M. PubNet: a flexible system for visualizing literature derived networks. Genome Biol. 2005;6 (9):R80. doi: 10.1186/gb-2005-6-9-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rebholz-Schuhmann D, Kirsch H, Arregui M, et al. EBIMed--text crunching to gather facts for proteins from Medline. Bioinformatics. 2007;23 (2):e237–244. doi: 10.1093/bioinformatics/btl302. [DOI] [PubMed] [Google Scholar]
- 51.Giglia E. Quertle and KNALIJ: searching PubMed has never been so easy and effective. European journal of physical and rehabilitation medicine. 2011;47 (4):687–690. [PubMed] [Google Scholar]
- 52.Coppernoll-Blach P. Quertle: The Conceptual Relationships Alternative Search Engine for PubMed. J Med Libr Assoc. 2011;99 (2):176–U159. doi: 10.3163/1536-5050.99.2.017. [DOI] [Google Scholar]
- 53.Wei CH, Kao HY, Lu Z. PubTator: a web-based text mining tool for assisting biocuration. Nucleic acids research. 2013;41 (Web Server issue):W518–522. doi: 10.1093/nar/gkt441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wei CH, Kao HY, Lu Z. PubTator: A PubMed-like interactive curation system for document triage and literature curation. Paper presented at the BioCreative Workshop; 2012; Washington DC. 2012. [Google Scholar]
- 55.Arighi CN, Carterette B, Cohen KB, et al. An overview of the BioCreative 2012 Workshop Track III: interactive text mining task. Database: the journal of biological databases and curation. 2013;2013:bas056. doi: 10.1093/database/bas056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Arighi CN, Roberts PM, Agarwal S, et al. BioCreative III interactive task: an overview. BMC bioinformatics. 2011;12(Suppl 8):S4. doi: 10.1186/1471-2105-12-S8-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lu Z, Hirschman L. Biocuration workflows and text mining: overview of the BioCreative 2012 Workshop Track II. Database: the journal of biological databases and curation. 2012;2012:bas043. doi: 10.1093/database/bas043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Neveol A, Wilbur WJ, Lu Z. Improving links between literature and biological data with text mining: a case study with GEO, PDB and MEDLINE. Database: the journal of biological databases and curation. 2012;2012:bas026. doi: 10.1093/database/bas026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lu Z, Kao HY, Wei CH, et al. The gene normalization task in BioCreative III. BMC bioinformatics. 2011;12(Suppl 8):S2. doi: 10.1186/1471-2105-12-S8-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Van Landeghem S, Bjorne J, Wei CH, et al. Large-scale event extraction from literature with multi-level gene normalization. PloS one. 2013;8 (4):e55814. doi: 10.1371/journal.pone.0055814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wei CH, Kao HY, Lu Z. SR4GN: a species recognition software tool for gene normalization. PloS one. 2012;7 (6):e38460. doi: 10.1371/journal.pone.0038460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wei CH, Harris BR, Kao HY, et al. tmVar: a text mining approach for extracting sequence variants in biomedical literature. Bioinformatics. 2013;29 (11):1433–1439. doi: 10.1093/bioinformatics/btt156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Leaman R, Dogan RI, Lu Z. DNorm: Disease Name Normalization with Pairwise Learning to Rank. Bioinformatics. doi: 10.1093/bioinformatics/btt474. (Submitted) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Leaman R, Khare R, Lu Z. NCBI at 2013 ShARe/CLEF eHealth Shared Task: Disorder Normalization in Clinical Notes with DNorm. Conference and Labs of the Evaluation Forum 2013 Working Notes. (In Press) [Google Scholar]
- 65.Ding J, Hughes LM, Berleant D, et al. PubMed Assistant: a biologist-friendly interface for enhanced PubMed search. Bioinformatics. 2006;22 (3):378–380. doi: 10.1093/bioinformatics/bti821. [DOI] [PubMed] [Google Scholar]
- 66.Schardt C, Adams MB, Owens T, et al. Utilization of the PICO framework to improve searching PubMed for clinical questions. BMC medical informatics and decision making. 2007;7:16. doi: 10.1186/1472-6947-7-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Richardson WS, Wilson MC, Nishikawa J, et al. The well-built clinical question: a key to evidence-based decisions. ACP Journal Club. 1995;123 (3):A 12–13. [PubMed] [Google Scholar]
- 68.Armstrong EC. The well-built clinical question: the key to finding the best evidence efficiently. WMJ: official publication of the State Medical Society of Wisconsin. 1999;98 (2):25–28. [PubMed] [Google Scholar]
- 69.Plikus MV, Zhang Z, Chuong CM. PubFocus: semantic MEDLINE/PubMed citations analytics through integration of controlled biomedical dictionaries and ranking algorithm. BMC Bioinformatics. 2006;7:424. doi: 10.1186/1471-2105-7-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bernstam EV, Herskovic JR, Aphinyanaphongs Y, et al. Using citation data to improve retrieval from MEDLINE. J Am Med Inform Assoc. 2006;13 (1):96–105. doi: 10.1197/jamia.M1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tanaka LY, Herskovic JR, Iyengar MS, et al. Sequential result refinement for searching the biomedical literature. J Biomed Inform. 2009;42 (4):678–684. doi: 10.1016/j.jbi.2009.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lin J. PageRank without hyperlinks: reranking with PubMed related article networks for biomedical text retrieval. BMC Bioinformatics. 2008;9:270. doi: 10.1186/1471-2105-9-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yeganova L, Comeau DC, Kim W, et al. How to Interpret PubMed Queries and Why It Matters. Journal of the American Society for Information Science and Technology. 2009;60 (2):264–274. doi: 10.1002/Asi.20979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yu H, Kim T, Oh J, et al. Enabling multi-level relevance feedback on PubMed by integrating rank learning into DBMS. BMC Bioinformatics. 2010;11(Suppl 2):S6. doi: 10.1186/1471-2105-11-S2-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.States DJ, Ade AS, Wright ZC, et al. MiSearch adaptive pubMed search tool. Bioinformatics. 2009;25 (7):974–976. doi: 10.1093/bioinformatics/btn033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Smalheiser NR, Zhou W, Torvik VI. Anne O’Tate: A tool to support user-driven summarization, drill-down and browsing of PubMed search results. Journal of biomedical discovery and collaboration. 2008;3:2. doi: 10.1186/1747-5333-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yamamoto Y, Takagi T. Biomedical knowledge navigation by literature clustering. J Biomed Inform. 2007;40 (2):114–130. doi: 10.1016/j.jbi.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 78.Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nature genetics. 2000;25 (1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Doms A, Schroeder M. GoPubMed: exploring PubMed with the Gene Ontology. Nucleic Acids Res. 2005;33 (Web Server issue):W783–786. doi: 10.1093/nar/gki470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Perez-Iratxeta C, Bork P, Andrade MA. XplorMed: a tool for exploring MEDLINE abstracts. Trends in biochemical sciences. 2001;26 (9):573–575. doi: 10.1016/s0968-0004(01)01926-0. [DOI] [PubMed] [Google Scholar]
- 81.Perez-Iratxeta C, Perez AJ, Bork P, et al. Update on XplorMed: A web server for exploring scientific literature. Nucleic Acids Res. 2003;31 (13):3866–3868. doi: 10.1093/nar/gkg538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lee EK, Lee HR, Quarshie A. SEACOIN--an investigative tool for biomedical informatics researchers. AMIA Annual Symposium proceedings/AMIA Symposium AMIA Symposium. 2011;2011:750–759. [PMC free article] [PubMed] [Google Scholar]
- 83.Mu X, Ryu H, Lu K. Supporting effective health and biomedical information retrieval and navigation: a novel facet view interface evaluation. J Biomed Inform. 2011;44 (4):576–586. doi: 10.1016/j.jbi.2011.01.008. [DOI] [PubMed] [Google Scholar]
- 84.Liu F, Yu C, Meng W. Personalized Web search for improving retrieval effectiveness. IEEE Transactions on Knowledge and Data Engineering. 2004;16 (1):28–40. [Google Scholar]


