Abstract
Recent studies have reported multiple cases of molecular adaptation in cetaceans related to their aquatic abilities. However, none of these has included the hippopotamus, precluding an understanding of whether molecular adaptations in cetaceans occurred before or after they split from their semi-aquatic sister taxa. Here, we obtained new transcriptomes from the hippopotamus and humpback whale, and analysed these together with available data from eight other cetaceans. We identified more than 11 000 orthologous genes and compiled a genome-wide dataset of 6845 coding DNA sequences among 23 mammals, to our knowledge the largest phylogenomic dataset to date for cetaceans. We found positive selection in nine genes on the branch leading to the common ancestor of hippopotamus and whales, and 461 genes in cetaceans compared to 64 in hippopotamus. Functional annotation revealed adaptations in diverse processes, including lipid metabolism, hypoxia, muscle and brain function. By combining these findings with data on protein–protein interactions, we found evidence suggesting clustering among gene products relating to nervous and muscular systems in cetaceans. We found little support for shared ancestral adaptations in the two taxa; most molecular adaptations in extant cetaceans occurred after their split with hippopotamids.
Keywords: RNA-sequencing, transcriptome, Cetartiodactyla, mammals
1. Introduction
Cetaceans are arguably the most specialized of all mammals, having evolved from a terrestrial ancestor to occupy an obligate aquatic niche [1]. Modern cetaceans show numerous phenotypic adaptations for life in the water; aside from the radical reorganization of their forelimbs into fins and loss of hindlimbs, they are able to dive and tolerate low oxygen, and possess modified circulatory and respiratory systems, large brains, hairlessness and transformations in sensory perception [1]. Some cetaceans show extreme longevity, as well as resistance to cancer, wound healing and insulin resistance [2,3]. Other major adaptations pertain to feeding ecology; indeed, modern cetaceans diverged approximately 34 Ma [4,5] into the toothed whales (suborder Odontoceti), which evolved echolocation to hunt using ultrasonic pulses and possess a highly specialized inner ear, and the baleen whales (suborder Mysticeti), which lost their teeth and instead evolved a novel keratinous material for filtering smaller prey [1].
Molecular evidence has revealed that the closest living relatives of the cetaceans are the two extant members of the family Hippopotamidae: the common and pygmy hippopotamus (‘hippo’) (e.g. [6,7]). Members of the Cetacea and Hippopotamidae are grouped together in the monophyletic clade Whippomorpha [8], which in turn is nested within the otherwise terrestrial mammalian order Cetartiodactyla that also includes the even-toed ungulates [9]. Recent molecular evidence suggests that the Whippomorpha diverged from other cetartiodactyls approximately 59 Ma and that the cetaceans and hippopotamids split approximately 55 Ma [5].
Both extant hippo species are adapted for spending long periods of time in water [10]. Like some early cetaceans, they can walk on the bottom of bodies of freshwater due to their thick, dense (pachyosteosclerotic) limb bones [11], and they are predominantly hairless, with thickened, lipid-rich skin that lacks sebaceous glands [12,13]. Hippo skin contains subepidermal capillaries with thickened walls to withstand high blood pressure, an adaptation for heat exchange that has also been reported in highly active species, such as cetaceans [13]. Hippos and cetaceans have some behavioural traits in common such as nursing underwater and subaquatic communication [1,14]. Furthermore, fossil cetaceans and hippos both possess a hyperinflated tegmen tympani of the petrosal bone, which may aid in interpreting the directionality of hearing [15]. Yet despite some shared specializations for life in the water, it is currently unclear to what extent cetaceans and hippos evolved these adaptations independently or whether they are ancestral traits, although some fossil evidence suggests the former [1,10,16]. Indeed, the question of whether the last common ancestor of the Whippomorpha exhibited a terrestrial, semi-aquatic or aquatic lifestyle remains unresolved, and there is particular interest in determining when in their evolutionary history cetaceans gained their specialized traits for living in water [1,10].
Genome-wide scans of selection can offer powerful insights into the evolutionary history of adaptive traits (e.g. [17]). However, all genome-scale studies of molecular adaptation in the Whippomorpha to date have been restricted to a few cetaceans [18–24]. Similarly, inferences of selection associated with the evolutionary transition from land to water have relied heavily on data from toothed whales, used in comparative evolutionary analyses together with the cow (Bos taurus) and other more distantly related terrestrial mammals [21,22,24].
To gain a better understanding of the timing and role of natural selection in the transition of cetartiodactyl mammals to a semi-aquatic/aquatic environment, we generated transcriptome data from the common hippopotamus (referred to as ‘hippo’ below) as well as from the humpback whale Megaptera novaeangliae. By analysing these together with existing data from two mysticetes and six odonotocetes, we conducted, to our knowledge, the most comprehensive genome-scale dataset of the group to date [18,23–26]. We reasoned that if adaptation to a semi-aquatic environment preceded the split between hippos and whales, then we would expect to see signatures of positive selection in multiple genes linked to an aquatic lifestyle on the ancestral branch of Whippomorpha. If, on the other hand, adaptation to a more aquatic way of life followed the split between these groups, then we might expect independent changes on each lineage. Under both scenarios, we also predicted a greater molecular signature of aquatic adaptation in cetaceans than in hippos, reflecting the more derived body plan in the former.
2. Material and methods
2.1. Taxon sampling, sequencing and RNA sequencing de novo assembly
New RNA sequencing data for the common hippo Hippopotamus amphibius and humpback whale M. novaeangliae were generated by pair-end Illumina HiSeq sequencing at BGI (electronic supplementary material, table S1), and combined with published sequence data from genomes or transcriptomes of eight other cetacean species: sperm whale (Physeter macrocephalus), Indo-Pacific humpback dolphin (Sousa chinensis), minke whale (Balaenoptera acutorostrata), fin whale (Balaenoptera physalus), finless porpoise (Neophocaena phocaenoides), bottlenose dolphin (Tursiops truncatus), killer whale (Orcinus orca) and Yangtze River dolphin (Lipotes vexillifer) [18,23–26].
2.2. Orthologue identification and dataset assembly
We obtained orthologous coding DNA sequences (CDSs) across cetaceans and the hippo using reciprocal blastx and tblastn searches, with T. truncatus and human as references. Orthologous sequences of 13 other laurasiatherian mammals were obtained from Ensembl [27]. CDS were aligned using PRANK v. 130820 [28] and filtered based on Guidance default parameters [29]. Sequences were further edited and trimmed to avoid problems with missing data and erroneous indels (electronic supplementary material, methods).
2.3. Natural selection analyses
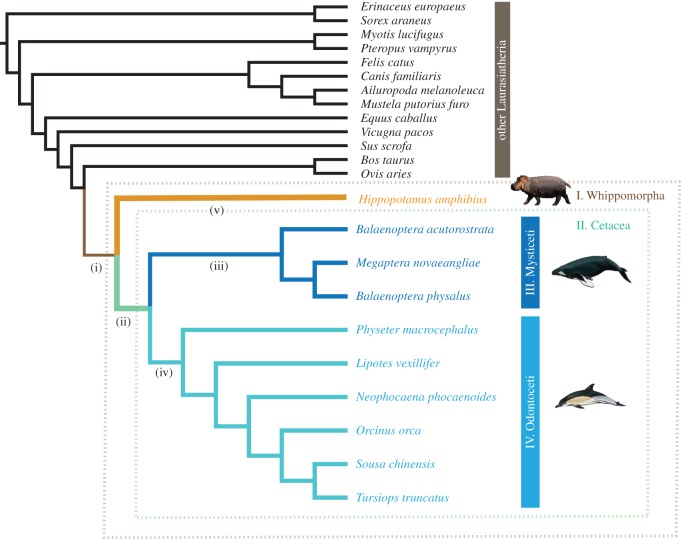
To identify episodes of positive selection, we used codon models in codeml of PAML v. 4.4 [30]. We first implemented branch-site model MA to identify sites under selection [30,31] on five focal branches: (i) Whippomorpha (Hippopotamidae + Cetacea); (ii) Cetacea; (iii) Mysticeti; (iv) Odontoceti; and (v) the terminal branch of H. amphibius (figure 1). Each branch-site model was compared to a null model using the likelihood ratio test (LRT) with 1 d.f., and sites with Bayes Empirical Bayes posterior probabilities of more than 0.50 were considered significant. To ensure that estimated positive ω-values represented genuine selection acting on genes, rather than alignment errors, we filtered out genes in which positively selected sites (PSSs) were found to be highly aggregated within the CDS alignment (median interval distance of PSSs ≤10 codons). The exact numbers of datasets remained in each clade or branch after filtering are given in table 1. Following this step, the associated p-values from the LRTs were corrected for multiple testing according to the Benjamini & Hochberg's [32] procedure that corrects the false discovery rate (FDR; Q-value) to Q<0.10. For further analyses using codon models, see the electronic supplementary material, methods, results and discussion, and table S3.
Figure 1.
Evolutionary relationships among laurasiatherian mammals as used in molecular evolution analyses. The four clades tested for divergent selection are shown in colour and numbered in uppercase: (I) Whippomorpha (Hippopotamidae + Cetacea); (II) Cetacea; (III) Mysticeti and (IV) Odontoceti. Branches tested for positive selection are numbered in lowercase: (i) Whippomorpha (Hippopotamidae + Cetacea); (ii) Cetacea; (iii) Mysticeti; (iv) Odontoceti and (v) hippo.
Table 1.
Genome-wide analysis for bursts of positive selection.
| initial screen for selection |
filtering | no. genes showing evidence of natural selection |
|||
|---|---|---|---|---|---|
| no. total datasets | p-value<0.05 | no. datasets excl. Med PSSs≥10 | p-value<0.05 | FDR q-value<0.10 | |
| branch-site model | ω0≤0, ω≥1 | ω0≤0, ω>1 | ω0≤0, ω>1 | ||
| ancestral Whippomorpha | 6845 | 106 | 4974 | 43 | 1 |
| hippo terminal branch | 6845 | 201 | 4974 | 64 | 2 |
| ancestral Cetacea | 11 925 | 391 | 8602 | 200 | 6 |
| ancestral Mysticeti | 11 925 | 439 | 8602 | 173 | 15 |
| ancestral Odontoceti | 11 925 | 335 | 8602 | 125 | 11 |
2.4. Network analysis of protein–protein interactions
For genes under positive selection, we built protein–protein interaction networks using Igraph v. 0.7.0 [33]. We interrogated the STRING database [34] with the human and dolphin Ensembl gene identifiers for combined interaction scores. To visualize proteins with common functions, we obtained gene ontology (GO) terms under the biological processes domain using topGO [35]. Corresponding GO term names were exported from Ensembl and AmiGO 2 v. 2.1.4, and GO terms were grouped into functional categories that we predicted to be important during the evolution of cetaceans and hippos (electronic supplementary material, methods). These grouped GO terms were then mapped onto each network.
2.5. Test of functional enrichment
For all genes under selection in our focal taxa, regardless of protein–protein interactions, we also tested for functional enrichment using the topGO package [35] (electronic supplementary material, methods, and results and discussion).
3. Results and discussion
3.1. RNA sequencing, assembly and orthologue identification
The number of candidate one-to-one orthologues across eight whippomorph species ranged from 6249 to 16 047 genes (electronic supplementary material, table S2). We combined annotated CDSs from bottlenose dolphin- and human-anchored blast searches (more than or equal to 50% coverage of the full gene length in the human genome), yielding 9267 and 14 234 one-to-one orthologues, respectively (electronic supplementary material, table S2). By sampling these sequences across additional laurasiatherian mammals, we built 11 925 gene alignments, each of which contained at least one member of each of the two extant cetacean suborders (Mysticeti and Odontoceti). Moreover, over half of these (n=6845) also contained the hippo sequence.
3.2. Scans for signatures of natural selection
To identify loci under positive selection at a genomic scale, we used branch-site codon models to estimate the ratio of non-synonymous to synonymous substitution rates (dN/dS) or omega (ω) on the ancestral branches of Whippomorpha, Cetacea, Mysticeti and Odontoceti, as well as the terminal hippo branch ((i), (ii), (iii), (iv) and (v), respectively; figure 1). Of 6845 genes containing the hippo sequence, we found 106 genes showing PSSs on the ancestral branch of Whippomorpha, as well as 201 genes on the hippo terminal branch (table 1). Across all 11 925 genes tested, signatures of molecular adaptation were detected in 391 genes on the ancestral branch of Cetacea, compared to 439 and 335 genes on the ancestral branches of Mysticeti and Odontoceti, respectively (table 1). Because signals of positive selection can sometimes arise from alignment errors, for each gene we inspected the distribution of sites with high omega values (ω>1) and filtered out genes (n=3323) in which such signals were highly aggregated (see Material and methods; table 1; electronic supplementary material, table S3). As a result, the numbers of genes retained as under positive selection decreased to 43 for the ancestral branch of Whippomorpha, and from 201 to 64 for the hippo branch (table 1; electronic supplementary material, table S5). Similarly, excluding genes with highly aggregated PSSs in the three ancestral cetacean branches, we retained 200 genes in the last common ancestor of Cetacea, compared to 173 and 125 genes on the ancestral branches of Mysticeti and Odontoceti, respectively (table 1; electronic supplementary material, table S5). Finally, we applied the FDR; Q<0.10) to correct for multiple testing, reducing further the numbers of genes to between one and 15 genes (table 1), although these numbers are likely to be underestimates given our large sample sizes and strict filtering regime.
3.3. Functional annotation of genes under selection
To compare whether sets of genes under selection in the hippo versus cetaceans show broad differences in both functional role and degree of interactions, we plotted interactions among protein products of genes found to be under positive selection (figure 2; electronic supplementary material, methods and figure S1). The hippo network consisted of 20 proteins with at least one interaction, and a key cluster was centred around serum albumin, ALB (see insets figure 2). In comparison, the network constructed for cetaceans comprised more proteins with a greater degree of connectedness; 105 proteins were connected to at least one other protein (figure 2), while several (e.g. GAPDH, EP300, CDH1) had high numbers of connections suggesting that they are important ‘hubs’. The network with the greatest concentration of connections centred around GMPS, a protein involved in guanine synthesis. By mapping GO terms onto these networks, we identified several instances in both taxa in which proteins associated with common tissue types and/or functions were clustered together. For example, in the hippo network, the linked proteins HMGCR and ALB are both associated with circulation (figure 2a), whereas in the cetacean network neighbouring proteins with functions related to the nervous system were centred around the hub of CDH1 (figure 2b). We also found clustering among proteins involved in other functions; for example, related to cell cycle and ageing in cetaceans (electronic supplementary material, figure S1A), and to lipids in both networks (electronic supplementary material, figure S1B). With one exception, proteins related to hypoxia and DNA repair occurred only in the cetacean network (RAD52, ERCC5 and SMC6), although clustering was limited (electronic supplementary material, figure S1C). No clustering was seen in proteins related to fluid, kidneys, lungs or sensory perception in either network (electronic supplementary material, figure S1D–G). Additional evidence of GO enrichment was restricted to cetaceans, and affected genes involved in brain, blood clotting and sensory perception (electronic supplementary material, results and discussion, tables S7–S9).
Figure 2.

Protein–protein interaction networks for 105 protein-coding gene products tested in both cetaceans and the hippo that were found to be under positive selection in cetaceans. Inset: protein–protein interaction networks for 20 protein-coding genes found to be under positive selection in the hippo. Nodes are labelled with the standard protein names, and the thickness of each connection is scaled to represent the strength of support, with thicker lines representing higher support. (a) Highlights proteins involved in the circulatory system (red) and (b) highlights proteins involved in muscle (yellow) the nervous system (blue) or both (yellow with blue).
3.4. Molecular adaptations in the hippo and whippomorph ancestors
Among the 64 genes that were found to have undergone positive selection along the hippo branch (electronic supplementary material, table S5), we found several associated with lipid metabolism, including those with key roles in the biosynthesis and absorption of cholesterol (i.e. HMGCR, CYP2J2 and CYP8B1; [36–38]) as well as genes linked to metabolic disorders and/or obesity, including CPXM1 and PON3[39–41]. Other genes seen to be under positive selection in the hippo branch alone are known to function in glucose regulation (e.g. PDK4; [42–44]). Interestingly, PDK4 has also been found to be under selection in another aquatic mammal, the walrus [18]. Another gene with a related metabolic function is AGL, involved in glycogen degradation [45,46]. Genes potentially linked to obesity may be related to the hippo's comparatively large size and capacity for fat storage, although the physiology of hippos in general remains little explored [47]. Other genes showing molecular adaptation in the hippo encode proteins that might relate to the unusual demands placed on its circulatory system; notably albumin, a constituent of blood plasma that helps regulate osmotic pressure [48]. Indeed, aside from the need to rapidly cool their skin, hippos appear to experience several circulatory changes during dives, including bradycardia (low heart rate), while at the same time maintaining their arterial blood pressure [49]. We also found positive selection in PER1, which encodes an essential component of the circadian clock [50], as well as genes associated with muscle function (e.g. CKMT2).
To identify genes important in the early evolution of the Whippomorpha, we examined the ancestral branch and recovered 43 genes under positive selection (electronic supplementary material, tables S5–S6). Of these, we were able to verify PSSs in nine genes for both hippo and cetaceans, as in the rest of the genes hippo sequences contained missing data for amino acid sites identified as being under positive selection. Positively selected genes included CPT1A, a gene associated with type II diabetes and involved in fatty acid oxidation, and XRCC6, which codes for a DNA repair protein.
3.5. Molecular adaptations during cetacean evolution
Like in the hippo, genes underpinning metabolism were also found to be under selection in the cetaceans. Indeed, our results indicate that as many as 25 positively selected genes in cetaceans are involved in sugar metabolism, insulin availability or lipid metabolism. For example, two solute-carrier genes (SLC5A10 and SLC9B2) together with LMF1 and MARCH6 have all been implicated in aspects of diabetes, obesity and/or body mass index (electronic supplementary material, table S9). In light of our results, it is noteworthy that the bottlenose dolphin has been proposed as an emerging model for studying type II diabetes based on reports that fasting individuals retain comparatively high glucose levels; this diabetic state may be related to the demand to provide glucose to the brain while diving [3].
Many of the other amino acid changes that we found in the cetacean branches also appear to correspond to their ability to dive and resist oxidative stress. Indeed, some species dive to extraordinary depths; for example, Cuvier's beaked whale (Ziphius cavirostris) can reach more than 1000 m [51]. To do this, cetaceans collapse lungs, sequester blood in retia mirabilia and maintain higher haemoglobin and myoglobin concentrations than terrestrial mammals [52]. Our genome-wide scans of nine cetacean species revealed selection in key hypoxia-related genes, including DDIT4, EP300 and MGEA5. MGEA5 interacts directly with the product of OGT, a hypoxia gene that has undergone massive gene copy number expansion in cetaceans [23]. We also found evidence for molecular adaptation in at least 13 genes involved in muscle and/or heart development and contraction in cetaceans (electronic supplementary material, table S9). Other studies have reported evidence that cetaceans have developed molecular adaptations to compensate for the lack of oxygen during dives. For example, in many diving aquatic mammals including cetaceans, myoglobin (a protein that stores oxygen in muscles) has evolved to have a greater charge, decreasing the tendency of molecules to clump and therefore increasing oxygen storage capacity in muscle cells [53].
Strikingly, we also found selection in eight genes related to blood clotting or platelet formation. For example, SERPINC1 produces the protein antithrombin, which interrupts the formation of blood clots [54]. It is notable that SERPINC1 was identified as being under positive selection in addition to containing a convergent amino acid change in two cetaceans (T. truncatus and O. orca), the walrus and manatee, in a recent study of marine mammal genomes [18]. Clotting in cetaceans differs from terrestrial mammals in that there is a relative lack of scab formation after wounding [55,56]. This reduced clotting has been attributed to the contact of blood with water rather than air, as well as the need to sequester blood in stagnant reservoirs while diving [56]. Moreover, cetaceans show accelerated wound healing, reducing infection and accelerating tissue repair [57].
We identified signatures of selection in at least 46 genes in the common ancestor of cetaceans associated with the nervous system and brain development, which is interesting given that cetaceans are characterized by large absolute brain sizes, large brain-to-body mass ratios [58], high numbers of neocortical neurons [59] and high cognitive capacity (e.g. [60]). Indeed, many of the genes we found have been implicated in neurological disorders in humans, such as microcephaly, mental retardation, major depressive disorder and Alzheimer's disease (electronic supplementary material). PSSs were also detected in genes involved in myelination [61], neural connectedness, axonal guidance, cognition, neuronal development and neural progenitor cell proliferation. Previous studies have also reported some nervous system genes to be under selection in the Tursiops genome [20–22]; however, owing to the number of cetacean taxa included, our study was able to localize positive selection to distinct branches within the tree. In addition, we found that 15 nervous system-related genes showed evidence of positive selection on the mysticete ancestral branch, while six genes contained PSSs on the odontocete ancestral branch. These results are contrary to expectations, as mysticetes, while possessing large absolute brain size in some species, have smaller brain to body size ratios than odontocetes and might be expected to have fewer nervous system genes under selection [58].
The transition from a terrestrial to a wholly aquatic environment means that cetaceans must depend on the properties of water for the transmission of light and sound. Adaptations for living in low light include a thickened cornea, spherical lens and reduced numbers of cones [62]. We determined whether cetaceans show positive selection in loci related to visual perception and found evidence in eight genes, some of which are known to be expressed in the cornea and/or retina, and are otherwise implicated in visual diseases (electronic supplementary material). These results complement earlier findings that cetaceans show several functional molecular changes in (or loss of) their opsin genes [63,64]. We also found molecular adaptation in five genes underpinning hearing; however, despite the fact that the toothed whales have evolved extremely high-frequency sound perception, none of these genes were exclusive to this group. Instead, the gene TNC was found to be under selection in the mysticetes only, while TECTA, JAG2 and USH1C were under selection in the ancestral branch of all cetaceans (although the latter also showed selection in odontocetes). These hearing genes add to the growing number that has been reported to be of potential importance in whales and dolphins [65–69].
In general, we find a much wider range of molecular adaptations in the cetaceans than hippos, probably reflecting their more derived body plan. For example, apart from those loci already discussed, molecular changes were also found in genes related to the kidneys (n=8) and skin/hair (n=8) (electronic supplementary material, table S9). Compared to terrestrial mammals, cetacean epidermis typically grows more quickly with fewer layers and is also less keratinized with increased cellular production and lifespan (e.g. [70,71]). Although other studies have identified the integument as a target of molecular evolution in whales and dolphins, these were unable to rule out the possibility that such changes occurred prior to the split with the hippos [20–22,24,72].
When not diving, cetaceans are exposed to the potentially harmful effects of solar radiation [73]. We found many genes under selection that are involved in DNA repair (n=5) and/or cancer suppression (n=13) in cetaceans, particularly in the mysticete lineage, where we discovered at least 12 of these genes under positive selection, including the RAD52, which is essential for double-strand DNA break repair and genomic maintenance of cancer prevention (electronic supplementary material, table S9). Aside from sun exposure, adaptive modification of genes involved in DNA damage repair and tumour suppression may serve to overcome the predicted 1000-fold increase in cancer risk thought to arise as a result of the increased number of cell divisions in these exceptionally large and long-lived mammals [2]. Indeed, Mysticeti contains both the largest (Balaenoptera musculus) and oldest recorded (Balaena mysticetus; more than 200 years) mammalian species, yet do not show elevated rates of cancer [2]. Our detection of positive selection in DNA damage-related genes in cetaceans augments the result of GO analyses (also see [23,24]).
Overall, we find little support for shared ancestral aquatic adaptations in hippos and cetaceans. In particular, while many molecular adaptations thought to be important for the aquatic environment were recorded on ancestral cetacean branches (mysticetes, odontocetes or both) a comparison of coding sequences that were available for all focal members of the Whippomorpha revealed only a few cases of positive selection along the ancestral branch of the entire clade. Explanations for these findings, apart from the greater degree of morphological adaptation for aquatic existence in cetaceans, include the fragmentary nature of the hippo RNA-sequencing data as well as the relatively short evolutionary time separating the split of Whippomorpha from Ruminantia and the subsequent divergence of hippos from cetaceans. Consequently, our results seem to suggest that cetaceans and hippos evolved most aquatic adaptations separately. On the other hand, we found similar selection pressures acting on genes implicated in lipids in both groups, and more work is needed to determine whether these signatures are related to specialized lipid-rich integuments that characterize semi-aquatic and aquatic animals [13].
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
We thank S. Bailey J. Parker, K. Warren and H. Oliveira for helpful advice and discussions. We are also grateful to C. Walker (QMUL GridPP High Throughput Cluster) for providing access to computing facilities. Analyses were performed with the assistance of SBCS-Informatics (http://informatics.sbcs.qmul.ac.uk) and the EPSRC-funded MidPlus cluster at Queen Mary University of London. Illustrations were by C. Buell and provided by J. Gatesy.
Data accessibility
The new short read data for the hippo and humpback whale have been deposited in the SRA of GenBank under the accession nos. SRR2183469 and SRR2183423. Alignments have been submitted to Dryad: http://dx.doi.org/10.5061/dryad.4cp98.
Authors' contributions
S.J.R. conceived the project, and together with G.T. designed the study. S.A. and A.P. collected the whale samples and performed the RNA extractions. M.F.B. provided the hippo samples. G.T. assembled the data and performed the molecular evolution analyses, with input from K.J.T.D., M.R.M. and S.J.R. K.J.T.D. performed the protein–protein interaction and network analyses, with input from G.T., M.R.M. and S.J.R. G.T., M.R.M. and S.J.R. drafted the manuscript. All other authors assisted in revising the manuscript. All authors read and approved the final manuscript.
Competing interests
We declare we have no competing interests.
Funding
This work was funded by the European Research Council (ERC 1076 Starting grant no. 310482) awarded to S.J.R. and a Newton International Fellowship awarded to M.R.M.
References
- 1.Gatesy J, Geisler JH, Chang J, Buell C, Berta A, Meredith RW, Springer MS, McGowen MR. 2013. A phylogenetic blueprint for a modern whale. Mol. Phyl. Evol. 66, 479–506. (doi:10.1016/j.ympev.2012.10.012) [DOI] [PubMed] [Google Scholar]
- 2.Caulin AF, Maley CC. 2011. Peto's paradox: evolution's prescription for cancer prevention. Trends Ecol. Evol. 26, 175–182. (doi:10.1016/j.tree.2011.01.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Venn-Watson SK, Ridgway SH. 2007. Big brains and blood glucose: common ground for diabetes mellitus in humans and healthy dolphins. Comp. Med. 57, 390–395. [PubMed] [Google Scholar]
- 4.McGowen MR, Spaulding M, Gatesy J. 2009. Divergence date estimation and a comprehensive molecular tree of extant cetaceans. Mol. Phyl. Evol. 53, 891–906. (doi:10.1016/J.Ympev.2009.08.018) [DOI] [PubMed] [Google Scholar]
- 5.Meredith RW. et al. 2011. Impacts of the Cretaceous terrestrial revolution and KPg extinction on mammal diversification. Science 334, 521–524. (doi:10.1126/science.1211028) [DOI] [PubMed] [Google Scholar]
- 6.Nikaido M, Rooney AP, Okada N. 1999. Phylogenetic relationships among cetartiodactyls based on insertions of short and long interspersed elements: hippopotamuses are the closest extant relatives of whales. Proc. Natl Acad. Sci. USA 96, 10 261–10 266. (doi:10.1073/pnas.96.18.10261) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Orliac M, Boisserie JR, Maclatchy L, Lihoreau F. 2010. Early Miocene hippopotamids (Cetartiodactyla) constrain the phylogenetic and spatiotemporal settings of hippopotamid origin. Proc. Natl Acad. Sci. USA 107, 11 871–11 876. (doi:10.1073/pnas.1001373107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Waddell PJ, Okada N, Hasegawa M. 1999. Towards resolving the interordinal relationships of placental mammals. Syst. Biol. 48, 1–5. (doi:10.1093/sysbio/48.1.1) [PubMed] [Google Scholar]
- 9.Montgelard C, Catzeflis FM, Douzery E. 1997. Phylogenetic relationships of artiodactyls and cetaceans as deduced from the comparison of cytochrome b and 12S rRNA mitochondrial sequences. Mol. Biol. Evol. 14, 550–559. (doi:10.1093/oxfordjournals.molbev.a025792) [DOI] [PubMed] [Google Scholar]
- 10.Boisserie JR, Fisher RE, Lihoreau F, Weston EM. 2011. Evolving between land and water: key questions on the emergence and history of the Hippopotamidae (Hippopotamoidea, Cetancodonta, Cetartiodactyla). Biol. Rev. 86, 601–625. (doi:10.1111/J.1469-185x.2010.00162.X) [DOI] [PubMed] [Google Scholar]
- 11.Coughlin BL, Fish FE. 2009. Hippopotamus underwater locomotion: reduced-gravity movements for a massive mammal. J. Mammal. 90, 675–679. (doi:10.1644/08-mamm-a-279r.1) [Google Scholar]
- 12.Luck CP, Wright PG. 1964. Aspects of the anatomy and physiology of the skin of the hippopotamus (H. amphibius). Exp. Physiol. 49, 1–14. (doi:0.1113/expphysiol.1964.sp001695) [DOI] [PubMed] [Google Scholar]
- 13.Meyer W, Schmidt J, Busche R, Jacob R, Naim HY. 2012. Demonstration of free fatty acids in the integument of semi-aquatic and aquatic mammals. Acta Histochem. 114, 145–150. (doi:10.1016/j.acthis.2011.03.011) [DOI] [PubMed] [Google Scholar]
- 14.Barklow WE. 2004. Amphibious communication with sound in hippos, Hippopotamus amphibius. Anim. Behav. 68, 1125–1132. (doi:10.1016/j.anbehav.2003.10.034) [Google Scholar]
- 15.O'Leary MA, Patel BA, Coleman MN. 2012. Endocranial petrosal anatomy of Bothriogenys (Mammalia, Artiodactyla, Anthracotheriidae), and petrosal volume and density comparisons among aquatic and terrestrial artiodactyls and outgroups. J. Paleontol. 86, 44–50. (doi:10.1666/10-091.1) [Google Scholar]
- 16.Gatesy J, O'Leary MA. 2001. Deciphering whale origins with molecules and fossils. Trends Ecol. Evol. 16, 562–570. (doi:10.1016/S0169-5347(01)02236-4) [Google Scholar]
- 17.Roux J, Privman E, Moretti S, Daub JT, Robinson-Rechavi M, Keller L. 2014. Patterns of positive selection in seven ant genomes. Mol. Biol. Evol. 31, 1661–1685. (doi:10.1093/Molbev/Msu141) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Foote AD. et al. 2015. Convergent evolution of the genomes of marine mammals. Nat. Genet. 47, 272–275. (doi:10.1038/ng.3198) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keane M. et al. 2015. Insights into the evolution of longevity from the bowhead whale genome. Cell Rep. 10, 112–122. (doi:10.1016/j.celrep.2014.12.008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McGowen MR, Grossman LI, Wildman DE. 2012. Dolphin genome provides evidence for adaptive evolution of nervous system genes and a molecular rate slowdown. Proc. R. Soc. B 279, 3643–3651. (doi:10.1098/rspb.2012.0869) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nery MF, Gonzalez DJ, Opazo JC. 2013. How to make a dolphin: molecular signature of positive selection in cetacean genome. PLoS ONE 8, e65491 (doi:10.1371/journal.pone.0065491) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun YB, Zhou WP, Liu HQ, Irwin DM, Shen YY, Zhang YP. 2013. Genome-wide scans for candidate genes involved in the aquatic adaptation of dolphins. Genome Biol. Evol. 5, 130–139. (doi:10.1093/gbe/evs123) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yim HS. et al. 2014. Minke whale genome and aquatic adaptation in cetaceans. Nat. Genet. 46, 88–92. (doi:10.1038/ng.2835) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou XM. et al. 2013. Baiji genomes reveal low genetic variability and new insights into secondary aquatic adaptations. Nat. Commun. 4, 2708 (doi:10.1038/ncomms3708) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gui D, Jia KT, Xia J, Yang LL, Chen JL, Wu YP, Yi MS. 2013. De novo assembly of the Indo-Pacific humpback dolphin leucocyte transcriptome to identify putative genes involved in the aquatic adaptation and immune response. PLoS ONE 8, e72417 (doi:10.1371/journal.pone.0072417) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lindblad-Toh K. et al. 2011. A high-resolution map of human evolutionary constraint using 29 mammals. Nature 478, 476–482. (doi:10.1038/nature10530) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kinsella RJ. et al. 2011. Ensembl BioMarts: a hub for data retrieval across taxonomic space. Database 2011, bar030. (doi:10.1093/database/bar030) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Loytynoja A, Goldman N. 2005. An algorithm for progressive multiple alignment of sequences with insertions. Proc. Natl Acad. Sci. USA 102, 10 557–10 562. (doi:10.1073/pnas.0409137102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Penn O, Privman E, Ashkenazy H, Landan G, Graur D, Pupko T. 2010. GUIDANCE: a web server for assessing alignment confidence scores. Nuclic Acid Res. 38, W23–W28. (doi:10.1093/nar/gkq443) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang ZH. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591. (doi:10.1093/Molbev/Msm088) [DOI] [PubMed] [Google Scholar]
- 31.Zhang JZ, Nielsen R, Yang ZH. 2005. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol. Biol. Evol. 22, 2472–2479. (doi:10.1093/Molbev/Msi237) [DOI] [PubMed] [Google Scholar]
- 32.Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57, 289–300. [Google Scholar]
- 33.Csardi G, Nepusz T. 2006. The igraph software package for complex network research. InterJournal Complex Systems, 1695 See http://igraph.org [Google Scholar]
- 34.Szklarczyk D. et al. 2015. STRING v10: protein–protein interaction networks, integrated over the tree of life. Nuclic Acid Res. 43, D447–D452. (doi:10.1093/nar/gku1003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alexa A, Rahnenfuhrer J, Lengauer T. 2006. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics 22, 1600–1607. (doi:10.1093/bioinformatics/btl140) [DOI] [PubMed] [Google Scholar]
- 36.Lindgren V, Luskey KL, Russell DW, Francke U. 1985. Human genes involved in cholesterol- metabolism: chromosomal mapping of the loci for the low-density-lipoprotein receptor and 3-hydroxy-3-methylglutaryl-coenzyme-A reductase with CDNA probes. Proc. Natl Acad. Sci. USA 82, 8567–8571. (doi:10.1073/pnas.82.24.8567) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang YZ, Eggertsen G, Gafvels M, Andersson U, Einarsson C, Bjorkhem I, Chiang JYL. 2004. Mechanisms of cholesterol and sterol regulatory element binding protein regulation of the sterol 12 α-hydroxylase gene (CYP8B1). Biochem. Biophys. Res. Commun. 320, 1204–1210. (doi:10.1016/j.bbrc.2004.06.069) [DOI] [PubMed] [Google Scholar]
- 38.Wu SN, Zhang Y, Gardner CO, Chen Q, Li Y, Wang GL, Gao PJ, Zhu DL. 2007. Evidence for association of polymorphisms in CYP2J2 and susceptibility to essential hypertension. Ann. Hum. Genet. 71, 519–525. (doi:10.1111/J.1469-1809.2007.00346.X) [DOI] [PubMed] [Google Scholar]
- 39.Maier EM. et al. 2005. Population spectrum of ACADM genotypes correlated to biochemical phenotypes in newborn screening for medium-chain acyl-CoA dehydrogenase deficiency. Hum. Mutat. 25, 443–452. (doi:10.1002/humu.20163) [DOI] [PubMed] [Google Scholar]
- 40.Perez-Montarelo D, Madsen O, Alves E, Rodriguez MC, Folch JM, Noguera JL, Groenen MAM, Fernandez AI. 2014. Identification of genes regulating growth and fatness traits in pig through hypothalamic transcriptome analysis. Phys. Genomics 46, 195–206. (doi:10.1152/physiolgenomics.00151.2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rai MF, Patra D, Sandell LJ, Brophy RH. 2014. Relationship of gene expression in the injured human meniscus to body mass index a biologic connection between obesity and osteoarthritis. Arthritis Rheumatol. 66, 2152–2164. (doi:10.1002/art.38643) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wende AR, Huss JM, Schaeffer PJ, Giguere V, Kelly DP. 2005. PGC-1 α coactivates PDK4 gene expression via the orphan nuclear receptor ERR α: a mechanism for transcriptional control of muscle glucose metabolism. Mol. Cell Biol. 25, 10 684–10 694. (doi:10.1128/mcb.25.24.10684-10694.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Connaughton S, Chowdhury F, Attia RR, Song SL, Zhang Y, Elam MB, Cook GA, Park EA. 2010. Regulation of pyruvate dehydrogenase kinase isoform 4 (PDK4) gene expression by glucocorticoids and insulin. Mol. Cell Endocrinol. 315, 159–167. (doi:10.1016/j.mce.2009.08.011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rodbell M, Birnbaum L, Pohl SL, Sundby F. 1971. Reaction of glucagon with its receptor: evidence for discrete regions of activity and binding in glucagon molecule. Proc. Natl Acad. Sci. USA 68, 909–913. (doi:10.1073/pnas.68.5.909) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Horinishi A, Okubo M, Tang NLS, Hui J, To KF, Mabuchi T, Okada T, Mabuchi H, Murase T. 2002. Mutational and haplotype analysis of AGL in patients with glycogen storage disease type III. J. Hum. Genet. 47, 55–59. (doi:10.1007/S100380200000) [DOI] [PubMed] [Google Scholar]
- 46.Fujita Y. et al. 2004. Hypercholesterolemia associated with splice-junction variation of inter-α-trypsin inhibitor heavy chain 4 (ITIH4) gene. J. Hum. Genet. 49, 24–28. (doi:10.1007/s10038-003-0101-8) [DOI] [PubMed] [Google Scholar]
- 47.Eltringham SK. 1999. The hippos: natural history and conservation. Princeton, NJ: Princeton University Press. [Google Scholar]
- 48.Ajioka RS, Phillips JD, Kushner JP. 2006. Biosynthesis of heme in mammals. Biochim.Biophys. Acta Mol. Cell. Res. 1763, 723–736. (doi:10.1016/J.Bbamcr.2006.05.005) [DOI] [PubMed] [Google Scholar]
- 49.Elsner R, Franklin DL, Van Citters RL, Kenney DW. 1966. Cardiovascular defense against asphyxia. Science 153, 941–949. (doi:10.1126/science.153.3739.941) [DOI] [PubMed] [Google Scholar]
- 50.Reppert SM, Weaver DR. 2002. Coordination of circadian timing in mammals. Nature 418, 935–941. (doi:10.1038/nature00965) [DOI] [PubMed] [Google Scholar]
- 51.Tyack PL, Johnson M, Soto NA, Sturlese A, Madsen PT. 2006. Extreme diving of beaked whales. J. Exp. Biol. 209, 4238–4253. (doi:10.1242/Jeb.02505) [DOI] [PubMed] [Google Scholar]
- 52.Kooyman GL, Ponganis PJ. 1998. The physiological basis of diving to depth: birds and mammals. Annu. Rev. Physiol. 60, 19–32. (doi:10.1146/Annurev.Physiol.60.1.19) [DOI] [PubMed] [Google Scholar]
- 53.Mirceta S, Signore AV, Burns JM, Cossins AR, Campbell KL, Berenbrink M. 2013. Evolution of mammalian diving capacity traced by myoglobin net surface charge. Science 340, 1234192 (doi:10.1126/science.1234192) [DOI] [PubMed] [Google Scholar]
- 54.Picard A, Oger PM, Daniel I, Cardon H, Montagnac G, Chervin JC. 2006. A sensitive pressure sensor for diamond anvil cell experiments up to 2 GPa: FluoSpheresr. J. Appl. Phys. 100, 034915, (doi:10.1063/1.2234821) [Google Scholar]
- 55.Bruceallen LJ, Geraci JR. 1985. Wound-healing in the bottlenose dolphin (Tursiops truncatus). Can. J. Fish Aquat. Sci. 42, 216–228. (doi:10.1139/F85-029) [Google Scholar]
- 56.Tibbs RF, Elghetany MT, Tran LT, Van Bonn W, Romano T, Cowan DF. 2005. Characterization of the coagulation system in healthy dolphins: the coagulation factors, natural anticoagulants, and fibrinolytic products. Comp. Clin. Pathol. 14, 95–98. (doi:10.1007/s00580-005-0567-1) [Google Scholar]
- 57.Griffeth RJ, Garcia-Parraga D, Mellado-Lopez M, Crespo-Picazo JL, Soriano-Navarro M, Martinez-Romero A, Moreno-Manzano V. 2014. Platelet-rich plasma and adipose-derived mesenchymal stem cells for regenerative medicine-associated treatments in bottlenose dolphins (Tursiops truncatus). PLoS ONE 9, e108439 (doi:10.1371/journal.pone.0108439) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Montgomery SH, Geisler JH, McGowen MR, Fox C, Marino L, Gatesy J. 2013. The evolutionary history of cetacean brain and body size. Evolution 67, 3339–3353. (doi:10.1111/evo.12197) [DOI] [PubMed] [Google Scholar]
- 59.Mortensen HS, Pakkenberg B, Dam M, Dietz R, Sonne C, Mikkelsen B, Eriksen N. 2014. Quantitative relationships in delphinid neocortex. Front. Neuroanat. 8, 132 (doi:10.3389/fnana.2014.00132) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Marino L. et al. 2007. Cetaceans have complex brains for complex cognition. PLoS Biol. 5, e139 (doi:10.1371/journal.pbio.0050139) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Noseda R. et al. 2013. Ddit4/Redd1/Rtp801 is a novel negative regulator of Schwann cell myelination.J. Neurosci. 33, 15 295–15 305. (doi:10.1523/JNEUROSCI.2408-13.2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mass AM, Supin AY. 2007. Adaptive features of aquatic mammals' eye. Anat. Rec. 290, 701–715. (doi:10.1002/ar.20529) [DOI] [PubMed] [Google Scholar]
- 63.Meredith RW, Gatesy J, Emerling CA, York VM, Springer MS. 2013. Rod monochromacy and the coevolution of cetacean retinal opsins. PLoS Genet. 9, e1003432 (doi:10.1371/journal.pgen.1003432) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhao H, Ru B, Teeling EC, Faulkes CG, Zhang S, Rossiter SJ. 2009. Rhodopsin molecular evolution in mammals inhabiting low light environments. PLoS ONE 4, e8326 (doi:10.1371/journal.pone.0008326) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Davies KT, Cotton JA, Kirwan JD, Teeling EC, Rossiter SJ. 2012. Parallel signatures of sequence evolution among hearing genes in echolocating mammals: an emerging model of genetic convergence. Heredity 108, 480–489. (doi:10.1038/hdy.2011.119) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li Y, Liu Z, Shi P, Zhang J. 2010. The hearing gene Prestin unites echolocating bats and whales. Curr. Biol. 20, R55–R56. (doi:10.1016/j.cub.2009.11.042) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liu Y, Rossiter SJ, Han X, Cotton JA, Zhang S. 2010. Cetaceans on a molecular fast track to ultrasonic hearing. Curr. Biol. 20, 1834–1839. (doi:10.1016/j.cub.2010.09.008) [DOI] [PubMed] [Google Scholar]
- 68.Parker J, Tsagkogeorga G, Cotton JA, Liu Y, Provero P, Stupka E, Rossiter SJ. 2013. Genome-wide signatures of convergent evolution in echolocating mammals. Nature 502, 228–231. (doi:10.1038/nature12511) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shen YY, Liang L, Li GS, Murphy RW, Zhang YP. 2012. Parallel evolution of auditory genes for echolocation in bats and toothed whales. PLoS Genet. 8, e1002788 (doi:10.1371/journal.pgen.1002788) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hicks BD, St Aubin DJ, Geraci JR, Brown WR. 1985. Epidermal growth in the bottlenose dolphin, Tursiops truncatus. J. Invest. Dermatol. 85, 60–63. (doi:10.1111/1523-1747.ep12275348) [DOI] [PubMed] [Google Scholar]
- 71.Reeb D, Best PB, Kidson SH. 2007. Structure of the integument of southern right whales, Eubalaena australis. Anat. Rec. 290, 596–613. (doi:10.1002/ar.20535) [DOI] [PubMed] [Google Scholar]
- 72.Chen Z, Wang Z, Xu S, Zhou K, Yang G. 2013. Characterization of hairless (Hr) and FGF5 genes provides insights into the molecular basis of hair loss in cetaceans. BMC Evol. Biol. 13, 34 (doi:10.1186/1471-2148-13-34) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Martinez-Levasseur LM, Gendron D, Knell RJ, O'Toole EA, Singh M, Acevedo-Whitehouse K. 2011. Acute sun damage and photoprotective responses in whales. Proc R. Soc. B. 278, 1581–1586. (doi:10.1098/rspb.2010.1903) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The new short read data for the hippo and humpback whale have been deposited in the SRA of GenBank under the accession nos. SRR2183469 and SRR2183423. Alignments have been submitted to Dryad: http://dx.doi.org/10.5061/dryad.4cp98.