Abstract
Increasingly sophisticated protein engineering efforts have been undertaken lately to generate protein therapeutics with desired properties. This has resulted in the discovery of the next generation of protein therapeutics, which include: engineered antibodies, immunoconjugates, bi/multi-specific proteins, antibody mimetic novel scaffolds, and engineered ligands/receptors. These novel protein therapeutics possess unique physicochemical properties and act via a unique mechanism-of-action, which collectively makes their pharmacokinetics (PK) and pharmacodynamics (PD) different than other established biological molecules. Consequently, in order to support the discovery and development of these next generation molecules, it becomes important to understand the determinants controlling their PK/PD. This review discusses the determinants that a PK/PD scientist should consider during the design and development of next generation protein therapeutics. In addition, the role of systems PK/PD models in enabling rational development of the next generation protein therapeutics is emphasized.
Keywords: Pharmacokinetic, Pharmacodynamic, Protein Therapeutics, Biologics, Novel Scaffold, ADME, Toxicity, Immunogenicity
INTRODUCTION
Protein-based therapeutics (biologics) have become an integral and significant part of current medical treatment. Since the approval of human insulin as the first protein therapeutic 3 decades ago, the field of protein therapeutics has matured tremendously with increasingly sophisticated engineering efforts being used to create protein molecules with desired pharmacologic, pharmacokinetic (PK), and pharmacodynamic (PD) properties. As a result, there are more than 130 protein and peptide based therapeutics approved by the Food and Drug Administration (FDA) for clinical as of 2015 [1]. From these approved therapeutics more than 30 are monoclonal antibody (mAb) based therapeutics [2], which are arguably the most successful and best-selling biotechnology drugs at present.
The clinical success of mAbs is mainly attributed to the following properties: high specificity of the molecules that help them selectively neutralize or inhibit a target; the presence of the Fc portion of the molecule that allows them to recruit the FcRn mediated salvage pathway to achieve an unusually long half-life (relative to most small molecules); and the potential to activate immune-mediated effector functions, e.g. antibody-dependent cellular cytotoxicity (ADCC) and complement-dependent cytotoxicity (CDC), via the interaction of Fc portion with Fc-gamma receptors. However, along with these desired properties, in recent years certain limitations have also appeared for this class of molecules. These limitations include: (a) large size and steric hindrance that restricts tissue penetration of mAbs into solid tumors and poorly vascularized tissues, (b) planar binding interfaces that makes the binding of mAbs to certain grooves and the catalytic sites difficult, (c) physical and chemical instability, (d) requirement for large doses, (e) potential immunogenicity, (f) costs-of-goods associated with production/purification that makes mAb therapies expensive, and (g) complex intellectual property issues associated with technological aspects [3,4]. To overcome these challenges protein engineers have been working relentlessly towards the discovery and development of the next generation of protein therapeutics/scaffolds. The pharmacologic, PK and PD properties of these novel scaffolds can be unique and different than first-generation mAbs. Therefore, it becomes important to get more insight into the determinants of the PK/PD of novel protein therapeutics. Here the determinants that one should consider to increase the probability of discovering and developing a successful drug molecule using the next generation of protein therapeutics is discussed.
NEXT GENERATION PROTEIN THERAPEUTICS
To develop the next generation of protein therapeutics having optimal target recognition, PK, biodistribution, and therapeutic function, the field has transitioned from naturally occurring proteins to the design of molecules using combinatorial protein engineering techniques. This next generation of protein therapeutics ranges from simply modified antibodies to non-antibody protein scaffolds, which are capable of engaging a broad range of targets. Table 1 provides a non-exhaustive list of different categories of the next generation protein therapeutics with examples from each category, along with their salient features, and the names of clinically approved drugs from that category.
Table 1.
Different categories of the next generation protein therapeutics.
| Category | Example(s) | Salient Feature(s) |
|---|---|---|
| Engineered mAb | Altered Fc domain with increased affinity to FcγIIIa receptors | Increased effector function (e.g. Antibody-dependent cell-mediated cytotoxicity (ADCC) and antibody-dependent cellular phagocytosis (ADCP)) |
| Altered Fc domain with decreased binding to Fcγ receptors and C1q | Decreased effector function | |
| Increased affinity of Fc domain with FcRn at pH 6.0 | Increased plasma half-life | |
| Addition of sialylated glycans in the Fc region | Enhanced anti-inflammatory properties | |
| Removal of recognized N-glycosylation sites in VH and VL | Prevents the formation of N-glycoforms | |
| N-glycosylation glyco-engineering | Decreased glycoforms | |
| Forced pyroglutamylation of the amino-terminal Gln residues | Decreased number of charge variants | |
| Deletion of carboxy-terminal Lys residues | Decreased number of charge variants | |
| IgG4 and IgG2 hinge engineering | Avoids ‘half’ IgGs and limits the scrambling of disulphide bonds | |
| Engineered variable domain to lower the isoelectric point | Decreased elimination of mAb | |
| Immunoconjugates | Antibody-Drug Conjugates: (1) cysteine or lysine based random conjugation of drugs on normal mAbs, or (2) site-specific conjugation of drugs on engineered antibodies. | Antibodies are used as targeted delivery agents for highly potent drug molecules. More than 50 in clinical trials. FDA approved drugs: Mylotarg®, Kadcyla®, Adcetris®. |
| Immunotoxins: Chimeric proteins containing antibody/fragment attached to a fragment of toxin (e.g. Resimmune, Moxetumomab pasudotox). | Antibody or fragments of it are used for targeted delivery of cytotoxic proteins derived from a bacteria or plant. | |
| Immunocytokines: recombinant antibody-cytokine fusion proteins (e.g. Teleukin, Darleukin, Fibromun). | Enhances the therapeutic index of cytokines by targeting them to the site of disease. | |
| Radioimmunoconjugates (RICs): Antibodies that are directly attached to radioactive isotopes. | Allows radiation therapy to be delivered directly to the surface of targeted cells. FDA approved drugs: Bexxar®, Zevalin®. | |
| Antibody-directed enzyme prodrug therapy (ADEPT): An antibody conjugated to an enzyme (e.g. A5CP, MFECP1). | Employs antibody-enzyme conjugates (AEC) to activate prodrugs selectively at the targeted cells. | |
| Bi/multi-specific | Asymmetric IgG like molecules: Triomab, CrossMAbs, Knobs-into-Holes, LUZ-Y, SEEDbody, Biclonic, and Fab-Exchanged antibody. | Retains the intact mAb structure, where each arm binds to a different antigen. FDA approved drug: Removab®. |
| Symmetric IgG like molecules: Dual-Targeting Ig, Two-in-one antibody, Crosslinked MAbs, mAb2, and CovX-body. | Antibodies whose each arm binds to two different antigens. Includes two antibodies cross-linked chemically, or Fc portion engineered to bind to a target. | |
| IgG Fusions: DVD-Ig, Ts2Ab, BsAb, TvAb, HERCULES, and IgG-like Bispecific. | Molecules bigger than normal mAb because of the added domains to achieve the bi/multi-specific binding. | |
| Fc Fusions: SCORPION, Fc-DART, Dual(ScFv)2-Fab, and ScFv/Fc Fusions. | Two to four ScFv molecules added to the Fc domain of IgG. | |
| Fab Fusions: F(ab)2, Tribody, Fab-Fv, Dock-and-Lock, Bis-Fab, Dual-Action, and Bivalent Bispecific. | Multiple combinations of Fab molecules with other Fab or ScFv molecules. | |
| ScFv and Diabody based: BiTE, DART, Tandab, COMBODY, Single-chain Diabody, HSA-ScFv fusion, and TCR-like antibodies. | Multiple combinations of ScFv/Diabody connected to each other, or non-IgG proteins. FDA approved drug: Blincyto®. | |
| Antibody mimetics/Novel scaffolds | Adhirons | A 100 amino-acid protein of plant origin, with four-strand antiparallel β-sheet core and a central helix. |
| AdNectin/Monobody | A 94 amino-acid protein inspired from the 10th domain of human fibronectin, which is a β-Sandwich of seven β-sheets. | |
| Affibody | A 58 amino-acid protein inspired from Z-domain of staphylococcal protein A, consisting of 3 α-helices. | |
| Affilin | A 176 amino-acid protein inspired from human γ-B-crystallin, which is a β-sheet. | |
| Affimer | A 98 amino-acid protein inspired from human protease inhibitor stefin-A. | |
| Affitin/Nanofitin | A 66 amino-acid protein inspired from DNA-binding protein Sac7d, which is a five-stranded incomplete β-barrel. | |
| Alphabody | 70–100 amino-acid artificial (de novo design) protein consisting triple antiparallel α-helices. | |
| Anticalin | A 160–180 amino-acid 8-stranded β-barrel protein inspired from human/insect lipocalins. | |
| Atrimer/Tetranectin | A 40 amino-acid protein with 5 flexible loops that is inspired from human C-type lectin domain. | |
| Avimer | A 43 amino-acid artificial protein inspired from human multimerized LDLR-A module. | |
| Centyrins | An 89 amino-acid β-Sheet protein inspired from Fn3 domains of human tenascin-C. | |
| DARPin | A 67 (+ multiples of 33) amino-acid artificial (consensus design) protein with α2/β2 repeats, inspired from human ankyrinrepeat proteins. | |
| Fynomer | A 63 amino-acid β-Sandwich protein inspired from SH3 domain of human Fyn tyrosine kinase. | |
| Kunitz domain | A 50–60 amino-acid, disulfide rich, protein consisting of α+β fold, which are inspired from the active domains of human protease inhibitors. FDA approved drug: Kalbitor®. | |
| Obody/OB-fold | A 111 amino-acid protein inspired from theOB-fold of the aspartyl tRNA synthetase that is usually a 5-stranded β-barrel. | |
| Pronectin | A 90–95 amino-acid protein with 2 β-sheets and 3 surface-exposed loops, which is inspired from the 14th extracellulardomain of human fibronectin-III. | |
| Repebody | An artificial (consensus design) 170 amino-acid protein with β-strand-turn-α-helix, which is inspired by the leucine-rich repeat (LRR) modules of the variable lymphocyte receptors (VLRs) from jawless vertebrates. | |
| Engineered ligands/receptors | Engineered soluble receptor that functions as a ‘ligand trap’ (e.g. TRAP-Fc, comprising ligand binding domains of EGFR and ErbB-4 fused to Fc domain). | Helps circumvent ligand multiplicity, where the targeted receptors often have multiple activating ligands. |
| ‘Receptor decoy’ strategy where binding domains of multiple receptors are expressed together to bind and inhibit multiple related ligands and their isoforms. | Helps block multiple ligand–receptor interactions driving independent pathways that synergizes by amplifying the same signal. FDA approved drugs: Eylea®, Zaltrap®. | |
| A ligand trap architecture consisting of receptors and their accessory proteins, which mimics multi-component receptor systems (e.g. IL-2 ‘superkine’). | Help exploit the avidity effects that drive the apparently high binding affinity of multimeric receptor complexes. FDA approved drug: Arcalyst®. | |
| Fc-fusion ligands: Therapeutics ligands (proteins) recombinantly fused with the Fc domain. | Provides enhanced exposure in vivo, leading to reduced dose and/or dosing frequency. FDA approved drug: Eloctate®, Alprolix®. | |
| Ligand-toxin conjugate | The ligand (e.g. cytokine) helps specifically deliver the toxin to the cells expressing the ligand specific receptors. FDA approved drug: Ontak®. |
Engineered mAb
The molecules that are very close to the naturally occurring antibodies are engineered mAbs. These molecules are designed to either enhance or abrogate already existing pharmacological or PK properties of mAbs, or to improve their production procedure and cost. This category includes: molecules whose Fc domain is altered to enhance the mAb binding to FcγIIIa receptor with the intent of enhancing effector function (e.g. antibodies with 3M i.e. S239D/A330L/I332E mutation [5]), molecules whose Fc domain shows reduced affinity to Fcγ receptors and C1q to reduce the effector function (e.g. antibodies with TM i.e. L234F/L235E/P331S mutation [6]), and molecules whose Fc domain is altered to increase the half-life of mAbs (e.g. antibodies with YTE i.e. M252Y/S254T/T256E mutation [7]). This category also includes mAbs whose glycosylation site/pattern has been changed to enhance anti-inflammatory properties, decrease the number of glycoforms, or to prevent the formation of N-glycoforms [2]. MAbs are also being engineered to lower the isoelectric point of the variable domain to decrease their elimination, and to modify the hinge region to avoid the formation of ‘half’ immunoglobulin-G (IgG) and scrambling of the inter-chain disulfide bonds.
Immunoconjugates
Another group of the next generation therapeutics can be categorized in to the immunoconjugates. Although these molecules have been under development for decades, recent advances in protein engineering and linker-chemistry have reinvigorated interest in the development of these molecules. These molecules are essentially conjugates of targeted proteins (e.g. mAb) and small molecules, which act as targeted delivery agents for their small molecule payloads. The protein helps target the conjugated potent small molecule to the desired site-of-action. Antibody-Drug Conjugates (ADCs) are one of the most prominent molecules of this category, with 2 ADCs approved by FDA in last 3 years and more than 50 ADCs under clinical development presently. The field of ADC is rapidly changing, where initially small molecules were randomly conjugated to lysine or cysteine amino-acids of a mAb to produce a heterogeneous formulation with a distribution of drug:antibody ratio (DAR). Lately, mAbs are specifically engineered to enable site-specific conjugation of small molecules to yield a homogenous product, which has been shown to demonstrate improved therapeutic index [8]. It is outside the scope of this manuscript to cover all the protein engineering approaches used to develop the next generation of site-specific ADCs (including the use of non-natural amino-acids); the reader is referred to references [9,10] for more details.
The immunoconjugate category also includes immunotoxins, wherein the antibody-based targeting domain is fused to a bacterial/plant toxin for targeted cell killing [11]. Immunoconjugates also include immunocytokines, where antibody or its fragments are fused with cytokines to enhance the therapeutic index of the cytokines by specifically targeting them to the site of disease [12]. Radioimmunoconjugates (RICs) that use an antibody or its fragments to specifically deliver radioisotopes to the site-of-action are also an old and successful member of this category. With the advent of new beta emitters with better physical properties (e.g. lutetium 177), new alpha emitters that can eradicate the microscopic clusters of tumor cells (e.g. bismuth 213 and astatine 211), and personalized treatments based on quantitative positron emission tomography (PET), the next generation of RICs are also under development [13].
Bi/multi-specific proteins
The next category of the next generation protein therapeutics is engineered bi-specific and multi-specific proteins. These proteins are designed to bind to two or more antigens/epitopes simultaneously in order to achieve improved binding, selectivity, and efficacy. This category include IgG-like molecules that are designed to have each of their arms bind to a different antigen (known as asymmetric IgG like molecules). Clinically approved drug catumaxomab (Removab®) is an example of this type, whose one arm binds to EpCAM and the other arm binds to the T-cell receptor CD3. Another type of molecule in this category is symmetric IgG-like molecules, whose each arm has the capability to bind to more than one antigen. Bi/multi-specific antibodies are also designed by fusion of IgG variable or Fc domain with extra binding domains (e.g. ScFv). These molecules are usually bigger in size than traditional mAbs, and Abbott’s Dual Variable Domain-Ig (DVD-Ig) format with ~200 kDa molecule weight is an example of such IgG fusion molecules [14]. Additionally, the Fc portion of IgG can be fused to two or more ScFv or Fab molecules to generate bi/multi-specific molecules like SCORPION™ and Fc-DART [15]. Similarly, Fab and ScFv molecules can also be fused to each other to make bi/multi-specific molecules (e.g. Bis-Fab, Fab-Fv, BiTE, and DART). Clinically approved drug blinatumomab (Blincyto®) is an example of bi-specific protein created in such way through the combination of two different ScFv domains, one that binds to CD19 receptors present on B-cells and the other that binds to CD3 receptors on the T-cells [16].
Antibody mimetics/novel scaffolds
Novel protein scaffolds (also known as antibody mimetics) are the newest and fastest growing class of next generation protein therapeutics. These molecules are developed based on the understanding that the biophysical and biochemical properties of proteins are very important to the design of an optimized pharmaceutical agent. These proteins are highly engineered and typically have more attractive physical and chemical properties than mAbs. They also demonstrate higher affinity per unit mass of the molecule because of their architecture [17]. About 50 different protein scaffolds have been discovered and documented during the past 20 years, where the term ‘scaffold’ is usually reserved for a single chain polypeptidic framework of reduced size (<200 amino-acids) that contains a highly structured core associated with variable portions that have high conformational tolerance, which allows for insertions, deletions, or other substitutions [4]. Table 1 features a few of these industrially-advanced protein scaffolds like AdNectin, Anticalin, Avimer, and DARPin. From these scaffolds one drug has been already approved based on the Kunitz domain, called ecallantide (Kalbitor®, DX-88). It is a 60 amino-acid inhibitor of the protein kallikrein developed through phage display to mimic antibodies inhibiting kallikrein, which is used for the treatment of hereditary angioedema and in the prevention of blood loss in cardiothoracic surgery [18]. Additionally, since 102 proteins have already been specifically targeted using 139 different non-IgG scaffold binders [3], it is just a matter of time before we see more successful drugs coming out of this category of proteins.
Engineered ligands/receptors
The last category of engineered proteins that have emerged as promising candidates for protein-based drug discovery efforts are natural ligands and receptors, which inherently modulate complex biological processes. Guided by biological principles, protein engineering strategies are allowing ligands and receptors to be developed as next-generation therapeutics with improved safety and efficacy [19]. This protein class can help overcome ligand multiplicity of the receptors, where a receptor have multiple activating ligands that complicates the design of therapeutics aimed to modulate the function of the receptor. For example, the ErbB family consists of four structurally related receptors that potentiate signaling through homodimerization and heterodimerization, and there are eleven distinct ligands that bind to and activate various ErbB receptor combinations. This diversity of the ErbB family and ligand multiplicity has represented an important challenge for drug development. This challenge can be addressed by creating a protein therapeutic capable of binding to and neutralizing the activity of multiple ligands (known as a ‘ligand trap’), like it was done for ErbB receptors by developing TRAP-Fc [20]. TRAP-Fc comprises the ligand binding domains of EGFR and ErbB-4 fused to the Fc domain of IgG, and is capable of antagonizing all ligand family members targeting ErbB receptors. Similarly, a ‘receptor decoy’ strategy has been developed to block multiple ligand–receptor interactions that drive independent pathways leading to the synergism in the same biological signal. For protein therapeutics designed to follow this strategy, binding domains of multiple receptors are expressed together to bind and inhibit multiple related ligands and their isoforms. A few examples of this kind of protein therapeutic are the clinically approved anti-angiogenic drugs Eylea® and Zaltrap® [21]. Ligand trap proteins can also be designed to take advantage of the avidity effects that drive high binding affinity of multimeric receptor complexes by developing therapeutic proteins consisting of receptors and their accessory proteins (e.g. FDA approved drug Rilonacept, also known as IL-1 Trap and Arcalyst®) [22]. The natural affinity of ligands for their receptors can also be exploited by employing them as targeted delivery agents. This is accomplished by the FDA approved drug denileukin-diftitox (Ontak®), which is an engineered protein combining IL-2 and Diphtheria toxin. This molecule binds to IL-2 receptors and introduces the diphtheria toxin into cells that express these receptors, leading to cell killing [23].
PHARMACOKINETIC CONSIDERATIONS
Table 2 summarizes the PK parameters of selected drug molecules approved by FDA. These molecules belong to various categories of the next generation protein therapeutics. While these molecules differ in many physicochemical properties, a few properties like their molecular weight stand out. As shown in Table 2, these molecules can range from <10 kDa to >200 kDa, and this diversity of size can bestow diverse ADME (absorption, distribution, metabolism, and elimination) characteristics to these molecules. In addition, there are several other molecular properties that can affect the ADME of the next generation molecules, which are discussed below.
Table 2.
PK parameters of selected protein therapeutics approved by FDA.
| Brand Name | Generic Name | M. Wt. (kDa) | Half-life | CL | Vss | F | Route | Category |
|---|---|---|---|---|---|---|---|---|
| Mylotarg® | Gemtuzumab Ozogamicin | ~150 | 67 h | 0.353 L/h | 20.6 L | - | IV | ADC |
| Kadcyla® | Trastuzumab emtansine | ~150 | 4.2 d | 0.68 L/d | 60 mL/kg | - | IV | ADC |
| Adcetris® | Brentuximab vedotin | ~150 | 4–6 d | 1.5–1.8 L/d | 6–10 L | - | IV | ADC |
| Bexxar® | Tositumomab | ~150 | 67 h | - | - | - | IV | RIC |
| Zevalin® | Ibritumomab tiuxetan | ~150 | 30 h | - | - | - | IV | RIC |
| Removab® | Catumaxomab | ~150 | 2.13 d | - | - | 0.82 | IP | Bi-specific |
| Blincyto® | Blinatumomab | ~54 | 2.11 h | 2.92 L/h | 4.52 L | - | IV | Bi-specific |
| Kalbitor® | Ecallantide | ~7 | 2 h | 153 mL/min | 26.4 L | 0.91 | SC | Scaffold |
| Eylea®/Zaltrap® | (Ziv)-Aflibercept | ~97 | 5–6 d | 0.88 L/d | 4 L | - | IV, IVT | Fc-fusion |
| Enbrel® | Etanercept | ~150 | 70 h | 0.11 L/h | 10.4 L | 0.76 | SC | Fc-fusion |
| Arcalyst® | Rilonacept | ~251 | 7.7 d | 0.8 L/d | 9.36 L | 0.5 | SC | Fc-fusion |
| Eloctate® | Antihemophilic-Fc Fusion | ~220 | 19.7 h | 2.06 mL/h/kg | 49.5 mL/kg | - | IV | Fc-fusion |
| Alprolix® | Factor IX-Fc Fusion Protein | ~98 | 86.5 h | 3.3 mL/h/kg | 327 mL/kg | - | IV | Fc-fusion |
| Ontak® | Denileukin Diftitox | ~58 | 70–80 m | 0.6–2 mL/min/kg | 0.06–0.09 L/kg | - | IV | Immunotoxin |
Absorption
Since most of the large peptides and proteins are unstable in the proteolytic environment of the gastrointestinal tract, these molecules are not usually delivered orally. Thus, oral absorption and oral bioavailability are not usually of concern for protein therapeutics. However, there is an increasing interest in using the Fc domain to transport orally delivered proteins into the systemic circulation [24,25], which might lead to the development of the next generation of protein therapeutics that can be delivered orally. At present, subcutaneous (SC) is the most widely used extravascular route of protein administration, followed by intramuscular (IM), intraperitoneal (IP), intravitreal (IVT), and other routes of administration.
Subcutaneous route
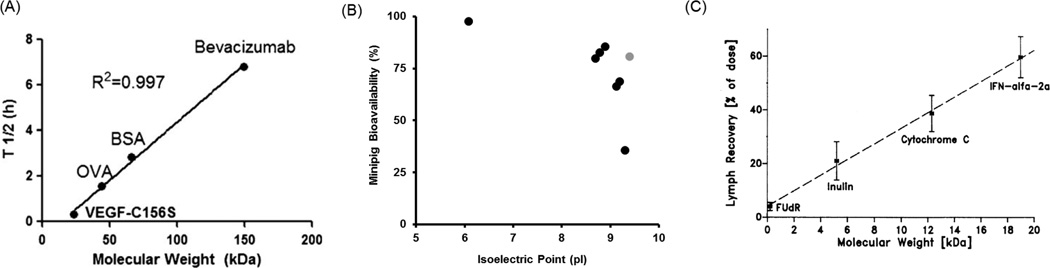
The ease of administration, relatively high bioavailability, and allowance for higher injection volume, has made SC the route of choice for the development of novel biologics (Table 2). Accordingly, there is increasing interest in understanding the determinants of the rate and extent of protein absorption from SC site of administration, in order to design next generation protein therapeutics with improved bioavailability. Protein molecular weight has been reported to be one of the most important factors that determine the rate of protein absorption from the SC site of administration. Protein molecular weight has also been found to correlate with the elimination half-life of proteins from the SC site of injection. Using 4 different proteins Wu et al. [26] have described that there is a proportional relationship between the molecular weight of a protein and its half-life of residence at the SC site of injection (Figure 1A). Affinity of the protein towards the salvage receptor FcRn has been shown to be another important determinant in the SC bioavailability of proteins. Deng et al. [27] have demonstrated that molecules with decreased affinity for FcRn show decreased bioavailability, because of reduced FcRn-mediated protection from catabolism at the absorption site. Thus, molecules with higher affinity for FcRn are expected to show greater SC bioavailability. The isoelectric point (pI) of proteins has also been found to correlate with their SC bioavailability. Using minipig as the animal model, Zheng et al. [28] have shown that proteins with high pI values exhibit lower bioavailability (Figure 1B), which was attributed to the nonspecific electrostatic interaction between the anionic cell membrane surface and the protein. Other molecular properties like the extent and type of glycosylation may also affect the SC bioavailability of proteins, however there is not enough data substantiating this relationship. As such, the published data indicate that molecules with smaller molecular weight (and by correlation, physical size), higher FcRn affinity, and lower pI values may be better candidates for designing the next generation protein therapeutics with higher SC bioavailability. Of note, the charge and solubility of the proteins may also be an important parameter to optimize SC bioavailability, since there is only a limited volume of fluid that can be injected at once into the dermis via the SC route. In addition, it has been described that a higher molecular weight may help deliver more amounts of the protein to the draining lymph node following SC delivery [29] (Figure 1C). So, if the protein is being developed for lymphoid disease, usage of a higher molecular weight protein may be advantageous. Recently, it is reported that formulation may also affect the bioavailability of SC administered protein therapeutics. Fathallah et al. [30] have described a significant increase in the bioavailability of SC administered rituximab when administered in a hypertonic buffer solution, because of an increase in the amount of protein that traffics through the lymph system.
Figure 1. Determinants for SC bioavailability of protein therapeutics.
(A) Relationship between half-life of protein therapeutics at the SC site-of-injection and molecular weight (Reprinted with the permission from [26]). (B) Effect of isoelectric point on the SC bioavailability of protein therapeutics in minipig (Reprinted with the permission from [28]). The gray dot represents an outlier. (C) Relationship between the percentages of SC injected dose of protein therapeutics found in the lymph and molecular weight (Reprinted with the permission from [29]).
Other routes
As the next generation of protein therapeutics are also being developed to treat tissue specific diseases (e.g. eye, lung, etc.) by local delivery, understanding the rate of absorption of proteins from these administration sites is also important. For example, following ocular delivery of proteins for macular degeneration, ideally one would like to achieve the maximum exposure of the therapeutic in the eye. Thus, it is important to understand and minimize the factors responsible for the efflux of the proteins from eye. It has been reported that FcRn may act as an efflux transporter for the proteins in the eye via transcytosis across blood-retinal barrier [31], and hence if the protein therapeutic is developed for local ocular delivery it may be preferable to avoid FcRn binding.
Distribution
The site-of-action for many protein therapeutics lies in the tissue compartments. Hence, understanding the determinants of tissue distribution for these molecules, following systemic or extravascular administration, is very important. In general, protein distribution is restricted to the vascular and interstitial space of the body (Table 2). Where tissue specific distribution depends on the physicochemical properties of the molecule, as well as physiological properties of a given tissue. Convection is one of the main processes responsible for the extravasation of proteins to tissues via paracellular pores of the vascular endothelium.
Molecular properties
The physical size of the protein and the size and distribution of vascular pores in a given tissue determines the extent of distribution of a given protein therapeutic in a particular tissue. This is evident by the fact that mAbs demonstrate different degrees of tissue distribution into different tissues based on the porosity of the tissues [32], and for a given tissue the extent of tissue distribution is inversely proportional to the size and molecular weight of the protein therapeutic (manuscript under review mAbs journal). Of note, diffusion can also contribute significantly towards the tissue distribution of smaller proteins [33]. The pI values of proteins have also been described as one of the determinant for their tissue distribution. Boswell et al. [34] have reported that the shifts in the pI value of ~1 unit or more can produce measurable changes in tissue distribution and kinetics of proteins. They have observed that an increase in net positive charge generally result in increased tissue retention, and a decrease in net positive charge generally result in decreased tissue retention. Thus, understanding electrostatic interactions between proteins and biological matrices is important for designing the next generation protein therapeutics with desired tissue distribution and retention properties. The role of FcRn in tissue distribution is still under debate. There are reports demonstrating that the tissue distribution of Fc-containing proteins is the same between wild-type and FcRn knockout mice [35,32], whereas there are also some recent data suggesting an important role of FcRn in biodistribution of antibodies [36]. In addition, there are reports hypothesizing the role of FcRn in limiting the distribution of Fc-containing proteins to specific tissues like brain [37] and eye [31] by acting as an efflux transporter. Thus, the decision to include an active Fc domain that binds to FcRn is an important one for the next generation protein therapeutics, as it may influence the whole body as well as tissue specific distribution of these molecules.
Binding to target
Since many of the protein therapeutics being developed bind specifically to a target, the expression profile of the target can also influence the tissue distribution of these molecules. In addition, novel and small proteins designed for tight binding to their antigen can also demonstrate off-target binding to unknown antigens, influencing their tissue distribution and PK [38]. However, just by analyzing the plasma PK of proteins it is difficult to realize this phenomena, which highlights the importance of whole body biodistribution studies that investigate tissue specific distribution of these molecules. For example, using preclinical biodistribution studies Vugmeyster et al. [38] have shown that antibodies with high affinity to tissue specific antigen showed selectively enhanced distribution to these organs, and also few antibodies demonstrated off-target biding to specific tissues. This phenomena was only observed when tissue to serum concentration ratios for these antibodies were compared to the ratios for the non-binding antibodies, since plasma PK profiles of these molecules were not altered significantly. Vugmeyster et al. have also demonstrated that the presence of a target antigen in a given tissue can alter the uptake rate of the protein in the tissue [38], suggesting target binding not only affects the extent but also rate of distribution of protein therapeutic into a specific tissue. Of note, protein therapeutics developed against a rapidly internalizing target could also experience selectively reduced concentrations at the target expressing tissue due to the target-mediated elimination of molecules locally. Thus, while designing the next generation protein therapeutics the contribution of target and possible off-target binding to tissue distribution of these molecules need to be considered and investigated.
Special tissues
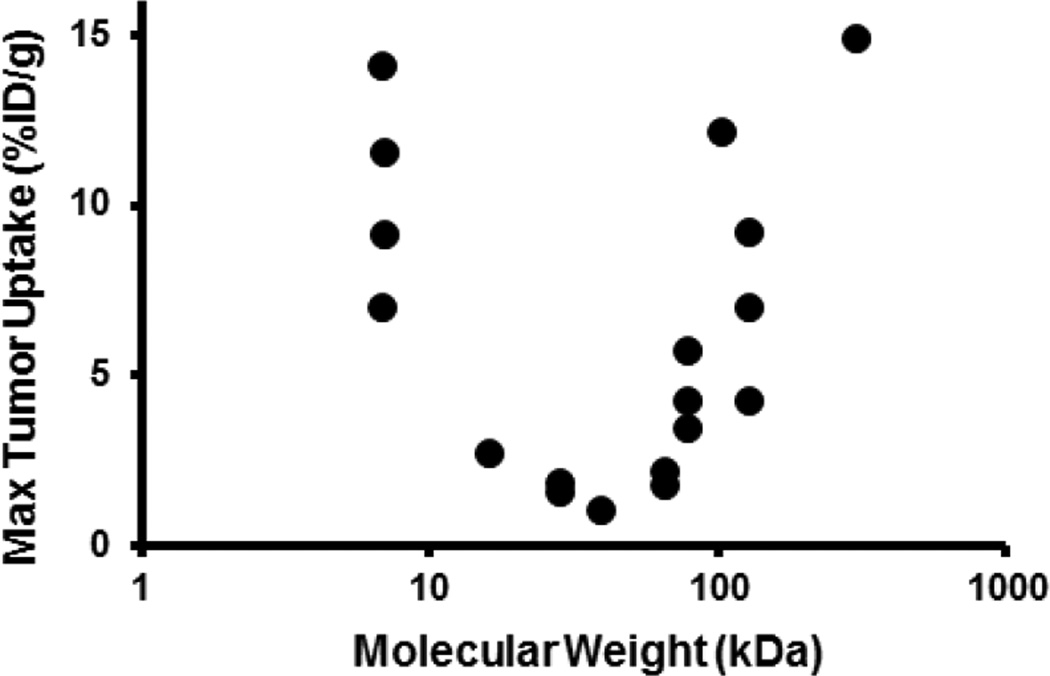
The unique properties of specific tissues is also worth considering. For example, solid tumors are one of the most complex and dynamic tissues and are the target of most novel protein therapeutics. It has been reported that there is a unique ‘U’ shape relationship between the molecular weight of the proteins and the extent of their distribution into solid tumor (Figure 2) [39]. This results from the effect of molecular weight on the complex interplay between systemic clearance and intratumor distribution of proteins. Thus, choosing the right molecular weight that can provide the desired tumor distribution is essential for the development of novel biologics. In addition, because of the presence of binding-site barrier around the tumor vasculature, the tightest binding protein molecules are not always appropriate for achieving homogenous tumor distribution [40]. Consequently, target antigen binding needs to be optimized while designing the next generation of protein therapeutics in order to achieve their homogenous tumor distribution. Brain is another organ that is very resistant to the distribution of protein therapeutics. Therefore, to develop a protein therapeutic for brain disorders it becomes essential to employ an enhanced permeation strategy (e.g. fusion with anti-TfR domain [41]) at the discovery stage itself. Kidney is an additional organ that demonstrates unique protein distribution propoerties. It is shown that for proteins like ScFv there is selective accumulation in the kidney [42], which is attributed to their reabsorption from the proximal tubule following glomerular filtration. While the exact location within the kidney for the distribution of these molecules, and the role of transporters like cubilin and megalin in their reabsorption is not clear, this observation can help design the next generation protein therapeutics for specifically targeting renal disorders.
Figure 2.
Relationship between the maximum uptake of protein therapeutics in solid tumor (represented as percentage of injected dose per gram of tumor) and molecular weight (Adapted from [39]).
Special modalities
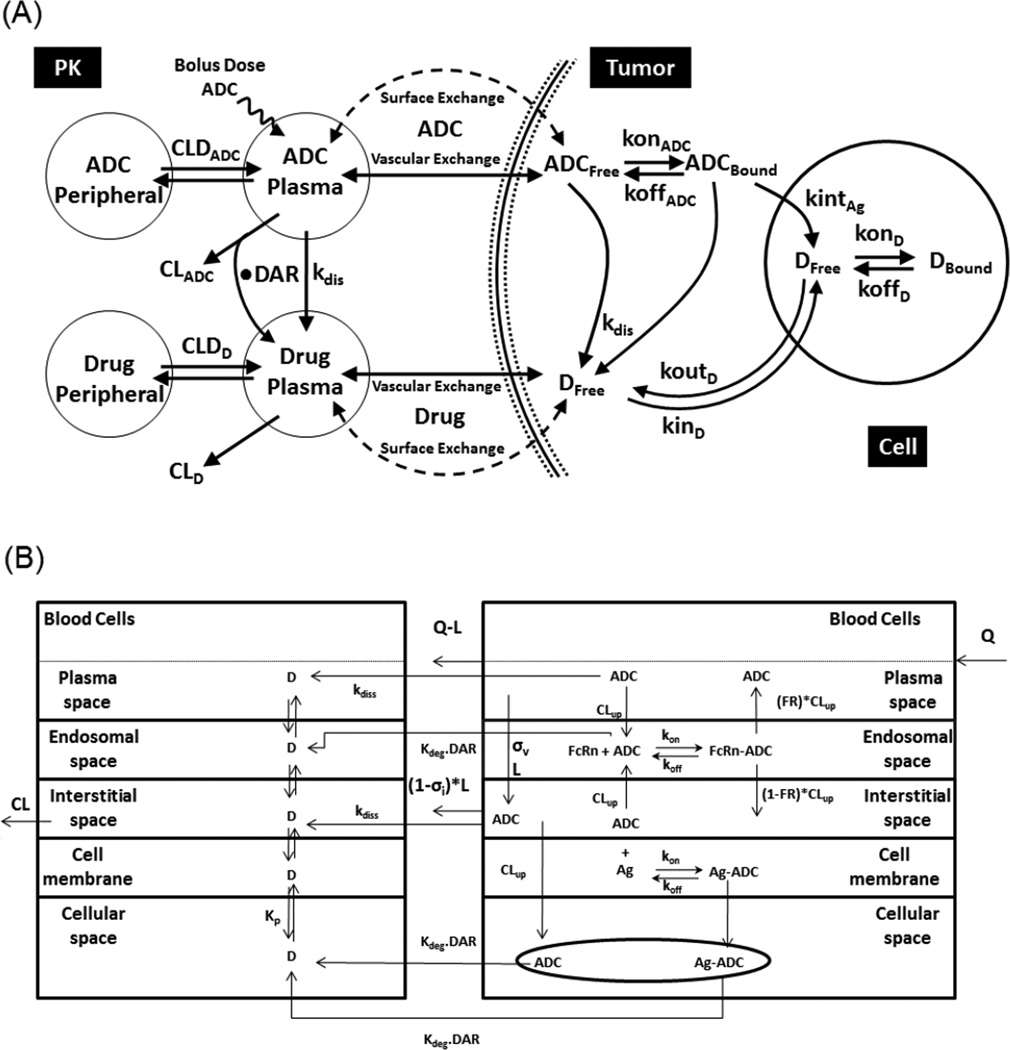
The tissue and tumor distribution of antibody-drug conjugates (ADCs) presents a unique challenge because understanding the distribution of these molecules not only requires a comprehensive understanding about the disposition of the conjugate, but also an understanding of the distribution of unconjugated drug and their active metabolites [43]. Our understanding about the determinants for the distribution of ADC and its components is very limited, and it is very resource intensive to experimentally evaluate their distribution routinely. Consequently, usage of systems PK models that can comprehensively integrate ADC tumor and tissue distribution data under a common quantitative framework becomes essential to design and develop the next generation of ADCs with optimal distribution properties [44,45] (Figure 3). In addition, for the development of next generation ADCs with desired distribution properties there is also a need to better understand cellular processing of ADCs.
Figure 3. Systems PK models developed for ADCs.
(A) A tumor disposition model developed to characterize and predict the concentrations of ADC and its components in the plasma and solid tumor [44]. (B) A PBPK model developed to characterize and predict the concentrations of ADC and its components throughout the body [45].
Understanding and predicting the distribution of bi/multi-specific proteins also presents another challenge. Since the distribution of these proteins not only depends on the expression and turnover rate of each target, but also on the relative affinity of the molecule to each target, and the turnover rate of each intermediate complex. Thus, without using a quantitative framework like a systems PK model [46] it would be very difficult to optimize the next generation bi/multi-specific molecules for desired distribution.
Metabolism and elimination
Protein therapeutics are generally catabolized to small peptides and amino acids. Hence, usually the metabolic product of these molecules are not considered pharmacologically active or toxic. Nonetheless, it is very important to understand the determinants of metabolism and elimination of protein therapeutics, since they control the clearance and exposure of these molecule throughout the body (including the site-of-action).
Molecular properties
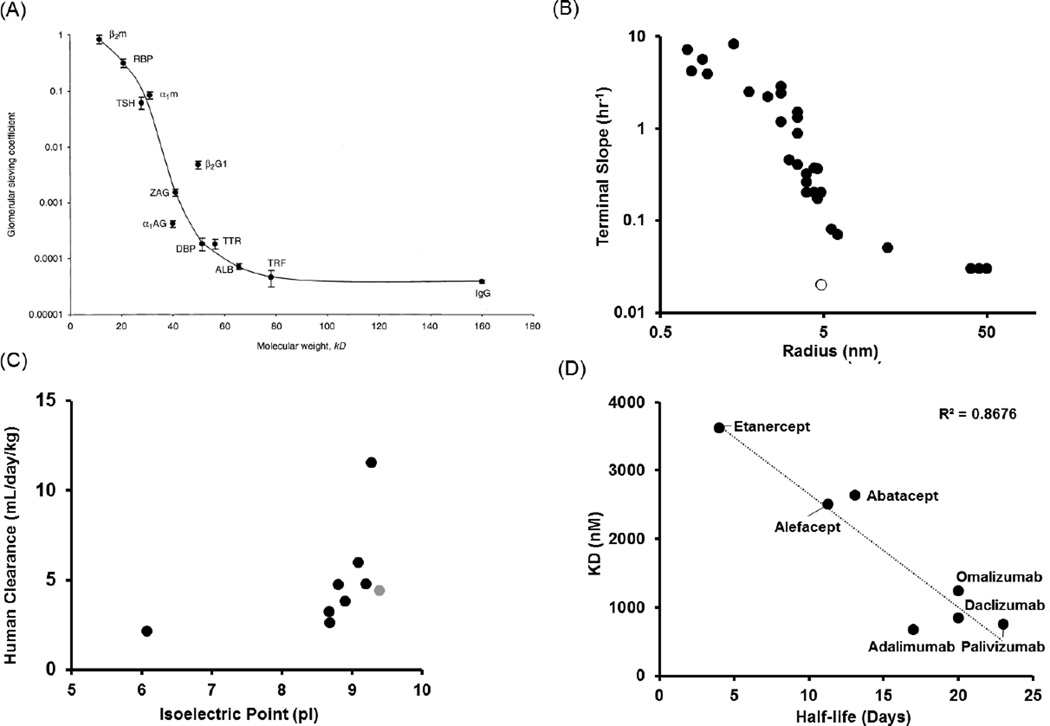
Nonspecific elimination of proteins through the kidney is one of the important determinants for their elimination. It is shown that there is an inverse sigmodal relationship between protein molecular weight and their glomerular sieving coefficient [47] (Figure 4A), where the glomerular filtration cut-off value (~60 kDa) serves as an inflection point. Consequently, one would expect a relationship between protein size and systemic clearance (especially renal clearance), which in fact has been described by Schmidt and Wittrup [39] (Figure 4B) in the form of ‘terminal-slope vs. protein radius’ relationship. Thus, in order to design the next generation protein therapeutics with longer systemic exposure, having a molecular weight above the glomerular filtration cut-off may be desirable. The pI value for proteins has also been shown to influence their clearance [34,28]. Boswell et al. [34] have described that an increase in net positive charge generally results in increased blood clearance and decreased net positive charge generally results in increased whole body clearance. Similarly, Zheng et al. [28] have also shown increases in the human clearance of proteins with an increase in pI value (Figure 4C). The importance of glycosylation in determining protein elimination is still under debate. Newkirk et al. have [48] described that glycosylation of proteins can affects their half-life, by showing that removal of the terminal sugars prolongs the half-life of IgGs due to the slower uptake and degradation by lectin receptors in the liver. Whereas, there are also publications claiming that mAb with different glycans and glycosylation levels have similar PK [49,50].
Figure 4. Determinants for the elimination of protein therapeutics.
(A) Relationship between glomerular sieving coefficient of proteins and their molecule weight (Reprinted with the permission from [47]). (B) Relationship between clearance (terminal slope of the PK profile) of protein therapeutics and their radius (Adapted from [39]). The open symbol represents IgG molecule that employs FcRn mediated salvage pathway. (C) Effect of isoelectric point on the clearance of protein therapeutics in human (Reprinted with the permission from [28]). The gray dot represents an outlier. (D) Relationship between the FcRn affinity (KD) of chosen protein therapeutics and their half-life in the clinic (Adapted from [52]).
FcRn mediated salvage
Another nonspecific pathway responsible for the clearance of protein therapeutics is their catabolism throughput the body following pinocytosis and lysosomal degradation in specific cells (e.g. vascular endothelial cells). However, proteins can escape this pathway by binding to FcRn salvage receptors which help them recycle back out of the cell. The unusually long half-life of IgG and albumin is indeed due to their ability to utilize this salvage mechanism by binding to FcRn. The next generation of protein therapeutics are being developed to exploit this escape mechanism. One strategy is to develop molecules with an already existing Fc domain (e.g. IgG) but increase its affinity towards FcRn at pH 6.0 (but not at pH 7.4). It has been shown that this can lead to further enhancement of the half-life for these molecules [51]. In fact, Suzuki et al. [52] have described an inversely proportional relationship between the Kd values of protein therapeutics to FcRn and the half-life of these molecules in human (Figure 4D). However, since this relationship was built using only 7 mAbs, one should be cautious before generalizing this relationship. Another strategy to employ FcRn is to fuse the Fc domain with proteins that do not naturally express it. These ‘Fc-fusion’ proteins have been commercially very successfully (e.g. etanercept), and do provide a significant enhancement in the half-life of short lived proteins in vivo [53]. Combining albumin or albumin-binding domain with protein therapeutics is also an efficient strategy to reduce the clearance of protein therapeutics, which has recently gained attention [54]. The next generation of protein therapeutics are sure to employ this strategy to enhance their half-lives.
Effect of target
The target mediated drug disposition (TMDD) pathway is a specific and significant pathway responsible for the elimination of protein therapeutics [55]. The fraction of total elimination that is contributed by TMDD depends on the non-specific clearance of the protein, affinity of the protein towards its antigen, antigen expression levels, internalization rate of cell membrane antigen, and the in vivo concentrations of the therapeutic protein [56]. It is usually observed that the influence of TMDD is more prominent when therapeutic drug concentrations are low, target antigen concentrations and internalization rates are high, and the affinity between the protein therapeutic and target is very high. In addition, when the target is expressed in blood, the influence of TMDD is more notable cf. the target expression on tissue cells that are not in rapid equilibrium with the systemic circulation. Thus, while designing the next generation protein therapeutics the possibility of TMDD needs to be explored early in the development, since significantly high TMDD in the clinic may be detrimental to the clinical success of some molecules.
Nonspecific binding
Although protein therapeutics are designed to bind to a specific target, there have been reports of off-target binding and its influence on their elimination. For example, Vugmeyster et al. have reported [57] unusually faster elimination of a humanized anti-Aβ antibody in cynomolgus monkeys due to its off-target binding to monkey fibrinogen. Similarly, Bumbaca et al. [58] have reported faster clearance a humanized anti-FGFR4 antibody in mice due to its off-target binding to mouse complement C3. Motavizumab, an affinity matured variant of palivizumab targeted against respiratory syncytial virus, has also been shown to exhibit faster elimination in rats and monkeys because of broad nonspecific tissue binding and sequestration. Thus, while designing novel protein therapeutics with novel protein scaffolds and very tight antigen affinity, the chances of increasing off-target binding should be considered. One can assess this off-target binding potential of drug candidates using commercially available protein chips (e.g. Protagen with ~400 different human proteins), or using ELISA or phage display based methods that can identify protein therapeutics with increased risk for fast clearance [59].
Immunogenicity
All protein therapeutics are potentially immunogenic, and development of anti-therapeutic antibodies (ATA, sometimes called anti-drug antibodies, ADAs) can lead to altered clearance of protein therapeutics. For example, it has been reported that immunogenic antibody can function as carrier proteins and prolong the circulatory half-life of protein therapeutics like insulin and various interleukins [60]. However in the majority of cases it has been reported that immunogenic antibodies lead to enhanced elimination of the therapeutic protein (e.g. adalimumab and infliximab [61]). Thus, in order to minimize the development of immunogenicity and the risk of enhanced elimination, the protein scaffolds for the next generation therapeutics need to be derived from naturally occurring human proteins with optimal glycosylation patterns. In addition, immunogenicity predictions can be carried out for drug candidates during early development stage using various in silico, in vitro, and preclinical tools [62,63].
Special modalities
The RICs and ADCs deserve special mention when it comes to metabolism, because unlike the rest of the protein therapeutics, the metabolites of these molecules are not innocuous. While the metabolism of RICs is unfortunately not well studied, ADCs have been relatively well studied [43,64]. For ADCs the formation of cytotoxic metabolites may occur via two simultaneous processes: deconjugation and catabolism. The deconjugation route includes release of the cytotoxic drug from an intact ADC via enzymatic/chemical processes that occur nonspecifically in plasma as well as the interstitial space. Catabolism includes proteolytic degradation of the conjugated antibody leading to the formation of cytotoxic drug-containing catabolites, and usually occurs following receptor-mediated endocytosis or fluid-phase pinocytosis of the ADC, via lysosomal trafficking and enzymatic degradation. The main determinants of ADC metabolism include linker stability, method and site of conjugation, total drug load, and the inherent PK of the parent antibody [64,43]. It has been reported that conventionally conjugated ADCs with higher drug load demonstrate higher elimination, and ADCs with unstable linker demonstrate higher deconjugation. Thus, the next generation of ADCs are being developed as site-specific ADCs that demonstrate drug load independent elimination [8] with relatively stable linkers [65]. It is also important to consider that the released drug itself has a unique PK profile, and the inherent clearance rate of this molecule will be an important determinant of ADC PK. In addition, there is a potential for ADC catabolites to engage in drug-drug interactions (DDIs) with other small molecule therapeutics, leading to alterations in serum/plasma concentrations of either the ADC catabolite or other co-administered medications [64]. Thus, optimizing the PK characteristics of the cytotoxic drug is equally important for developing the next generation ADCs.
Other factors affecting ADME of protein therapeutics
The analytical methods employed to determine the concentrations of the protein therapeutic are an important factor to consider while interpreting PK data. Most protein therapeutics are analyzed using ligand-binding immunoassays like ELISA. While this method is sensitive and versatile, there are a few limitations: (a) they lack specificity and ability to distinguish between active and inactive forms of macromolecules, (b) a variety of endogenous and exogenous substances (e.g. binding proteins and antibodies) can interfere with these assays, (c) formation of ATA can lead to measurement of artificially lower concentrations through competition at the assay binding site, (d) they are matrix specific and difficult to apply to tissue samples, and (e) development of reagents for these assays can take months.
For next generation protein therapeutics like Fc-fusion proteins it is very important that the analytical method is capable of detecting the Fc-clipping of the fusion protein, in order to capture the correct PK of the fusion molecule. However, as demonstrated by the case study with FGF21-Fc fusion proteins [66], a single ELISA is not capable of providing a detailed and accurate picture of the PK. Consequently, multiple ELISA formats or more sophisticated methods like LB-MS or LC/MS/MS are necessary to accurately analyze these next generation protein therapeutics. ADCs are another class of next generation molecules that require both small and large molecule analytical methods to capture the comprehensive PK of these conjugate. Kaur et al. [67] have elaborated on this subject matter and indicated that both a ligand-binding assay that is insensitive to drug loading and a LC/MS assay are required to quantify ADC and its components accurately. As such, it seems that in order to analyze the next generation of protein therapeutics, ligand-binding assays may not be sufficient, and mass spectrometry (MS) will emerge as the standard technique for in vivo characterization of the biotransformation products of these biologics [68]. The superiority of MS based methods over ELISA is also apparent when analyzing the protein therapeutic PK in the presence of ATA. For example, Wang et al. [69] have demonstrated that in the presence of the ATA the ELISA method was only capable of measuring the free circulating drug concentrations whereas the LC-MS/MS method was able to measure the total circulating drug concentrations.
Formulation is another factor that can affect the PK of protein therapeutics, and should be considered while developing the next generation molecules. For instance, it is generally assumed that the presence of a higher percentage of aggregates will lead to a higher probability of immunogenicity, resulting in rapid clearance of the protein therapeutics. The nature of the biological sample used for analyzing protein therapeutic concentrations is also believed to affect the PK. For example, there are unpublished reports suggesting the selection of plasma vs. serum as a sample matrix can lead to differences in the measured protein concentrations. The presence of disease in the patient population could also affect the PK of protein therapeutics. For example, patients with renal failure may demonstrate reduced clearance of smaller protein therapeutics that are significantly eliminated into urine. In addition, altered FcRn function or target expression profile due to the disease can lead to altered nonspecific or specific clearance of protein therapeutics. Lastly, the potential for drug interactions, where the protein therapeutic may be a victim, should also be considered while developing the next generation molecules, as it can notably affect the PK of these molecules. For example, using xenografts bearing mice it has been demonstrated that concomitant administration of an anti-VEGF antibody leads to reduced solid tumor exposure of a therapeutic antibody because of reduced blood flow to the tumor [70].
PHARMACODYNAMIC CONSIDERATIONS
Therapeutic target
Protein therapeutics are very versatile molecules that are able to interact with a broad range of targets ranging from small organic molecules (e.g. carbohydrates and nucleic acids) to viral, bacterial, animal, and human proteins. Most of the time though, therapeutic proteins are developed to target human proteins, which can be classified into cytokines, receptors, enzymes, blood proteins, hormones, growth factors, signaling proteins, regulatory proteins, and immunoglobulins [3]. Selection of the right target is crucial for the clinical success of a protein therapeutic since the PD effect depends on the inherent properties of the target (e.g. expression profile, turnover rate), the mechanism of therapeutic interaction with the target (stable vs. transient, covalent vs. non-covalent, and homo- vs. hetero-oligomerization), and the pharmacological consequences of the interaction (agonism, antagonism, neutralization, signal transduction, internalization).
Target selection
Most of the novel protein therapeutics are currently being developed to treat oncology and immunology disorders. While the variety of targets that can be employed to ameliorate these diseases are plenty, only few of them have gained popularity as targets of choice for the development of next generation protein therapeutics, mainly because it is very risky to choose clinically unestablished targets for the development of novel scaffolds whose clinical PK and behavior is not known. This is proven by the fact that TNF-α is the most frequently targeted molecule for treating immunology indications with 8 different protein scaffold, and it is also the target for the top three best-selling biopharmaceutical products. Similarly, the surface receptors EGFR, HER2, and fibronectin extra-domain B are the most popular targets for treating oncology indications, since each has been targeted by at least three different scaffolds [71]. As such, it seems that targeting clinically validated targets maybe a safe way to develop novel protein therapeutics. However, the clinical success of the Kunitz domain-based kallikrein inhibitor Ecallantide clearly shows that a combination of novel target and novel scaffold can still be developed into a drug.
Target evaluation
Before choosing the novel target for a protein therapeutic it is very important to understand the disease physiology and system biology of potential targets. Once the target/targets are chosen it is equally important to evaluate the therapeutic potential using systems pharmacology approaches in order to validate the target/targets [72]. Based on the systems pharmacology analysis one can decide whether it is necessary to modulate multiple targets in order to efficiently treat the disease or whether a single target is sufficient. Subsequently, an appropriate protein scaffold should be chosen that is capable of archiving desired modulation of validated target/targets in an in vivo setting. As such, for protein therapeutics the decision about which molecule(s) to target and which scaffold to use for targeting is inter-dependent. The discovery of aflibercept is a befitting example of this practice [19]. Based the systems biology of angiogenesis it was deemed important to neutralize all the ligands involved in this process in order to discover an anti-angiogenic therapeutic that is superior to bevacizumab and achieves complete physiological inhibition of angiogenesis. Therefore, a decoy receptor scaffold was designed by combining different VEGF-R fragments to generate a broadly neutralizing biologic capable of binding to and inhibiting multiple VEGF isoforms and placental growth factor. The discovery of bi/multi-specific protein therapeutics also hinges on a similar procedure, where it is important to justify the need for integrating multiple binding domain in one scaffold for a superior efficacy cf. using a combination of therapeutics that binds to each of the target in a monovalent manner.
The criteria for choosing a target for developing immunoconjugates like ADCs is slightly different, since here the target merely acts as a vehicle for the small molecule conjugate to get inside the cell. Thus, it is possible to choose a target that may be physiologically inert, as long as it is capable of delivering the extracellular cargo to the desired location inside the cell. Desirable properties for such targets are: (a) differentially high expression on the diseased cells, (b) ability to internalize and regenerate, and (c) traffic bound molecules to the lysosome.
Preclinical evaluation
While the use of in silico and in vitro systems to evaluate the PD effect of protein therapeutics is important, in vivo evaluations are irreplaceable because the PK of protein therapeutics at the site-of-action, the nature and severity of the disease, and the model system employed to evaluate the PD effect are all important determinants of the PD effect.
Animal model
In order to evaluate the PD effect preclinically the choice of the right animal model is very important. Ideally the animal model should be disease-bearing to mimic the physiology and pathological condition similar to the clinical condition. In order to properly evaluate the therapeutic index the target expression profile in normal and diseased tissues should also match the target profile in the clinic. If a relevant animal model is not available or the protein therapeutic is not cross-reactive with the target in the animal model, use of a surrogate molecule to evaluate the preclinical PD effect is warranted. For example, in order to evaluate the PD effect of novel immunotherapy agents, one needs to either use an animal model with active human immune system or develop a surrogate that can interact with the immune system of the animal model.
Dosing regimen
Choosing the correct dosing regimen to evaluate preclinical PD of protein therapeutics is also important. Based on the PK of the protein therapeutic in the animal model, one needs to choose a dosing regimen that provides sufficient drug exposure at the site-of-action. If the compound is efficacious preclinically, before moving forward to the clinic, it is also important to verify that the systemic exposures achieved at the preclinically efficacious dosing regimen are achievable and tolerable in the clinical setting. As mentioned later, usually PK/PD models are used to achieve these goals.
Toxicity
Along with evaluating the potential of efficacy, it is also important to evaluate the toxicity of protein therapeutics early in the development.
On-target
Protein therapeutics mainly demonstrate on-target toxicity in the form of exaggerated or undesired pharmacology [73]. For example, Yan et al. [74] have shown that anti-DLL4 antibody manifested proliferative vascular lesions in various tissues and liver toxicity with increased transaminase levels in rats and monkeys, due to DLL4/NOTCH1 pathway blockade in these tissues. The famous case of TGN1412 [75], which manifested a cytokine storm in healthy volunteers due to the exaggerated pharmacology, is another example for on-target toxicity of protein therapeutics.
Off-target
Since protein therapeutics are designed for highly specific binding to their targets, off target toxicity due to their binding to unrelated targets is not common. Nonetheless, there are few case studies that showcase off-target toxic effects of these molecules. For example, Santostefano et al. [76] have shown that an antibody developed against a soluble human protein (AMG X) that was not expressed on platelets, manifested thrombocytopenia, platelet activation, reduced mean arterial pressure, and transient loss of consciousness in cynomolgus monkeys after the first dose. Later it was found that the platelet activation occurred through an off-target binding of AMG X to cynomolgus platelets, and this antibody did not bind to humans or baboon platelets.
Immunogenicity
Since protein therapeutics can induce an immunogenic response, the toxicity related to ATA formation is also important to consider. Holland et al. [77] have provided a nice case study to emphasize this point using a novel scaffold: VH domain antibody (VH) developed against human TNF-α receptor 1 (TNFR1). They observed human anti-VH autoantibodies (HAVH) in approximately 50% of drug-naïve healthy human subjects. First, they demonstrated in vitro that HAVH autoantibody/VH complexes are capable of activating TNFR1 and cause cytokine release in human cell types. And, subsequently when VH was administered to healthy subjects, clinical and physiological signs of cytokine release were observed in HAVH autoantibody-positive subjects.
PK/PD relationship
Development of a reliable PK/PD relationship is essential for preclinical and clinical development, as well as preclinical-to-clinical translation, of protein therapeutics. Two of the most important factors required to develop a reliable PK/PD relationship are: (i) the PK of an analyte at the site-of-action, which is responsible for the pharmacological effect, and (ii) a PD endpoint that accurately represents the pharmacology of the drug.
PK at the site-of-action and predictive models
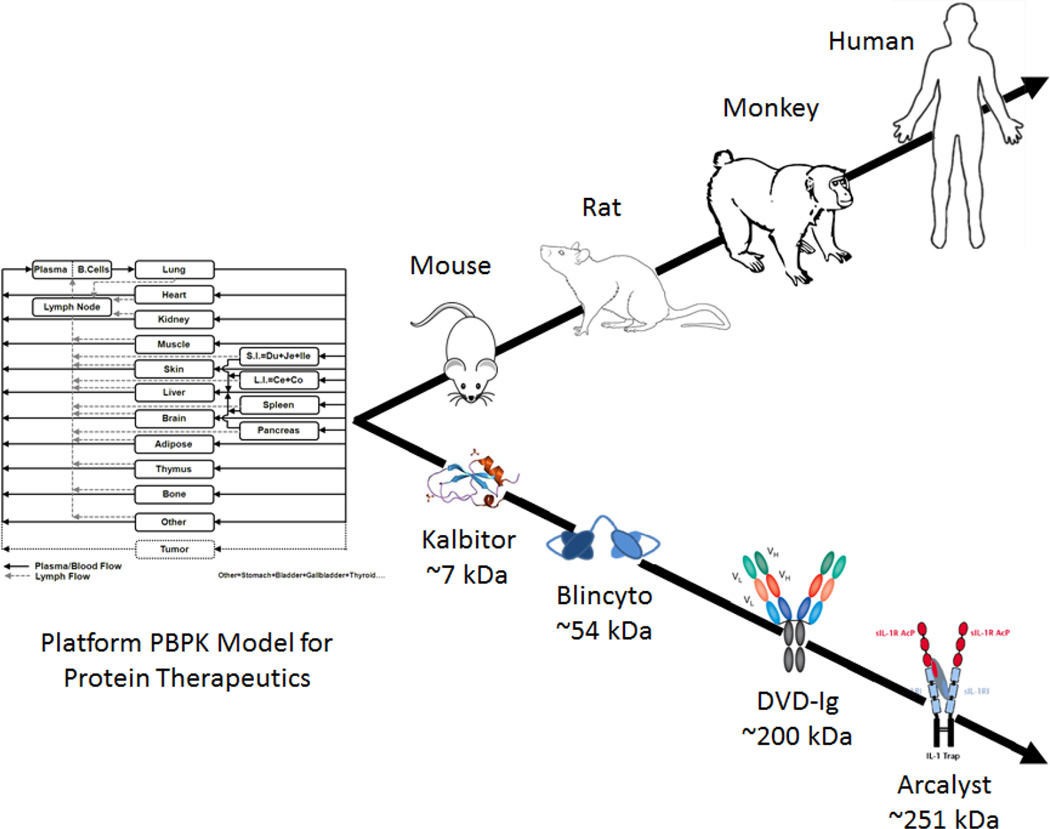
Depending on the mechanism-of-action of the protein therapeutic and the disease being treated, the analyte and the site-of-action for the measurement of the analyte changes. For example, for a protein therapeutic that neutralizes inflammatory cytokines to treat rheumatoid arthritis, the concentration of the unbound protein therapeutic at the inflammatory joint/synovium may be the most appropriate measurement to correlate with the efficacy [78,79]. For an anticancer therapeutic that activates the tyrosine kinase mediated apoptosis signal via occupancy of a membrane bound receptor, the concentration of the therapeutic protein in the tumor interstitium may be the most appropriate measurement [80]. Whereas for a solid tumor targeting ADC, the concentration of unconjugated drug molecules inside the cancer cell would be the most appropriate measurement to correlate with the PD endpoint [44]. However, it is very difficult to measure the concentrations of protein therapeutics at the site-of-action. Therefore, most often the plasma/serum concentration of the protein therapeutics is used as a surrogate. Since protein therapeutics in plasma/serum are usually not in rapid equilibrium with the site-of-action in tissues, a PK/PD relationship built based on plasma PK of protein therapeutics may not be ideal. A solution to this issue is to use a systems or mechanistic PK model that can accurately predict the concentrations of protein therapeutics at the site-of-action, based on the plasma/serum PK. Figure 5 showcases the schematic of one such platform PBPK model that is capable of predicting the site-of-action concentration of a protein therapeutics in several preclinical species and humans [81]. Figure 3 displays similar systems PK models specifically developed for ADCs. These kind of models are ideal for developing a reliable PK/PD relationships for protein therapeutics, which can be further used for their preclinical/clinical development and preclinical-to-clinical translation.
Figure 5.
Schematic of a platform PBPK model that is envisioned to encompass several animal species and human, and protein therapeutics with a wide range of molecular weight [81].
PD endpoint
The availability of a PD endpoint that can accurately represent target modulation and pharmacology of a protein therapeutic is another hurdle for the development of a reliable PK/PD relationship. For most diseases it is relatively easy to monitor the PD effect preclinically, however finding the PD endpoint that can be translated into the clinic is a major challenge. For example, one can routinely monitor changes in solid tumor preclinically, but monitoring the same in the clinic is challenging. A solution this problem is to find a biomarker in blood that closely resemble the changes in the PD endpoint. For example, circulating levels of free IgE have been used as a biomarker for the treatment of asthma with omalizumab in both preclinical and clinical settings. It was observed in the clinic that patients with asthma who responded better to omalizumab treatment had free IgE serum levels reduced in a dose-dependent manner. However, the science of biomarkers has come under question recently, because of the ubiquitous failure of biomarkers [82]. For example, less than 1% of published cancer biomarkers have actually entered into the clinical practice. Nonetheless, irrespective of what is the nature of the PD endpoint, the PD models used to characterize the efficacy/toxicity data for protein therapeutics are similar to the ones used for small molecules [72,55]. Of note, the PK model used to characterize the TMDD of protein therapeutics is also capable of being used to characterize both PK and PD of protein therapeutics [83].
PK/PD modeling and simulation
PK/PD models are usually first built during the preclinical stage by integrating exposure and efficacy/toxicity data. As mentioned earlier, it is very important that the PK and PD data used to build preclinical PK/PD models are generated using the right molecule in an appropriate animal model. It is usually advisable to monitor the concentration-time profile of proteins at 3 or more doses for at least 5 half-lives, in order to adequately characterize the PK (including TMDD). In addition, the best PK/PD relationships are built when PD studies are conducted using diverse dosing regimens. The duration of PD studies should be chosen based on the nature of the PD endpoint. For example, for protein therapeutics being developed for bone indications, the PD study may have to be continued for months. The preclinical PK/PD models are very useful in optimizing the experimental designs for further preclinical experiments. In addition, these models also provide the structure for a base PK/PD model that could be employed in the clinic, following simplification and addition of statistical models.
Another important use of preclinical PK/PD models is to enable preclinical-to-clinical translation of protein therapeutics. If appropriately translated these models can provide a priori estimates regarding First-in-Human dose and pharmacologically equivalent dose (PED) of protein therapeutics. For example, Luu et al. [84] have shown that the TMDD model was successfully able to predict the human PK of an ALK1 targeting IgG2 antibody exhibiting nonlinearity in animal models. PK/PD models are also capable of translating the efficacy of novel protein therapeutics. We have shown that a systems PK/PD model developed for ADCs was able to a priori predict the progression-free survival (PFS) and objective-response rates (ORR) of brentuximab-vedotin in Hodgkin's lymphoma patients. The important thing to consider while translating the PD using preclinical PK/PD models is to account for the differences in the target profile, protein therapeutic PK, and disease severity between the preclinical model and clinical trial population. Of note, PK/PD models can also be used to characterize the clinical exposure-response relationships for the efficacy and toxicity of protein therapeutics in the clinic. The use of these models in the clinic may help design better Phase-III trials and enable informed go/no-go decisions [85].
Immunogenicity
Finally, immunogenicity is also an important factor to consider regarding the PD of protein therapeutics, since neutralizing antibodies can abrogate the efficacy of these drugs. However, in the absence of any promising predictive models of immunogenicity its effect on the PD of protein therapeutics is hard to envision before going to the clinic.
SUMMARY
Protein-based molecules are very promising candidates for the development of the next generation therapeutics. There are a large number of protein therapeutics and novel scaffolds already in the development for the treatment of a variety of disorders. The PK of these molecules is unique and challenging to comprehend for certain therapeutics like ADCs and bi/multi-specific molecules. Thus, it is important to understand the determinants controlling the PK of protein therapeutics, and consider them during the design and development of next generation therapeutics. The PD of protein therapeutics depends on their exposure at the site-of-action and the property of the target. In addition, off-target binding and immunogenicity can also significantly affect the efficacy and toxicity of protein therapeutics. It is generally difficult to develop a reliable PK/PD relationship for protein therapeutics that can be translated to the clinic. However, the use of systems PK/PD models can facilitate the development of these relationships, which can further aid in the discovery, development, and preclinical-to-clinical translation of the next generation protein therapeutics.
ACKNOWLEDGEMENTS
This work was supported by the Center for Protein Therapeutics at the University at Buffalo, and NIH grant GM114179 to DKS.
REFERENCES
- 1.Leader B, Baca QJ, Golan DE. Protein therapeutics: a summary and pharmacological classification. Nature reviews drug discovery. 2008;7(1):21–39. doi: 10.1038/nrd2399. [DOI] [PubMed] [Google Scholar]
- 2.Beck A, Wurch T, Bailly C, Corvaia N. Strategies and challenges for the next generation of therapeutic antibodies. Nature reviews immunology. 2010;10(5):345–352. doi: 10.1038/nri2747. [DOI] [PubMed] [Google Scholar]
- 3.Skrlec K, Strukelj B, Berlec A. Non-immunoglobulin scaffolds: a focus on their targets. Trends in biotechnology. 2015;33(7):408–418. doi: 10.1016/j.tibtech.2015.03.012. [DOI] [PubMed] [Google Scholar]
- 4.Wurch T, Pierre A, Depil S. Novel protein scaffolds as emerging therapeutic proteins: from discovery to clinical proof-of-concept. Trends in biotechnology. 2012;30(11):575–582. doi: 10.1016/j.tibtech.2012.07.006. [DOI] [PubMed] [Google Scholar]
- 5.Shields RL, Namenuk AK, Hong K, Meng YG, Rae J, Briggs J, Xie D, Lai J, Stadlen A, Li B, Fox JA, Presta LG. High resolution mapping of the binding site on human IgG1 for Fc gamma RI, Fc gamma RII, Fc gamma RIII, and FcRn and design of IgG1 variants with improved binding to the Fc gamma R. The Journal of biological chemistry. 2001;276(9):6591–6604. doi: 10.1074/jbc.M009483200. [DOI] [PubMed] [Google Scholar]
- 6.Oganesyan V, Gao C, Shirinian L, Wu H, Dall'Acqua WF. Structural characterization of a human Fc fragment engineered for lack of effector functions. Acta crystallographica Section D, Biological crystallography. 2008;64(Pt 6):700–704. doi: 10.1107/S0907444908007877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dall'Acqua WF, Kiener PA, Wu H. Properties of human IgG1s engineered for enhanced binding to the neonatal Fc receptor (FcRn) The Journal of biological chemistry. 2006;281(33):23514–23524. doi: 10.1074/jbc.M604292200. [DOI] [PubMed] [Google Scholar]
- 8.Strop P, Delaria K, Foletti D, Witt JM, Hasa-Moreno A, Poulsen K, Casas MG, Dorywalska M, Farias S, Pios A, Lui V, Dushin R, Zhou D, Navaratnam T, Tran TT, Sutton J, Lindquist KC, Han B, Liu SH, Shelton DL, Pons J, Rajpal A. Site-specific conjugation improves therapeutic index of antibody drug conjugates with high drug loading. Nature biotechnology. 2015;33(7):694–696. doi: 10.1038/nbt.3274. [DOI] [PubMed] [Google Scholar]
- 9.Agarwal P, Bertozzi CR. Site-specific antibody-drug conjugates: the nexus of bioorthogonal chemistry, protein engineering, and drug development. Bioconjugate chemistry. 2015;26(2):176–192. doi: 10.1021/bc5004982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou Q, Kim J. Advances in the development of site-specific antibody-drug conjugation. Anti-cancer agents in medicinal chemistry. 2015 doi: 10.2174/1871520615666150302125448. [DOI] [PubMed] [Google Scholar]
- 11.Alewine C, Hassan R, Pastan I. Advances in anticancer immunotoxin therapy. The oncologist. 2015;20(2):176–185. doi: 10.1634/theoncologist.2014-0358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.List T, Neri D. Immunocytokines: a review of molecules in clinical development for cancer therapy. Clinical pharmacology : advances and applications. 2013;5:29–45. doi: 10.2147/CPAA.S49231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kraeber-Bodere F, Bodet-Milin C, Rousseau C, Eugene T, Pallardy A, Frampas E, Carlier T, Ferrer L, Gaschet J, Davodeau F, Gestin JF, Faivre-Chauvet A, Barbet J, Cherel M. Radioimmunoconjugates for the treatment of cancer. Seminars in oncology. 2014;41(5):613–622. doi: 10.1053/j.seminoncol.2014.07.004. [DOI] [PubMed] [Google Scholar]
- 14.DiGiammarino E, Ghayur T, Liu J. Design and generation of DVD-Ig molecules for dual-specific targeting. Methods in molecular biology. 2012;899:145–156. doi: 10.1007/978-1-61779-921-1_9. [DOI] [PubMed] [Google Scholar]
- 15.Zanin M, Keck ZY, Rainey GJ, Lam CY, Boon AC, Rubrum A, Darnell D, Wong SS, Griffin Y, Xia J, Webster RG, Webby R, Johnson S, Foung S. An anti-H5N1 influenza virus FcDART antibody is a highly efficacious therapeutic agent and prophylactic against H5N1 influenza virus infection. Journal of virology. 2015;89(8):4549–4561. doi: 10.1128/JVI.00078-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Buie LW, Pecoraro JJ, Horvat TZ, Daley RJ. Blinatumomab: A First-in-Class Bispecific T-Cell Engager for Precursor B-Cell Acute Lymphoblastic Leukemia. The Annals of pharmacotherapy. 2015 doi: 10.1177/1060028015588555. [DOI] [PubMed] [Google Scholar]
- 17.Caravella J, Lugovskoy A. Design of next-generation protein therapeutics. Current opinion in chemical biology. 2010;14(4):520–528. doi: 10.1016/j.cbpa.2010.06.175. [DOI] [PubMed] [Google Scholar]
- 18.Lehmann A. Ecallantide (DX-88), a plasma kallikrein inhibitor for the treatment of hereditary angioedema and the prevention of blood loss in on-pump cardiothoracic surgery. Expert opinion on biological therapy. 2008;8(8):1187–1199. doi: 10.1517/14712598.8.8.1187. [DOI] [PubMed] [Google Scholar]
- 19.Kariolis MS, Kapur S, Cochran JR. Beyond antibodies: using biological principles to guide the development of next-generation protein therapeutics. Current opinion in biotechnology. 2013;24(6):1072–1077. doi: 10.1016/j.copbio.2013.03.017. [DOI] [PubMed] [Google Scholar]
- 20.Lindzen M, Carvalho S, Starr A, Ben-Chetrit N, Pradeep CR, Kostler WJ, Rabinkov A, Lavi S, Bacus SS, Yarden Y. A recombinant decoy comprising EGFR and ErbB-4 inhibits tumor growth and metastasis. Oncogene. 2012;31(30):3505–3515. doi: 10.1038/onc.2011.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Al-Halafi AM. Vascular endothelial growth factor trap-eye and trap technology: Aflibercept from bench to bedside. Oman journal of ophthalmology. 2014;7(3):112–115. doi: 10.4103/0974-620X.142591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gabay C. IL-1 trap. Regeneron/Novartis. Current opinion in investigational drugs. 2003;4(5):593–597. [PubMed] [Google Scholar]
- 23.Foss FM. DAB(389)IL-2 (denileukin diftitox, ONTAK): a new fusion protein technology. Clinical lymphoma. 2000;1(Suppl 1):S27–S31. [PubMed] [Google Scholar]
- 24.Low SC, Nunes SL, Bitonti AJ, Dumont JA. Oral and pulmonary delivery of FSH-Fc fusion proteins via neonatal Fc receptor-mediated transcytosis. Human reproduction. 2005;20(7):1805–1813. doi: 10.1093/humrep/deh896. [DOI] [PubMed] [Google Scholar]
- 25.Pridgen EM, Alexis F, Kuo TT, Levy-Nissenbaum E, Karnik R, Blumberg RS, Langer R, Farokhzad OC. Transepithelial transport of Fc-targeted nanoparticles by the neonatal fc receptor for oral delivery. Science translational medicine. 2013;5(213):213ra167. doi: 10.1126/scitranslmed.3007049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu F, Bhansali SG, Law WC, Bergey EJ, Prasad PN, Morris ME. Fluorescence imaging of the lymph node uptake of proteins in mice after subcutaneous injection: molecular weight dependence. Pharmaceutical research. 2012;29(7):1843–1853. doi: 10.1007/s11095-012-0708-6. [DOI] [PubMed] [Google Scholar]
- 27.Deng R, Meng YG, Hoyte K, Lutman J, Lu Y, Iyer S, DeForge LE, Theil FP, Fielder PJ, Prabhu S. Subcutaneous bioavailability of therapeutic antibodies as a function of FcRn binding affinity in mice. mAbs. 2012;4(1):101–109. doi: 10.4161/mabs.4.1.18543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zheng Y, Tesar DB, Benincosa L, Birnbock H, Boswell CA, Bumbaca D, Cowan KJ, Danilenko DM, Daugherty AL, Fielder PJ, Grimm HP, Joshi A, Justies N, Kolaitis G, Lewin-Koh N, Li J, McVay S, O'Mahony J, Otteneder M, Pantze M, Putnam WS, Qiu ZJ, Ruppel J, Singer T, Stauch O, Theil FP, Visich J, Yang J, Ying Y, Khawli LA, Richter WF. Minipig as a potential translatable model for monoclonal antibody pharmacokinetics after intravenous and subcutaneous administration. mAbs. 2012;4(2):243–255. doi: 10.4161/mabs.4.2.19387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Supersaxo A, Hein WR, Steffen H. Effect of molecular weight on the lymphatic absorption of water-soluble compounds following subcutaneous administration. Pharmaceutical research. 1990;7(2):167–169. doi: 10.1023/a:1015880819328. [DOI] [PubMed] [Google Scholar]
- 30.Fathallah AM, Turner MR, Mager DE, Balu-Iyer SV. Effects of hypertonic buffer composition on lymph node uptake and bioavailability of rituximab, after subcutaneous administration. Biopharmaceutics & drug disposition. 2015;36(2):115–125. doi: 10.1002/bdd.1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim H, Robinson SB, Csaky KG. FcRn receptor-mediated pharmacokinetics of therapeutic IgG in the eye. Molecular vision. 2009;15:2803–2812. [PMC free article] [PubMed] [Google Scholar]
- 32.Shah DK, Betts AM. Antibody biodistribution coefficients: inferring tissue concentrations of monoclonal antibodies based on the plasma concentrations in several preclinical species and human. mAbs. 2013;5(2):297–305. doi: 10.4161/mabs.23684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rippe B, Haraldsson B. Fluid and protein fluxes across small and large pores in the microvasculature. Application of two-pore equations. Acta physiologica Scandinavica. 1987;131(3):411–428. doi: 10.1111/j.1748-1716.1987.tb08257.x. [DOI] [PubMed] [Google Scholar]
- 34.Boswell CA, Tesar DB, Mukhyala K, Theil FP, Fielder PJ, Khawli LA. Effects of charge on antibody tissue distribution and pharmacokinetics. Bioconjugate chemistry. 2010;21(12):2153–2163. doi: 10.1021/bc100261d. [DOI] [PubMed] [Google Scholar]
- 35.Garg A, Balthasar JP. Physiologically-based pharmacokinetic (PBPK) model to predict IgG tissue kinetics in wild-type and FcRn-knockout mice. Journal of pharmacokinetics and pharmacodynamics. 2007;34(5):687–709. doi: 10.1007/s10928-007-9065-1. [DOI] [PubMed] [Google Scholar]
- 36.Chen N, Wang W, Fauty S, Fang Y, Hamuro L, Hussain A, Prueksaritanont T. The effect of the neonatal Fc receptor on human IgG biodistribution in mice. mAbs. 2014;6(2):502–508. doi: 10.4161/mabs.27765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cooper PR, Ciambrone GJ, Kliwinski CM, Maze E, Johnson L, Li Q, Feng Y, Hornby PJ. Efflux of monoclonal antibodies from rat brain by neonatal Fc receptor, FcRn. Brain research. 2013;1534:13–21. doi: 10.1016/j.brainres.2013.08.035. [DOI] [PubMed] [Google Scholar]
- 38.Vugmeyster Y, DeFranco D, Szklut P, Wang Q, Xu X. Biodistribution of [125I]-labeled therapeutic proteins: application in protein drug development beyond oncology. Journal of pharmaceutical sciences. 2010;99(2):1028–1045. doi: 10.1002/jps.21855. [DOI] [PubMed] [Google Scholar]
- 39.Schmidt MM, Wittrup KD. A modeling analysis of the effects of molecular size and binding affinity on tumor targeting. Molecular cancer therapeutics. 2009;8(10):2861–2871. doi: 10.1158/1535-7163.MCT-09-0195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weinstein JN, van Osdol W. Early intervention in cancer using monoclonal antibodies and other biological ligands: micropharmacology and the "binding site barrier". Cancer research. 1992;52(9 Suppl):2747s–2751s. [PubMed] [Google Scholar]
- 41.Yu YJ, Zhang Y, Kenrick M, Hoyte K, Luk W, Lu Y, Atwal J, Elliott JM, Prabhu S, Watts RJ, Dennis MS. Boosting brain uptake of a therapeutic antibody by reducing its affinity for a transcytosis target. Science translational medicine. 2011;3(84):84ra44. doi: 10.1126/scitranslmed.3002230. [DOI] [PubMed] [Google Scholar]
- 42.Hamilton S, Odili J, Wilson GD, Kupsch JM. Reducing renal accumulation of single-chain Fv against melanoma-associated proteoglycan by coadministration of L-lysine. Melanoma research. 2002;12(4):373–379. doi: 10.1097/00008390-200208000-00010. [DOI] [PubMed] [Google Scholar]
- 43.Lin K, Tibbitts J. Pharmacokinetic considerations for antibody drug conjugates. Pharmaceutical research. 2012;29(9):2354–2366. doi: 10.1007/s11095-012-0800-y. [DOI] [PubMed] [Google Scholar]
- 44.Shah DK, Haddish-Berhane N, Betts A. Bench to bedside translation of antibody drug conjugates using a multiscale mechanistic PK/PD model: a case study with brentuximab-vedotin. Journal of pharmacokinetics and pharmacodynamics. 2012;39(6):643–659. doi: 10.1007/s10928-012-9276-y. [DOI] [PubMed] [Google Scholar]
- 45.Singh AP, Shin YG, Shah DK. Application of Pharmacokinetic-Pharmacodynamic Modeling and Simulation for Antibody-Drug Conjugate Development. Pharmaceutical research. 2015 doi: 10.1007/s11095-015-1626-1. [DOI] [PubMed] [Google Scholar]
- 46.Chudasama VL, Zutshi A, Singh P, Abraham AK, Mager DE, Harrold JM. Simulations of site-specific target-mediated pharmacokinetic models for guiding the development of bispecific antibodies. Journal of pharmacokinetics and pharmacodynamics. 2015;42(1):1–18. doi: 10.1007/s10928-014-9401-1. [DOI] [PubMed] [Google Scholar]
- 47.Norden AG, Lapsley M, Lee PJ, Pusey CD, Scheinman SJ, Tam FW, Thakker RV, Unwin RJ, Wrong O. Glomerular protein sieving and implications for renal failure in Fanconi syndrome. Kidney international. 2001;60(5):1885–1892. doi: 10.1046/j.1523-1755.2001.00016.x. [DOI] [PubMed] [Google Scholar]
- 48.Newkirk MM, Novick J, Stevenson MM, Fournier MJ, Apostolakos P. Differential clearance of glycoforms of IgG in normal and autoimmune-prone mice. Clinical and experimental immunology. 1996;106(2):259–264. doi: 10.1046/j.1365-2249.1996.d01-847.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Deng R, Jin F, Prabhu S, Iyer S. Monoclonal antibodies: what are the pharmacokinetic and pharmacodynamic considerations for drug development? Expert opinion on drug metabolism & toxicology. 2012;8(2):141–160. doi: 10.1517/17425255.2012.643868. [DOI] [PubMed] [Google Scholar]
- 50.Huang L, Biolsi S, Bales KR, Kuchibhotla U. Impact of variable domain glycosylation on antibody clearance: an LC/MS characterization. Analytical biochemistry. 2006;349(2):197–207. doi: 10.1016/j.ab.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 51.Yeung YA, Leabman MK, Marvin JS, Qiu J, Adams CW, Lien S, Starovasnik MA, Lowman HB. Engineering human IgG1 affinity to human neonatal Fc receptor: impact of affinity improvement on pharmacokinetics in primates. Journal of immunology. 2009;182(12):7663–7671. doi: 10.4049/jimmunol.0804182. [DOI] [PubMed] [Google Scholar]
- 52.Suzuki T, Ishii-Watabe A, Tada M, Kobayashi T, Kanayasu-Toyoda T, Kawanishi T, Yamaguchi T. Importance of neonatal FcR in regulating the serum half-life of therapeutic proteins containing the Fc domain of human IgG1: a comparative study of the affinity of monoclonal antibodies and Fc-fusion proteins to human neonatal FcR. Journal of immunology. 2010;184(4):1968–1976. doi: 10.4049/jimmunol.0903296. [DOI] [PubMed] [Google Scholar]
- 53.Czajkowsky DM, Hu J, Shao Z, Pleass RJ. Fc-fusion proteins: new developments and future perspectives. EMBO molecular medicine. 2012;4(10):1015–1028. doi: 10.1002/emmm.201201379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sand KM, Bern M, Nilsen J, Noordzij HT, Sandlie I, Andersen JT. Unraveling the Interaction between FcRn and Albumin: Opportunities for Design of Albumin-Based Therapeutics. Frontiers in immunology. 2014;5:682. doi: 10.3389/fimmu.2014.00682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mager DE, Jusko WJ. General pharmacokinetic model for drugs exhibiting target-mediated drug disposition. Journal of pharmacokinetics and pharmacodynamics. 2001;28(6):507–532. doi: 10.1023/a:1014414520282. [DOI] [PubMed] [Google Scholar]
- 56.Peletier LA, Gabrielsson J. Dynamics of target-mediated drug disposition: characteristic profiles and parameter identification. Journal of pharmacokinetics and pharmacodynamics. 2012;39(5):429–451. doi: 10.1007/s10928-012-9260-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vugmeyster Y, Szklut P, Wensel D, Ross J, Xu X, Awwad M, Gill D, Tchistiakov L, Warner G. Complex pharmacokinetics of a humanized antibody against human amyloid beta peptide, anti-abeta Ab2, in nonclinical species. Pharmaceutical research. 2011;28(7):1696–1706. doi: 10.1007/s11095-011-0405-x. [DOI] [PubMed] [Google Scholar]
- 58.Bumbaca D, Wong A, Drake E, Reyes AE, 2nd, Lin BC, Stephan JP, Desnoyers L, Shen BQ, Dennis MS. Highly specific off-target binding identified and eliminated during the humanization of an antibody against FGF receptor 4. mAbs. 2011;3(4):376–386. doi: 10.4161/mabs.3.4.15786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hotzel I, Theil FP, Bernstein LJ, Prabhu S, Deng R, Quintana L, Lutman J, Sibia R, Chan P, Bumbaca D, Fielder P, Carter PJ, Kelley RF. A strategy for risk mitigation of antibodies with fast clearance. mAbs. 2012;4(6):753–760. doi: 10.4161/mabs.22189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rehlaender BN, Cho MJ. Antibodies as carrier proteins. Pharmaceutical research. 1998;15(11):1652–1656. doi: 10.1023/a:1011936007457. [DOI] [PubMed] [Google Scholar]
- 61.Ducourau E, Mulleman D, Paintaud G, Miow Lin DC, Lauferon F, Ternant D, Watier H, Goupille P. Antibodies toward infliximab are associated with low infliximab concentration at treatment initiation and poor infliximab maintenance in rheumatic diseases. Arthritis research & therapy. 2011;13(3):R105. doi: 10.1186/ar3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brinks V, Weinbuch D, Baker M, Dean Y, Stas P, Kostense S, Rup B, Jiskoot W. Preclinical models used for immunogenicity prediction of therapeutic proteins. Pharmaceutical research. 2013;30(7):1719–1728. doi: 10.1007/s11095-013-1062-z. [DOI] [PubMed] [Google Scholar]
- 63.Bryson CJ, Jones TD, Baker MP. Prediction of immunogenicity of therapeutic proteins: validity of computational tools. BioDrugs : clinical immunotherapeutics, biopharmaceuticals and gene therapy. 2010;24(1):1–8. doi: 10.2165/11318560-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 64.Han TH, Zhao B. Absorption, distribution, metabolism, and excretion considerations for the development of antibody-drug conjugates. Drug metabolism and disposition: the biological fate of chemicals. 2014;42(11):1914–1920. doi: 10.1124/dmd.114.058586. [DOI] [PubMed] [Google Scholar]
- 65.Mack F, Ritchie M, Sapra P. The next generation of antibody drug conjugates. Seminars in oncology. 2014;41(5):637–652. doi: 10.1053/j.seminoncol.2014.08.001. [DOI] [PubMed] [Google Scholar]
- 66.Hager T, Spahr C, Xu J, Salimi-Moosavi H, Hall M. Differential enzyme-linked immunosorbent assay and ligand-binding mass spectrometry for analysis of biotransformation of protein therapeutics: application to various FGF21 modalities. Analytical chemistry. 2013;85(5):2731–2738. doi: 10.1021/ac303203y. [DOI] [PubMed] [Google Scholar]
- 67.Kaur S, Xu K, Saad OM, Dere RC, Carrasco-Triguero M. Bioanalytical assay strategies for the development of antibody-drug conjugate biotherapeutics. Bioanalysis. 2013;5(2):201–226. doi: 10.4155/bio.12.299. [DOI] [PubMed] [Google Scholar]
- 68.Ezan E, Becher F, Fenaille F. Assessment of the metabolism of therapeutic proteins and antibodies. Expert opinion on drug metabolism & toxicology. 2014;10(8):1079–1091. doi: 10.1517/17425255.2014.925878. [DOI] [PubMed] [Google Scholar]
- 69.Wang SJ, Wu ST, Gokemeijer J, Fura A, Krishna M, Morin P, Chen G, Price K, Wang-Iverson D, Olah T, Weiner R, Tymiak A, Jemal M. Attribution of the discrepancy between ELISA and LC-MS/MS assay results of a PEGylated scaffold protein in post-dose monkey plasma samples due to the presence of anti-drug antibodies. Analytical and bioanalytical chemistry. 2012;402(3):1229–1239. doi: 10.1007/s00216-011-5527-9. [DOI] [PubMed] [Google Scholar]
- 70.Abuqayyas L, Balthasar JP. Pharmacokinetic mAb-mAb interaction: anti-VEGF mAb decreases the distribution of anti-CEA mAb into colorectal tumor xenografts. The AAPS journal. 2012;14(3):445–455. doi: 10.1208/s12248-012-9357-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Walsh G. Biopharmaceutical benchmarks 2014. Nature biotechnology. 2014;32(10):992–1000. doi: 10.1038/nbt.3040. [DOI] [PubMed] [Google Scholar]
- 72.Jusko WJ. Moving from basic toward systems pharmacodynamic models. Journal of pharmaceutical sciences. 2013;102(9):2930–2940. doi: 10.1002/jps.23590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hansel TT, Kropshofer H, Singer T, Mitchell JA, George AJ. The safety and side effects of monoclonal antibodies. Nature reviews Drug discovery. 2010;9(4):325–338. doi: 10.1038/nrd3003. [DOI] [PubMed] [Google Scholar]
- 74.Yan M. Therapeutic promise and challenges of targeting DLL4/NOTCH1. Vascular cell. 2011;3:17. doi: 10.1186/2045-824X-3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Goodyear M. Learning from the TGN1412 trial. Bmj. 2006;332(7543):677–678. doi: 10.1136/bmj.38797.635012.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Santostefano MJ, Kirchner J, Vissinga C, Fort M, Lear S, Pan WJ, Prince PJ, Hensley KM, Tran D, Rock D, Vargas HM, Narayanan P, Jawando R, Rees W, Reindel JF, Reynhardt K, Everds N. Off-target platelet activation in macaques unique to a therapeutic monoclonal antibody. Toxicologic pathology. 2012;40(6):899–917. doi: 10.1177/0192623312444029. [DOI] [PubMed] [Google Scholar]
- 77.Holland MC, Wurthner JU, Morley PJ, Birchler MA, Lambert J, Albayaty M, Serone AP, Wilson R, Chen Y, Forrest RM, Cordy JC, Lipson DA, Bayliffe AI. Autoantibodies to variable heavy (VH) chain Ig sequences in humans impact the safety and clinical pharmacology of a VH domain antibody antagonist of TNF-alpha receptor 1. Journal of clinical immunology. 2013;33(7):1192–1203. doi: 10.1007/s10875-013-9915-0. [DOI] [PubMed] [Google Scholar]
- 78.Lon HK, Liu D, Jusko WJ. Pharmacokinetic/pharmacodynamic modeling in inflammation. Critical reviews in biomedical engineering. 2012;40(4):295–312. doi: 10.1615/critrevbiomedeng.v40.i4.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tabrizi M, Bornstein GG, Suria H. Biodistribution mechanisms of therapeutic monoclonal antibodies in health and disease. The AAPS journal. 2010;12(1):33–43. doi: 10.1208/s12248-009-9157-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lammerts van Bueren JJ, Bleeker WK, Bogh HO, Houtkamp M, Schuurman J, van de Winkel JG, Parren PW. Effect of target dynamics on pharmacokinetics of a novel therapeutic antibody against the epidermal growth factor receptor: implications for the mechanisms of action. Cancer research. 2006;66(15):7630–7638. doi: 10.1158/0008-5472.CAN-05-4010. [DOI] [PubMed] [Google Scholar]
- 81.Shah DK, Betts AM. Towards a platform PBPK model to characterize the plasma and tissue disposition of monoclonal antibodies in preclinical species and human. Journal of pharmacokinetics and pharmacodynamics. 2012;39(1):67–86. doi: 10.1007/s10928-011-9232-2. [DOI] [PubMed] [Google Scholar]
- 82.Kern SE. Why your new cancer biomarker may never work: recurrent patterns and remarkable diversity in biomarker failures. Cancer research. 2012;72(23):6097–6101. doi: 10.1158/0008-5472.CAN-12-3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Betts AM, Clark TH, Yang J, Treadway JL, Li M, Giovanelli MA, Abdiche Y, Stone DM, Paralkar VM. The application of target information and preclinical pharmacokinetic/pharmacodynamic modeling in predicting clinical doses of a Dickkopf-1 antibody for osteoporosis. The Journal of pharmacology and experimental therapeutics. 2010;333(1):2–13. doi: 10.1124/jpet.109.164129. [DOI] [PubMed] [Google Scholar]
- 84.Luu KT, Bergqvist S, Chen E, Hu-Lowe D, Kraynov E. A model-based approach to predicting the human pharmacokinetics of a monoclonal antibody exhibiting target-mediated drug disposition. The Journal of pharmacology and experimental therapeutics. 2012;341(3):702–708. doi: 10.1124/jpet.112.191999. [DOI] [PubMed] [Google Scholar]
- 85.Bender BC, Schaedeli-Stark F, Koch R, Joshi A, Chu YW, Rugo H, Krop IE, Girish S, Friberg LE, Gupta M. A population pharmacokinetic/pharmacodynamic model of thrombocytopenia characterizing the effect of trastuzumab emtansine (T-DM1) on platelet counts in patients with HER2-positive metastatic breast cancer. Cancer chemotherapy and pharmacology. 2012;70(4):591–601. doi: 10.1007/s00280-012-1934-7. [DOI] [PubMed] [Google Scholar]