Abstract
How embryonic stem cells (ESC) commit to specific cell lineages and ultimately yield all cell types of a fully formed organism remains a major question. ESC differentiation is accompanied by large-scale histone and DNA modifications, but the relations between these two categories of epigenetic changes are not understood. Here we demonstrate the hierarchical interplay between the histone deacetylase, sirtuin 6 (Sirt6), which targets acetylated histone H3 at lysines 9 and 56 (H3K9ac and H3K56ac), and the Tet (Ten-eleven translocation) enzymes, which convert 5-methylcytosine (5mC) into 5-hydroxymethylcytosine (5hmC). ESCs derived from Sirt6 knockout (S6KO) mice are skewed towards neuroectoderm development. This phenotype is associated with derepression of Oct4, Sox2 and Nanog, which in turn causes an upregulation of Tet enzymes and elevated production of 5hmC. Genome-wide analysis revealed an upregulation of neuroectoderm genes marked with 5hmC in S6KO ESCs, thereby implicating Tet enzymes in the neuroectoderm-skewed differentiation phenotype of S6KO ESCs, which is fully rescued upon knockdown of Tets. We demonstrate a new role for Sirt6 as a chromatin regulator safeguarding the balance between pluripotency and differentiation through Tet-dependent regulation of 5hmC levels.
INTRODUCTION
During early stages of development, embryonic stem cells (ESCs) proliferate and differentiate into all somatic cell types. ESC differentiation requires global changes of chromatin architecture to elicit specific epigenetic programs of gene expression associated to each somatic cell type. Chromatin alterations including changes in histone modifications and DNA methylation patterns play a critical role during the commitment, establishment and maintenance of a particular cell lineage during early embryogenesis (Chen and Dent, 2014). Notably, the interplay between these chromatin alterations, and how they execute epigenetic programs of gene expression during ESC differentiation remain largely unknown.
DNA methylation is usually linked to chromatin compaction and gene inactivation, which constitutes a critical process to establish cell lineage specification during ESC differentiation (Smith and Meissner, 2013). DNA methylation is a reversible process catalyzed by the Fe2+ and α-ketoglutarate-dependent dioxygenases, Tet enzymes (Iyer et al., 2009; Tahiliani et al., 2009). There are three Tet orthologues in the mouse, Tet1, Tet2 and Tet3. These enzymes revert the methylation status of DNA by successive oxidation of 5mC into 5hmC, 5-carboxycytosine (5caC) and 5-formylcytosine (5fC), which are intermediates of an active DNA demethylation mechanism (He et al., 2011; Ito et al., 2011). Increased levels in 5hmC are tightly associated to the maintenance of the pluripotency state of ESCs (Ficz et al., 2011; Williams et al., 2011; Wu et al., 2011). The expression of Tet1 and Tet2, maintained at high levels in ESCs, diminishes during differentiation, which correlates with repression of pluripotent genes and activation of developmental genes (Kriaoucionis and Heintz, 2009; Tahiliani et al., 2009; Ito et al., 2010; Ko et al., 2010; Szwagierczak et al., 2010). The Tet-dependent production of 5hmC has been implicated in cell lineage specification of ESCs (Koh et al., 2011). However, upstream regulatory mechanisms underlying the participation of Tet enzymes and the potential role of 5hmC as a direct epigenetic component regulating specific genes during ESC differentiation remain undetermined.
One of the histone modifications involved in ESC function is acetylation of lysine 56 in histone H3 (H3K56ac), which has been linked to the pluripotent transcriptional network in human ESCs (Xie et al., 2009). More specifically, H3K56ac levels correlates with the transcriptional activation of pluripotent genes, its levels diminishing significantly on those genes during ESC differentiation (Xie et al., 2009). How this mark is regulated during ESCs differentiation remained unclear. The NAD-dependent histone deacetylase Sirt6 was shown to target H3K56ac in mouse ESCs (Yang et al., 2009; Michishita et al., 2009), and is one of seven mammalian members of the sirtuin protein network, with roles in genome stability, glucose metabolism and tumor suppression (Mostoslavsky et al., 2006; Michishita et al., 2009; Yang et al., 2009; Zhong et al., 2010; Sebastian et al., 2012; Toiber et al., 2013). Because of its ability to specifically target H3K56ac, we investigated the potential participation of Sirt6 in ESC differentiation. Our results demonstrate that Sirt6 directly regulates the expression of the core pluripotent genes Oct4, Sox2 and Nanog, via deacetylation of H3K56ac, which in turn controls ESC differentiation through Tet-mediated oxidation of 5mC into 5hmC.
RESULTS
Sirt6 Deletion Skews Embryonic Stem Cell Differentiation Potential Towards Neuroectoderm
ESCs derived from S6KO mouse embryos showed skewed differentiation compared to those derived from their WT littermates. When cultured to form embryoid bodies (EBs), S6KO EBs from three different ESC lines were significantly smaller in size compared to their WT counterparts (Figure 1A). Immunofluorescence (IF) analysis showed expression of the endoderm marker Gata4 to be downregulated in S6KO EBs, while expression of the neuroectoderm marker Gfap was upregulated (Figure 1B). Gfap is also upregulated in EBs derived from S6KO iPSCs (Figure S1A). The skewing of S6KO EBs towards neuroectoderm and away from endoderm, mesoderm and trophoectoderm was confirmed by examining the expression of additional markers (Figures 1C and S1B). Moreover, when we subjected ESCs to an in vitro neurogenesis protocol, we found a striking increase in the number of Nestin and β-III-Tubulin expressing neurons in S6KO versus WT controls (Figures 1D, S1C and S1D). The expression on Nestin was upregulated in S6KO EBs even under normal culturing conditions (Figure S1E). Notably, even prior to differentiation, S6KO ESCs exhibited a downregulation of genes associated with endoderm, mesoderm and trophectoderm, while neuroectoderm related genes were upregulated, consistent with a primed differentiation state in the absence of Sirt6 (Figure 1E and S1F). These results point towards a previously unidentified role for Sirt6 in regulating cell lineage specification during ESC differentiation.
Figure 1. Sirt6 deficiency skews ESC differentiation towards neuroectoderm and promotes expression of Oct4, Sox2, Nanog.
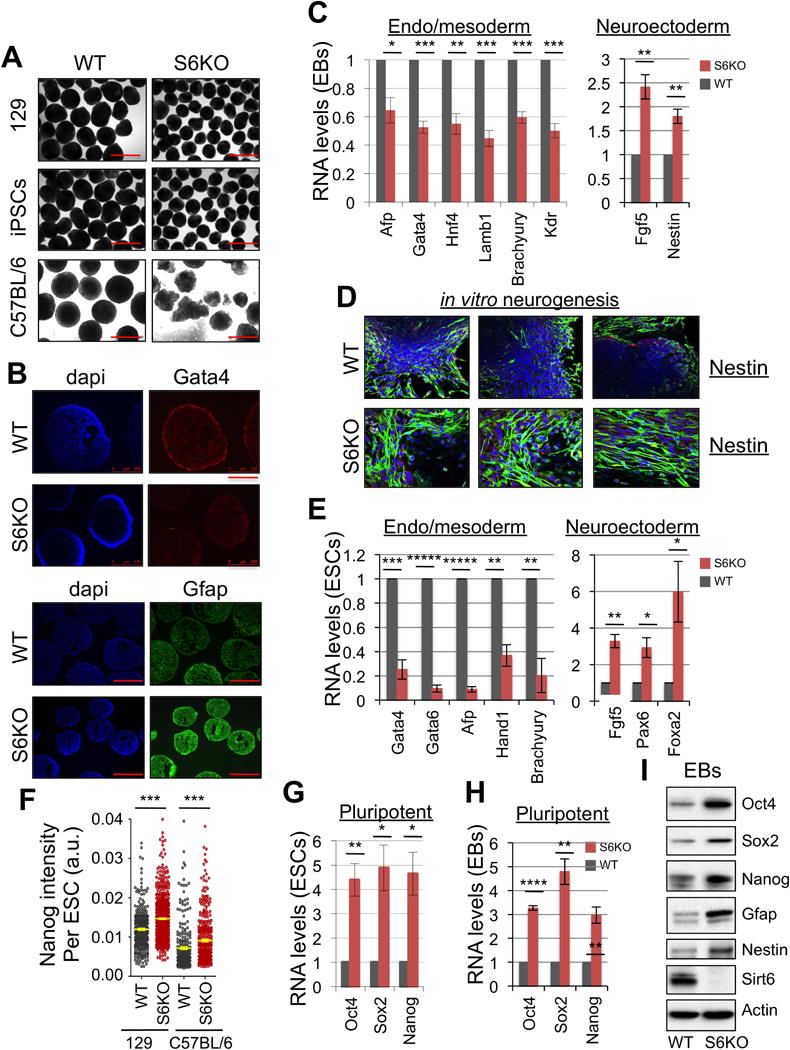
(A) EBs derived from WT and S6KO ESCs (129, iPSCs from 129 and C57BL/6 mouse strain). Scale bar, 250 μm.
(B) Immunofluorescence of EBs from WT and S6KO (129 mouse strain) for Gata4 (scale bar, 250 μm) and Gfap (scale bar 500 μm).
(C) Expression of endoderm, mesoderm and neuroectoderm genes in WT versus S6KO EBs. qRT-PCR data is expressed relative to WT EBs.
(D) Immunofluorescence of in vitro generated neurons from WT and S6KO EBs for Nestin (green). Nuclei were stained with DAPI.
(E) Expression of endoderm, mesoderm and neuroectoderm genes in WT versus S6KO ESCs. qRT-PCR data is expressed relative to WT EBs.
(F) Quantification of Nanog levels (mean intensity, a.u.) per cell by immunostaining in WT and S6KO ESCs from 129 and C57BL/6 genetic backgrounds. Yellow bars represent mean +/− s.e.m. *** P < 0.001 by 1 way Anova followed by Tukey test analysis. (a.u., arbitrary units).
(G) Core pluripotent gene expression in WT versus S6KO ESCs assessed by qRT-PCR analysis.
(H) Core pluripotent gene expression in WT versus S6KO EBs assessed by qRT-PCR analysis.
(I) Western blot analysis for the core pluripotent factors on WT versus S6KO EBs.
These are representatives of at least n = 3 experimental replicates.
The data on panels (C), (E), (F), (G) and (H) are at least n = 3 experimental replicates, values are mean +/− s.e.m. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, *****P < 0.00001, by t-test analysis.
Pluripotent genes are not Repressed During Differentiation of ESCs Lacking Sirt6
Persistent expression of Oct4, Sox2 and Nanog is critical to maintain the pluripotency state, but it needs to be silenced upon ESC differentiation (Young, 2011). However, during early stages of ESC differentiation, Oct4 and Sox2 were shown to orchestrate germ layer fate decisions. Oct4 was found to suppress neuroectoderm differentiation while promoting development of the mesoderm. Divergently, Sox2 inhibits mesodermal differentiation, but promotes the development of neuroectoderm. An overall downregulation of Nanog is a causal event to impel the differentiation state (Thomson et al., 2011). Thus, we assessed whether lack of Sirt6 could alter expression of these pluripotent genes following differentiation. Notably, Sirt6 deficiency was characterized by persistent expression of the core pluripotency genes Oct4, Sox2 and Nanog and their protein products in both ESCs and EBs (Figures 1F–1I). Using an alternative differentiation protocol (retinoic acid) we find that even though Oct4 was normally repressed, the expression of Sox2 and Nanog persisted upon forced differentiation (Figure S1G). These data suggest that Sirt6 negatively regulates the expression of these core pluripotent genes to achieve proper ESC differentiation.
Sirt6 Regulates Levels of H3K56ac and H3K9ac at the Promoters of Oct4, Sox2 and Nanog
To determine the mechanism by which Sirt6 regulates expression of the core pluripotent genes, we subjected ESCs from both S6KO and WT to chromatin immunoprecipitation (ChIP) before and after differentiation. We first assessed binding of SIRT6 to the pluripotent genes. Notably, Sirt6 was found at the promoter regions of Oct4, Sox2 and Nanog both before and after differentiation (Figure 2A). We then tested for the presence of H3K56Ac, one of the SIRT6 substrates previously linked to pluripotent gene expression (Xie et al., 2009). Consistently, the levels of H3K56ac at these promoters were increased in S6KO compared to WT ESCs and EBs (Figure 2B). Additionally, another Sirt6 substrate, H3K9ac, was also increased at the promoter regions of these core pluripotent genes in S6KO versus WT ESCs (Figure 2C). Furthermore, the recruitment of Sirt6 was extended inside the Oct4 locus, showing maximum binding at the promoter (primers A and B, at −195 and −13 positions, respectively) and towards the 3′ end of exon 1 (primer C at position +719) before and after differentiation (Figures 2D and 2E), which is paralleled by an increase in H3K56ac in S6KO versus WT EBs (Figure 2F). The recruitment of Sirt6 was not apparent at position +4220 (primer D, between exons 4 and 5) towards the end of Oct4 paralleling the unchanged levels of H3K56ac between WT and S6KO EBs. Moreover, Sirt6-dependent deacetylation of both H3K9ac and H3K56ac on the Oct4, Sox2 and Nanog loci was confirmed by genome wide analyses (Figures 2G and 2H). Notably, an increase on these histone modifications is retained after RA-dependent differentiation, particularly at the Sox2 locus (Figure S1H). These results support the idea that Sirt6 negatively regulates expression of the core pluripotency genes Oct4, Sox2 and Nanog, thereby emphasizing a new role for Sirt6 in stem cell function. The inability to suppress these core pluripotent genes might in part account for the skewed differentiation towards the neural lineage in S6KO compared to WT ESCs and iPSCs.
Figure 2. Sirt6-dependent regulation of core pluripotent genes.
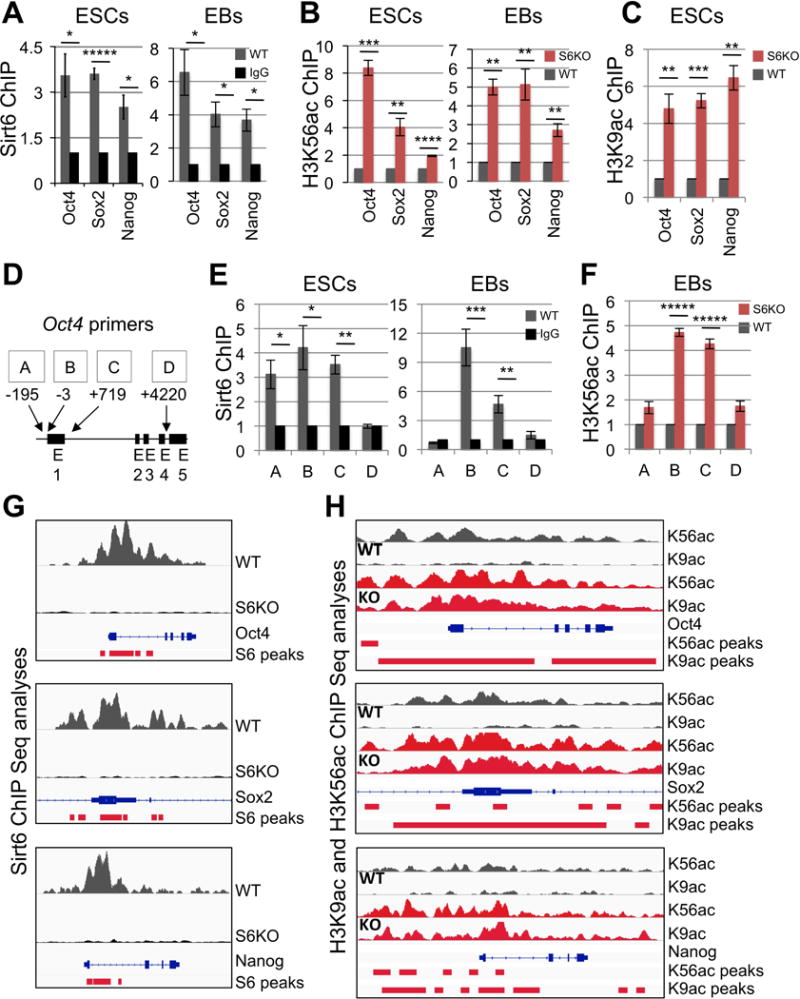
(A) Chromatin immunoprecipitation (ChIP) analysis for Sirt6 on core pluripotent gene promoters in WT ESCs and EBs. Data is expressed relative to IgG-ChIP control.
(B) ChIP analysis for H3K56ac on core pluripotent gene promoters in both ESCs and EBs from WT and S6KO. Data is expressed relative to WT values.
(C) ChIP analysis for H3K9ac on core pluripotent gene promoters in EBs from WT and S6KO. Data is expressed relative to WT values.
(D) Schematic diagram of the Oct4 locus depicting primers used for ChIP assays in panels (E) and (F).
(E) ChIP analysis for Sirt6 on the Oct4 locus in WT ESCs and EBs. Data is expressed relative to IgG-ChIP control.
(F) ChIP analysis for H3K56ac on the Oct4 locus in EBs from WT and S6KO. Data is expressed relative to WT values.
(G) Sirt6 ChIP-Seq binding profiles on Oct4, Sox2, and Nanog genes in WT and S6KO ESCs. Images were created with the Integrative Genomic Viewer (IGV) (Robinson et al., 2011). Data are normalized to total counts, and the scale rage is 0.0 – 7.0.
(H) ChIP-Seq binding profiles of histone marks H3K56ac and H3K9ac on Oct4, Sox2 and Nanog genes in WT and S6KO ESCs. Images were created with the Integrative Genomic Viewer (IGV) (Robinson et al., 2011). Data are normalized to total counts, and the scale rage is 0.0 – 2.0.
The red bars under each plot in panels (G) and (H) represent statistically significant peaks for each ChIP seq analysis.
Global increase of Tet expression and 5hmC in S6KO versus WT ESCs
Tet1 and Tet2 genes are postulated targets of Oct4 and Sox2 (Koh et al., 2011; Wu et al., 2013). Strikingly, we observed significant upregulation of Tet1 and Tet2 expression in S6KO compared to WT ESCs, at both mRNA and protein levels (Figures 3A, 3B). Slot blot analysis showed a striking global increase of 5hmC in S6KO compared to WT ESCs, without a global alteration in the levels of 5mC (Figures 3C, 3D). Concordantly to the upregulation of Tet genes in S6KO, we found an increased recruitment of Oct4 and Sox2 to both Tet1 and Tet2 genes in S6KO versus WT ESCs by ChIP analysis, using primers for previously identified Oct4:Sox2 consensus binding sites (Koh et al., 2011; Figures 3E, 3F). Additionally, Oct4 and Sox2 binding to their own genes were enhanced in S6KO versus WT ESCs (Figure 3G). These data support a positive role for Oct4:Sox2-dependent regulation of Tet expression, which is enhanced in S6KO compared to WT ESCs, and thereby suggest a novel function for Sirt6, as a potential regulator of a Tet-dependent mechanism associated with ESC differentiation.
Figure 3. Oct4:Sox2-dependent upregulation of Tets in S6KO versus WT ESCs and EBs.
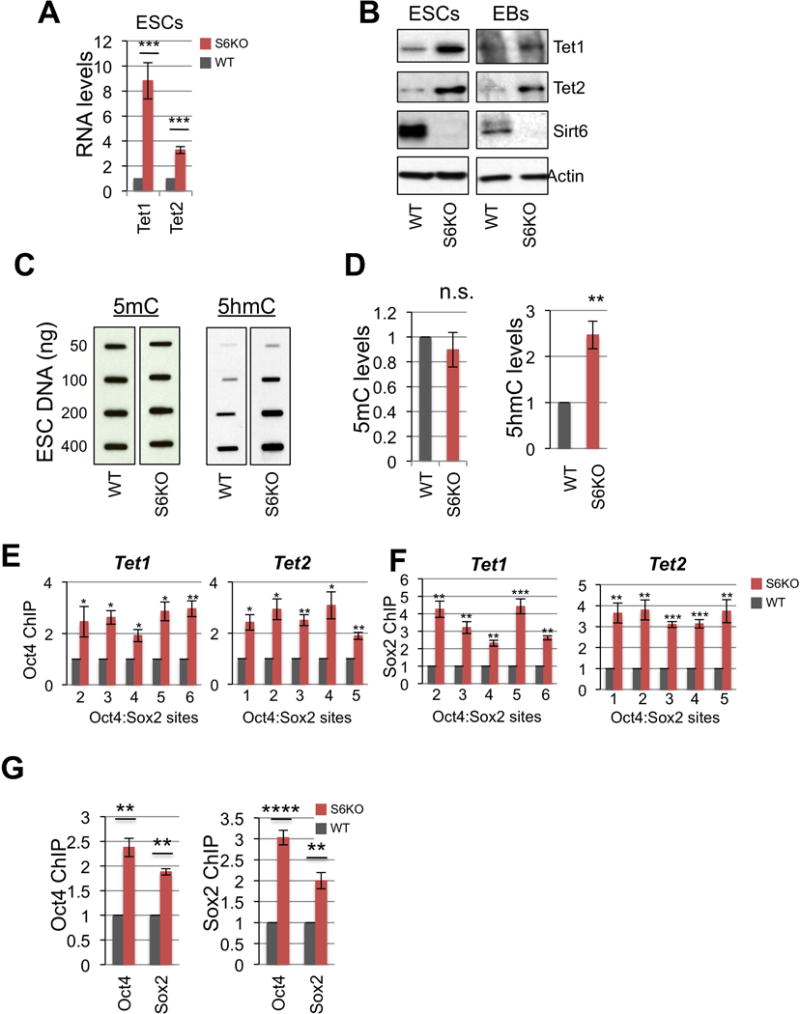
(A) Tet1 and Tet2 gene expression in WT versus S6KO ESCs. qRT-PCR data is expressed relative to WT ESCs.
(B) Western blot analysis for Tet1 and Tet2 in both ESCs and EBs. A representative of n = 3 biological replicates is shown.
(C) Global 5mC and 5hmC levels assayed by slot blot analysis in WT versus S6KO ESCs.
(D) Graphs showing fold change of 5mC and 5hmC from panel (H).
(E) ChIP analysis for Oct4 on both Tet1 and Tet2 at Oct4:Sox2 predicted binding sites (Koh et al., 2011) in WT versus S6KO ESCs. Data is expressed relative to WT values.
(F) ChIP analysis for Sox2 on both Tet1 and Tet2 at Oct4:Sox2 predicted binding sites (Koh et al., 2011) in WT versus S6KO ESCs. Data is expressed relative to WT values. The data are n = 3 experimental replicates. Values are mean +/− s.e.m. *P < 0.05, **P < 0.01, ***P < 0.001, by t-test analysis.
Depletion of Tet1 or Tet2 Rescues the Differentiation Phenotype in S6KO ESCs
The upregulation of Tets and 5hmC production in S6KO led us to examine the genetic interaction between Sirt6 and Tets in ESCs. Strikingly, shRNA-mediated depletion of either Tet1 or Tet2 (Koh et al., 2011) fully rescued not only the abnormal morphology of EBs derived from S6KO ESCs (Figure 4A), but also the skewed differentiation, as demonstrated by the normalized expression of the neuroectoderm marker Gfap (Figures 4B and 4C). Furthermore, the expression of additional germ layer markers – the neuroectoderm markers Fgf4 and Nestin and the endoderm markers Gata4 and Gata6 was restored upon Tet knockdown (Figure 4D). As expected, the levels of Tet1 and Tet2 transcripts, as well as Oct4 and Nanog transcripts, were brought back to near WT levels upon Tet knockdown in S6KO EBs (Figure 4E), as were the elevated levels of 5hmC in S6KO ESCs (Figure 4F, 4G). Interestingly, knockdown of either Tet1 or Tet2 caused downregulation of both Tet genes (Figure 4E), thereby implicating both Tets in the S6KO differentiation phenotype. These results further support a role for Sirt6 in suppressing expression of the core pluripotent genes in both pluripotent and differentiating ESC, thus indirectly, controlling the levels of Tet enzymes to facilitate proper differentiation.
Figure 4. Tet knockdown rescues the differentiation phenotype of S6KO ESCs and global levels of 5hmC.
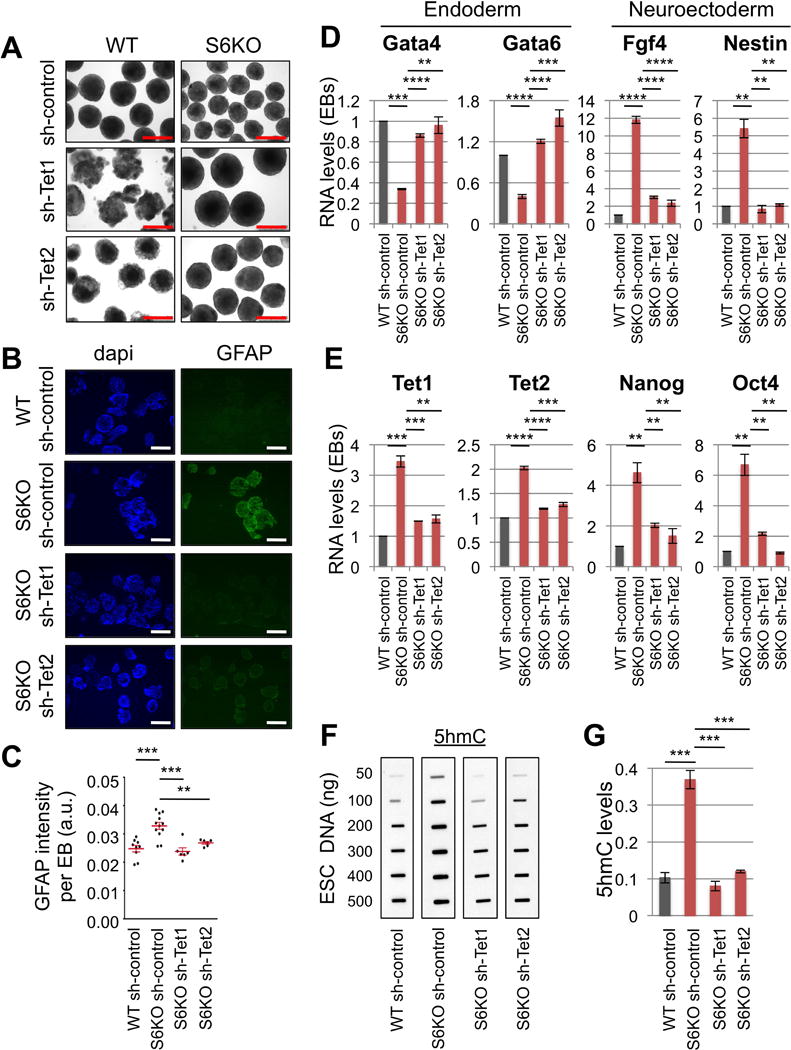
(A) EBs derived from WT and S6KO ESCs stably infected with shRNA targeting Tet1 or Tet2. Scale bar, 500 μm.
(B) Immunofluorescence of Tet knockdown EBs for Gfap. Scale bar, 500 μm.
(C) Graph showing quantification of Gfap intensity (mean intensity, a.u.) per EB on panel (B). Red bars represent mean +/− s.e.m. ** P < 0.01, *** P < 0.001 by 1 way Anova followed by Tukey test analysis. (a.u., arbitrary units). Data are represented as detection of Gfap per each EB. Values are mean +/− s.e.m. **P < 0.01, ***P < 0.001 by ttest analysis.
(D) Gene expression of endoderm, and neuroectoderm genes in Tet knockdown EBs. qRT-PCR data is expressed relative to WT EBs stably transfected with shRNA control.
(E) Expression of Tet and core pluripotent genes in Tet knockdown EBs analyzed as described above.
(F) Global 5hmC levels assayed by slot blot analysis in Tet knockdown ESCs.
(G) Graphs show fold change of 5hmC from panel (F).
The data on panels (D), (E) and (G) are n = 3 experimental replicates, values are mean +/− s.e.m. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 by t-test analysis.
SIRT6 Controls Levels of 5hmC of Genes Associated with Neuroectoderm
To elucidate the role of Tet proteins in WT and S6KO ESCs, we analyzed the genome-wide distribution of 5hmC in WT and S6KO ESCs by CMS-IP (Pastor et al., 2011). We then compared 5hmC levels in adjacent, non-overlapping 500-bp windows genome-wide (Lienhard et al., 2014). S6KO ESCs showed significant (p<1.e-3) gain of 5hmC at more windows (2,218) than lost 5hmC (1,562 windows; (Supplemental Table S1), confirming at a genome-wide level the overall gain of 5hmC observed by dot blot analysis (Figures 3C, 3D). Differentially hydroxymethylated regions (DHMRs) with gain of 5hmC were enriched over DHMRs with loss of 5hmC at promoters (p<1.3e-252 and p<6.3e-16, respectively) as well as CpG islands (p<1.4e-269 and p<1.2e-48, respectively) (Figure 5A). By grouping gene promoters according to their CpG densities (+/− 1kb around their transcription start sites), we found DHMRs to be equally enriched in low (LCP) as well as high (HCP) CpG density promoters (Figure 5A). However, gain of 5hmC within exons was preferentially observed at genes important for regulation of transcription and neuronal differentiation (Figure 5B, Supplemental Table S2) (Figures S2, S3). Notably, the presence of 5hmC at exons was recently shown to positively correlate with gene expression (Huang et al., 2014). Particularly, the Hoxa gene cluster, which is implicated in neural crest development, displays a significant enrichment of 5hmC, along with Gata2 and Pax6, also implicated in neurogenesis (Figure 5C). However, the housekeeping gene β-glucuronidase (Gusb) shows no difference in 5hmC levels between WT and S6KO ESCs (Figure 5C), which ensures specificity on this analysis. By correlating gain or loss of 5hmC with publically available data on histone marks, bisulfite derived methylation, and transcription factor binding data (Supplemental Table S3), we found that regions that gained 5hmC in S6KO compared to WT ESCs were also enriched in H3K4me2, an epigenetic mark associated with both promoters and enhancers that is involved in transcriptional activation and the binding of Tet1 (Figure 5D). Interestingly, the gain of 5hmC in S6KO, compared to WT ESCs, occurs at regions with low 5mC in WT ESC (Figure 5D), confirming an important role of Sirt6 in regulating 5hmC at low methylated regions (LMRs) which have been previously associated with distal regulatory elements (Stadler et al., 2011) (Figure 5D). Concomitant to the enrichment of 5hmC and H3K4me2, we found the expression of the Hoxa gene cluster together with Gata2 and Pax6, along with other genes of the neural lineage to be upregulated in S6KO versus WT ESCs (Figures 6A, S4A). Importantly, the upregulation of these genes was rescued upon Tet1 or Tet2 knockdown (Figures 6A, S4A). The association between 5hmC and H3K4me2 was further evaluated by correlation with public data as described above, on a set of genes from the neural lineage whose expression is upregulated in S6KO and rescued upon Tet knockdown (Figure 6B). This analysis shows a strong association between 5hmC with H3K4me2 within neuroectoderm genes whose expression is increased in S6KO versus WT ESCs (Figures 6A, 6B). We also address any potential interplay between 5hmC with H3K9ac and/or H3K56ac by genome wide analyses. Consistent with our previous work, we found a global elevation of both H3K56ac and H3K9ac at various genomic regions in ESCs, which is maintained after RA-dependent differentiation (Figure S7). We found Sirt6 and its histone targets, H3K56ac and H3K9ac, to mark genes involved in transcription, metabolism, RNA processing, cell cycle, chromatin organization, DNA repair as well as ESC marker genes associated with the maintenance of pluripotency (Tables S4–8). Notably, we found no significant correlation on the enrichments of 5hmC, H3K56ac, H3K9ac, as well as SIRT6 binding in S6KO versus WT ESCs (Figures 6C, 6D, S8A–D). Additionally, we evaluated the role of Sox2, which was shown to promote expression of the neural lineage (Thomson et al., 2011), by comparing the genomic regions enriched for 5hmC with the Sox2 ChIP-seq data set from Lodato and colleagues (Lodato et al., 2013), and found no significant correlation with the gain of 5hmC in S6KO versus WT ESCs (Figures 6C, 6D). Additionally, we found no correlation between H3K56ac and H3K9ac with Sox2 targets (Figure S8, Table S9). Overall, these data indicate that enrichment of 5hmC in S6KO is not directly interlocked with H3K56ac and/or H3K9ac, as well as Sirt6 or Sox2 targeted genomic regions. Therefore, Sirt6 plays a hierarchical role in regulating ESC differentiation by modulating Tet-dependent production of 5hmC through direct repression of the core pluripotent gene network via deacetylation of H3K56ac and H3K9ac. Furthermore, these data strongly suggest that 5hmC may functions as a positive transcriptional determinant to control the expression of genes associated with neuroectoderm development.
Figure 5. Characterization of genomic regions with change of 5hmC in S6KO compared to WT ESCs.
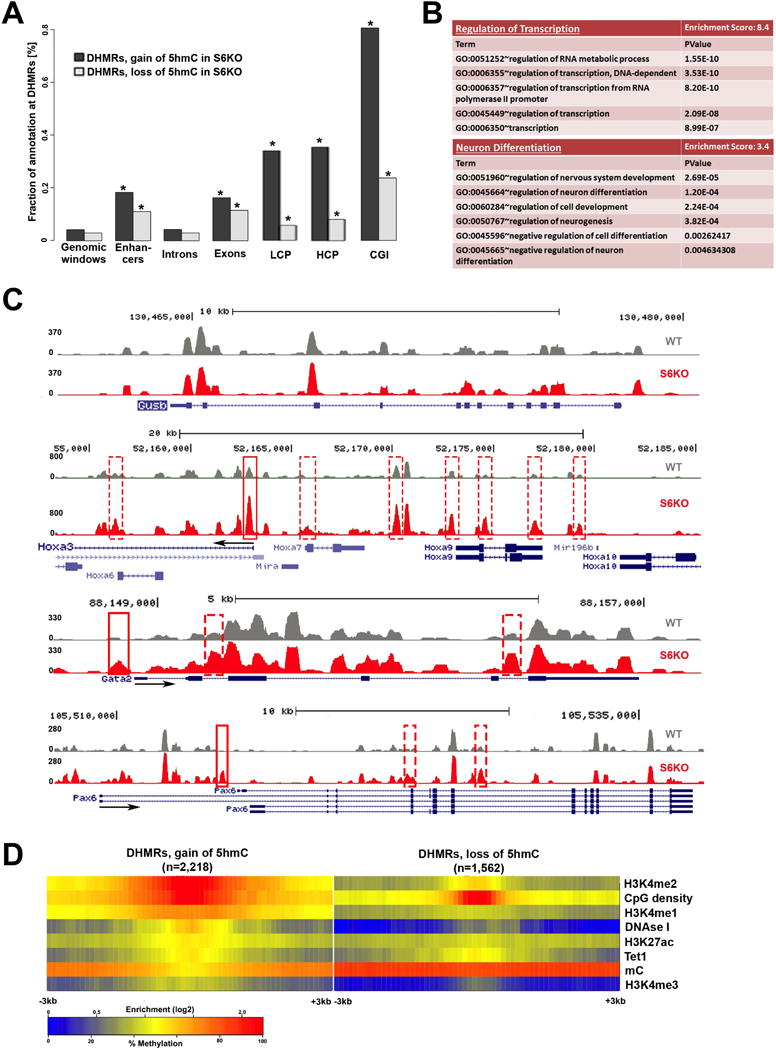
(A) Total DHMRs with gain or loss of 5hmC in S6KO represent 0.04% or 0.03% of the genome, respectively (genomic windows). Both classes of DHMRs are significantly enriched in exons, promoters, and CpG islands, where DHMRs with gain or loss of 5hmC are similarly enriched at exons (p value ≤ 6.3e-155 and p value ≤ 1.5e-110, respectively), whereas DHMRs with gain of 5hmC are much stronger enriched than DHMRs with loss of 5hmC at promoters (p value ≤ 1.3e-252 and p value ≤ 6.3e-16, respectively), and at CpG islands (p value ≤ 1.4e-269 and p value ≤ 1.2e-48, respectively) (Fisher’s exact test).
(B) Functional annotation (Huang et al., 2009) of genes with change of 5hmC (gain or loss) at exons, reveals significantly enriched gene ontology clusters associated with regulation of transcription and neuron differentiation.
(C) UCSC browser visualization of 5hmC of gain of 5hmC in S6KO (red) versus WT (gray) at the promoter of Hoxa3, Gata2 and Pax6 genes, and at multiple other genomic regions in the vicinity. No changes in 5hmC levels on the Gusb gene, is shown as an analytical control.
(D) Enrichment analysis of histone H3 modifications strongly connects H3K4me2 and low methylated regions to DHMRs with gain of 5hmC in S6KO. Both, gain and loss of 5hmC occurs at regions with elevated levels of CpG density and Tet1 binding. The marks are sorted by high (top) to low (bottom) enrichment at the center of DHMRs with gain of 5hmC in S6KO.
Figure 6. Association of 5hmC with H3K4me2, but not H3K9ac and/or H3K56ac in neuroectoderm genes upregulated in S6KO compared to WT ESCs.
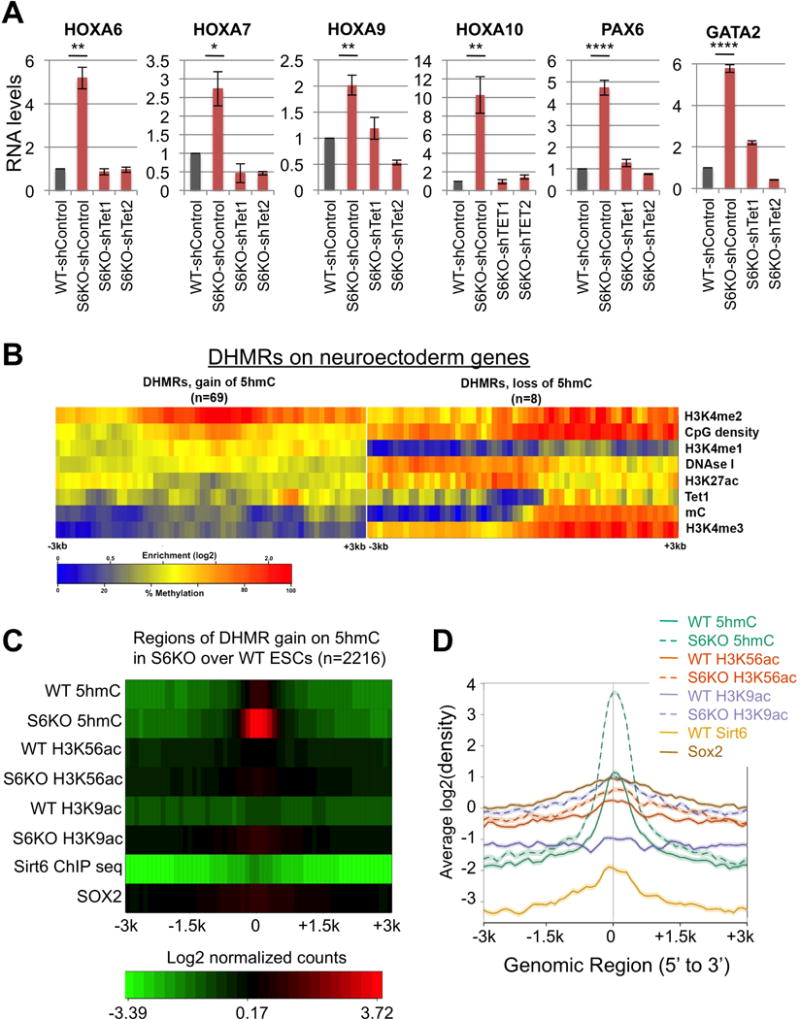
(A) Expression of neuroectoderm genes with gain of 5hmC in Tet knockdown ESCs. qRT-PCR data is expressed relative to WT-shControl. Data are n = 3 experimental replicates, values are mean +/− s.e.m. *P < 0.05, **P < 0.01, ****P < 0.0001 by t-test analysis.
(B) Enrichment analysis of histone H3 modifications strongly connects H3K4me2 to upregulated neuroectoderm genes with gain of 5hmC in S6KO. The marks are sorted by high (top) to low (bottom) enrichment at the center of DHMRs with gain of 5hmC in S6KO.
(C) Heat map plot of regions of DHMRs gain on 5hmC in S6KO over WT in ESCs showing average profile for +/− 3k centered around the 2216 regions with DHMR gains for the factors 5hmC, H3K56ac, H3K9ac, Sirt6, and Sox2 in WT and S6KO ESCs. Each row of the heat map represents mean values of enrichment z-scores in 100 bp windows in the +/−3kb region.
(D) Enrichment line plot of average profile for regions of DHMRs gain on 5hmC in S6KO over WT mouse ESCs (n=2216) for data of panel (C). Semi-transparent band behind line shows standard error of the mean for each average profile.
SIRT6 Regulates Developmental Programs In Vivo
To further evaluate the role of Sirt6 in ESC differentiation in vivo, we injected S6KO and WT ESCs subcutaneously into immunodeficient mice and followed teratoma formation. Teratomas derived from S6KO were significantly smaller compared to their WT counterparts (Figure 7A). The neuroectoderm marker β-III Tubulin as well as Oct4 were expressed at elevated levels in S6KO-derived teratomas (Figure 7B). Remarkably, knocking down either Tet1 or Tet2 rescued the smaller size of teratomas derived from S6KO ESCs (Figures 7C). Additionally, the upregulation of Oct4, β-III Tubulin and Gfap expression was also rescued upon either Tet1 or Tet2 knockdown (Figures 7D, 7E). To explore this phenotype further, we integrated a GFP-encoding gene within the Rosa locus in both WT and S6KO iPSCs. These cells were then injected into mouse blastocysts and chimerism was determined in mid-gestation (E12.5) embryos by immunohistochemistry using an anti-GFP antibody. As expected, WT GFP-iPSCs gave raise to most if not all tissues of the GFP chimeras (Figure 8A, Table 8B). However, mice originated from blastocysts injected with S6KO iPSCs exhibited a weak GFP staining and in some cases almost exclusively in tissues of the central nervous system, such as diencephalon and partial regions of the neural cord (marked by yellow arrows) (Figure 8A, Table 8B).
Figure 7. Sirt6 deficiency triggers an in vivo differentiation defect in mouse and in human EBs, which is rescued by Tet knockdown on mouse teratomas.
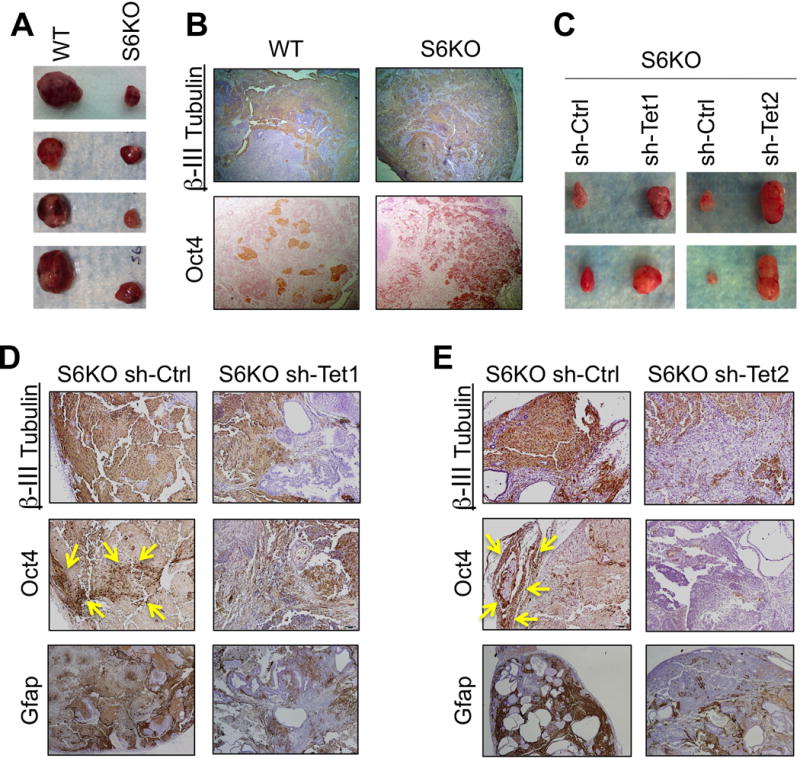
(A) Teratomas from C57BL/6 WT and S6KO ESCs. Data are shown as n = 5 biological replicates.
(B) IHC analysis for β-III Tubulin and Oct4 on WT versus S6KO teratomas. Pictures are taken at 5×. IHC of one representative from n=3 experimental replicates is shown.
(C) Teratomas derived from stably infected S6KO ESCs with shRNA targeting either Tet1 or Tet2 versus control shRNA.
(D) and (E) IHC staining for β-III Tubulin, Oct4 and Gfap of teratomas from panel (C). One representative from n=3 experimental replicates is shown. Areas of Oct4 positive nuclear staining are demarked by the yellow arrows.
Figure 8. Sirt6 deficiency triggers an in vivo differentiation defect in mouse and in human EBs.
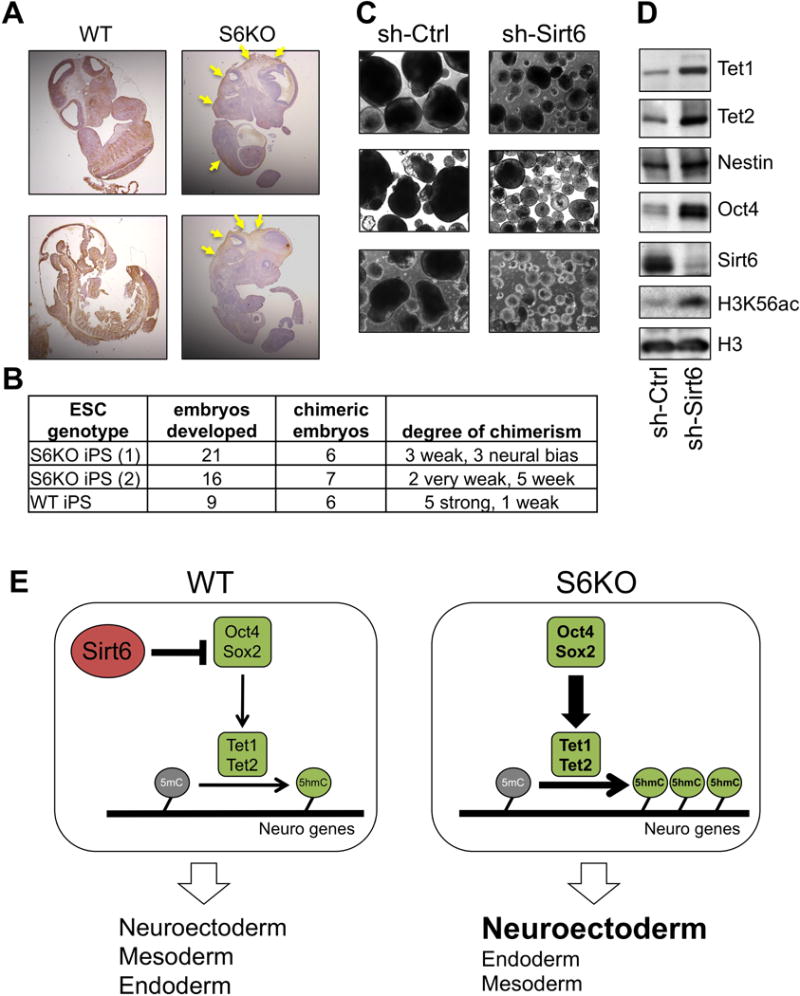
(A) IHC analysis for GFP on chimeric mice (E12.5) derived from WT or S6KO iPSCs. Note that contribution of S6KO iPSCs is restricted to some neural tissues (yellow arrows).
(B) Table showing the numbers of embryos developed and the degree of chimerism based on IHC with anti-GFP antibody. Note that S6KO iPSCs exhibit a week degree of chimerism or is restricted to the neural tissue in three embryos.
(C) Human EBs (hEBs) stably infected with an shRNA control or shRNA targeting Sirt6. Scale bar, 300 μm. A representative of n = 5 biological replicates is shown.
(D) Western blot analysis for Tet enzymes, Oct4 and the neuroectoderm marker Nestin on hEBs stably infected with an shRNA control or shRNA targeting Sirt6. A representative from n = 3 experimental replicates is shown.
(E) Schematic representation depicting the role of Sirt6 as a regulator of ESC differentiation via repression of Oct4 and Sox2 gene expression which in turn controls Tet-dependent oxidation of 5mC into 5hmC, which is needed to achieve proper development of the germ layers. Sirt6 depletion causes a derepression of Oct4 and Sox2 triggering an upregulation of Tet-dependent 5hmC production that results in skewed development towards neuroectoderm.
Depletion of Sirt6 in hESCs Leads to a Differentiation Defect Similarly to Mouse S6KO ESCs
To assess whether this Sirt6-dependent developmental mechanism is evolutionarily conserved, we explored the differentiation capacity of human ESCs (hESCs) upon shRNA-mediated depletion of Sirt6, and found that Sirt6-depleted human EBs (hEBs) were significantly smaller compared to their controls (Figure 8C). Furthermore, the expression of Tet1, Tet2 and Oct4 was elevated in Sirt6-depleted hESCs (Figure 8D). Paralleling the role of Sirt6 in mouse ESC, we found from genome-wide analysis by Ram and colleagues (Ram et al., 2011) that human Sirt6 is recruited at human Oct4 and Sox2 genes (Figure S4B), supporting an evolutionarily conserved role for Sirt6 in regulating the core pluripotent genes. Consistently, we found minimal or no binding of SIRT6 to the Tet genes (Figure S4C), again suggesting that SIRT6 modulates Tet-dependent 5hmC indirectly. Additionally, the neuroectoderm marker Nestin was upregulated in Sirt6-deficient hESCs (Figure 8D), supporting the predisposition towards the neural cell lineage in the absence of Sirt6. Hence, we propose a model whereby Sirt6 controls ESC differentiation by repressing the expression of Oct4 and Sox2, consequently diminishing the Oct4:Sox2-activated expression of Tet enzymes, and thereby limiting the levels of 5hmC at specific genomic regions to allow balanced transcription of developmentally regulated genes (Figure 8E).
DISCUSSION
A critical step during ESC differentiation involves the silencing of the pluripotent gene network to allow expression of cell lineage specific genes. The core pluripotent genes Oct4 and Nanog undergo transcriptional silencing through DNA methylation at their regulatory regions, which is maintained in differentiated somatic cells (Epsztejn-Litman et al., 2008; Li et al., 2007). Our work demonstrates an additional mechanism to repress expression of the core pluripotent genes during ESC differentiation, which involves the histone deacetylase Sirt6. Previous studies showed that Sirt6 is a critical modulator of glycolytic metabolism, DNA repair and cancer (Mostoslavsky et al., 2006; Zhong et al., 2010; Sebastian et al., 2012; Toiber et al., 2013). Our work determines a novel function for Sirt6 as a key regulator of ESC differentiation, by repressing the expression of Oct4, Sox2 and Nanog. Importantly, derepression of these core pluripotent genes in S6KO during ESC differentiation suggests that lack of Sirt6 could potentially increase efficiency of reprogramming. Indeed, we found a ~10-fold increase in iPSC formation during reprogramming of mouse neural progenitor cells (NPCs) derived from S6KO mice compared to WT controls (Figure S5F–H).
Somatic cells switch from an oxidative metabolic state to a glycolytic state during reprogramming to iPSCs (Folmes et al., 2011; Panopoulos et al., 2012; Prigione and Adjaye, 2012; Varum et al., 2011; Zhou et al., 2012). Furthermore glycolysis is critical for the maintenance of pluripotency (Zhang et al., 2012). Both MEFs and ESCs lacking Sirt6 exhibit a higher rate of glycolysis compared to WT (Zhong et al., 2009). However, the differentiation phenotype of S6KO EBs was not rescued upon glycolysis inhibition, by knocking down Pdk1 (Figure S5A). These data suggest a predominant role for Sirt6 in regulating expression of pluripotent genes and 5hmC levels, independent of its function in metabolism, during ESC differentiation. Importantly, ectopic expression of human Sirt6 rescues the differentiation phenotype of S6KO EBs, thereby establishing specificity for the role of Sirt6 on the S6KO differentiation phenotype as well as its conserved role between mouse and human (Figure S6A–C).
The biological significance of Tet-mediated 5mC oxidations in epigenetic regulation remains poorly understood, especially with regard to its relevance during ESC differentiation, whereby pluripotent cells commit to specific cell lineages. In addition to be a DNA demethylation intermediate, 5hmC is recognized by chromatin regulatory proteins and therefore postulated to function as an epigenetic mark (Spruijt et al., 2013). However, specific target genes, whose expression is associated to 5hmC as an epigenetic determinant of cell lineage specification during ESC differentiation, are unknown. We identified genes associated to the neural lineage whose expression is directly correlated with an enrichment of 5hmC at promoters and exons in S6KO ESCs (Figures 5A–C). Furthermore, the enrichment of 5hmC in these genes occurs near H3K4me2 mark, an epigenetic signature involved in transcription activation (Figures 5D and 6B). The upregulated expression of Tet enzymes in S6KO ESCs suggests a potential increase in further oxidized forms of 5mC. Indeed, we detected elevated levels of 5caC in S6KO versus WT ESCs by slot blot analysis, which are rescued upon Tet1 or Tet2 knockdown (Figures S5B–E). Therefore, S6KO ESCs represents a relevant biological system to further analyze the importance of Tet-mediated DNA oxidation during cell fate choices.
Recent studies have shown that Tet1/Tet2 double knockout ESCs are depleted of 5hmC, which correlates with developmental defects in teratomas and chimeric embryos (Dawlaty et al., 2013). Tet1/Tet2-deficient mice were obtained at lower frequency indicating a phenotype of partial lethality and thereby supporting the critical role of Tet enzymes in embryonic development (Dawlaty et al., 2013). In this context, Sirt6 deficiency in specific genetic backgrounds also causes partial embryonic lethality (data not shown), further supporting that imbalanced levels of Tet proteins impair embryonic development.
Changes in overall chromatin architecture are required during the transition from pluripotency to differentiated states. However, the interplay between chromatin regulators and epigenetic determinants associated with the establishment of transcriptional programs during cell fate choices remain poorly understood. Our work identify the chromatin enzyme Sirt6 as a key regulator of ESC differentiation, acting through sequential regulation of the core pluripotent genes and Tet-mediated production of 5hmC to control expression of genes involved in neural cell fate. Thus, in the absence of Sirt6, neural-related genes are marked with elevated levels of 5hmC, suggesting that this modification might function as either an epigenetic determinant or a facilitator of local DNA demethylation that channels ESC commitment to the neural cell lineage. Interestingly, Tet2-dependent hydroxylation of 5mC was also found to be required for the transcriptional activation of the Hoxa cluster, which is critical for cell lineage specification in NT2 cells, an embryonic carcinoma cell line that can be differentiated with retinoic acid (RA) into neural cells (Bocker et al., 2012). Together, our data argue that the differentiation defect we observe in S6KO EBs is linked to Tet function, resulting in a predisposition towards a neuroectoderm developmental pathway.
Collectively, our studies unravel a molecular mechanism implicating Sirt6 as a critical regulator of ESC differentiation that involves the core pluripotent genes and Tet-dependent production of 5hmC. Future studies will determine, at a gene-by-gene level, whether 5hmC and its further oxidized forms 5fC and 5caC participates in gene expression and ESC differentiation as molecular intermediates in the process of DNA demethylation, as epigenetic marks that recruits chromatin and transcriptional regulators to gene regulatory regions, or whether both mechanisms apply.
Supplementary Material
Acknowledgments
This work was supported in part by NIH grants GM093072-01, DK088190-01A1 (R.M.), HD065812 and CA151535 (to A.R). R.M. is the Kristine and Bob Higgins MGH Research Scholar, the Warshaw Institute Fellow, and a Howard Goodman Awardee. L.C. is the recipient of a Feodor Lynen Research Fellowship from the Alexander von Humboldt Foundation. Y.H. is supported by a postdoctoral fellowship from the Leukemia and Lymphoma Society. C.A.S. is the recipient of the Evans Center Fellow Award.
References
- Bocker MT, Tuorto F, Raddatz G, Musch T, Yang FC, Xu M, Lyko F, Breiling A. Hydroxylation of 5-methylcytosine by TET2 maintains the active state of the mammalian HOXA cluster. Nature Communications. 2012;3.818:1–12. doi: 10.1038/ncomms1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Dent SY. Chromatin modifiers and remodellers: regulators of cellular differentiation. Nat Rev Genet. 2014;15:93–106. doi: 10.1038/nrg3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawlaty MM, Breiling A, Le T, Raddatz G, Barrasa MI, Cheng AW, Gao Q, Powell BE, Li Z, Xu M, et al. Combined deficiency of Tet1 and Tet2 causes epigenetic abnormalities but its compatible with postnatal development. Cell. 2013;24:310–323. doi: 10.1016/j.devcel.2012.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epsztejn-Litman S, Feldman N, Abu-Remaileh M, Shufaro Y, Gerson A, Ueda J, Deplus R, Fuks F, Shinkai Y, Cedar H, et al. De novo DNA methylation prmoted by G9a prevents reprogramming of embryonically silenced genes. Nat Struct Mol Biol. 2008;15:1176–1183. doi: 10.1038/nsmb.1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ficz G, Branco MR, Seisenberger S, Santos F, Krueger F, Hore TA, Marques CJ, Andrews S, Reik W. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature. 2011;473:398–402. doi: 10.1038/nature10008. [DOI] [PubMed] [Google Scholar]
- Folmes CD, Nelson TJ, Martinez-Fernandez A, Arrell DK, Lindor JZ, Dzeja PP, Ikeda Y, Perez-Terzic C, Terzic A. Somatic oxidative bioenergetics transitions into pluripotency-dependent glycolysis to facilitate nuclear reprogramming. Cell Metab. 2011;14:264–271. doi: 10.1016/j.cmet.2011.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar-Maia A, Alajem A, Polesso F, Sridharan R, Mason MJ, Heidersbach A, Ramalho-Santos J, McManus MT, Plath K, Meshorer E, et al. Chd1 regulates open chromatin and pluripotency of embryonic stem cells. Nature. 2013;460:863–868. doi: 10.1038/nature08212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes IC, Acquarone M, Maciel Rde M, Erlich RB, Rehen SK. Analysis of pluripotent stem cells by using cryosections of embryoid bodies. Journal of Visualized Experiments. 2014 doi: 10.3791/2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, Ding J, Jia Y, Chen Z, Li L, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–1306. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Chavez L, Chang X, Wang X, Pastor WA, Kang J, Zepeda-Martinez JA, Pape UJ, Jacobsen SE, Peters B, et al. Proc Natl Acad Sci USA. 2014;111:1361–1366. doi: 10.1073/pnas.1322921111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Ito S, D’Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y. Tet proteins can convert 5-Methycytosine to 5-Formylcytosine and 5-Carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer LM, Tahiliani M, Rao A, Aravind L. Prediction of novel family of enzymes involved in oxidative and other complex modifications of bases in nucleic acids. Cell Cycle. 2009;11:1698–1710. doi: 10.4161/cc.8.11.8580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko M, Huang Y, Jankowska AM, Pape UJ, Tahiliani M, Bandukwala HS, An J, Lamperti ED, Koh KP, Ganetzky R, et al. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature. 2010;468:839–843. doi: 10.1038/nature09586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh KP, Yabuuchi A, Rao S, Huang Y, Cunniff K, Nardone J, Laiho A, Tahiliani M, Sommer CA, Mostoslavsky G, et al. Tet1 and tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell. 2011;8:200–213. doi: 10.1016/j.stem.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory- efficient alignment of short DNA sequences to the human genome. Genome Biology. 2009;10(3):R25. doi: 10.1186/gb-2009-10-3-r25. 10. Doi. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JY, Pu MT, Hirasawa R, Li BZ, Huang YN, Zeng R, Jing NH, Chen T, Li E, Sasaki H, et al. Synergistic function of DNA methyltransferases Dnmt3a and Dnmt3b in the methylation of Oct4 and Nanog. Mol Cell Biol. 2007;27:8748–8759. doi: 10.1128/MCB.01380-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup The sequence alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lienhard M, Grimm C, Morkel M, Herwig R, Chavez L. MEDIPS: genome-wide differential coverage analysis of sequencing data derived from DNA enrichment experiments. Bioinformatics. 2014;30:284–286. doi: 10.1093/bioinformatics/btt650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leinonen R, Sugawara H, Shumway M. The sequence read archive. Nucleic Acid Research. 2011;39:19–21. doi: 10.1093/nar/gkq1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodato MA, Ng CW, Wamstad JA, Cheng AW, Thai KK, Fraenkel E, Jaenisch R, Boyer LA. SOX2 co-occupies distal enhancer elements with distinct POU factors in ESCs and NPCs to specific cell state. PLOS Genetics. 9:e1003288. doi: 10.1371/journal.pgen.1003288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellen M, Mellén M, Ayata P, Dewell S, Kriaucionis S, Heintz N. MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell. 2012;151:1417–1430. doi: 10.1016/j.cell.2012.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michishita E, McCord RA, Boxer LD, Barber MF, Hong T, Gozani O, Chua KF. Cell cycle-dependent deacetylation of telomeric histone H3 lysine K56 by human Sirt6. Cell Cycle. 2009;16:2664–2666. doi: 10.4161/cc.8.16.9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostoslavsky R, Chua KF, Lombard DB, Pang WW, Fischer MR, Gellon L, Liu P, Mostoslavsky G, Franco S, Murphy MM, et al. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell. 2006;124:315–329. doi: 10.1016/j.cell.2005.11.044. [DOI] [PubMed] [Google Scholar]
- Panopoulos AD, Yanes O, Ruiz S, Kida YS, Diep D, Tautenhahn R, Herrerías A, Batchelder EM, Plongthongkum N, Lutz M, et al. The metabolome of induced pluripotent stem cells reveals metabolic changes occurring in somatic cell reprogramming. Cell Res. 2012;22:168–177. doi: 10.1038/cr.2011.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastor WA, Pape UJ, Huang Y, Henderson HR, Lister R, Ko M, McLoughlin EM, Brudno Y, Mahapatra S, Kapranov P, et al. Genome-wide mapping of 5-hydroximethylcytosine in embryonic stem cells. Nature. 2011;473:394–397. doi: 10.1038/nature10102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastor WA, Aravind L, Rao A. TETonic shift: biological roles of TET proteins in DNA demethylation and transcription. Nature Reviews. 2013;14:341–356. doi: 10.1038/nrm3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigione A, Adjaye J. Modulation of mitochondrial biogenesis and bioenergetic metabolism upon in vitro and in vivo differentiation of human ES and iPS cells. Int J Dev Biol. 2010;54:1729–1741. doi: 10.1387/ijdb.103198ap. [DOI] [PubMed] [Google Scholar]
- Ram O, Goren A, Amit I, Shoresh N, Yosef N, Ernst J, Kellis M, Gymrek M, Issner R, Coyne M, et al. Combinatorial patterning of chromatin regulators uncovered by genome-wide location analysis in human cells. Cell. 2011;147:1628–1639. doi: 10.1016/j.cell.2011.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastián C, Zwaans BM, Silberman DM, Gymrek M, Goren A, Zhong L, Ram O, Truelove J, Guimaraes AR, Toiber D, et al. Cell. 2012;151:1185–1199. doi: 10.1016/j.cell.2012.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Yue F, McCleary DF, Ye Z, Edsall L, Kuan S, Wagner U, Dixon J, Lee L, Lobanenkov VV, et al. A map of the cis-regulatory sequence in the mouse genome. Nature. 2012;488:116–120. doi: 10.1038/nature11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ZD, Meissner A. DNA methylation: roles in mammalian development. Nat Rev Genet. 2013;14:204–220. doi: 10.1038/nrg3354. [DOI] [PubMed] [Google Scholar]
- Spruijt CG, Gnerlich F, Smits AH, Pfaffeneder T, Jansen PW, Bauer C, Münzel M, Wagner M, Müller M, Khan F, et al. Dynamic readers for 5-(hydroxy)Methylcytosine and its oxidized derivatives. Cell. 2013;152:1146–1159. doi: 10.1016/j.cell.2013.02.004. [DOI] [PubMed] [Google Scholar]
- Stadler MB, Murr R, Burger L, Ivanek R, Lienert F, Schöler A, van Nimwegen E, Wirbelauer C, Oakeley EJ, Gaidatzis D, et al. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature. 2011;480:490–495. doi: 10.1038/nature10716. [DOI] [PubMed] [Google Scholar]
- Suva ML, Riggi N, Bernstein BE. Epigenetic reprogramming in cancer. Science. 2013;339:1567–1570. doi: 10.1126/science.1230184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szwagierczak A, Bultmann S, Schmidt CS, Spada F, Leonhardt H. Sensitive enzymatic quantification of 5-hydroxymethylcytosine in genomic DNA. Nucleic Acids Res. 2010;38(19):e181. doi: 10.1093/nar/gkq684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et al. Conversion of 5-methylcitosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson M, Liu SJ, Zou LN, Smith Z, Meissner A, Ramanathan S. Pluripotency factors in embryonic stem cells regulate differentiation into germ layers. Cell. 2011;145:875–889. doi: 10.1016/j.cell.2011.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toiber D, Erdel F, Bouazoune K, Silberman DM, Zhong L, Mulligan P, Sebastian C, Cosentino C, Martinez-Pastor B, Giacosa S, et al. SIRT6 recruits SNF2H to DNA break sites, preventing instability through chromatin remodeling. Mol Cell. 2013;51:454–468. doi: 10.1016/j.molcel.2013.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varum S, Rodrigues AS, Moura MB, Momcilovic O, Easley CA, 4th, Ramalho-Santos J, Van Houten B, Schatten G. Energy metabolism in human pluripotent stem cells and their differentiated counterparts. PLoS One. 2011;6(6):e20914. doi: 10.1371/journal.pone.0020914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams K, Christensen J, Pedersen MT, Johansen JV, Cloos PA, Rappsilber J, Helin K. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature. 2011;473:343–348. doi: 10.1038/nature10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, D’Alessio AC, Ito S, Xia K, Wang Z, Cui K, Zhao K, Sun YE, Wu H, Zhang Y. Reversing DNA methylation: mechanisms, genomics, and biological functions. Cell. 2014;156:45–63. doi: 10.1016/j.cell.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Guo Z, Liu Y, Tang B, Wang Y, Yang L, Du J, Zhang Y. Oct4 and the small molecule inhibitor, SC1, regulates Tet2 expression in mouse embryonic stem cells. Mol Biol Rep. 2013;40:2897–2906. doi: 10.1007/s11033-012-2305-5. [DOI] [PubMed] [Google Scholar]
- Xie W, Song C, Young NL, Sperling AS, Xu F, Sridharan R, Conway AE, Garcia BA, Plath K, Clark AT, et al. Histone H3 lysine 56 acetylation is linked to the core transcriptional network in human embryonic stem cells. Molecular Cell. 2009;33:417–427. doi: 10.1016/j.molcel.2009.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B, Zwaans BMM, Eckersdoroff M, Lombard DB. The sirtuin Sirt6 deacetylates H3K56Ac in vivo to promote genomic stability. Cell Cycle. 2009;16:2662–2663. doi: 10.4161/cc.8.16.9329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young RA. Control of the embryonic stem cell state. Cell. 2011;144:940–954. doi: 10.1016/j.cell.2011.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. Dual functions of Tet1 in transcription regulation in mouse embryonic stem cells. Nature. 2011;473:389–93. doi: 10.1038/nature09934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Nuebel E, Daley GQ, Koehler CM, Teitell MA. Metabolic regulation in pluripotent stem cells during reprogramming and slef-renewal. Cell Stem Cell. 2012;11:589–595. doi: 10.1016/j.stem.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong L, D’Urso A, Toiber D, Sebastian C, Henry RE, Vadysirisack DD, Guimaraes A, Marinelli B, Wikstrom JD, Nir T, et al. The histone deacetylase SIRT6 regulates glucose homeostasis via Hif1α. Cell. 2010;140:280–293. doi: 10.1016/j.cell.2009.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Choi M, Margineantu D, Margaretha L, Hesson J, Cavanaugh C, Blau CA, Horwitz MS, Hockenbery D, Ware C, et al. HIF1α induced switch from bivalent to exclusively glycolytic matabolism during ESC-to-EpiSC/hESC transitions. EMBO J. 2012;31:2103–2116. doi: 10.1038/emboj.2012.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


