Abstract
Cells’ ability to sense and interpret mechanical signals from the extracellular milieu modulates the degree of cell spreading. Yet how cells detect such signals and activate downstream signaling at the molecular level remain elusive. Herein, we utilize tension gauge tether (TGT) platform to investigate underlying molecular mechanism of cell spreading. Our data from both differentiated cells of cancerous and non-cancerous origin show that for the same stiff underlying glass substrates and for same ligand density it is the molecular forces across single integrins that ultimately determine cell spreading responses. Furthermore, by decoupling molecular stiffness and molecular tension we demonstrate that molecular stiffness has little influence on cell spreading. Our data provide strong evidence that links molecular forces at the cell-substrate interface to the degree of cell spreading.
Keywords: Cell adhesion, spreading, tension gauge tether (TGT) technology
INTRODUCTION
Cells are sensitive to mechanical and physical cues from the extracellular matrix (ECM). This exceptional ability of sensing regulates cellular events that dictate cell fate decisions1, 2, proliferation3, regeneration4, and cancer metastasis5. At the core of such regulation lies the degree of cell spreading which is controlled by the bulk properties of the underlying substrates such as stiffness1, 6-8. For example, naïve human mesenchymal stem cells spread differentially on substrates of different stiffness to commit to neurogenic, myogenic or osteogenic lineages1. Previously, we demonstrated that stress-induced cell spreading of mouse embryonic stem cells elicits differentiation9. Changes in cell shape and efficient motor neuron differentiation from human pluripotent cells can be achieved by simply tuning the mechanical stiffness of the microposts10. However, the molecular mechanism by which cells sense stiffness and control spreading that ultimately influences cell differentiation remains poorly understood. Although several studies reported contractile elements to be responsible for rigidity sensing, cells spreading, and differentiation1, 11-15, they fail to capture the molecular mechanism related to early events in cell spreading. A recent report proposed that it is the collagen tethering density, not the physical bulk stiffness of the substrates that determine cellular morphology and gene expression change during stem cell differentiation16. On the contrary, a subsequent study suggested that differentiation of stem cells is possible even in the absence of such tethering17. Therefore, the underlying molecular mechanisms connecting the substrate stiffness to cell spreading, and subsequently fate regulation remain unresolved. In this study, to investigate cell spreading mechanism of differentiated cells of cancerous and non-cancerous origin, we employed our recently developed tension gauge tether (TGT)18 platform that can determine single molecular forces required to activate signaling through ligand-receptor bonds and cap the force through a single bond at a defined value.
RESULTS AND DISCUSSION
Molecular tension driven cell spreading
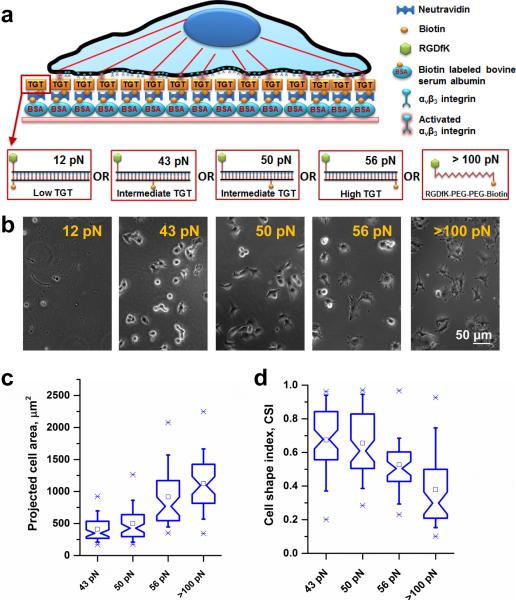
We conjugated cyclic-RGDfK peptide, a ligand specific to αvβ3 integrins19-21, to TGTs of four different values of calculated tension tolerance, Ttol (12, 43, 50, 56 pN). Ttol is the lowest when the biotin is on the same end of the duplex DNA tether as the peptide ligand because the force between the ligand and biotin is applied in the unzipping direction (Fig. 1a). Ttol increases progressively as the biotin is moved toward the other end of the DNA because the force is now applied in a shear configuration that requires much higher forces for DNA rupture (Fig. 1a). Passivated surfaces were prepared by incubating biotinylated bovine serum albumin (BSA) on glass surfaces followed by incubation of neutravidin as described before22. The RGDfK conjugated TGTs were then immobilized through a neutravidin-biotin linker (Fig. 1a). Biotinylated cyclic-RGDfK peptide which has much larger rupture force (>100 pN)23 was also directly immobilized on the surface as a control. B16-F1 melanoma cells cultured on rigid plastic dishes were plated on the surface presenting different Ttol values 12, 43, 50, 56, and >100 pN (Fig. 1b). As reported for five different cell lines previously18, B16-F1 cells did not adhere on surfaces with Ttol < 40 pN but adhered well to surfaces with higher Ttol suggesting that they apply about 40 pN peak force during initial cell adhesion.
Figure 1.
Cell spreading increases with increasing tension tolerance surface cues. (a) A schematic representation of a cell on a TGT surface. TGTs with different tension tolerances were immobilized through biotin-neutravidin interactions on biotinylated BSA passivated rigid glass surfaces. (b) Cells were plated on different tension tolerance surfaces. The measured force requirement during initial cell adhesion process was ~40 pN. Cell spreading increases with increasing tension tolerance. Tension tolerance >100 pN represents biotinylated RGDfK ligand immobilized directly on to the surface, not through TGTs. (c) Projected cell area of cells are presented as box-and-whisker plots. Cells (n=64, 63, 46, 48 for 43 pN, 50 pN, 56 pN, and >100 pN surface respectively) increased cell spreading area with increasing tension tolerances. There were significant differences in cell area change between 43 pN and 50 pN (p<0.03), 50 pN and 56 pN (p<1.38×10−8), and 56 pN and >100 pN (p<0.02) tension tolerance surfaces. (d) A box-and-whisker plot shows a dimensionless parameter, measuring geometric circularity called cell shape index (CSI). CSI varies from 1 for a perfect circular shape to 0 for a highly irregular shape. No significant differences were observed between 43 pN and 50 pN TGT surface (p>0.58). However, CSI values between 50 pN and 56 pN, and 56 and <100 pN surfaces showed significant changes (p<4.38×10−4, 3.51 ×10−4 respectively).
For the four high-tension surfaces supporting cell adhesion, the projected cell area of the cells increased with increasing Ttol (Fig. 1c). Although hydrogel studies have shown that cells spread more on stiffer surfaces1, 6, 8, our data show that the molecular forces across single receptor-ligand bonds can determine cell spreading responses. Three additional cancer cell lines (Supplementary Fig. 1) and non-cancerous mouse embryonic fibroblasts (Supplementary Fig. 2) we tested, all showed Ttol-dependent cell spreading, suggesting the universality of our finding.
To further quantify the differences in cell morphology, we computed cell shape index (CSI), a dimensionless parameter for geometric circularity measurement. CSI of 1 represents a perfect circle while any deviation from 1 indicates departure from a perfect circle. Fig. 1d shows that as the tension tolerance increases cells exhibit progressively lower CSI values indicating more extensions leading to a complex cell spreading pattern.
Molecular tension, but not stiffness, controls cell spreading
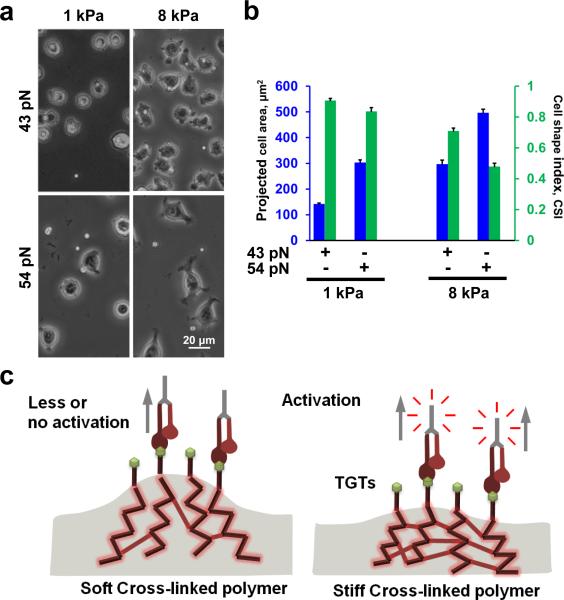
Next, we evaluated whether cell spreading is still affected by the underlying substrate stiffness when the cells are tethered through TGTs. If Ttol is solely responsible for the regulation of cell spreading we expect to see similar cell spreading response regardless of the substrate stiffness. When we plated cells on polyacrylamide gel substrates, through TGTs, we observed that as the substrate stiffness increased from 1 kPa to 8 kPa, cell spreading was also increased (Fig. 2a). In addition, for the same substrate stiffness, as the molecular tension increases from 43 pN to 54 pN, cell spreading was also increased. The Ttol dependent cell spreading was more pronounced on 8 kPa gel substrates (Fig. 2a).
Figure 2.
Increasing molecular tension increases cell spreading on same substrate stiffness. (a) Cells were plated on two different substrate stiffness, namely 1 and 8 kPa. For same substrate stiffness, increasing molecular tension tolerance increased degree of cell spreading. (b) Cell spreading significantly increased from 43 pN TGT to 54 pN TGT engineered 1 kPa substrates (n= 35 and 46 for 43 pN and 54 pN respectively; p<9.85×10−20). CSI values also showed significant changes (p<0.01). Similarly significant difference in cell spreading is also observed on 43 pN TGT and 54 pN TGT engineered 8 kPa substrates (n= 41 and 40 for 43 pN and 54 pN respectively; p<2.41×10−14). Pronounced difference in CSI values is observed between 43 pN and 54 pN engineered 8 kPa substrate compared to 1 kPa substrate (p<6.66×10−12). (c) Confounding effects of cross-linked substrates. On softer substrates, the degree of cross-linking is much lower allowing larger substrate deformation compared to rigid substrates where higher degree of cross-linking yields smaller deformation for the same applied forces. Data represent mean± s.e.m.
We next quantified projected cell area and CSI for 1 and 8 kPa gels functionalized with 43 pN or 54 pN TGTs (Fig. 2b). For same gel stiffness, cell spreading was significantly increased for 54 pN TGTs compared to 43 pN TGTs with concomitant decrease in CSI values (Fig. 2b). We also observed a significant increase in cell spreading for 8 kPa gel substrate compared to 1 kPa gel substrate with concomitant drop in CSI values for either 43 pN or 54 pN TGTs (Fig. 2b).
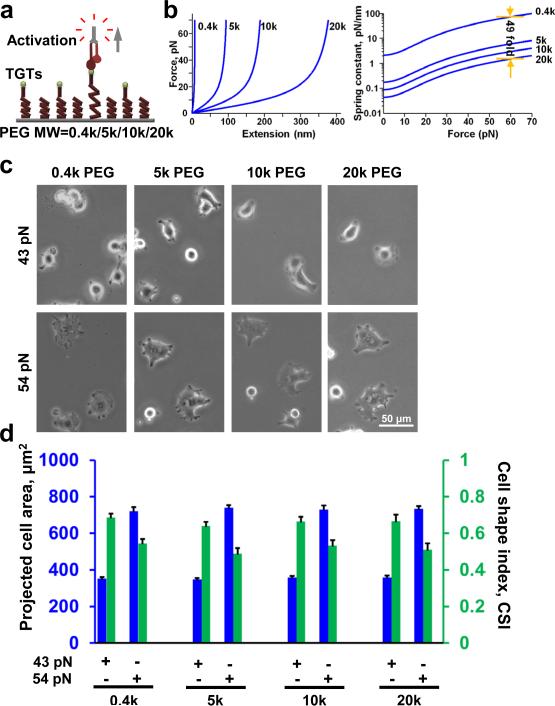
Because we observed that cell spreading is dependent on both the substrate stiffness and Ttol of TGTs we wondered whether substrate stiffness dependent cell spreading is due to the difference in cross-linking density of the gel substrates, where softer gel promotes a larger deformation when force is applied by the cells via integrins (Fig. 2c) which may lead to rapid dissociation of integrins from TGTs. Others have suggested similar mechanism of dissociation of integrins from ECM proteins on soft substrates24. In order to rule out the confounding effects of gel crosslinking, we tested cell spreading on a platform where we can immobilize TGTs through independent (noncross-linked) tethers of different molecular stiffness. We linked TGTs to a surface via linear polyethylene glycol (PEG) tethers (non-crosslinked) of four different molecular weights 0.4kD, 5kD, 10kD, and 20kD (Fig. 3a). A single non-crosslinked PEG molecule when stretched end-to-end exhibits a non-linear elasticity as shown from single molecule force spectroscopy25. We approximated the low force (< 100 pN) response of PEG using the extended freely-jointed chain model (Fig. 3b, left). Taking the derivative of force with respect to extension, which gives us the molecular stiffness at a given force (Fig. 3b, left), we can estimate a 49 fold difference in molecular stiffness between the lowest (0.4 kD) and highest (20 kD) molecular weight PEG at the same tension (Fig. 3b, right). If molecular stiffness has any influence on cell spreading, one may expect to see an increase in cell spreading with an increase in molecular stiffness. Instead, we observed similar cell spreading across all molecular stiffness surfaces for both 43 and 54 pN TGTs (Fig.3c, d). Comparing cell spreading between 43 and 54 pN, we observed increase in cell spreading with increase in Ttol across all molecular stiffness surfaces (Fig.3c, d). These results suggest that cell spreading is mainly controlled by molecular tension, and not by molecular stiffness.
Figure 3.
Molecular stiffness does not affect cell spreading. (a) Highly passivated mPEG surfaces with different molecular weight biotinylated PEGs were prepared to immobilize TGTs with different tension tolerances. PEGs with higher molecular weight are more elastic with lower spring constants. (b) Extended freely-jointed chain model is used to predict force deformation characteristics of PEGs with different molecular weights. Computing the slope at different points provides molecular stiffness information for a given force range in different molecular weight PEGs. (c) Cells were plated on different molecular weight PEGs with two different TGTs. For same tension tolerance, cell spreading is similar in different molecular weight PEG suggesting that molecular stiffness do not control cell spreading. (d) No significant changes in cell spreading was observed on different molecular weight surfaces engineered with 43 pN TGTs (n=32, 27, 29, 23 for 0.4k, 5k, 10k, 20k PEG; one way ANOVA, p>0.82) and 54 pN TGTs (n=28, 24, 28, 22 for 0.4k, 5k, 10k, 20k PEG; one way ANOVA, p>0.92). Similarly, no significant changes in CSI values were found (one way ANOVA, p>0.64 for 43 pN and p>0.53 for 54 pN). Data represent mean± s.e.m.
At present, the underlying molecular mechanism of stiffness sensing, and resulting changes in cell spreading and gene expression remains elusive. Previous studies suggest that cell spreading and gene regulation are dictated by the bulk stiffness of the underlying substrates1, 6-8. However, the molecular mechanism of sensing bulk material stiffness at the cell level is not clear. Many studies have been devoted to understand this sensing mechanism. One study proposes that the difference in collagen tethering density, based on the gel porosity, between soft vs. stiff substrates determines cellular morphology and gene expression change during stem cell differentiation16 but this was questioned by a recent report suggesting that such tethering is not necessary for differentiation17. Another study suggests that enhanced internalization of integrins on soft substrates results in diminished cell spreading and gene expression24. Most recently, it was proposed that the kinetics of integrin-ECM ligand bond is involved in rigidity sensing through the differential rate of loading of integrin bonds for different substrate stiffness26. Many other efforts have been made to study and limit average molecular forces that alter cellular behavior in a force dependent manner27, 28. But the TGT technology with its autonomous gauge can precisely cap the molecular forces that can act on single integrins. According to our data, it is ultimately the tension tolerance of single bonds that dictates such differential spreading response. We showed for the same stiff glass substrates and the same ligand density, the amount of cell spreading is profoundly affected by the single molecular forces that hold the ligand to the substrate. Moreover, we showed by changing the molecular stiffness of the individual tethers that it is the molecular tension, not molecular stiffness that controls cell spreading. Collectively, we demonstrate that the stiffness sensing and resultant changes in cell spreading can occur by sensing molecular forces across ligand-receptor bonds.
Recent development of single molecule tools including our own TGT platform is offering us an exceptional opportunity to interrogate many critical cellular processes at the molecular level. One of the earlier studies reported a Fluorescence Resonance Energy Transfer (FRET) based DNA force sensor with a goal to measure cellular adhesion forces29. Although there is a significant challenge in measuring FRET signals due to signal quenching by the cells, several other groups later demonstrated feasibility of such measurements30-33. Another approach was successfully implemented using genetically encoded tension probes to investigate the mechanobiology of cell surface receptors including vinculin34 and E-cadherin35, 36. Analysis of such data sets requires considerable care and sophisticated data analysis algorithm to remove background noise especially in the low signal areas. Moreover, the major limitation of these genetically encoded force sensors are detectable force not exceeding 5-6 pN. Our TGT platform provides an alternative approach to FRET based signal where we utilize loading configuration of a double-stranded DNA. These rupturable tethers are weak in an unzipping mode but offer more resistance in shear mode Fig. 1. Nevertheless, the reported force values for many cellular functions from any of these single molecule measurement techniques heavily rely on cellular loading rate. TGTs with different Ttol were immobilized on the surface using a widely used strategy thorough neutravidin-biotin association. A recent study reported that streptavidin-biotin bonds rupture due to cell-generated forces possibly because of low loading rates37. Therefore, even for the large force supporting tether reported in this manuscript (>100 pN, immobilizing biotinylated cyclic-RGDfK peptide directly onto the surface) can possibly rupture at a lower force level depending on the loading rate. The big challenge now is to determine in precise manner how forces are applied to single ligand receptor bonds in living cells, including the effective loading rates and maximum forces applied. In the future, a robust covalently linked scheme will be adopted to immobilize TGTs.
We engineered our TGT constructs to offer ligands (cyclic-RGDfK peptide) specific to αvβ3 integrins19-21 to measure molecular forces exerted during cell adhesion and capture their cell spreading response. Because the ligand density can affect cell spreading, the cyclic-RGDfK ligand density (~ 600 ligands/μm2) on the surface was kept constant for all TGT constructs so that that there is at least one ligand every 40 nm. As a reference, an earlier study showed that cell spreading and FA formations are not supported when spacing between integrin binding sites was more than 58 nm38. In the future, experiments can be designed to explore differences in cell adhesion and spreading through non-RGD integrins (e.g., LVD-binding integrins, αA domain integrins, and non- αA domain integrins)39 in 3D.
MATERIALS AND METHODS
Cell culture
Cells were routinely maintained in rigid culture dishes with high-glucose Dulbecco's Modified Eagle's Medium (DMEM) (Invitrogen) cell culture medium containing 10% fetal bovine serum (Hyclone, Thermo Fisher Scientific) at 37 °C with 5% CO2. The growth medium was supplemented with 2 mM L-glutamine, 1 mM sodium pyruvate, and 50 μg/ml penicillin-streptomycin. All experiments were carried out with low serum (1%) culture medium.
Surface modification and functionalization
Glass bottom dishes were passivated and functionalized with an array of tension gauge tethers described elsewhere18. In short, the glass surface was incubated with biotin labeled BSA (Sigma) for 20 min in room temperature. Following the incubation process, the glass surface was washed with PBS and further incubated with neutravidin (Thermo Fisher Scientific) for 10 min at room temperature. It was washed again with PBS and incubated with either TGT constructs or biotin labeled RGDfK (Peptides International)
Cell area measurements
ImageJ (NIH) was used to measure projected cell spreading area and perimeter. Cell shape index (CSI), a measure of circularity, is a non-dimensional parameter calculated based on the projected cell area and perimeter using the relation, . CSI values ranges from 0 to 1. For a perfect circle the value of CSI is equal to 1. Any value less than 1 indicates cell protrusions which deviate from a perfect circle.
Statistical testing
All statistical analysis was carried out using a two-tailed Student's t-test unless mentioned otherwise.
CONCLUSIONS
In summary, our findings provide a fundamental understanding of cell spreading mechanism by establishing a link between molecular forces across integrins and cell spreading behavior. We successfully decoupled molecular stiffness and molecular tension to conclusively provide a novel insight that it is the molecular tension rather than molecular stiffness that controls cell spreading.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Arash Tajik and Youhua Tan for technical help and discussion, and Qian Xu for carefully reading the manuscript. This work was partly supported by NIH GM072744 (N.W.). F.C. acknowledges IGB Fellowship from Carl R. Woese Institute for Genomic Biology, University of Illinois at Urbana-Champaign. T.H. is an investigator with the Howard Hughes Medical Institute.
Footnotes
Author contributions
T.H., F.C., and N.W. conceived the project and designed the experiments. F.C., I.L., B.L., S.D., R.S., S.L., X.W., S.P., J.S. performed experiments and/ or analyzed data. T.H., F.C., and N.W. wrote the manuscript with feedback from all.
Supplemental Information
Supplemental information includes supplementary experimental procedures and two supplementary figures.
REFERENCES
- 1.Engler AJ, Sen S, Sweeney HL, Discher DE. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 2.Chowdhury F, Li Y, Poh YC, Yokohama-Tamaki T, Wang N, Tanaka TS. PLoS ONE. 2010;5:e15655. doi: 10.1371/journal.pone.0015655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Winer JP, Janmey PA, McCormick ME, Funaki M. Tissue Eng Part A. 2009;15:147–154. doi: 10.1089/ten.tea.2007.0388. [DOI] [PubMed] [Google Scholar]
- 4.Cosgrove BD, Gilbert PM, Porpiglia E, Mourkioti F, Lee SP, Corbel SY, Llewellyn ME, Delp SL, Blau HM. Nat Med. 2014;20:255–264. doi: 10.1038/nm.3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paszek MJ, Zahir N, Johnson KR, Lakins JN, Rozenberg GI, Gefen A, Reinhart-King CA, Margulies SS, Dembo M, Boettiger D, Hammer DA, Weaver VM. Cancer Cell. 2005;8:241–254. doi: 10.1016/j.ccr.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 6.Discher DE, Janmey P, Wang YL. Science. 2005;310:1139–1143. doi: 10.1126/science.1116995. [DOI] [PubMed] [Google Scholar]
- 7.Vogel V, Sheetz M. Nat Rev Mol Cell Biol. 2006;7:265–275. doi: 10.1038/nrm1890. [DOI] [PubMed] [Google Scholar]
- 8.Solon J, Levental I, Sengupta K, Georges PC, Janmey PA. Biophys J. 2007;93:4453–4461. doi: 10.1529/biophysj.106.101386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chowdhury F, Na S, Li D, Poh YC, Tanaka TS, Wang F, Wang N. Nat Mater. 2010;9:82–88. doi: 10.1038/nmat2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun Y, Yong KM, Villa-Diaz LG, Zhang X, Chen W, Philson R, Weng S, Xu H, Krebsbach PH, Fu J. Nat Mater. 2014 doi: 10.1038/nmat3945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ghassemi S, Meacci G, Liu S, Gondarenko AA, Mathur A, Roca-Cusachs P, Sheetz MP, Hone J. Proc Natl Acad Sci U S A. 2012;109:5328–5333. doi: 10.1073/pnas.1119886109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Plotnikov SV, Pasapera AM, Sabass B, Waterman CM. Cell. 2012;151:1513–1527. doi: 10.1016/j.cell.2012.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chan CE, Odde DJ. Science. 2008;322:1687–1691. doi: 10.1126/science.1163595. [DOI] [PubMed] [Google Scholar]
- 14.McBeath R, Pirone DM, Nelson CM, Bhadriraju K, Chen CS. Dev Cell. 2004;6:483–495. doi: 10.1016/s1534-5807(04)00075-9. [DOI] [PubMed] [Google Scholar]
- 15.Schwartz MA. Cold Spring Harb Perspect Biol. 2010;2:a005066. doi: 10.1101/cshperspect.a005066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Trappmann B, Gautrot JE, Connelly JT, Strange DG, Li Y, Oyen ML, Cohen Stuart MA, Boehm H, Li B, Vogel V, Spatz JP, Watt FM, Huck WT. Nat Mater. 2012;11:642–649. doi: 10.1038/nmat3339. [DOI] [PubMed] [Google Scholar]
- 17.Wen JH, Vincent LG, Fuhrmann A, Choi YS, Hribar KC, Taylor-Weiner H, Chen S, Engler AJ. Nat Mater. 2014;13:979–987. doi: 10.1038/nmat4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang X, Ha T. Science. 2013;340:991–994. doi: 10.1126/science.1231041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aumailley M, Gurrath M, Muller G, Calvete J, Timpl R, Kessler H. FEBS Lett. 1991;291:50–54. doi: 10.1016/0014-5793(91)81101-d. [DOI] [PubMed] [Google Scholar]
- 20.Gurrath M, Muller G, Kessler H, Aumailley M, Timpl R. Eur J Biochem. 1992;210:911–921. doi: 10.1111/j.1432-1033.1992.tb17495.x. [DOI] [PubMed] [Google Scholar]
- 21.Pfaff M, Tangemann K, Muller B, Gurrath M, Muller G, Kessler H, Timpl R, Engel J. J Biol Chem. 1994;269:20233–20238. [PubMed] [Google Scholar]
- 22.Roy R, Hohng S, Ha T. Nat Methods. 2008;5:507–516. doi: 10.1038/nmeth.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moy VT, Florin EL, Gaub HE. Science. 1994;266:257–259. doi: 10.1126/science.7939660. [DOI] [PubMed] [Google Scholar]
- 24.Du J, Chen X, Liang X, Zhang G, Xu J, He L, Zhan Q, Feng XQ, Chien S, Yang C. Proc Natl Acad Sci U S A. 2011;108:9466–9471. doi: 10.1073/pnas.1106467108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oesterhelt F, Rief M, Gaub HE. New J Phys. 1999;1 [Google Scholar]
- 26.Elosegui-Artola A, Bazellieres E, Allen MD, Andreu I, Oria R, Sunyer R, Gomm JJ, Marshall JF, Jones JL, Trepat X, Roca-Cusachs P. Nat Mater. 2014;13:631–637. doi: 10.1038/nmat3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu CH, Law JB, Suryana M, Low HY, Sheetz MP. Proc Natl Acad Sci U S A. 2011;108:20585–20590. doi: 10.1073/pnas.1109485108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu CH, Rafiq NB, Krishnasamy A, Hartman KL, Jones GE, Bershadsky AD, Sheetz MP. Cell Rep. 2013;5:1456–1468. doi: 10.1016/j.celrep.2013.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tarsa PB, Brau RR, Barch M, Ferrer JM, Freyzon Y, Matsudaira P, Lang MJ. Angew Chem Int Ed Engl. 2007;46:1999–2001. doi: 10.1002/anie.200604546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blakely BL, Dumelin CE, Trappmann B, McGregor LM, Choi CK, Anthony PC, Duesterberg VK, Baker BM, Block SM, Liu DR, Chen CS. Nat Methods. 2014;11:1229–1232. doi: 10.1038/nmeth.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y, Ge C, Zhu C, Salaita K. Nat Commun. 2014;5:5167. doi: 10.1038/ncomms6167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morimatsu M, Mekhdjian AH, Adhikari AS, Dunn AR. Nano Lett. 2013;13:3985–3989. doi: 10.1021/nl4005145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morimatsu M, Mekhdjian AH, Chang AC, Tan SJ, Dunn AR. Nano Lett. 2015;15:2220–2228. doi: 10.1021/nl5047335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grashoff C, Hoffman BD, Brenner MD, Zhou R, Parsons M, Yang MT, McLean MA, Sligar SG, Chen CS, Ha T, Schwartz MA. Nature. 2010;466:263–266. doi: 10.1038/nature09198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cai D, Chen SC, Prasad M, He L, Wang X, Choesmel-Cadamuro V, Sawyer JK, Danuser G, Montell DJ. Cell. 2014;157:1146–1159. doi: 10.1016/j.cell.2014.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Borghi N, Sorokina M, Shcherbakova OG, Weis WI, Pruitt BL, Nelson WJ, Dunn AR. Proc Natl Acad Sci U S A. 2012;109:12568–12573. doi: 10.1073/pnas.1204390109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jurchenko C, Chang Y, Narui Y, Zhang Y, Salaita KS. Biophys J. 2014;106:1436–1446. doi: 10.1016/j.bpj.2014.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arnold M, Cavalcanti-Adam EA, Glass R, Blummel J, Eck W, Kantlehner M, Kessler H, Spatz JP. Chemphyschem. 2004;5:383–388. doi: 10.1002/cphc.200301014. [DOI] [PubMed] [Google Scholar]
- 39.Humphries JD, Byron A, Humphries MJ. J Cell Sci. 2006;119:3901–3903. doi: 10.1242/jcs.03098. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.