Abstract
Objective
The baboon provides a natural model of genetic generalized epilepsy. This study compares the intrinsic connectivity networks of epileptic and healthy control baboons using resting-state fMRI and data-driven functional connectivity mapping.
Methods
Twenty baboons, matched for gender, age, and weight were classified into two groups (10 epileptic, 10 control) on the basis of scalp EEG findings. Each animal underwent one MRI session which acquired one 5-minute resting state fMRI scan and one anatomical MRI scan—used for registration and spatial normalization. Using independent component analysis, we identified 14 unique components/networks, which were then used to characterize each group’s functional connectivity maps of each brain network.
Results
The epileptic group demonstrated network-specific differences in functional connectivity when compared to the control animals. The sensitivity and specificity of the two groups’ functional connectivity maps were significantly different in the visual, motor, amygdala, insular, and default mode networks. Significant increases were found in the occipital gyri of the epileptic group’s functional connectivity map for the default mode, cingulate, intraparietal, motor, visual, amygdala, and thalamic regions.
Significance
This is the first study using resting-state fMRI to demonstrate intrinsic functional connectivity differences between epileptic and control non-human primates. These results are consistent with seed-based genetic generalized epilepsy studies in humans; however, our use of a data-driven approach expands the scope of functional connectivity mapping to include brain regions/networks comprising the whole brain.
Keywords: resting-state fMRI, functional connectivity, genetic generalized epilepsy, non-human primates
INTRODUCTION
Although almost 40% of individuals with recurrent unprovoked seizures have genetic generalized epilepsy (GGE),1 little is known about the underlying mechanisms. GGE is electroclinically characterized by generalized seizures, associated with synchronization of the ictal discharge over both hemispheres on scalp electroencephalography (EEG), and by interictal epileptic discharges, which are predominantly generalized. Routine neuroimaging is typically unremarkable. Focal epilepsies, on the other hand, tend to be symptomatic of an underlying developmental abnormality or injury of the brain, and there is a better understanding of mechanisms due to routine investigations using intracranial electrodes and access to pathology, providing an important correlation with neuroimaging. Voxel-based morphometry and functional neuroimaging have been adapted to study possible structural or functional abnormalities underlying GGE,2–4 and EEG-fMRI has been useful to map blood-oxygen-level dependent (BOLD) signal during epileptic discharges.5, 6 While many of the morphometric studies demonstrate subtle structural alterations in the frontoparietal cortices and thalami,2, 3 resting-state fMRI (rs-fMRI) studies often require hypothesis-driven analyses to uncover differences in functional connectivity.4, 7, 8 So far, EEG-fMRI has been the most informative resource, demonstrating that regional BOLD increases occur several seconds before ictal or interictal generalized spike-and-wave discharges, and are decreased for several seconds after the discharges.5–7 The BOLD increases during the ictal discharge are brief, and often limited to the medial frontal lobe and anteromedial thalamic nuclei, but little is known about the cause for the more substantial peri-ictal BOLD changes. Unfortunately, these BOLD changes cannot be correlated with intracranial recordings in humans, and therefore animal models of GGE are needed. As a first step, however, it is important to demonstrate the translatability of the fMRI changes from the animal models to humans with GGE.
The epileptic baboon represents a natural animal model of photosensitive, generalized epilepsy. Epileptic baboons exhibit spontaneous myoclonic, absence and generalized tonic-clonic seizures mainly in the morning hours with a peak incidence in adolescence.9 Scalp EEG demonstrates a generalized ictal onset, as well as (predominantly) 4–6 Hz generalized interictal epileptic discharges.10 The electroclinical features of the epileptic baboon are similar to juvenile myoclonic epilepsy—the most common form of GGE in humans. In order to better understand the epileptic networks in the epileptic baboon, our group evaluated cerebral blood flow changes in response to intermittent light stimulation in photosensitive baboons using H215O positron emission tomography (PET),11–13 and recorded ictal and interictal discharges with continuous intracranial EEG.9 Both modalities demonstrated regional activations and ictal and interictal epileptic discharges in the parietal and premotor, mainly orbitofrontal, cortices. However, there were no differences in cerebral blood flow of the resting PET scans between the epileptic and control animals.14
In the present study, we compare the intrinsic brain networks of epileptic, photosensitive and control baboons using rs-fMRI. In contrast to human studies—which usually employ a hypothesis-based, region seeded approach—we utilized a data-driven approach to evaluate the functional connectivity of all of the baboon’s (data-derived) brain networks so that our results would not be limited by seed selection. The goal of this research was to demonstrate the translatability of the epileptic baboon’s intrinsic brain networks to human GGE.
METHODS
Animal subjects
Twenty, adult baboons (Papio hamadryas anubis; 10 controls (CTL)/10 epileptic (EPI)) with a mean age of 11.52 ± 4.50 yrs (mean ± standard deviation) and mean body weight of 17.34 ± 4.95 kg were obtained from the Texas Biomedical Research Institute (TBRI; San Antonio, Texas). The demographics of each animal group are shown in Supplementary Table 1. Before imaging, each animal was screened for neurological disorders (i.e. epilepsy) using scalp electroencephalographic (EEG) techniques described by Szabó et al..10 All ten baboons in the EPI group had generalized interictal epileptic discharges, 5 of whom were also photosensitive. Six of the baboons had witnessed seizures; a seventh baboon had myoclonic seizures recorded during the EEG. The anesthetized animal preparation—for optimized physiological stability and functional imaging responses—has been described in previous studies.15, 16 Briefly, for each rs-fMRI scan, the animal received an injection of intramuscular ketamine (5 mg/kg) to facilitate oral intubation and catheterization of a venous delivery line; intramuscular atropine (0.3 mg) was administered to reduce oropharyngeal secretions. During each imaging session, we maintained sedation with continuous i.v. administration of ketamine (5–6 mg/kg/hr) and vecuronium (0.25 mg/kg/hr)—a paralytic that acts at the neuromuscular junction. This dosage range provided us with a stable level of sedation throughout the MRI session in the EPI and CTL baboons. Upon conclusion of the imaging session, we administered atropine (0.6–1.2 mg, i.v.) and neostigmine (0.5–2.0 mg, i.v.) to reverse muscle paralysis. During the entire procedure, the animals’ respiration, heart rate and oxygenation were monitored. No complications were noted during or after any of the procedures. All animals were studied in accordance with the policies of the Institutional Animal Care and Use Committee of the University of Texas Health Science Center at San Antonio; this study fully complied with U.S. Public Health Service’s Guide for the Care and Use of Laboratory Animals17 and the Animal Welfare Act.18
Data acquisition
The MRI scans were performed on a 3T Siemens TIM Trio clinical MRI scanner using a body radio-frequency transmission coil with a 12-channel head RF receiver coil (Siemens, Erlangen, Germany). Blood-oxygen-level dependent (BOLD) rs-fMRI was acquired using a gradient echo-planar imaging pulse sequence. Whole brain coverage rs-fMRI was acquired parallel to the anterior commissure - posterior commissure line with the following scanning parameters: repetition time (TR) = 3,000 msec, echo time (TE) = 30 msec, acquisition matrix = 124 × 124 mm, spatial resolutions of 1.048 mm × 1.048 mm, and 27 axial slices (slice thickness = 1.9 mm) were acquired for at least five minutes. In the same session, high-resolution anatomical images were acquired using a magnetization-prepared, rapid gradient-echo (MP-RAGE) sequence (TR/TE/flip angle = 2100 ms/3.1 ms/12°), non-selective inversion pulse, TI = 1100 ms, FOV = 160 × 192 × 192 mm, 1.0 mm isotropic spatial resolution. We used the anatomical MRIs for within modality co-registration and spatial normalization.
Data preprocessing
All MRIs were processed using tools developed by the University of Oxford’s Functional Magnetic Resonance Imaging of the Brain (FMRIB) division. Anatomic MRIs were preprocessed with the Brain Extraction Tool19 in the FMRIB’s Software Library (FSL), to remove nonbrain tissues. FSL’s Fully Automated Segmentation Tool20 was used to segment the brain into white matter, gray matter and ventricular masks; a global (whole-brain) mask was created by combining each of these masks. The resulting images were used during the rs-fMRI preprocessing stages described below.
All rs-fMRIs were preprocessed using FSL’s FMRI Expert Analysis Tool (FEAT).21 The initial rs-fMRI preprocessing steps include: removal of the first 4 volumes of rs-fMRI data for T1-equilibration, motion correction (using standard and extended motion parameters), slice timing correction, and rs-fMRI brain extraction. The white matter and ventricular masks were eroded to 65% of their original volume to ensure that each mask was only sampling data from the respective brain region;22 the global, white matter, and ventricular masks were then used to determine each subject’s nuisance signals. Initially, each animal’s preprocessed dataset underwent nuisance regression using only the white matter and ventricular signal regressors (i.e. 2-covariates).23 To determine whether or not to include the mean global signal in nuisance regression, we implemented the global negative index (GNI) proposed by Chen et al.23. In their article, Chen and colleagues empirically demonstrated that a numerical relationship (i.e. the GNI) exists between functional connectivity errors and global signal noise and that this value can indicate (a priori) whether or not mean global signal regression induces more error in functional connectivity estimates. Chen et al.23 determined the criterial GNI value to be 3.0%. Thus, datasets with GNI values below this criterial GNI should include the mean global signal in nuisance regression, whereas datasets with GNIs greater than or equal to 3.0% should not. We used each animal’s 2-covariate image to calculate each animal’s GNI using in-house computer programs developed in MATLAB (Natick, MA, USA). The GNI-informed version of each animal’s rs-fMRI image then underwent isotropic spatial smoothing (FWHM = 6.0 mm), co-registration to a representative baboon brain, and band-pass filtering (0.008 < f < 0.1 Hz). Co-registration consisted of multiple steps using FSL’s FLIRT and FNIRT tools: 1) linear, affine registration with 7 degrees-of-freedom (DOF) between each subject’s rs-fMRI and 3D anatomical image, 2) linear, affine registration with 12 DOF between each subject’s 3D anatomical image and a representative baboon brain, 3) non-linear registration between each subject’s 3D anatomical image and the representative baboons brain. These transformation matrices were then used to transform each animal’s rs-fMRI dataset into the representative baboon’s brain space. Henceforth, we refer to the GNI-informed version of each image as an animal’s “preprocessed rs-fMRI image”.
Intrinsic connectivity networks
We performed independent component analysis (ICA) on all animals (N = 20) using FSL’s MELODIC program.24 The ICA was performed using temporal concatenation and an automatic estimation of the number of components to determine the baboon’s major brain networks. MELODIC determines the number of independent components in the ICA using Bayesian dimensionality estimation techniques.24 Once determined, each ICA-derived brain network was thresholded (z > 4.0) to create brain masks representing the major components of each network; the maximum ICA brain regions were identified using homologous brain areas listed in Saleem & Logothetis’ rhesus brain atlas.25 The thresholded mask of each ICA-derived brain network was then used to calculate each animal’s network-specific time-series for functional connectivity analysis.
Functional connectivity analysis
Each animal’s preprocessed rs-fMRI dataset underwent voxel-wise, “First-level” analysis for each ICA-derived brain network—using FSL’s FEAT program—to determine the functional connectivity of each brain network. In this step, the time-course of each subject’s ICA-derived brain networks were correlated with all other voxels in the brain to demonstrate which brain regions were functionally connected in each brain network. Next, each subject was assigned to a group and each brain network was analyzed using FEAT’s “Higher-level” (i.e. group-wise) analysis using the FMRIB’s Local Analysis of Mixed Effects—also in FEAT; the resulting images were thresholded (|z| > 2.3; p < 0.05, family-wise error (FWE) corrected) using FSL’s “randomise” command with threshold-free cluster enhancement. These images were then overlaid onto our representative baboon brain to calculate the descriptive statistics of each group’s functional connectivity map for each brain network; voxels within 1 mm of the white matter were excluded from each animal’s functional connectivity map for this calculation. The full range of each group’s respective functional connectivity map was used to determine the maximum and minimum z-scores for each brain network. Each group’s thresholded (|z| > 2.3; p < 0.05, FWE corrected) functional connectivity map was used to determine the total number of voxels and mean z-scores of each brain network. Student’s t-tests were used to assess any group differences in the total number of significant voxels in each network’s functional connectivity map. In addition, cluster analysis was performed on the contrast images between the CTL and EPI groups’ network responses. Only significant clusters (p < 0.05, FWE corrected)—with their maximum cluster location located in the gray matter—were reported in the cluster analysis results. These brain regions were identified using homologous regions in the rhesus brain.25
RESULTS
Subjects
There were no significant (p < 0.05) differences in the gender, age, or weight between the EPI and CTL animals (demographics were reported in Supplementary Table 1). The EPI animals tended to be younger and larger in size than the CTL group, though these differences were not statistically significant. These differences were due to a selection bias inherent to our methodology—i.e. preferentially performing EEG studies in seizure animals soon after their first witnessed seizure and in older asymptomatic animals before they died or otherwise exited the colony. The greater prevalence of females in the asymptomatic group is due to colony management policies at the TBRI that maintain harem groups for the purposes of breeding. In this study, all animals had GNIs greater than the critical GNI value of 3.0% (mean = 11.23%, SD = 4.33%), therefore, excluding the global signal from nuisance regression induced less correlation error and only the white matter and ventricular nuisance signals were regressed in each of these animals’ rs-fMRI scans.
Intrinsic connectivity networks
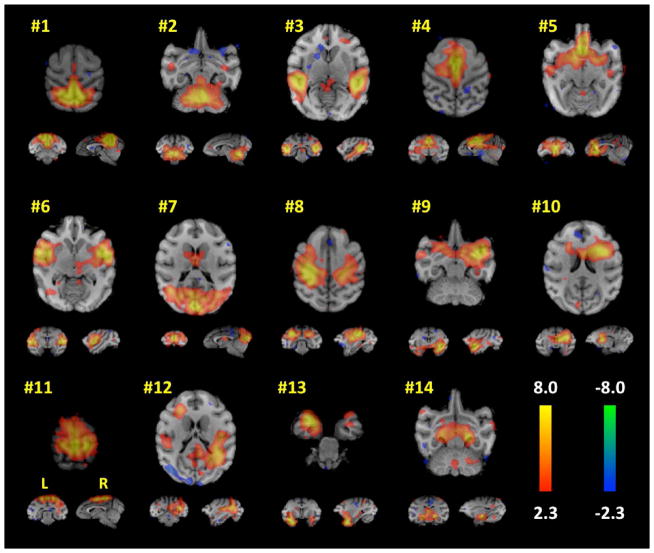
The whole-group ICA produced 14 meaningful brain components (i.e. networks), shown in Figure 1. The largest cluster of each of these brain networks was identified and labeled using homologous region in a rhesus brain atlas;25 these regions are listed in Table 1. The 14 ICA-derived brain networks spanned the whole brain and included the following brain regions: visual, default mode network (DMN), cingulate, auditory, motor, amygdala, insula, basal ganglia, lateral geniculate nucleus, and cerebellum. These regions were used to assess group-wise differences between the EPI and CTL animals. Interestingly, the thalamus did not have a unique ICA component. This result may be due to the fact that the thalamus is intricately connected to many brain regions, thus certain brain networks may have merged specific thalamic nuclei into their ICA components. The parcellation of the thalamus into several distinct ICA components has been shown by Kim et al.26
Figure 1. Concordant resting-state network maps determined by the spatial ICA of all subjects.
Each network number corresponds to the Intrinsic Connectivity Networks listed in Table 1. Orthogonal slices of the most representative points in space are shown. Each ICA map was thresholded at |z| > 2.3.
Table 1.
Intrinsic connectivity networks derived using an independent component analysis of all subjects.
| IC Seed Region (#) | Intrinsic Connectivity Network | Maximum Network Location | Primate Brain Label† |
|---|---|---|---|
| 1 | Default Mode Network | Medial Parietal Cortex of Superior Parietal Lobule | Area 7m |
| 2 | Cerebellum | Cerebellum | cb |
| 3 | Auditory | R. Fundus of the Superior Temporal Sulcus | Area PGa |
| 4 | Cingulate | Midcingulate Cortex | Area 24b |
| 5 | Anterior Cingulate | Subcallosal Gyrus/Anterior Cingulate Cortex | Area 25 |
| 6 | Insula | R. Dysgranular Insular Cortex | Id |
| 7 | Visual | Occipital Gyrus | V1 |
| 8 | Anterior Intraparietal | L. Inferior Parietal Lobule | AIP |
| 9 | Parainsula | R. Insular Proisocortex | Pi |
| 10 | Basal Ganglia | R. Caudate | cd |
| 11 | Motor | Dorsal Precentral Gyrus | F1 |
| 12 | Visual Association | R. Fundus of the Superior Temporal Sulcus | Area FST |
| 13 | Amygdala | L. Amygdala | amy |
| 14 | Thalamus | R. Lateral Geniculate Nucleus | LGN |
Labels correspond to the homologous areas listed in the rhesus25
R = Right; L = Left
Descriptive statistics
Cluster analysis of the functional connectivity of each ICA-derived brain network was used to determine the descriptive statistics for each animal group (Figure 2). The maximum strength (i.e. sensitivity) of each network’s functional connectivity map was stronger in the EPI group than in the CTL group when assessing the following brain networks: DMN (#1), cerebellum (#2), insula (#6), visual (#7), intraparietal (#8), motor (#11), visual association (#12), and the thalamus (#14). However, there were no significant (p < 0.05) differences in the mean values between the CTL and EPI groups (Figure 2A). The size (i.e. specificity) of each network’s functional connectivity map was shown in Figure 2B. The EPI group had significantly (p < 0.05) more voxels above threshold than the CTL group in all ICA-derived brain regions except the anterior cingulate (#5) and parainsular (#9) cortices. These data indicate that the intrinsic brain networks of the EPI group were consistently more widespread than the CTL group even though their mean z-scores were similar. Therefore, we are confident in stating that the EPI brain networks were functionally connected to a larger extent of the same and/or more brain regions than the CTL group in each brain network.
Figure 2. Descriptive statistics of the functional connectivity of the Epileptic (EPI) and Control (CTL) group’s network responses.
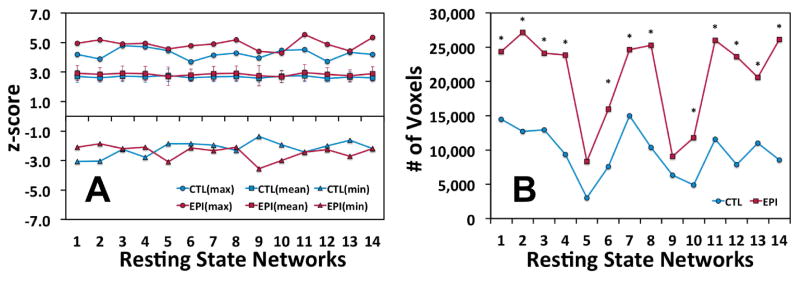
(A) The maximum and mean z-scores of voxels above threshold (|z| > 2.3; p < 0.05, FWE corrected) for each respective brain network in the EPI and CTL groups. The error bars in the mean z-score graph represent the standard deviation of the mean. (B) The total number of voxels above threshold for each group’s functional connectivity map. Significant (p < 0.05) differences between the groups were identified with an asterisk. The “Resting State Network” numbers in this figure refer to the list of networks shown in Table 1.
Functional connectivity
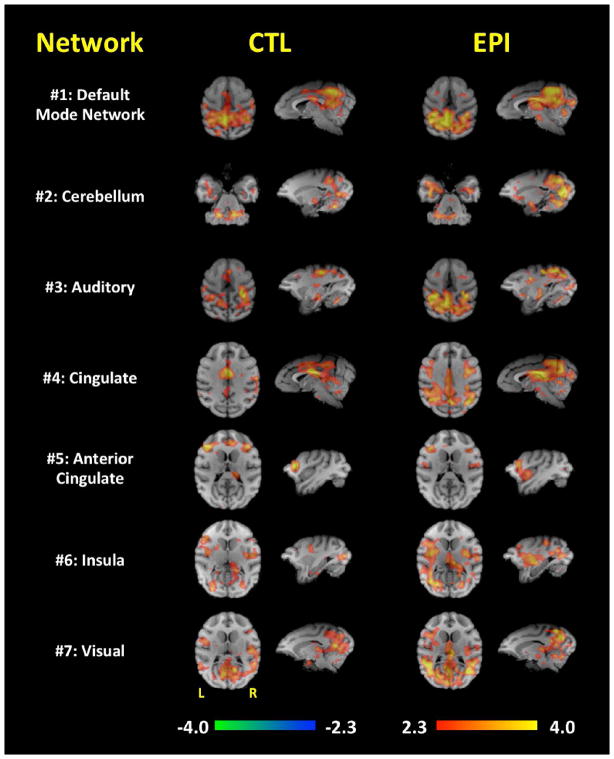
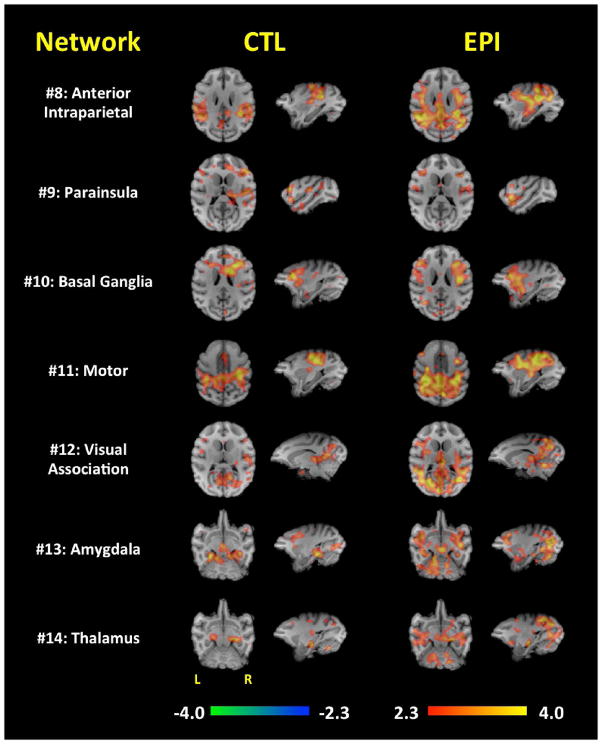
Figures 3 and 4 represent each network’s major brain activations produced when assessing each ICA-derived brain network. In general, the EPI group had larger (i.e. widespread) functional connectivity maps compared to the CTL group. Because of this, there were several regions in which the EPI group demonstrated functional connections, whereas the CTL group did not. For example, the EPI group’s functional connectivity was more widespread in the DMN (#1), cerebellum (#2), cingulate (#4), insula (#6), visual (#7), anterior intraparietal (#8), basal ganglia (#10), motor (#11), visual association (#12), amygdala (#13), and thalamic (#14) regions. The vast majority of the network-specific, group-wise functional connectivity differences were in the occipital gyrus; The extent of these network-specific differences were distributed throughout the primary and secondary visual cortices. Additionally, the parietal operculum appears to be more functionally connected to the motor, inferior parietal lobule, and visual networks in the EPI group. The DMN (#1), cingulate (#4), and basal ganglia (#10) brain regions also demonstrated increased functional connectivity to the precentral gyrus in the EPI group. Interestingly, the EPI group’s functional connectivity of the cingulate, anterior intraparietal, visual, and visual association brain regions very much resemble the functional connectivity map of the DMN suggesting a possible linking of these brain regions in the EPI group. These functional connectivity differences are summarized in Table 2.
Figure 3. Functional connectivity of networks #1–7.
Each row demonstrates the functional connectivity of the EPI and CTL groups for the DMN (#1), cerebellum (#2), auditory (#3), cingulate (#4), anterior cingulate (#5), insula (#6), and visual (#7) brain regions. Orthogonal slices of the most representative points in space are shown.
Figure 4. Functional connectivity of networks #8–14.
Each row demonstrates the functional connectivity of the EPI and CTL groups for the anterior intraparietal (#8), parainsular (#9), basal ganglia (#10), motor (#11), visual association (#12), amygdala (#13), and thalamic (#14) brain regions. Orthogonal slices of the most representative points in space are shown.
Table 2.
Maximum cluster locations of the functional connectivity differences between the EPI and CTL groups.
| IC # | Intrinsic Connectivity Network | Maximum Cluster Location | Label† | z-score | Size (voxels) | Location (mm)
|
|||
|---|---|---|---|---|---|---|---|---|---|
| X | Y | Z | |||||||
| EPI > CTL | 1 | Default Mode Network | Dorsal precentral gyrus | F1 | 3.18 | 57 | −22 | 5 | 20 |
| Occipital gyrus | V1 | 2.96 | 134 | 13 | −48 | 16 | |||
|
| |||||||||
| 2 | Cerebellum | Occipital gyrus | V1 | 4.09 | 121 | 14 | −48 | 16 | |
| Occipital gyrus | V2 | 2.87 | 159 | −8 | −43 | 8 | |||
| Agranular insular cortex | Ial | 2.80 | 97 | 18 | 6 | 0 | |||
| Occipital gyrus | V2 | 2.78 | 51 | −24 | −42 | 6 | |||
|
| |||||||||
| 3 | Auditory | Occipital gyrus | V2 | 2.99 | 58 | 3 | −39 | 15 | |
|
| |||||||||
| 4 | Cingulate | Occipital gyrus | V1 | 3.68 | 255 | −3 | −47 | 9 | |
| Occipital gyrus | V3d | 3.61 | 272 | 18 | −39 | 18 | |||
| Dorsal precentral gyrus | F1 | 3.52 | 105 | −22 | 5 | 20 | |||
| Occipital gyrus | V2 | 3.39 | 482 | −14 | −39 | 8 | |||
| Occipital gyrus | V2 | 3.12 | 110 | 3 | −39 | 17 | |||
| Occipital gyrus | V2 | 2.93 | 117 | −25 | −42 | 6 | |||
|
| |||||||||
| 5 | Anterior Cingulate | Globus pallidus | GPe | 3.13 | 104 | 13 | −6 | 3 | |
|
| |||||||||
| 6 | Insula | Occipital gyrus | V3v | 3.14 | 335 | −25 | −29 | 2 | |
|
| |||||||||
| 7 | Visual | Parietal operculum | 7op | 3.06 | 80 | 18 | −7 | 12 | |
|
| |||||||||
| 8 | Anterior Intraparietal | Occipital gyrus | V2 | 3.24 | 75 | −3 | −46 | 9 | |
| Occipital gyrus | V2 | 3.21 | 293 | −9 | −37 | 6 | |||
| Parietal operculum | 7op | 2.97 | 85 | 18 | −8 | 12 | |||
|
| |||||||||
| 10 | Basal Ganglia | Dorsal precentral gyrus | F1 | 2.83 | 77 | −22 | 6 | 20 | |
| Occipital gyrus | V2 | 2.62 | 54 | −13 | −37 | 9 | |||
|
| |||||||||
| 11 | Motor | Occipital gyrus | V2 | 3.53 | 140 | 4 | −39 | 19 | |
| Dorsal precentral gyrus | F1 | 3.47 | 61 | −22 | 5 | 20 | |||
| Occipital gyrus | V2 | 3.45 | 128 | −9 | −34 | 4 | |||
| Occipital gyrus | V1 | 3.33 | 74 | 11 | −50 | 9 | |||
| Dorsal bank of the superior temporal sulcus | MST | 3.29 | 121 | −16 | −23 | 19 | |||
| Occipital gyrus | V1 | 3.23 | 127 | −3 | −47 | 8 | |||
| Occipital gyrus | V2 | 3.18 | 89 | −23 | −44 | 7 | |||
| Dorsal bank of the superior temporal sulcus | MST | 3.05 | 64 | 14 | −25 | 23 | |||
| Parietal operculum | 7op | 2.84 | 90 | 18 | −9 | 12 | |||
|
| |||||||||
| 12 | Visual Association | Superior parietal lobule | Area 5 | 3.14 | 92 | −13 | −20 | 23 | |
| Occipital gyrus | V1 | 3.08 | 115 | −13 | −37 | 8 | |||
|
| |||||||||
| 13 | Amygdala | Occipital gyrus | V1 | 3.04 | 100 | −14 | −34 | 11 | |
| Occipital gyrus | V1 | 2.90 | 68 | 15 | −47 | 15 | |||
|
| |||||||||
| 14 | Thalamus | Occipital gyrus | V3A | 3.96 | 252 | 18 | −38 | 19 | |
| Occipital gyrus | V3d | 3.36 | 393 | −20 | −35 | 15 | |||
| Occipital gyrus | V1 | 3.36 | 107 | −3 | −47 | 9 | |||
|
| |||||||||
| CTL > EPI | 1 | Default Mode Network | Postcentral gyrus | Areas 3a/b | 3.00 | 57 | 27 | −13 | 19 |
|
| |||||||||
| 2 | Cerebellum | Postcentral gyrus | Areas 3a/b | 2.82 | 59 | 26 | −12 | 19 | |
|
| |||||||||
| 5 | Anterior Cingulate | Perigenual anterior cingulate cortex | Area 24c | 3.11 | 73 | 7 | 17 | 13 | |
|
| |||||||||
| 9 | Parainsula | Thalamus | - | 3.46 | 83 | −9 | −11 | −1 | |
| Dorsal bank of the superior temporal sulcus | Area TPO | 2.51 | 62 | 26 | −25 | 13 | |||
|
| |||||||||
| 13 | Amygdala | Parietal operculum | 7op | 2.69 | 54 | 26 | −16 | 15 | |
Labels correspond to the homologous areas listed in the rhesus25
DISCUSSION
In this study we examined the rs-fMRI functional connectivity differences between EPI and CTL non-human primates in fourteen ICA-derived brain networks. Using voxel-wise analysis of each brain network’s functional connectivity, we were able to produce functional connectivity maps for each group, then assess any significant differences between their network-specific functional connectivity maps. To the best of our knowledge, this is the first study to use a data-driven neuroimaging approach (e.g. ICA) to identify multiple brain networks involved in GGE.
Functional connectivity of GGE
Several previous studies7, 8, 27–29 have utilized seed-based, voxel-wise approaches (using rs-fMRI) to investigate functional connectivity in generalized epilepsy patients. Many of these studies8, 28, 29 have focused on sub-cortical regions—such as the thalamus—to demonstrate functional connectivity differences in epileptic patients. Kim et al.,29 seeded the anteromedial thalamus and found that thalamocortical functional connectivity in the medial prefrontal and the posterior cingulate cortices—one of the main brain regions associated with the DMN—were negatively correlated with disease duration. The authors suggested that the aberrant functional connectivity in the DMN was possibly due to long-standing effects of the disease and that these effects may be used to track the severity/stage of generalized epilepsies over time.
Other studies,7, 29 specifically investigated the DMN, finding decreases in the functional connectivity between the frontal and parietal brain regions of the DMN. Luo et al.,27 reported that although the voxel-wise representation of the DMN appeared to be similar between groups, however, graph theoretical analysis revealed that the node-to-node relationships within the DMN were significantly weaker in the epileptic group; McGill et al.,7 also found decreased functional connectivity between the DMN brain regions in the epileptic group. Kay et al.30 used ICA (and confirmatory seed-based approaches) to quantify DMN connectivity in genetic generalized epilepsy (GGE) and reported no significant differences in the functional connectivity of the DMN between healthy controls and GGE patients. However, they did find significant decreases in the mean functional connectivity of the DMN in the GGE patients when correcting for treatment-resistance. In our study, we found a strong representation of the DMN in both the CTL and EPI groups, with DMN brain regions in the frontal and parietal regions of the brain—similar to the human DMN. We also did not find significant differences between the CTL and EPI groups in the mean functional connectivity of DMN. However, since our baboon model of GGE is medication-naïve, we did not have to adjust for medication effects and the lack of response to medications.
In addition, our results are also consistent with a study by Kay et al.,31 which found that the paracingulate cortex demonstrated increased functional connectivity to the anterior thalamus and the DMN in GGE patients with frequent generalized spike and wave discharges. Our intrinsic connectivity network associated with the cingulate cortex (#4 in Figure 3) demonstrated an increased functional connectivity map in the EPI group, which resembled that of the DMN (#1 in Figure 3). These similarities between brain networks suggest that these networks have been linked—possibly in the posterior cingulate cortex and/or the precuneus—in the EPI baboons. These similarities between the cingulate and DMN networks of humans with GGE and epileptic baboons support the translatability of our baboon model of GGE.
The occipital lobes represent one region with increased connectivity to all of the networks in the epileptic compared to healthy control baboons. This was contrary to expectations, as the occipital lobes were not thought to participate in generalized epileptic networks, except in the setting of photosensitivity.32, 33 Although we did have five photosensitive baboons in our epileptic group, we did not have enough subjects—photosensitive epileptic baboons vs. non-photosensitive epileptic baboons—to definitively show whether or not the occipital differences in functional connectivity were due (explicitly) to the five photosensitive subjects included in the epilepsy group. Early studies did not find the occipital lobe to be spontaneously epileptogenic, but instead merely capable of eliciting ictal and interictal discharges in the motor and premotor cortices. On the other hand, similar to the resting-state fMRI data presented in this study, PET studies correlating cerebral blood flow with the rate of interictal epileptic discharges in the baboon demonstrated activation of primary and secondary visual areas.14 More recent intracranial EEG recordings demonstrated frequent parieto-occipital interictal—and at times (even) ictal—discharges, suggesting that the occipital lobes are also part of the epileptic network in the baboon model of GGE.9
The prominence of parietal and occipital lobe networks diverge from resting-state fMRI studies in humans.7, 8, 26–28, 31 There is ample structural and functional neuroimaging data indicating increased thalamocortical connectivity within the frontal lobes, the brain regions believed to be harboring the epileptic network in GGE.4, 34, 35 However, many of the resting-state fMRI studies utilized seed-based analyses or correlated network connectivity with other clinical parameters in order to demonstrate differences between people with GGE and healthy controls.4 The data-driven approach utilized in this study is likely to yield a more extensive connectivity map by avoiding bias. Based upon voxel-based morphometry studies in humans with juvenile myoclonic epilepsy—indicating structural abnormalities in temporal, parietal and occipital lobes—we would expect to find differences in functional connectivity as well.2, 3 Intracranial EEG recordings of ictal epileptic discharges in rodent models of absence epilepsy indicate the primary epileptogenic zone to be in the primary somatosensory areas,36 and multiregionally in the epileptic baboon, including a prominent role of the parietal and occipital cortices.9 To some extent, EEG-fMRI studies in human absence epilepsies, have confirmed an important pre-ictal contribution of the parietal lobes.5, 6 Some factors favoring the frontal lobe bias in the resting-state connectivity studies in humans are their larger and more highly-developed frontal lobe networks and increased variability of cortical structures.16
We were surprised to find increases in the functional connectivity of the motor cortex in our EPI baboon group, specifically in light of a recent study by Young et al.,37 which found that epileptic baboons have a lower cell density in the motor cortex. We would have expected a decrease in the motor network’s functional connectivity commensurate with the lower number of neurons in the EPI group’s sensorimotor cortex. However, based upon the increased motor excitability demonstrated in drug-naïve JME patients,38 one possible explanation may be that those missing neurons would usually inhibit output from the sensorimotor network, allowing increased corticocortical connectivity mediated by pyramidal cell projections. This hypothesis may be tested using EEG-fMRI; however, this is beyond the scope of the current study and should be investigated in future research projects.
Limitations and future directions
One major limitation of our study is the lack of EEG monitoring during the rs-fMRI studies. EEG-fMRI would not only elucidate BOLD changes around the time of ictal or interictal epileptic discharges,5, 6 but more importantly, it could help to correlate connectivity patterns with ictal or interictal discharge rates during the resting period to evaluate their contributions to the resting-state BOLD signal.14 Furthermore, the ability to identify sequences contaminated by ictal or interictal epileptic discharges would better provide a more accurate comparison of the resting-state networks between EPI and CTL groups.39 In our previous baboon EEG studies,11–14 we have reported that the baboon model of GGE has between 0 and 5 interictal epileptic discharges per 90 second period (average = 1.33 discharges/minute). Thus, in a 5-minute rs-fMRI scan, the EPI baboons may have 6–7 interictal epileptic discharges during the scan. We have not observed ictal discharges in our prior EEG/PET studies (at rest), but without EEG-fMRI we cannot rule out that our rs-fMRI results are not influenced by either ictal or interictal epileptic discharges.
A second disadvantage of baboon neuroimaging studies is the need for anesthesia. We have investigated the effects of ketamine on regional and global cerebral blood flow measurements using PET12–15 and BOLD fMRI.40 At the (relatively) low doses used in this study, we have not found any anesthetic effects on cerebral blood flow (or the BOLD signal) either globally or locally. By maintaining the relatively low dosing of ketamine (5–6 mg/kg/hr) throughout each rs-fMRI session, we are confident that we have assuaged any possible anesthesia-induced BOLD signal effects and that the brain network differences reported here are valid and not due to anesthesia effects.
One major advantage of using a baboon model of GGE is that the baboons are medication-naïve, and therefore will demonstrate a more natural disease state. Additionally, once we have thoroughly mapped the brain networks (and differences) of the baboon model of GGE, we can then utilize these fMRI findings as potential biomarkers for epilepsy. These brain regions may then be used to evaluate whether medical therapies—such as anti-epileptic drugs or neurostimulation—can alter or “normalize” resting-state brain activity in these disease-specific brain networks. Furthermore, functional connectivity maps in the baboon model of GGE may also inform intracranial electrode placement for further electrophysiological monitoring and/or neurostimulation studies, thereby increasing the fidelity of our EEG recordings.
CONCLUSION
This is the first study using rs-fMRI to demonstrate intrinsic functional connectivity differences between EPI and CTL non-human primates. These results are consistent with seed-based GGE studies in humans in the cingulate and DMN regions, however, our use of a data-driven approach expands the scope of functional connectivity mapping to include brain regions/networks comprising the whole brain. This data-driven approach in a medication-naïve, natural animal model of GGE may enable the utilization of fMRI findings as potential biomarkers for epilepsy research and treatments.
Supplementary Material
Key Points.
This is the first study using rs-fMRI to demonstrate intrinsic functional connectivity differences between EPI and CTL non-human primates.
The baboon is a medication-naïve, natural animal model of GGE which demonstrates results consistent with seed-based studies in humans.
Our data-driven approach expands the scope of functional connectivity mapping in GGE to include brain regions throughout the brain.
Our results may enable the utilization of fMRI findings as potential biomarkers for epilepsy research and treatments.
Acknowledgments
This study was supported by National Institutes of Health (R21-NS065431; R21-NS084198) and utilized the primate resources of the Texas Biomedical Research Institute in San Antonio, Texas, which is supported by the Research Facilities Improvement Grants (C06-RR013556; C06-RR014578; C06-RR015456). We would like to thank 1) Dr. Jinqi Li from UTHSCSA’s Research Imaging Institute for MRI acquisition and 2) Katie Strychalski and Angelique Colby from UTHSCSA’s Department of Laboratory Animal Resources for animal support throughout the study.
Footnotes
DISCLOSURE
Dr. Salinas does not have any conflicts of interest to disclose. Dr. Szabó is a speaker for UCB Pharma and Lundbeck, however these affiliations did not influence his contributions to this study. We confirm that we have read the Journal’s position on issues involved in ethical publication and affirm that this report is consistent with those guidelines.
References
- 1.http://www.epilepsyfoundation.org/about/statistics.cfm.
- 2.Lin K, Jackowski AP, Carrete H, Jr, et al. Voxel-based morphometry evaluation of patients with photosensitive juvenile myoclonic epilepsy. Epilepsy Res. 2009;86:138–145. doi: 10.1016/j.eplepsyres.2009.05.016. [DOI] [PubMed] [Google Scholar]
- 3.Ronan L, Alhusaini S, Scanlon C, et al. Widespread cortical morphologic changes in juvenile myoclonic epilepsy: evidence from structural MRI. Epilepsia. 2012;53:651–658. doi: 10.1111/j.1528-1167.2012.03413.x. [DOI] [PubMed] [Google Scholar]
- 4.Vollmar C, O’Muircheartaigh J, Symms MR, et al. Altered microstructural connectivity in juvenile myoclonic epilepsy: the missing link. Neurol. 2012;78:1555–1559. doi: 10.1212/WNL.0b013e3182563b44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bai X, Vestal M, Berman R, et al. Dynamic time course of typical childhood absence seizures: EEG, behavior, and functional magnetic resonance imaging. J Neurosci. 2010;30:5884–5893. doi: 10.1523/JNEUROSCI.5101-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benuzzi F, Mirandola L, Pugnaghi M, et al. Increased cortical BOLD signal anticipates generalized spike and wave discharges in adolescents and adults with idiopathic generalized epilepsies. Epilepsia. 2012;53:622–630. doi: 10.1111/j.1528-1167.2011.03385.x. [DOI] [PubMed] [Google Scholar]
- 7.McGill ML, Devinsky O, Kelly C, et al. Default mode network abnormalities in idiopathic generalized epilepsy. Epilepsy Behav. 2012;23:353–359. doi: 10.1016/j.yebeh.2012.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Z, Zhang Z, Jiao Q, et al. Impairments of thalamic nuclei in idiopathic generalized epilepsy revealed by a study combining morphological and functional connectivity MRI. PLoS One. 2012;7:e39701. doi: 10.1371/journal.pone.0039701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Szabó CÁ, Salinas FS, Leland MM, et al. Baboon model of generalized epilepsy: Continuous intracranial video-EEG monitoring with subdural electrodes. Epilepsy Res. 2012;101(1-2):46–55. doi: 10.1016/j.eplepsyres.2012.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Szabó CA, Leland MM, Knape K, et al. Clinical and EEG phenotypes of epilepsy in the baboon (Papio hamadryas spp. ) Epilepsy Res. 2005;65:71–80. doi: 10.1016/j.eplepsyres.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 11.Szabó CÁ, Narayana S, Kochunov PV, et al. PET imaging in the photosensitive baboon: case-controlled study. Epilepsia. 2007;48:245–253. doi: 10.1111/j.1528-1167.2006.00949.x. [DOI] [PubMed] [Google Scholar]
- 12.Szabó CA, Salinas FS, Narayana S. Functional PET evaluation of the photosensitive baboon. Open Neurosci J. 2011;5:206–215. doi: 10.2174/1874440001105010206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Szabó CÁ, Salinas FS, Li K, et al. Modeling the effective connectivity of the visual network in healthy and photosensitive, epileptic baboons. Brain Struct Funct. 2015 doi: 10.1007/s00429-015-1022-y. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Szabó CA, Narayana S, Franklin C, et al. “Resting” CBF in the epileptic baboon: correlation with ketamine dose and interictal epileptic discharges. Epilepsy Res. 2008;82:57–63. doi: 10.1016/j.eplepsyres.2008.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Salinas FS, Szabó CÁ, Zhang W, et al. Functional neuroimaging of the baboon during concurrent image-guided transcranial magnetic stimulation. Neuroimage. 2011;57:1393–1401. doi: 10.1016/j.neuroimage.2011.05.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wey H-Y, Phillips KA, McKay DR, et al. Multi-region hemispheric specialization differentiates human from nonhuman primate brain function. Brain Struct Funct. 2014;219:2187–2194. doi: 10.1007/s00429-013-0620-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Book Guide for the Care and Use of Laboratory Animals. The National Academies Press; Washington, D.C: 2011. Guide for the Care and Use of Laboratory Animals. [Google Scholar]
- 18.Animal Welfare Act 7 U.S.C. 54 § 2143, 2009.
- 19.Smith SM. Fast robust automated brain extraction. Hum Brain Mapp. 2002;17:143–155. doi: 10.1002/hbm.10062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y, Brady M, Smith S. Segmentation of brain MR images through a hidden Markov random field model and the expectation-maximization algorithm. IEEE Trans Med Imaging. 2001;20:45–57. doi: 10.1109/42.906424. [DOI] [PubMed] [Google Scholar]
- 21.Woolrich MW, Jbabdi S, Patenaude B, et al. Bayesian analysis of neuroimaging data in FSL. Neuroimage. 2009;45:S173–186. doi: 10.1016/j.neuroimage.2008.10.055. [DOI] [PubMed] [Google Scholar]
- 22.Chai XJ, Castanon AN, Ongur D, et al. Anticorrelations in resting state networks without global signal regression. Neuroimage. 2012;59:1420–1428. doi: 10.1016/j.neuroimage.2011.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen G, Xie C, Ward BD, et al. A method to determine the necessity for global signal regression in resting-state fMRI studies. Magn Reson Med. 2012;68:1828–1835. doi: 10.1002/mrm.24201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beckmann CF, DeLuca M, Devlin JT, et al. Investigations into Resting-State Connectivity Using Independent Component Analysis. Philos Trans R Soc Lond B Biol Sci. 2005;360:1001–1013. doi: 10.1098/rstb.2005.1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saleem KS, Logothetis NK. A combined MRI and histology atlas of the rhesus monkey brain in stereotaxic coordinates. Academic Press; London: 2007. [Google Scholar]
- 26.Kim D-J, Park B, Park H-J. Functional connectivity-based identification of subdivisions of the basal ganglia and thalamus using multilevel independent component analysis of resting state fMRI. Hum Brain Mapp. 2013;34:1371–1385. doi: 10.1002/hbm.21517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luo C, Li Q, Lai Y, et al. Altered functional connectivity in default mode network in absence epilepsy: a resting state fMRI study. Hum Brain Mapp. 2011;32:438–449. doi: 10.1002/hbm.21034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Masterton RA, Carney PW, Jackson GD. Cortical and thalamic resting-state functional connectivity is altered in childhood absence epilepsy. Epilepsy Res. 2012;99:327–334. doi: 10.1016/j.eplepsyres.2011.12.014. [DOI] [PubMed] [Google Scholar]
- 29.Kim JB, Suh S-i, Seo W-K, et al. Altered thalamocortical functional connectivity in idiopathic generalized epilepsy. Epilepsia. 2014;55:592–600. doi: 10.1111/epi.12580. [DOI] [PubMed] [Google Scholar]
- 30.Kay BP, DiFrancesco MW, Privitera MD, et al. Reduced default mode network connectivity in treatment-resistant idiopathic generalized epilepsy. Epilepsia. 2013;54:461–470. doi: 10.1111/epi.12057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kay BP, Holland SK, Privitera MD, et al. Differences in paracingulate connectivity associated with epileptiform discharges and uncontrolled seizures in genetic generalized epilepsy. Epilepsia. 2014;55:256–263. doi: 10.1111/epi.12486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fischer-Williams M, Poncet M, Riche D, et al. Light-induced epilepsy in the baboon, Papio papio: cortical and depth recordings. Electroencephalogr Clin Neurophysiol. 1968;25:557–569. doi: 10.1016/0013-4694(68)90235-6. [DOI] [PubMed] [Google Scholar]
- 33.Naquet R, Catier J, Menini C. Neurophysiology of photically induced epilepsy in Papio papio. Adv Neurol. 1975;10:107–118. [PubMed] [Google Scholar]
- 34.Woermann FG, Free SL, Koepp MJ, et al. Voxel-by-voxel comparison of automatically segmented cerebral gray matter--A rater-independent comparison of structural MRI in patients with epilepsy. Neuroimage. 1999;10:373–384. doi: 10.1006/nimg.1999.0481. [DOI] [PubMed] [Google Scholar]
- 35.Gotman J, Grova C, Bagshaw A, et al. Generalized epileptic discharges show thalamocortical activation and suspension of the default state of the brain. Proc Natl Acad Sci U S A. 2005;102:15236–15240. doi: 10.1073/pnas.0504935102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Luijtelaar G, Sitnikova E. Global and focal aspects of absence epilepsy: the contribution of genetic models. Neurosci Biobehav Rev. 2006;30:983–1003. doi: 10.1016/j.neubiorev.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 37.Young NA, Szabó CÅ, Phelix CF, et al. Epileptic baboons have lower numbers of neurons in specific areas of cortex. Proc Natl Acad Sci U S A. 2013;110:19107–19112. doi: 10.1073/pnas.1318894110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Puri V, Sajan P, Chowdury V, et al. Cortical excitability in drug naïve juvenile myoclonic epilepsy. Seizure. 2013;22(8):662–9. doi: 10.1016/j.seizure.2013.05.001. [DOI] [PubMed] [Google Scholar]
- 39.Moeller F, Maneshi M, Pittau F, et al. Functional connectivity in patients with idiopathic generalized epilepsy. Epilepsia. 2011;52:515–522. doi: 10.1111/j.1528-1167.2010.02938.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wey H-Y, Wang DJ, Duong TQ. Baseline CBF, and BOLD, CBF, and CMRO(2) fMRI of visual and vibrotactile stimulations in baboons. J Cereb Blood Flow Metab. 2011;31:715–724. doi: 10.1038/jcbfm.2010.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.