Abstract
Atopic dermatitis (AD) is a waxing and waning illness of childhood that is likely caused by interactions between an altered skin barrier and immune dysregulation. The goal of our study was to evaluate the association of DRB1 genetic variants and the persistence of AD using whole exome sequencing and high resolution typing. DRB1 was interrogated based on previous reports that utilized high throughput techniques. We evaluated an ongoing nation-wide long-term cohort of children with AD in which patients are asked every 6 months about their medication use and their AD symptoms. In total, 87 African-American and 50 European-American children were evaluated. Genetic association analysis was performed using a software tool focusing on amino acid variable positions shared by HLA-DRB1 alleles covering the antigen presenting domain. Amino acid variations at position 9 (pocket 9), position 26, and position 78 (pocket 4) were marginally associated with the prevalence of AD. However, the odds ratio was 0.30 (0.14, 0.68; p=0.003) for residue 78, 0.27 (0.10, 0.69; p=0.006) for residue 26 and not significant for residue 9 with respect to the persistence of AD. In conclusion, amino acid variations at peptide-binding pockets of HLA-DRB1 were associated with the persistence of AD in African-American children.
1. Introduction
Atopic dermatitis (AD) is a common chronic episodic ailment manifested by itchy red patches that most frequently occur in the flexural areas of the elbows and knees.(1;2) The acute cutaneous appearance of AD can vary from red patches with fine scales and/or vesicles, to large areas of decimated, cracked, swollen, and crusted skin. The terms AD, eczema, and flexural dermatitis are often used interchangeably especially in children. AD often has its onset prior to 2 years of age. The lifetime prevalence of AD is about 5–20%.(3;4) AD has profound effects on a child’s quality of life as well as that of their caregivers.(5) With respect to disability adjusted life years, it is one of the most debilitating skin diseases.(6) In the US the yearly prevalence of AD is about 10% in both children and adults regardless of race.(3) Children with more persistent disease tend to have been diagnosed earlier, and usually have histories of asthma as well as seasonal allergies and family histories of eczema.(7) While AD was once believed to resolve by late childhood, recent studies have shown that it can be a persistent and life-long disease.(8;9)
The etiology of AD and the manifestation of its classic skin findings are often explained based on alterations in skin barrier function, immune dysregulation, and presence of environmental exposures. These factors are conceptually modeled as either “outside-in” or “inside-out”. (10) The outside-in model states that the transfer of external exposures occurs across a dysfunctional skin barrier eliciting AD symptoms due to immunologic activation. In this context, a model antigen passes through a constitutively defective skin barrier. The resulting allergic sensitization is partially manifested by high total serum IgE levels (which while often elevated in atopic disease is inconsistently associated with AD) and the presence of IgE for specific environmental allergens. In contrast, the inside-out conceptual model is based on the notion that the classic epidermal changes noted in AD result from constitutive immunologic abnormalities that in turn alter the skin barrier causing the skin findings of AD.(10;11) In this model, immunologic activation is proposed to be due to T helper type (Th2) immune-cell associated cytokine responses that directly promote the defective skin barrier thereby allowing the passage of antigen resulting in an even brisker immunologic response.(10–12)
AD is a complex disease with a strong genetic component. The most commonly reported genetic variant associated with skin barrier dysfunction is filaggrin (FLG), which codes for a skin barrier protein and has been associated with the prevalence and persistence of AD.(13;14) However, while FLG loss of function (LOF) mutations are quite common in individuals of European and Asian ancestry with AD, individuals of African ancestry with AD rarely have these mutations.(15–18) Previously, we found variants of FLG2, another skin barrier gene related to FLG, may be associated with the persistence of AD in African-Americans.(16;19) Several genes that have a role in the immune response have also been associated with AD.(20) These include thymic stromal lymphopoietin (TSLP), which is a keratinocyte-derived protein that initiates Th2 immune responses in the skin. We have recently demonstrated that TSLP variation is associated with the persistence of AD in both African-Americans and European-Americans.(21) Eosinophilic esophagitis and food allergy, which are diseases associated with AD, have recently been shown to be associated with TSLP variation.(22;23) Also recent clinical trials using anti-TSLP antibody in patients with asthma demonstrated improvements in parameters of bronchoconstriction and airway inflammation.(24) Further, we recently have shown that variation in the TSLP gene can markedly reduce the association of FLG with the persistence of AD.(21)
In order for TSLP protein to prime the immunologic response to external antigen, antigen presentation must occur. This process is guided by human leukocyte antigen (HLA) molecules, which are encoded within the major histocompatibility complex (MHC) located on chromosome 6.(25) Hundreds of genes have been identified in this region.(26) These genes affect immune system function by coding for cell-surface antigens.(25) More specifically, in the presence of TSLP, MHC II expressing antigen-presenting cells (APCs) present antigens to T helper (Th) cells thereby stimulating and perhaps eliciting the cutaneous symptoms of AD.(27;28) Six large high throughput genetic studies (e.g., genome wide association studies (GWAS)) of European and/or Asian children with AD have been published.(29–34) In one study the loci that includes HLA-DRB1 region was identified and in two of these studies, the G-protein signaling modulator 3 (GPSM3) gene in the 6p21.3 cytoband, which is the same cytoband region as the MHC loci and structurally very close to HLA-DRB1 of the MHC class II region, was identified as a potential genetic loci associated with AD.(29–32;35;36)
Our recent interest has focused on the persistence of AD, while others have focused on evaluations of factors that increase the risk of a child developing AD.(14;19;21) We base this interest on the notion because prevention efforts have not been successful (e.g., the elimination of risk factors) and since AD is a prevalent lifelong illness that waxes and wanes by evaluating persistence we hope to better understand therapeutic targets for future study. (5;37;38) The goal of our study was to specifically evaluate the HLA-DRB1 genetic variants for its association with the persistence of AD in children. We are particularly interested in African-American children who have been understudied in candidate gene studies and have not been included in any of the GWAS studies of AD.
2. Materials and Methods
2.1 Cohort
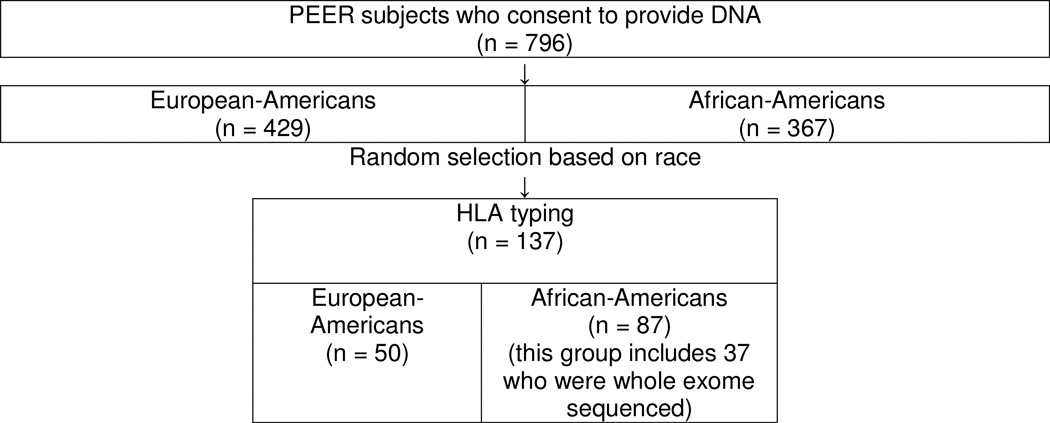
Subjects for this study were enrolled in the Pediatric Eczema Elective Registry (PEER), www.thepeerprogram.com, which is an ongoing prospective 10-year observational registry that is part of a post-marketing commitment to the Food and Drug Administration and the European Medicines Agency. The enrollment criteria and goals of the PEER study are described in detail elsewhere.(14) The diagnosis of AD for each child was made by the enrolling physicians, the majority of whom were pediatricians, allergists, and dermatologists.(14) The diagnosis was confirmed based on the UK working party criteria.(14) Children/parents in the PEER study who also enrolled in our current study completed an additional informed consent approved by the Institutional Review Board of the University of Pennsylvania. These children subsequently provided a saliva sample from which DNA was extracted. In total, 796 children (367 African Americans and 429 European-Americans) enrolled in this part of the PEER study (Figure 1). We randomly selected DNA for 137 children enrolled in the PEER study for HLA genotyping. In total, 87 African-Americans (including 37 that had whole exome sequencing) and 50 European – Americans were HLA-DRB1 genotyped. The cohort for each evaluation is described in Figure 1. A child’s race was self-described as African-American or European-American (white) and the reliability of this determination was previously genetically confirmed using ancestry informative markers.(14)
Figure 1.
Flow chart for selection of study subjects
2.2 Clinical Outcome
As in previous reports, we investigated longitudinally the self-reported outcome of whether or not a child’s skin was reported to be AD symptom-free during multiple six-month periods while not requiring the use of medication.(14;21) Information on these outcomes was collected longitudinally every six months thus allowing for repeated assessments over time thereby mimicking the natural waxing and waning nature of AD.
2.3 HLA genotyping
For this investigation, we focused only on the HLA-DRB1 locus based on the previous GWAS findings as described above.(29–32) HLA-DRB1 genotyping was performed on 87 African-American (including 37 who were also analyzed by whole exome sequencing) and 50 European-American samples by the Histocompatibility and Immunogenetics Laboratory in the Hospital of the University of Pennsylvania. Typing was conducted using the standard operating techniques of this clinical reference laboratory using sequence based HLA-DRB1 typing with SeCore® sequencing kits via fluorescent dye terminator sequencing chemistry with a Big Dye Terminator Chemistry/3500xL Genetic Analyzer. For many cases (30%) this approach did not yield unambiguous two-field typing and we defaulted to the most probable second field. This choice was supplemented and confirmed by using our exome data (described below) that provide DRB1 typing independently.
2.4 Whole exome sequencing
As part of our broader interest to identify variants associated with AD or its progression, we generated whole exome sequencing data for a subset of subjects. This data was used to supplement our HLA genotyping efforts. For this purpose the Omixon “Target” software was used. This software can analyze whole exome or whole genome sequencing data and provide HLA genotyping. Exome sequencing was performed by Ambry Genetics (Aliso Viejo, California) utilizing whole exome targeted enrichment by Agilent SureSelect technology.(16) The libraries for the whole exome study were indexed, 100 bp paired end and processed using Illumina HiSeq2000 at 100x coverage per exome.
2.5 Analysis of HLA-DRB1 amino acid variations
HLA-Amino acid sequence alignment data was obtained using HLA-DRB1 allele sequences contained in the IMGT/HLA database (Release 3.16.0, April 14, 2014). Analysis of amino acid variations was focused on the antigen presenting domain (beta 1 domain) of the HLA-DRB1 molecule. We used the SKDM software tool to analyze the association of amino acid sequence variations with disease risk as previously described. (39)
2.6 Genetic association analysis
Data were summarized by means, medians or percentages as appropriate. Initial assessments of basic demographic and health related data were based on the individual. Individuals in this study were followed longitudinally and surveyed every six months regarding the status of their AD. We assessed the statistical association between repeated measures of the binary outcome (six-month symptom free and not using topical steroids) and specific amino acid residue changes among HLA-DRB1 alleles using generalize linear latent and mixed models with a binary logistic link function and binomial family. These models include both random-effects and fixed-effects terms, allowing for subject specific estimates of the association of a risk factor with a repeated outcome in longitudinal data. In other words, this statistical technique allows for the proper estimation of the likelihood at any given 6-month evaluation that an individual has less persistent disease based on the variables in the model. We compared the prevalence of specific amino acid residues in peptide-binding pockets of HLA-DRB1 alleles in our AD cohort to a control group collected by author DSM. The DRB1 allelic frequencies of this group were similar to the NDMP control cohort (https://bioinformatics.bethematchclinical.org/WorkArea/DownloadAsset.aspx?id=6398) (Table 2).(40)
Table 2.
Two-field DRB1 allelic frequency for the study cohort, a control group, and NMDP control group.
| DRB1 | European American AD Cohort N=50 |
European American Controls N=109 |
European American Controls NMDP |
African American Cohort N=86 |
African American Controls N=59 |
African American NMDP |
|---|---|---|---|---|---|---|
| 0101 | 0.0700 | 0.0963 | 0.09149 | 0.0085 | 0.02599 | |
| 0102 | 0.0400 | 0.0183 | 0.01703 | 0.0523 | 0.0169 | 0.03992 |
| 0103 | 0.0100 | 0.0138 | 0.00889 | 0.0085 | 0.00229 | |
| 0301 | 0.0900 | 0.1376 | 0.12916 | 0.0756 | 0.0339 | 0.07069 |
| 0302 | 0.00006 | 0.0465 | 0.0678 | 0.06528 | ||
| 0305 | 0.00000 | 0.00021 | ||||
| 0306 | 0.00000 | 0.00021 | ||||
| 0401 | 0.0700 | 0.0688 | 0.09111 | 0.0291 | 0.0085 | 0.02287 |
| 0402 | 0.0275 | 0.00972 | 0.00042 | |||
| 0403 | 0.0200 | 0.0138 | 0.00572 | 0.0085 | 0.00229 | |
| 0404 | 0.0200 | 0.0413 | 0.03634 | 0.0058 | 0.00686 | |
| 0405 | 0.0046 | 0.00368 | 0.0116 | 0.0339 | 0.00956 | |
| 0406 | 0.0100 | 0.00025 | 0.00062 | |||
| 0407 | 0.0100 | 0.0138 | 0.00947 | 0.0058 | 0.0085 | 0.00395 |
| 0408 | 0.0092 | 0.00248 | 0.0085 | 0.00062 | ||
| 0409 | 0.00000 | 0.00021 | ||||
| 0410 | 0.00000 | 0.00062 | ||||
| 0411 | 0.00000 | 0.00083 | ||||
| 0414 | 0.00000 | 0.00000 | ||||
| 0417 | 0.00000 | 0.00000 | ||||
| 0418 | 0.00000 | 0.00000 | ||||
| 0701 | 0.1100 | 0.1330 | 0.13767 | 0.1512 | 0.0847 | 0.09771 |
| 0703 | 0.00000 | 0.00000 | ||||
| 0801 | 0.0200 | 0.0275 | 0.02363 | 0.0058 | 0.00457 | |
| 0802 | 0.0100 | 0.0092 | 0.00025 | 0.00104 | ||
| 0803 | 0.0100 | 0.00133 | 0.00042 | |||
| 0804 | 0.00089 | 0. 0756 | 0.0678 | 0.05052 | ||
| 0805 | 0.00000 | 0.00000 | ||||
| 0806 | 0.00006 | 0.0169 | 0.00520 | |||
| 0809 | 0.00000 | 0.00000 | ||||
| 0810 | 0.00000 | 0.00000 | ||||
| 0811 | 0.00006 | 0.00104 | ||||
| 0901 | 0.0300 | 0.0138 | 0.00820 | 0.0349 | 0.03160 | |
| 1001 | 0.0046 | 0.00826 | 0.0763 | 0.01850 | ||
| 1101 | 0.0600 | 0.0321 | 0.05654 | 0. 0814 | 0.1102 | 0.08711 |
| 1102 | 0.0100 | 0.0046 | 0.00152 | 0.0174 | 0.0593 | 0.03909 |
| 1103 | 0.0100 | 0.0046 | 0.00483 | 0.00062 | ||
| 1104 | 0.0400 | 0.0505 | 0.03189 | 0.00561 | ||
| 1106 | 0.00000 | 0.00000 | ||||
| 1108 | 0.00000 | 0.00000 | ||||
| 1109 | 0.00006 | 0.00000 | ||||
| 1110 | 0.00000 | 0.00166 | ||||
| 1111 | 0.00000 | 0.00000 | ||||
| 1115 | 0.00000 | 0.00000 | ||||
| 1117 | 0.00000 | 0.00021 | ||||
| 1139 | 0.00006 | 0.00000 | ||||
| 1201g | 0.0300 | 0.0275 | 0.01468 | 0.0465 | 0.0169 | 0.03950 |
| 1202 | 0.00000 | 0.0169 | 0.00270 | |||
| 1208 | 0.00000 | 0.00000 | ||||
| 1301 | 0.0600 | 0.0229 | 0.06283 | 0.0465 | 0.0763 | 0.05551 |
| 1302 | 0.0600 | 0.0459 | 0.04015 | 0.0640 | 0.0508 | 0.06445 |
| 1303 | 0.0200 | 0.0092 | 0.00991 | 0.0233 | 0.0085 | 0.03701 |
| 1304 | 0.00006 | 0.0174 | 0.0085 | 0.01310 | ||
| 1305 | 0.00235 | 0.00042 | ||||
| 1306 | 0.00000 | 0.00000 | ||||
| 1309 | 0.00000 | 0.00000 | ||||
| 1311 | 0.00006 | 0.00021 | ||||
| 1312 | 0.00000 | 0.00000 | ||||
| 1316 | 0.00000 | 0.00042 | ||||
| 1320 | 0.00000 | 0.00021 | ||||
| 1331 | 0.00000 | 0.00062 | ||||
| 1336 | 0.00000 | 0.00021 | ||||
| 1340 | 0.00000 | 0.00000 | ||||
| 1350 | 0.00000 | 0.00000 | ||||
| 1401g | 0.02459 | 0.0058 | 0.0085 | 0.02141 | ||
| 1402 | 0.00000 | 0.00062 | ||||
| 1403 | 0.00000 | 0.00000 | ||||
| 1404 | 0.0100 | 0.00032 | 0.00042 | |||
| 1405 | 0.00000 | 0.00000 | ||||
| 1406 | 0.00006 | 0.00000 | ||||
| 1407 | 0.00006 | 0.00000 | ||||
| 1408 | 0.00000 | 0.00000 | ||||
| 1412 | 0.00000 | 0.00000 | ||||
| 1418 | 0.00000 | 0.00000 | ||||
| 1419 | 0.00000 | 0.00000 | ||||
| 1422 | 0.00000 | 0.00000 | ||||
| 1425 | 0.00000 | 0.00000 | ||||
| 1432 | 0.0100 | |||||
| 1454 | 0.0300 | 0.0138 | 0.0174 | 0.0254 | ||
| 1501 | 0.1200 | 0.1239 | 0.14441 | 0.0233 | 0.0085 | 0.02931 |
| 1502 | 0.0092 | 0.00775 | 0.0116 | 0.00166 | ||
| 1503 | 0.0138 | 0.00019 | 0.1337 | 0.1441 | 0.11746 | |
| 1504 | 0.00000 | 0.00000 | ||||
| 1506 | 0.00000 | 0.00000 | ||||
| 1507 | 0.00000 | 0.00000 | ||||
| 1514 | 0.00000 | 0.00000 | ||||
| 1601 | 0.0200 | 0.0046 | 0.01061 | 0.0085 | 0.00104 | |
| 1602 | 0.0046 | 0.00127 | 0.0233 | 0.0085 | 0.01538 |
3. Results
3.1 Characteristics of the study subjects
The mean and median age (in years) at onset of these subjects was 2.5 years (SD 3.11) and 0.75 years (interquartile range (IQR) = 0.5, 7), respectively (Table 1). Male subjects accounted for 57.5% (n = 95) of the cohort. At enrollment, 60.0% (N=99) had a history of asthma or wheezing, 66.7% (N=110) had a history of seasonal allergies, and 28.5% (N=47) reported food allergies (N=13). These characteristics were similar to the full PEER cohort (Table 1). At the time of this evaluation, the participants had been followed for an average of 6.38 years (SD: 2.03) and with a median follow up of 7 years (interquartile range (IQR) = 5, 8). The median number of six month intervals evaluated was 8 (IQR: 4–12).
Table 1.
Characteristics of the PEER cohort that provided DNA and the cohort analyzed in this study.
| PEER Cohort (N=796) |
Current Study Cohort (N=160) |
|
|---|---|---|
| age at onset (years) | 0.75 (SD: 2.0) | 0.75 (SD 2.5) |
| gender (male) | 51.8% | 57.5% |
| uncontrolled disease at enrollment |
10.2% | 13.9% |
| asthma | 52.4% | 60.0% |
| seasonal allergies | 67.7% | 66.6% |
| food allergies | 31.6% | 28.5% |
| medication allergies | 13.6% | 13.3% |
| pet allergies | 30.7% | 24.8% |
3.2 Analysis of Exome sequencing data
Sequencing data was analyzed using the Omixon “Target” computational pipeline. We had exome sequencing data for 37 individuals (or 74 genomes) that also had high resolution HLA-DRB1 typing (two-field). 32 of the 37 (64 of 74 alleles) had concordant DRB1 allele assignments. A discordant allele assignment (for the 2 alleles) was observed in one subject; and in four subjects, exome sequencing data was not adequate for this evaluation. Overall, the concordance rate was 83.7% for all samples or 97.0% for samples of sufficient quality (i.e. poor extent of exons covered) for analysis using the Omixon software tool.(41) All (100%) of the imputed second fields based on the HLA genotyping were confirmed by the use of Omixon software.
3.3 HLA genotyping
The HLA-DRB1 genotyped allelic frequency results for 137 children are presented in Table 2. These results were compared to a control panel of children from the Children’s Hospital of Philadelphia. The SKDM pipeline was used to analyze these findings based on biologically relevant protein residue changes in the binding pockets of HLA-DRB1. This analysis revealed that amino acid residues at position 78 (DRB1*07:01 and *09:01) of pocket 4, position 26 (DRB1*03:01 and *09:01) pocket 4 and position 9 (DRB1*07:01, *01:02, *15:01, *16:02, *15:02, *15:03, *01:03; *16:01) of pocket 9 were more prevalent in the PEER cohort then the control cohort. The odds ratio (OR) of association comparing the PEER African-American children to the control group was 2.59 (p=0.02) for position 78, 3.11 (p=0.03) for position 26, and 2.27 (p=0.02) for position 9. No associations were noted for the European-American cohort.
Based on these findings we evaluated whether African-American children with these residue variations had more persistent disease (i.e., more six-month periods with active AD symptoms requiring the use of topical medication) (Table 3). More specifically children with substitution at position 78 were more likely to not (i.e. more persistent) have a six-month period of disease free skin without using medication (OR: 0.30 (0.14, 0.88; p=0.003)) when compared to children with the baseline receptor residues (DRB1-01:01). Similar findings were noted for amino acid variation at position 26 (OR: 0.27 (0.10, 0.69; p=0.006) but not for position 9 (0.66 (0.27, 1.61; p=0.36). Interestingly, the findings for residues at position 78 were not significantly affected by adjustment (i.e., not confounded) by the FLG2 gender, age of onset, and the presence of seasonal allergies, food allergies, pet allergies, and asthma but position 26 was confounded. However, after adjustment the association with residue 9 became significant (OR: 0.34 (0.18, 0.66; p=0.001)).
Table 3.
The Persistence of AD and DRB1 protein residues (Odds ratios and 95% confidence intervals). Fully adjusted, FLG2 gender, age of onset, and the presence of seasonal allergies, food allergies, pet allergies, and asthma.
| Symptom Free without need for Medication | |||
|---|---|---|---|
| Pocket/ Position |
Full cohort | African-Americans | Fully Adjust African-Americans |
| Pocket 4 78 |
0.31 (0.18,0.52) <0.0001 |
0.30 (0.14, 0.68) 0.003 |
0.22 (0.04,1.12) 0.06 |
| Pocket 4 26 |
0.32 (0.15,0.69) 0.004 |
0.27 (0.10,0.69) 0.006 |
1.98 (0.53,7.40) 0.31 |
| Pocket 9 9 |
0.46 (0.27, 0.78) 0.004 |
0.66 (0.27,1.61) 0.36 |
0.34 (0.18,0.66) 0.001 |
4. Discussion
In our set of investigations, we interrogated the DRB1 gene, part of the HLA class II region, which was identified as a region of interest for AD in previous large genetic studies.(29–32) Amino acid variable positions shared by HLA-DRB1 alleles in antigen presenting domains as identified by a software tool were found to be nominally associated with an increased prevalence of AD. The specific amino acid variants where localized to three peptide-binding pockets 4 and 9. It is important to realize that we account for residue 26 among residues associated with pocket 4. The residue 26 was not originally associated with pocket 4 residues.(42;43) This is due to the fact that only residues in the 4 Angstrom radius of the incoming side chain of the side chain of an amino acid were accounted as pocket 4 residues.(42;43) However in a subsequent study of ours we have shown that upon extending the radius to 4.5 Angstroms residue 26 is part of pocket 4.(44) In fact, residue 26 has been independently shown to influence peptide binding so we feel that residue 26 is a pocket 4 residue.(43) We then evaluated these residues with respect to a clinically relevant outcome based on the persistence of AD. We were able to further confirm that two of three of these protein residues are likely associated with more persistent AD and have effect estimates similar to those we had previously found for mutations of the skin barrier, FLG or FLG2 mutations.(14;19) We were not able to find similar abnormalities in those of European-American ancestry; however, our sample size for this group was very small.
This study is one of the first examples where GWAS data helped to guide investigators to evaluate an HLA region resulting in an association of an illness with a receptor phenotype. Even though the reported SNP association in the GWAS study was not with persistence of AD but rather with AD susceptibility, the designation of the DRB1 region as being implicated in the disease was suggestive. Interestingly, mostly GWAS SNPs findings are to alleles already known within the MHC to be disease associated such as type 1 diabetes or Multiple Sclerosis. In our study the opposite is true in that GWAS findings helped guide us to the DRB1 gene and we subsequently identified the exact elements possibly related to the persistence of AD.
In addition, we showed excellent concordance between Omixon “Target” software tool genotyping of the MHC II region using whole exome sequencing data and the HLA-DRB1 typing using sequence based typing. Our evaluation showed that when using high quality whole exome sequencing data the concordance between the methods is greater than 95%.
MHC class II molecules are transmembrane glycoproteins found primarily on the surface of specialized cells of the immune system.(25;26) These proteins are important for the presentation of antigens by APCs to CD 4+ T cells. MHC class II expression is critical for the control of normal immune response and its variation is often associated with pathologic immune responses.(25;26) In the current study we used HLA genotyping to precisely define DRB1 alleles. We then grouped the allelic variation to characterized amino acid sequence substitutions that may result in structural changes in the molecule likely affecting peptide binding and antigen presentation by this MHC II molecule thereby yielding potential targets for further exploration. Previous studies in autoimmune disease revealed that combinations of variable amino acid sites shared by several HLA alleles (shared epitopes) are better descriptors of actual causative genetic variants implicating specific amino acid residues in peptide binding pockets of HLA-DRB1 that account for major disease risk attributable to HLA-DRB1.(45)
AD is a disease that has long been thought to have an immunologic component indicating that an evaluation of the HLA-MHC region of the genome could be fruitful to study.(32) Several studies have explored the relationship between HLA genes and atopic sensitization disorders. For example, Aron et al. found a strong positive relationship between HLA-DR4 and DR7 alleles with atopy and asthma in a Finnish cohort.(46) HLA-DRB1, -DQB1 and -DPB1 genotypes were associated with peanut allergy in a British study.(47) In a small Japanese study, HLA-DRB1 and -DQB1 alleles were found to be associated with severe atopic dermatitis with high IgE levels.(48) The GPSM3 gene in the MHC region (6p21.3) and likely in linkage disequilibrium with HLA-DRB1 was implicated in two AD GWAS, one performed in a Japanese cohort and the other in a German population using the Immunochip.(29;30) Park et al. showed an association of HLA-DRB1 with AD in Korean children.(49) Madore et al. reported an association between HLA-DQB1 and peanut allergy.(50) In an AD GWAS performed on European subjects, Weidinger et al. imputed the classical HLA alleles using the SNP data and observed a protective effect of HLA-DRB1 whereas HLA-B conferred increased risk of AD.(32) We know of no previous studies that analyzed protein residue variation with respect to AD.
Our study has a few weaknesses. First, this is a small first study that may not have been sufficiently powered for all analyses. Larger studies will be needed to further replicate and validate our findings. It is likely that we did not find any associations in the European-American cohort due to its small size. Second, we did not correct for multiple testing. However, it is important to note that our hypothesis was based on previous studies and focused solely on DRB1. Our final analyses of persistence were all guided by the preliminary observations and not based on an agnostic approach that could result in multiple testing concerns. Furthermore our findings are consistent with many hypothetical models of AD that describe a process that begins with external antigen penetrating a defective skin barrier triggering APCs that in turn activate the Th2 cell-associated cytokine response. A well-known function of MHC class II molecules is the presentation of antigen by APCs to Th2 cells. In addition, given that AD is a common disease in all races and ethnicities, it might be expected that a common mechanism (e.g. MHC class II-mediated immune dysregulation) might exist. Finally, it is possible that in some of our samples the HLA-DRB1 allele assignment was not accurate since for several of our samples all allele ambiguities were not resolved. In these cases we defaulted to the most probable alleles. However, 14 of the subjects (representing 23 alleles) that were not fully resolved were also part of the group evaluated by the Omixon software and for these the Omixon approach was in 100% agreement with this “default” approach. Thus it is unlikely that this was a source of important error.
In conclusion, GWAS evaluation of the complex diversity of the MHC has yielded few potentially causal associations with illness in the MHC II region. We used previous GWAS studies to help inform our investigation. In the PEER cohort, specific residue variation of binding pockets of the HLA-DRB1 locus was marginally associated with had an increased prevalence of AD and significantly associated with the persistence of AD. This association was noted for those of African ancestry and was not influenced by skin barrier protein variation (FLG2 status). This is the first report associating MHC II variation with the persistence of AD. Future studies need to replicate and validate the findings from our study, include larger samples sizes of children from different ancestries, investigate functional aspects of DRB1 variation, as well associations of DRB1 within multi-loci MHC haplotypes with respect to AD.
Acknowledgement
Role of funding source
This study was supported by R01-AR0056755 from the National Institutes of Arthritis Musculoskeletal and Skin Disease, Arguild Foundation, and a grant from Valeant Pharmaceuticals for the Pediatric Eczema Elective Registry (PEER) study. The sponsors had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors have no financial relationships relevant to this manuscript.
Reference List
- 1.Akdis CA, Akdis M, Bieber T, Bindslev-Jensen C, Boguniewicz M, Eigenmann P, et al. Diagnosis and treatment of atopic dermatitis in children and adults: European Academy of Allergology and Clinical Immunology/American Academy of Allergy, Asthma and Immunology/PRACTALL Consensus Report. Allergy. 2006;61(8):969–987. doi: 10.1111/j.1398-9995.2006.01153.x. [DOI] [PubMed] [Google Scholar]
- 2.Charman CR, Williams HC. Epidemiology. In: Bieber T, Leung DYM, editors. Atopic Dermatitis. 1 ed. New York: Marcel Dekker, Inc; 2002. pp. 21–42. [Google Scholar]
- 3.Hanifin JM, Reed ML. the Eczema Prevalence Impact Working Group, A population-based survey of eczema prevalence in the United States. Dermatitis. 2007;82(2):82–91. doi: 10.2310/6620.2007.06034. [DOI] [PubMed] [Google Scholar]
- 4.Silverberg JI, Hanifin JM. Adult eczema prevalence and associations with asthma and other variables: A US population-based study. Journal of Allergy & Clinical Immunology. 2013;132(5):1132–1138. doi: 10.1016/j.jaci.2013.08.031. [DOI] [PubMed] [Google Scholar]
- 5.Eichenfield LF, Tom WL, Chamlin SL, Feldman SR, Hanifin JM, Simpson EL, et al. Guidelines of care for the management of atopic dermatitis: section 1. Diagnosis assessment of atopic dermatitis[Review] Journal of the American Academy of Dermatology. 2014;70(2):338–351. doi: 10.1016/j.jaad.2013.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Murray CJ, Vos T, Lozano R, Naghavi M, Flaxman AD, Michaud C, et al. Disability-adjusted life years (DALYs) for 291 diseases and injuries in 21 regions, 1990–2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380(9859):2197–2223. doi: 10.1016/S0140-6736(12)61689-4. [DOI] [PubMed] [Google Scholar]
- 7.Hoare C, Li Wan Po A, Williams H. Systemic review of treatments for atopic eczema. Health Technology Assessment. 2002;4:1–191. [PMC free article] [PubMed] [Google Scholar]
- 8.Silverberg JI, Simpson EL. Association between severe eczema in children and multiple comorbid conditions and increased healthcare utilization. Pediatric Allergy & Immunology. 2013;24(5):476–486. doi: 10.1111/pai.12095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Margolis JS, Abuabrar K, Bilker W, Hoffstad O, Margolis DJ. Persistance of mild of mild to moderate atopic dermatitis. JAMA Dermatology. 2014;150(6):593–600. doi: 10.1001/jamadermatol.2013.10271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dhinga N, Gulati N, Guttman-Yassky E. Mechanisms of contact sensitization offer insights into roll of barrier defects vs intrinsic immune abnormalities as drivers of atopic dermatitis. Journal of Investigative Dermatology. 2013;133:2311–2314. doi: 10.1038/jid.2013.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Newell L, Polak ME, Perera J, Owen C, Boyd P, Pickard C, et al. Sensitization via healthy skin programs Th2 responses in indivduals with atopic dermatitis. Journal of Investigative Dermatology. 2013;133:2372–2380. doi: 10.1038/jid.2013.148. [DOI] [PubMed] [Google Scholar]
- 12.Guttman-Yassky E, Nograles KE, Krueger JG. Contrasting pathogenesis of atopic dermatitis, psoriasis--part I: clinical, pathologic concepts [Review] Journal of Allergy & Clinical Immunology. 2011;127(5):1110–1118. doi: 10.1016/j.jaci.2011.01.053. [DOI] [PubMed] [Google Scholar]
- 13.Palmer CN, Irvine AD, Terron-Kwiatkowski A, Zhao Y, Liao H, Lee SP, et al. Common loss-of-function variants of the epidermal barrier protein filaggrin are a major predisposing factor for atopic dermatitis. Nature Genetics. 2006;38(4):441–446. doi: 10.1038/ng1767. [DOI] [PubMed] [Google Scholar]
- 14.Margolis DJ, Apter AJ, Gupta J, Hoffstad O, Papadopoulos M, Campbell LE, et al. The persistence of atopic dermatitis and Filaggrin mutations in a US longitudinal cohort. Journal of Allergy & Clinical Immunology. 2012;130(4):912–917. doi: 10.1016/j.jaci.2012.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Winge MC, Bilcha KD, Lieden A, Shibeshi D, Sandilands A, Wahlgren CF, et al. Novel filaggrin mutation but no other loss-of-function variants found in Ethiopian patients with atopic dermatitis. British Journal of Dermatology. 2011;165(5):1074–1080. doi: 10.1111/j.1365-2133.2011.10475.x. [DOI] [PubMed] [Google Scholar]
- 16.Margolis DJ, Gupta J, Apter A, Hoffstad O, Papdopoulos M, Rebbeck TR, et al. Exome sequencing of Filaggrin and related genes in African-American children with atopic dermatitis. Journal of Investigative Dermatology. 2014;134(8):2272–2274. doi: 10.1038/jid.2014.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thaswer-Esmail F, Jakasa I, Todd G, Wen Y, Brown SJ, Krobach K, et al. South African amaXhosa patient with atopic dermatitis have decreased levels of filaggrin breakdown products but no loss-of-function mutations in filaggrin. Journal of Allergy & Clinical Immunology. 2014;133(1):280–282. doi: 10.1016/j.jaci.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Polican I, Becker L, Stein SL, Smith MS, aller AS. Filaggrin gene mutations in African Americans with both ichthyosis vulgaris and atopic dermatitis. Pediatric Dermatology. 2014;31(4):489–492. doi: 10.1111/pde.12355. [DOI] [PubMed] [Google Scholar]
- 19.Margolis DJ, Gupta J, Apter AJ, Ganguly T, Hoffstad O, Papadopoulos M, et al. Filaggrin-2 variation is associated wtih more persistent atopic dermatitis in Arican Americans. Journal of Allergy & Clinical Immunology. 2014;133(3):784–789. doi: 10.1016/j.jaci.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barnes KC. An update on the genetics of atopic dermatitis: scratching the surface in 2009[Review] [219 refs] Journal of Allergy & Clinical Immunology. 2010;125(1):16–29. doi: 10.1016/j.jaci.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Margolis DJ, Kim B, Apter AJ, Gupta J, Hoffstad O, Papadopoulos M, et al. Thymic stromal lymphopoietin variation, filaggrin loss-of-function, and persistence of atopic dermatitis. JAMA Dermatology. 2013;150(3):254–259. doi: 10.1001/jamadermatol.2013.7954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Noti M, Wojno ED, Kim BS, Siracusa MC, Giacomin PR, Nair MG, et al. Thymic stromal lymphopoietin-elicited basophil responses promote eosinophilic esophagitis. Nature Medicine. 2013;19(8):1005–1013. doi: 10.1038/nm.3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rothenberg ME, Spergel JM, Sherrill JD, Annaiah K, Martin LJ, Cianferoni A, et al. Common variants at 5q22 associate with pediatric eosinophilic esophagitis. Nature Genetics. 2010;42(4):289–291. doi: 10.1038/ng.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gauvreau GM, O’Bryne PM, Boulet LP, Wang Y, Cockcroft D, Bigler J, et al. Effects of anti-TSLP antibody on allergin-induced asthmatic reponses. New England Journal of Medicine. 2014;370:2102–2110. doi: 10.1056/NEJMoa1402895. [DOI] [PubMed] [Google Scholar]
- 25.Trowsdale J, Knight JC. Major histocompatibility compex genomics and human disease. Annu Rev Genomics Hum Genet. 2013;14:301–323. doi: 10.1146/annurev-genom-091212-153455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krawczyk M, Reith W. Regulation of MHC class II expression, a unique regulatory system identified by the study of primary immunodeficiency disease. Tissue Antigens. 2006;67:183–197. doi: 10.1111/j.1399-0039.2006.00557.x. [DOI] [PubMed] [Google Scholar]
- 27.Kim BS, Wang K, Siracusa MC, Saenz SA, Brestoff JR, Monticelli LA, et al. Basophils promote innnate lymphoid cell responses in inflammed skin. Journal of Immunology. 2014;193:3717–3725. doi: 10.4049/jimmunol.1401307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mirchandani AS, Besnard AG, Yip E, Scott C, Bain CC, Cerovic V, et al. Type 2 innate lymphoid cells drive CD4+ Th2 cell responses. Journal of Immunology. 2014;192(5):2442–2448. doi: 10.4049/jimmunol.1300974. [DOI] [PubMed] [Google Scholar]
- 29.Ellinghaus D, Baurecht H, Esparza-Gordillo J, Rodriguez E, Matanovic A, Marenholz I, et al. High-density genotyping study indentifies four new susceptibility loci for atopic dermatitis. Nature Genetics. 2013;45:808–812. doi: 10.1038/ng.2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hirota T, Takahashi A, Kubo M, Tsunoda T, Tomita K, Sakashita M, et al. Genome-wide association study identifies eight new susceptibility loci for atopic dermatitis in the Japanese population. Nature Genetics. 2012;44(11):1222–1226. doi: 10.1038/ng.2438. [DOI] [PubMed] [Google Scholar]
- 31.Paternoster L, Standl M, Chen CM, Ramasamy A, Bonnelykke K, Duijts L, et al. Meta-analysis of genome-wide association studies identifies three new risk loci for atopic dermatitis. Nature Genetics. 2012;44(2):187–192. doi: 10.1038/ng.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weidinger S, Willis-Owen SA, Kamatani Y, Baurecht H, Morar N, Liang L, et al. A genome-wide association study of atopic dermatitis identifies loci with overlapping effects on asthma and psoriasis. Human Molecular Genetics. 2013;22(23):4841–4856. doi: 10.1093/hmg/ddt317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Esparza-Gordillo J, Weidinger S, Folster-Holst R, Bauerfeind A, Ruschendorf F, Patone G, et al. A common variant on chromosome 11q13 is associated with atopic dermatitis. Nature Genetics. 2009;41(5):596–601. doi: 10.1038/ng.347. [DOI] [PubMed] [Google Scholar]
- 34.Sun LD, Xiao FL, Li Y, Zhou WM, Tang HY, Tang XF, et al. Genome-wide association study identifies two new susceptibility loci for atopic dermatitis in the Chinese Han population. Nature Genetics. 2011;43(7):690–694. doi: 10.1038/ng.851. [DOI] [PubMed] [Google Scholar]
- 35.Bussman C, Weidinger S, Novak N. Genetics of atopic dermatitis. JDDG: Journal of German Society of Dermatology. 2011;9:670–679. doi: 10.1111/j.1610-0387.2011.07656.x. [DOI] [PubMed] [Google Scholar]
- 36.Kendall E, Sargent CA, Campbel RD. Human major histocompatibility complex contains a new cluster of genes between HLA-D and complement C4 loci. Nucleic Acids Res. 1990;18(24):7251–7257. doi: 10.1093/nar/18.24.7251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eichenfield LF, Tom WL, Berger TG, Krol A, Paller AS, Schwarzenberger K, et al. Guidelines of care for the management of atopic dermatitis: section 2, Management and treatment of atopic dermatitis with topical therapies. Journal of the American Academy of Dermatology. 2014;71(1):116–132. doi: 10.1016/j.jaad.2014.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sidbury R, Tom WL, Bergman JN, Cooper KD, Silverman RA, Berger TG, et al. Guidelines of care for the management of atopic dermatitis: Section 4: Prevention of disease flares and use of adjunctive therapies and approaches. J Am Acad Dermatol. 2014;71:1218–33. doi: 10.1016/j.jaad.2014.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kanterakis S, Magira E, Rosenman KD, Rossman M, Talsznia K, Monos DS. SKDM human leukocyte antigen (HLA) tool: A comprehensive HLA and disease associations analysis software. Human Immunology. 2008;69:522–525. doi: 10.1016/j.humimm.2008.05.011. [DOI] [PubMed] [Google Scholar]
- 40.Maiers M, Gragert L, Klitz W. High resolution HLA alleles and haplotypes in the US population. Human Immunology. 2007;68:779–788. doi: 10.1016/j.humimm.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 41.Major E, Rigo K, Hague T, Berces A, Juhos S. HLA typing from 1000 genomes whole genome and whole exome illumina data. PLOS One. 2013;8(11):e78410. doi: 10.1371/journal.pone.0078410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stern LJ, Brown JH, Jardetzky TS, Gorga JC, Urban RG, Strominger JL, et al. Crystal structure of the human class II MHC protein HLA-DR1 complexed with an influenza virus peptide. Nature. 1994;368:215–221. doi: 10.1038/368215a0. [DOI] [PubMed] [Google Scholar]
- 43.Reche PA, Reinherz EL. Sequence variability analysis of human class I and class II MHC molecules: Functional and structural correlates of amino acid polymorphisms. J Mol Biol. 2003;331:623–641. doi: 10.1016/s0022-2836(03)00750-2. [DOI] [PubMed] [Google Scholar]
- 44.Androulakis IP, Nayak NN, Ierapetritou MG, Mono DS, Floudas CA. A predictive method for th evaluation of peptide binding in pocket 1 of HLA-DRB1 via global minimization of energy interactions. Proteins: Structures, function, and Genetics. 1997;29:87–102. [PubMed] [Google Scholar]
- 45.Thomson G, Marthandan N, Hollenbach JA, Mack SJ, Erlich HA, Single RM, et al. Sequence feature variant type (SFVT) analysis of the HLA genetic association in juvenile idiopathic arthritis. Pacific Symposium on Biocomputing. 2010:359–370. doi: 10.1142/9789814295291_0038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aron Y, Desmazes-Dufeu N, Matran R, Polla BS, Dusser D, Lockhart A, et al. Evidence of a strong, positive association between atopy and the HLA class II alleles DR4 and DR7. Clinical and Experimental Allergy. 1996;26:821–828. [PubMed] [Google Scholar]
- 47.Howell WM, Standring P, Warner JA, Warner JO. HLA class II genotype, HLA-DR B cell surface expression and allergan specific IgE production in atopic and non-atopic members of asthmatic family pedigrees. Clinical and Experimental Allergy. 1999;29(Suppl 4):35–38. [PubMed] [Google Scholar]
- 48.Saeki H, Kuwata S, Nakagawa H, Etoh T, Yanagisawa M, Miyamoto M, et al. Analysis of diseae-associated amino acid epitopes on HLA class II moleculres in atopic dermatitis. J Allergy Clin Immunol. 1995;96:1061–1068. doi: 10.1016/s0091-6749(95)70191-5. [DOI] [PubMed] [Google Scholar]
- 49.Park H, Ahn K, Park MH, Lee SI. The HLA-DRB1 polymorphism is associated with atopic dermatitis, but not egg allergy in Korean children. Allergy Asthma Immunol Res. 2012;4(3):143–149. doi: 10.4168/aair.2012.4.3.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Madore AM, Vaillancourt VT, Asi Y, Alizadehfar R, Ben-Shoshan M, Michel DL, et al. HLA-DQB1*02 and DQB1*06:03P are associated with peanut allergy. European Journal of Human Genetics. 2013;21:1181–1184. doi: 10.1038/ejhg.2013.13. [DOI] [PMC free article] [PubMed] [Google Scholar]