Abstract
Cows exposed to short day photoperiod (SD, 8L:16D) during the 60-day nonlactating period prior to parturition produce more milk in their subsequent lactation compared with cows exposed to long day photoperiod (LD, 16L:8D). Although this response is well established in dairy cows, the underlying mechanisms are not understood. We hypothesized that differential gene expression in cows exposed to SD or LD photoperiods during the dry period could be used to identify the functional basis for the subsequent increase in milk production during lactation. Pregnant, multiparous cows were maintained on an SD or LD photoperiod for 60 days prior to parturition. Mammary biopsies were obtained on days −24 and −9 relative to parturition and Affymetrix GeneChip Bovine Genome Arrays were used to quantify gene expression. Sixty-four genes were differentially expressed (P ≤ 0.05 and fold-change ≥ |1.5|) between SD and LD treatments. Many of these genes were associated with cell growth and proliferation, or immune function. Ingenuity Pathway Analysis predicted upstream regulators to include TNF, TGF-β1, interferon-γ, and several interleukins. In addition, expression of 125 genes was significantly different between day −24 and day −9; those genes were associated with milk component metabolism and immune function. The interaction of photoperiod and time affected 32 genes associated with insulin-like growth factor I signaling. Genes differentially expressed in response to photoperiod were associated with mammary development and immune function consistent with the enhancement of milk yield in the ensuing lactation. Our results provide insight into the mechanisms by which photoperiod affects the mammary gland and subsequently lactation.
Keywords: photoperiod, mammary gland, gene expression, dairy cows
the typical lactation cycle of a dairy cow includes a nonlactating, or dry, period of 40–60 days between successive lactations. This dry period promotes remodeling and restoration of mammary secretory tissue prior to the next lactation (30). Manipulation of photoperiod, or the duration of daily light exposure, during the dry period is a management tool to increase milk yield in the subsequent lactation. Specifically, cows exposed to a short day (SD; 8 h light-16 h dark) photoperiod during the dry period produce about 3 kg/day (∼5–10%) more milk in the subsequent lactation than cows exposed to long day (LD; 16 h light-8 h dark) photoperiod (4). This photoperiodic effect on milk production is substantial and consistent across multiple studies and species, including sheep and goats (21). The increased milk yield must result from enhanced function of the mammary gland; yet the underlying mechanisms have not been elucidated.
Milk production is fundamentally determined by the number and activity of secretory cells in the mammary gland (12), suggesting that photoperiod manipulation must affect one, or both, of those factors. Indeed, mammary epithelial cells of cows exposed to SD during the dry period had higher proliferation rates at 3 wk prior to parturition and reduced apoptosis rates overall, relative to cows on LD (53). Photoperiod manipulation also affects mammary parenchymal growth in prepubertal calves, such that mammogenesis is enhanced by LD treatment (21). Together, these studies clearly demonstrate that photoperiod can influence mammary development and function; however, data on the effects of photoperiod on mammary gene expression are limited.
To identify potential mediators of photoperiodic effects on mammary gene expression and function most studies have focused on investigating the role of two hormones, insulin-like growth factor I (IGF-I) and prolactin. Relative to LD, exposure to SD photoperiod, during either the dry period or lactation, decreases circulating prolactin (21). Prepartum SD exposure is also accompanied by an increase in mammary expression of prolactin receptor (4, 21). The increase in receptor abundance could promote growth through signaling pathways that include members of the suppressor of cytokine signaling gene family (53), although additional transcriptional regulation seems likely. Circulation of IGF-I, a growth factor known to have galactopoietic effects in goats (47), increases early in the dry period (1), potentially inhibiting mammary cell apoptosis (39). Exposure to LD photoperiod during lactation also increases circulating IGF-I concentrations in cows concurrently producing more milk than their SD counterparts (18). These findings suggest that circulating IGF-I may mediate the galactopoietic response to photoperiod in lactating dairy cows. To the contrary, Miller and coworkers (41) reported only a nonsignificant increase in blood IGF-I concentrations in cows exposed to SD, compared with those on LD, while dry. This led Wall and coworkers (53) to hypothesize that local expression of IGF-I in the mammary gland of dry cows might replace the blood-borne supply. However, they reported a significant increase in mammary expression of IGF-II but not IGF-I in cows on SD relative to those on LD photoperiod (53).
As indicated above, very few genes have been investigated as potential mediators of the milk yield response to photoperiod treatment in dairy cows (21). Recent transcriptomic studies of peripartum mammary development (13, 25, 26), milking frequency (15, 38, 54), environmental stress (14), bacterial invasion (50), and lactation (7, 8, 29, 33) have identified changes in gene expression related to functional outcomes in the bovine mammary gland. Genes identified in those studies are good candidates to be modulators of mammary function in response to external cues. To identify photoperiod-responsive genes, we have compared the mammary transcriptome of dairy cows exposed to LD or SD photoperiod during the dry period. We also compared gene expression at two time points to identify genes differentially expressed during mammary differentiation prepartum. We hypothesized that differential gene expression in cows exposed to LD or SD photoperiods during the dry period could be used to identify the functional basis for the subsequent difference in milk production during lactation. Specifically, we hypothesized that exposure to SD photoperiod would alter the expression of genes associated with enhanced mammary development and functional support of milk production. The objectives of this study were to identify genes differentially expressed between SD and LD photoperiod, and between 24 and 9 days prepartum, to determine associated signaling pathways, biofunctions, and upstream regulators and to draw functional associations to explain how their differential expression may influence subsequent milk production.
MATERIALS AND METHODS
Animals, photoperiod treatment, mammary biopsies.
All animal procedures were approved by the Animal Care and Use Committees of the University of Vermont and the University of Illinois. Mammary gland samples used in this experiment were derived from a study described previously (4, 52, 53). In brief, multiparous Holstein cows were dried off ∼60 days before their predicted parturition date and randomly assigned to LD or SD photoperiod treatment for the duration of the dry period. Lights were turned on at 0800 for both groups, and off at 1600 for SD or at 2400 for LD. Mammary biopsies were collected on day −24 ± 2 and −9 ± 2 relative to calving, as described by Farr and coworkers (23). Biopsies were trimmed of nonparenchymal tissue, then immediately snap-frozen in liquid nitrogen, and stored at −80°C pending subsequent RNA isolation.
RNA isolation and reverse transcription.
RNA was isolated from biopsied mammary tissues as previously described (53) using TRIzol Reagent (Invitrogen) according to the manufacturer's instructions. Total RNA was further purified using the RNeasy Mini Kit (Qiagen). Nucleic acid concentration was quantified using a NanoDrop ND1000 spectrophotometer and RNA quality was assessed using an Agilent 2100 Bioanalyzer (Agilent Technologies). Samples meeting the criteria of RNA integrity number > 7.0 were used for microarray analysis.
Microarray analysis - Affymetrix GeneChip bovine genome arrays.
Microarray analysis was conducted on samples from four cows for each photoperiod treatment and time (total n = 16 cows). Preparation of RNA and microarray procedures were performed at the University of Vermont Microarray Core Facility using previously described protocols (2). In brief, 2 μg of total RNA were reverse transcribed to single-stranded cDNA using T7-oligo (dT) primer. T4 DNA polymerase was used to synthesize double-stranded cDNA, which served as a template for in vitro transcription using T7 RNA polymerase to produce biotinylated cRNA. The biotinylated cRNAs were fragmented into 50- to 200-base fragments and then hybridized to GeneChip Bovine Genome Arrays (Affymetrix) for 16 h at 45°C in a rotating Affymetrix GeneChip Hybridization Oven 320. After hybridization, arrays were washed and stained with streptavidin-phycoerythrin on an automated Affymetrix GeneChip Fluidic Station F450. The arrays were scanned with an Affymetrix GeneChip Scanner 2700, and the images quantified using Affymetrix GeneChip Operating Software.
Data and statistical analysis.
The signal intensity for each probe was calculated from scanned images using GeneChip Operating Software (Affymetrix). Signal intensities were analyzed using BioConductor (http://www.bioconductor.org), background corrected, normalized by the loess method, and summarized as robust multichip averages (RMA) (9, 31). Two samples, one from each photoperiod treatment at the day −24 time point, were excluded from further analysis due to large variation in average signal intensities. This resulted in n = 3/treatment for day −24 and n = 4/treatment for day −9 (total n = 14). Data were analyzed for the effect of photoperiod treatment (LD − SD), time relative to parturition (day −24 − day −9) and the interaction of photoperiod treatment and time. Individual probes meeting the criteria fold-change ≥ |1.5| and P value ≤ 0.05, were considered differentially expressed (45). Expression data were also analyzed with the Benjamini and Hochberg test for multiple comparisons [false discovery rate (FDR)] (6). The adjusted P value ≤ 0.2 greatly restricted the number of genes available for functional analysis and was not used as a filter (32, 43, 46). To visualize the effects of photoperiod and time relative to parturition on specific genes, we generated heat maps from average RMA values using JMP 10 Pro.
Gene function and pathway analysis.
Probe set information and Gene Ontology biological and molecular functions were obtained on Affymetrix NetAffx Analysis Center (http://www.affymetrix.com). Ingenuity Pathway Analysis (IPA; Ingenuity Systems, http://www.ingenuity.com) was used to identify biofunctions, canonical pathways, and upstream regulators enriched in our data sets. Differentially expressed probes (fold-change ≥ |1.5|, P ≤ 0.05) for each effect (treatment: n = 131, time: n = 177, interaction: n = 956) were imported to IPA for analysis. When multiple probes for a single gene were differentially expressed, data were consolidated within IPA; probes with no current annotation were excluded from further analysis. Mapped IDs (treatment: n = 64, time: n = 125, interaction: n = 601), were identified from the Ingenuity Knowledge Base at the time of analysis (December 2013). “Analysis ready IDs” (treatment: n = 44, time: n = 104, interaction: n = 556) were subjected to Core Analysis. The Affymetrix GeneChip Bovine Genome Array was identified as a reference platform in IPA. We excluded biofunctions and canonical pathways specifically relating to diseases, disorders, and cancer from the analyses to obtain results most applicable to the bovine system. All other default settings were maintained. Statistical significance of enriched biofunctions and pathways was corrected for multiple comparisons within IPA using Benjamini-Hochberg (B-H) P value test correction threshold of P ≤ 0.05 (6). The z-score of activation, generated in IPA, provided insight into the functional effects of differentially expressed genes. The z-score denotes the predicted relationship between experimentally observed gene expression that is either activating (z > 0) or inhibiting (z < 0), as compiled in the Ingenuity Knowledge Base. The two functional effects with the largest |z-score| were considered representative of the biofunctional category. Canonical pathway ratios represent the number of differentially expressed genes relative to total number of genes in the IPA pathway. Comparison of our dataset to gene lists reported by Lemay and coworkers (bovine lactation genome: milk protein, pregnancy, lactation, and involution) (33) was conducted within IPA.
Analysis of upstream regulators included all IPA-defined molecule types. Two metrics, P value of overlap and predicted activation state, were used to understand the relationship among upstream regulators and differentially expressed genes. Fisher's exact P value of overlap (P ≤ 0.05) assesses the significance of the expression data for genes downstream of an upstream regulator. Given the fold-change of the differentially expressed genes, the predicted activation state indicates whether the upstream regulator would have activated, inhibited, or affected (unknown direction) the differentially expressed gene. In the analysis of photoperiod, upstream regulators affecting > 5 genes are presented, whereas in the analysis of time relative to parturition, the top five upstream regulators by lowest P value of overlap are shown.
RESULTS
The physiological responses to photoperiod treatment of cows used in this study were reported previously (4, 52). In brief, cows exposed to SD photoperiod during the dry period produced significantly more milk (∼38 kg/day) compared with cows exposed to LD photoperiod (∼35 kg/day) for the first 16 wk of lactation (4, 52).
Photoperiod induces differential expression of genes in the mammary gland.
Microarray analysis was used to identify differentially expressed genes in response to photoperiod and time relative to parturition and the interaction of photoperiod and time. For complete lists of differentially expressed probes, see Supplemental Table S1.1 Among the three effects analyzed, 757 mapped genes (1,264 probes) met our criteria for differential expression.
In response to photoperiod, 131 probes were differentially expressed (LD − SD; Supplemental Table S1). Of the 131 differentially expressed probes, 64 unique genes were annotated by Affymetrix, 35 of which had lower expression in cows exposed to SD, relative to LD photoperiod (Table 1). Probes (n = 19) representing eight known genes met the criteria of FDR ≤ 0.2 (Table 1), in total (n = 53) probes met the FDR ≤ 0.2 criteria (Supplemental Table S1). The top biofunctions enriched by photoperiod-responsive genes were cell-to-cell interactions, small molecule biochemistry, cell movement, hematological system development, and immune cell trafficking (Table 1, Supplemental Table S2). Notably, nine members of the bovine lymphocyte antigen (BoLA) family were differentially expressed, including BoLA-DQA5, which underwent the largest (28.5) fold-change. Genes associated with immune cell trafficking (CCR1, S100A12, and S100A8) were primarily decreased in response to SD photoperiod. Of the differentially expressed genes in response to photoperiod, 15 were in common with the bovine lactation genome (33) (Table 1).
Table 1.
Effect of photoperiod on differentially expressed genes in the mammary gland of dry cows
| Gene Symbol | Gene Name | Fold Change | P Value | Probe Set ID | IPA Biofunctions | GO Molecular Function | GO Biological Function |
|---|---|---|---|---|---|---|---|
| Expression Higher in Short Day Photoperiod | |||||||
| ADAM1B | A disintegrin and metalloproteinase 1B | −1.54 | 0.0382 | Bt.12799.1.S1_at | CI, CM | oxidoreductase activity | |
| AGA | aspartylglucosaminidase | −1.85 | < 0.0001 | Bt.3115.1.A1_at | |||
| ALDH5A1 | aldehyde dehydrogenase 5 member A1 | −1.91 | 0.0025 | Bt.2173.1.S1_at | CI, SMB | nucleic acid/amino acid metabolic process | |
| AKR1C3‡ | aldo-keto reductase family 1, member C3 | −1.61 | 0.0028 | Bt.23094.4.S1_at | SMB | proliferation, differentiation | |
| BCHE‡ | butyrylcholinesterase | −1.85 | 0.0494 | Bt.28385.1.A1_at | CI, SMB | hydrolase activity | |
| BoLA†‡ | major histocompatibility complex (MHC) I | −5.15 | 0.0001* | Bt.29823.1.S1_x_at | receptor activity | antigen processing/presentation, cellular defense | |
| BoLA-DQB1†‡ | MHC class II, DQ β1 | −3.48 | < 0.0001* | Bt.350.1.S1_at | CI, HSD, ICT | antigen processing/presentation, cellular defense response | |
| BoLA-DQA1† | MHC class II DQ α1 | −7.07 | < 0.0001* | Bt.22867.2.A1_at | CI, HSD, ICT | ||
| BoLA-DQA2† | MHC, class II, DQ α2 | −6.29 | < 0.0001* | Bt.4751.2.S1_at | |||
| BoLA-DQA3† | MHC, class II DQ α3 | −3.20 | < 0.0001 | Bt.4751.2.S1_at | |||
| BoLA-N† | MHC class I antigen | −7.95 | 0.0003 | Bt.5324.1.S1_s_at | |||
| BoLA-NC1 | MHC class I antigen | −3.72 | < 0.0001* | Bt.28022.1.A1_at | |||
| BTN3A2 | butyrophilin, subfamily 3A | −2.17 | 0.0021 | Bt.28475.1.A1_at | ubiquitin-protein ligase activity | immune system process | |
| DMP1 | dentin matrix acidic phosphoprotein 1 | −2.27 | 0.0284 | Bt.554.1.S1_at | CM | ||
| GBP1‡ | guanylate binding protein 1 | −1.54 | 0.0006 | Bt.24012.1.A1_at | invasion, tubulation, proliferation | ||
| gzmA | granzyme A | −1.58 | 0.0227 | Bt.29672.1.S1_at | serine-type peptidase activity | ||
| IgCgamma | IgG2a heavy chain constant region | −1.61 | 0.013 | Bt.12490.2.A1_x_at | |||
| KERA | Keratocan | −1.98 | 0.0022 | Bt.5391.1.S1_at | HSD | receptor activity | visual perception; GCPR/cytokine-mediated signaling |
| KIR2DS1 | killer cell immunoglobulin-like receptor 2DS1 | −1.56 | 0.0027 | Bt.11174.2.S1_at | |||
| MAL | mal, T-cell differentiation protein | −1.53 | 0.0100 | Bt.4060.1.S1_at | |||
| MAN1C1 | mannosidase, alpha, class 1C, member 1 | −1.52 | 0.0191 | Bt.10587.1.S1_at | hydrolase activity | metabolic process, glycosylation, proteolysis | |
| MAP1LC3C | microtubule-associated protein 1 light chain 3 γ | −1.63 | 0.0062 | Bt.3000.2.S1_a_at | cytoskeleton, microtubule binding | ||
| MCPH1 | microcephalin 1 | −1.56 | 0.0021 | Bt.11907.1.A1_at | |||
| MOXD1 | monooxygenase, DBH-like 1 | −1.56 | 0.0253 | Bt.20519.1.A1_at | oxidoreductase activity | neurological system, metabolic process | |
| OAS1X | 2′,5′-oligoadenylate synthetase 1, 40/46 kDa | −1.75 | 0.0001 | Bt.20922.1.S1_at | |||
| PRSS2 | protease, serine, 2 | −11.1 | < 0.0001* | Bt.4404.1.A1_at | |||
| SEPT2 | septin 2 | −1.81 | 0.0001 | Bt.20352.1.S1_at | |||
| SLC27A6‡ | solute carrier family 27 (fatty acid transporter), member 6 | −1.63 | 0.0104 | Bt.26921.1.A1_at | SMB | ligase activity, transporter activity | immune system process, fatty acid metabolic process |
| SLPI | secretory leukocyte peptidase inhibitor | −1.61 | 0.0183 | Bt.9736.1.S1_at | protein binding, endopeptidase inhibitor | proteolysis | |
| Expression Lower in Short Day Photoperiod | |||||||
| ABCC4 | ATP-binding cassette, subfamily C 4 | 2.10 | 0.0001 | Bt.27292.1.S1_at | |||
| BEX4 | brain expressed, X-linked 4 | 1.65 | 0.0034 | Bt.3698.1.S1_at | |||
| BoLA-DQA5‡ | MHC class II, DQ α5 | 28.5 | < 0.0001* | Bt.215.1.S1_at | CI, HSD, ICT | ||
| BoLA-DRB3‡ | MHC class II, DRB3 | 8.38 | < 0.0027* | Bt.20925.1.S1_at | |||
| CCR1‡ | chemokine (C-C motif) receptor 1 | 1.58 | 0.0013 | Bt.1377.1.S1_at | CI,SMB, CM, HSD, ICT | G-protein coupled receptor activity | immune/stimulus response, cytokine-mediated signaling |
| CD68‡ | CD68 molecule | 1.65 | 0.0106 | Bt.2334.1.S1_at | CI | lysosomal transport, proteolysis | |
| CLEC4E | C-type lectin domain family 4, member E | 1.86 | 0.0156 | Bt.16271.2.A1_at | CI, CM, SD, ICT | ||
| CSTB | cystatin B (stefin B) | 1.54 | 0.0068 | Bt.24354.1.S1_at | protein binding, endopeptidase inhibitor activity | proteolysis | |
| DDC | dopa decarboxylase | 1.75 | 0.003 | Bt.115.1.S1_at | CI, SMB | ||
| DEFB1 | defensin, beta 1 | 2.14 | 0.0007 | Bt.13125.1.S1_s_at | immune system, metabolic process, response to stress | ||
| DEFB5† | defensin beta 5 | 4.53 | 0.0005 | Bt.13125.1.S1_at | immune system. metabolic process, response to stress | ||
| DRD1 | dopamine receptor D1 | 1.6 | 0.0173 | Bt.9029.1.S1_at | CI, SMB, CM | G protein-coupled receptor activity | neurological system, GPCR/cell-cell signaling |
| F13A1† | coagulation factor XIII, A1 polypeptide | 1.64 | 0.0123 | Bt.19845.2.A1_at | CI,CM, HSD, ICT | acyltransferase activity | immune system, protein modification, blood coagulation |
| FXYD3 | FXYD domain containing ion transport regulator 3 | 1.53 | 0.0272 | Bt.9573.1.S1_a_at | ion channel activity, protein binding | ion transport, signal transduction | |
| GIMAP8 | GTPase IMAP family member 8-like | 1.81 | 0.0001 | Bt.26769.1.S1_at | |||
| GPNMB†‡ | glycoprotein (transmembrane) nmb | 1.94 | 0.0108 | Bt.9807.1.S1_at | |||
| IGL@ | immunoglobulin light chain, λ gene cluster | 1.52 | 0.0078 | ||||
| IL17RB‡ | interleukin 17 receptor B | 1.64 | 0.0118 | Bt.24532.1.A1_at | CM, HSD, ICT | ||
| IL8 | interleukin 8 | 1.67 | 0.002 | Bt.155.1.S1_at | CI,SMB, CM,HSD, ICT | chemokine activity | macrophage activation, cytokine-mediated signaling |
| KIR2DL5A | killer cell immunoglobulin-like receptor, two domains, long cytoplasmic tail, 5A | 1.89 | 0.002 | Bt.11174.1.S1_at | |||
| NPL | N-acetylneuraminate pyruvate lyase | 1.64 | 0.0006 | Bt.26155.1.A1_at | lyase activity | cellular amino acid biosynthetic process | |
| PEBP4 | phosphatidylethanolamine-binding protein 4 | 2.93 | 0.0002 | Bt.14398.1.S1_at | kinase regulator activity | signal transduction | |
| PTI† | pancreatic trypsin inhibitor | 2.96 | 0.0111 | Bt.28518.1.S1_at | |||
| RARRES1 | retinoic acid receptor responder | 1.61 | 0.0386 | Bt.24933.1.S1_at | CM | ||
| S100A12‡ | S100 calcium binding protein A12 | 1.91 | 0.019 | Bt.357.1.S1_at | CI, ICT | calcium ion, calmodulin binding | immune response, cell cycle |
| S100A8 | S100 calcium binding protein A8 | 1.91 | 0.0142 | Bt.9360.1.S1_at | CI,CM, ICT | calcium ion/calmodulin binding | macrophage activation, cell cycle |
| SDS | serine dehydratase | 1.61 | 0.0391 | Bt.5878.1.A1_at | SMB | ||
| SERPINA3 | serpin peptidase inhibitor A3 | 1.56 | 0.0036 | Bt.5362.1.S1_at | CM, HSD, ICT | protein binding, endopeptidase inhibitor activity | proteolysis |
| SFRP1 | secreted frizzled-related protein 1 | 1.55 | 0.0276 | Bt.5226.1.S1_at | CI CM, HSD, ICT | ||
| SPADH1 | spermadhesin 1 | 2.24 | 0.0295 | Bt.457.1.S1_at | |||
| TMEM183A‡ | transmembrane protein 183A | 1.55 | 0.0004 | Bt.17595.3.A1_at | |||
| TRAF3IP3‡ | TRAF3 interacting protein 3 | 1.56 | 0.0242 | Bt.2266.3.S1_at | |||
| VNN1 | vanin 1 | 1.84 | 0.0446 | Bt.28243.1.S1_a_at | CI | hydrolase activity | signal transduction, adhesion, vitamin metab |
| VNN2 | vanin 2 | 1.72 | 0.0061 | Bt.19160.1.A1_at | |||
| VSTM1 | V-set and transmembrane domain 1 | 1.59 | 0.0071 | Bt.9131.1.S1_at | |||
Biopsies were obtained from cows exposed to long or short day photoperiod. Microarray analysis was conducted on purified RNA using Affymetrix GeneChip Bovine Genome Arrays. Genes identified in response to photoperiod treatment (long day-short day). Differential expression was attributed to genes meeting the criteria of P ≤ 0.05 and fold-change ≥ |1.5|.
Multiple probes were differentially expressed; average fold-change and P values are shown. Top 5 Ingenuity Pathway Analysis (IPA) biofunctions and corresponding genes identified as differentially expressed in response to photoperiod.
CI, cell-to-cell interactions; SMB, small molecule biochemistry; CM, cell movement; HSD, hematological system development; ICT, immune cell trafficking.
Gene Ontology (GO) molecular and biological functions were assigned using Affymetrix NetAffx Analysis Center (http://www.affymetrix.com).
Genes in common with the bovine lactation genome (35).
Genes meeting the criteria of false discovery rate ≤ 0.2.
To gain further insight into the functions of these genes, we examined the top five canonical pathways enriched by genes differentially expressed in response to photoperiod (Table 2). Four of the top five pathways were associated with immune function, largely due to the high fold-change of three BoLA genes. Genes associated with fatty acid oxidation (Table 2, Supplemental Table S3) enriched the fifth pathway. The ratios of differentially expressed genes to total number of genes in these canonical pathways were generally low (average: 0.043), which can be attributed to the relatively small number of differentially expressed genes in response to photoperiod.
Table 2.
Top five canonical pathways enriched by genes differentially expressed in response to photoperiod, time relative to parturition and the interaction
| Effect | Canonical Pathway | B-H P Value | Ratio | Differentially Expressed Genes |
|---|---|---|---|---|
| Photoperiod | cytotoxic T lymphocyte mediated apoptosis of cells | 0.008 | 0.034 | ↓BoLA, ↑BoLA-DQA5, BoLA-DRB3 |
| OX40 signaling | 0.010 | 0.031 | ↓BoLA, ↑BoLA-DQA5, BoLA-DRB3 | |
| B cell development | 0.036 | 0.056 | ↑BoLA-DQA5, BoLA-DRB3 | |
| antigen presentation | 0.055 | 0.048 | ↓BoLA, ↑BoLA-DQA5 | |
| fatty acid beta oxidation I | 0.059 | 0.044 | ↑SDS, ↓SLC27A6 | |
| Time | acute phase response signaling | < 0.0001 | 0.043 | ↑C3, C4BPA, CFB, CP, HP, IL6, IL33, LBP, RBP1 |
| LXR/RXR activation | 0.004 | 0.043 | ↑C3, CD14, IL6, IL33, LBP, LPL | |
| VDR/RXR activation | 0.004 | 0.057 | ↓CAMP, CXCL10, ↑CD14, IGFBP5, THBD | |
| granulocyte adhesion and diapedesis | 0.016 | 0.033 | ↓CXCL10, CXCL13, CLDN1, ↑IL33, SDC2, CXCL2 | |
| IL-10 | 0.019 | 0.051 | ↓CAMP, CXCL10, ↑CD14, IGFBP5, THBD | |
| Interaction | IGF-I signaling | 0.014 | 0.103 | ↑NOV, SOCS1, ↓FOS, IGF1R, JUN, KRAS, NEDD4, PRKACB, PRKAR2B, PTK2, YWHAG |
| CDK5 signaling | 0.064* | 0.093 | ↓ITGA2, KRAS, PPM1L, PPP1CB, PRKACB, PRKAR2B ↑ADCY3, PPP1R10, PPP1R3C | |
| dopamine receptor signaling | 0.077* | 0.083 | ↓PPM1L, PPP1CB, PRKACB, PRKAR2B ↑ADCY3, GCH1, PPP1R10, PPP1R3C | |
| sertoli cell junction signaling | 0.077* | 0.051 | ↓ACTN4, ATF2, CDH1, CLDN1, ITGA2 JUN, KRAS, PRKACB, PRKAR2B, PVRL3, SPTBN1 ↑ACTA1, ACTG2, TGFB3 | |
| protein kinase A signaling | 0.09* | 0.051 | ↓PPP1R3C, ADCY3, PPP1R10, TGFB3, HIST1H1C, TTN, PYGM,↑PRKACB, PPP1CB, PDE1A, AKAP5, DUSP10, PRKAR2B, ATF2, TGFBR1, YWHAG, BRAF, PTK2, EYA3, PTPLA, PTPN2 |
Biopsies were obtained from cows exposed to either long or short day photoperiod on days −24 and −9 relative to parturition. Microarray analysis was conducted on purified RNA using Affymetrix GeneChip Bovine Genome Arrays. Genes were identified in the effect of photoperiod treatment, time relative to parturition and the interaction. Differential expression was attributed to genes meeting the criteria of P ≤ 0.05 and fold-change ≥ |1.5|. IPA was used to identify canonical pathways. Benjamini-Hochberg (B-H) P value test correction to identify the probability of gene associations by chance (6). Ratio of differentially expressed genes to the total number of genes in the canonical pathway as defined by IPA.
Canonical pathways that did not meet the B-H threshold criteria of P ≤ 0.5. ↓Negative fold-change; ↑positive fold-change, within respective comparisons.
Of the upstream regulators identified (n = 368, Supplemental Table S4), nearly all (97%) were predicted to affect ≤ 5 of the photoperiod-responsive genes. Upstream regulators predicted to affect > 5 differentially expressed genes were selected for further investigation and included 2 factors involved in tumor necrosis factor (TNF) signaling (TNF and FAS, a member of the TNF receptor superfamily), four interleukins (IL), interferon-γ (IFNG), transforming growth factor-β1 (TGF-β1), oncostatin M (OSM), and the hormone β-estradiol (Table 3). Prolactin was predicted to regulate three genes and was included as a reference upstream regulator. Together, these upstream regulators were predicted to affect 26 of the photoperiod-responsive genes, the majority of which were activated. Complete lists of upstream regulators are available in Supplemental Table S4.
Table 3.
Upstream regulators predicted to affect >5 genes differentially expressed in the mammary gland of dairy cows in response to photoperiod treatment during the dry period
| Upstream Regulators |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene Symbol | PRL* | DEX | IFNG | LPS | TGF-β1 | TNF | E2 | OSM | IL-13 | FAS | IL-6 | IL-1B | IL-10 |
| AKR1C3 | ↑ | ↓ | ↑ | ||||||||||
| ALDH5A1 | ↔ | ||||||||||||
| BoLA | ↓ | ↓ | ↓ | ||||||||||
| BoLA-DQA5 | ↑ | ↓ | ↓ | ↑ | ↔ | ↑ | |||||||
| BoLA-DRB3 | ↓ | ↑ | ↔ | ↓ | |||||||||
| CCR1 | ↑ | ↑ | ↓ | ↑ | ↓ | ↑ | ↔ | ↑ | ↔ | ↑ | |||
| CD68 | ↔ | ↑ | ↑ | ||||||||||
| CLEC4E | ↑ | ↑ | ↑ | ↓ | ↑ | ||||||||
| CSTB | ↑ | ||||||||||||
| DMP1 | ↓ | ||||||||||||
| DRD1 | |||||||||||||
| F13A1 | ↑ | ↑ | ↑ | ||||||||||
| GBP1 | ↑ | ↓ | ↓ | ↑ | ↑ | ↓ | ↓ | ↓ | |||||
| GPNMB | ↑ | ↓ | ↓ | ↑ | |||||||||
| IL17RB | ↓ | ↓ | ↑ | ↑ | ↑ | ↑ | |||||||
| IL8 | ↓ | ↑ | ↑ | ↓ | ↑ | ↑ | ↑ | ↓ | ↑ | ↑ | ↑ | ↓ | |
| MAL | ↓ | ||||||||||||
| MAN1C1 | |||||||||||||
| PEBP4 | |||||||||||||
| RARRES1 | ↔ | ↑ | |||||||||||
| S100A12 | ↔ | ↑ | ↔ | ||||||||||
| S100A8 | ↑ | ↑ | ↑ | ↑ | ↓ | ↑ | ↑ | ||||||
| SDS | ↑ | ||||||||||||
| SERPINA3 | ↑ | ↑ | ↑ | ↑ | ↑ | ↑ | ↑ | ↑ | ↑ | ||||
| SFRP1 | ↑ | ↑ | ↔ | ↑ | ↑ | ||||||||
| VNN1 | ↑ | ||||||||||||
| Genes affected, n | 3 | 11 | 11 | 11 | 11 | 10 | 8 | 8 | 7 | 6 | 6 | 6 | 6 |
Biopsies were obtained from cows exposed to long or short day photoperiod. Microarray analysis was conducted on purified RNA using Affymetrix GeneChip Bovine Genome Arrays. Genes were identified in the effect of photoperiod treatment (long day-short day). Differential expression was attributed to genes meeting the criteria of P ≤ 0.05 and fold-change ≥ |1.5|. Predicted upstream regulators - PRL, prolactin; DEX, dexamethasone; IFNG, interferon γ; LPS, lipopolysaccharide; TGF-β1, transforming growth factor-β1; TNF, tumor necrosis factor; E2, beta-estradiol; OSM, oncostatin M; IL, interleukin members 1B, 6, 10, 13; FAS, TNF receptor superfamily member 6. Upstream regulators were identified by IPA upstream regulator function. IPA predictions of the upstream regulators' activation state are based on the direction of gene expression change. ↑ Predicted activation, ↔ affected, ↓ predicted inhibition.
Prolactin was included based on its role in lactation and photoperiod biology.
Expression of genes related to milk synthesis increases with approaching parturition.
Time relative to parturition significantly affected the expression of 125 mapped genes (171 probes, Supplemental Table S1). Of the probe sets, 146 were more highly expressed on day −9 than day −24. These differentially expressed genes aligned with 28 unique (38 total) significantly enriched (B-H P ≤ 0.05) biofunctions (Table 4). Biofunctions predicted to increase (activation z-score ≥ 2.0) include lipid synthesis, migration of granulocytes and neutrophils, and metabolism of carbohydrate. The functional effect, organismal death, had a z-score of activation < −2.0 and thereby was predicted to decrease. Genes associated with all biofunctional categories are available in Supplemental Table S2.
Table 4.
IPA biofunctions enriched by genes differentially expressed in the mammary gland of cows in response to time (day −9 minus day −24) relative to parturition
| IPA Biofunction | Genes, n | Functional Effect | z-Score of Activation | P Value | Associated Biofunctions |
|---|---|---|---|---|---|
| Lipid metabolism | 32 | ↑synthesis of lipid | 3.4 | 0.003 | small molecule biochemistry |
| ↑fatty acid metabolism | 2.6 | 0.027 | |||
| Molecular transport | 42 | ↑transport of molecules | 2.4 | 0.019 | |
| secretion of molecules | 1.7 | 0.023 | |||
| Cellular movement | 38 | ↑migration of granulocytes | 2.8 | 0.000 | hematological sys. devel. and funct., immune cell trafficking, tissue devel. |
| ↑migration of neutrophils | 2.2 | 0.005 | |||
| Cell-to-cell signaling and interaction | 34 | ↑binding of phagocytes | 2.6 | 0.000 | hematological sys. devel. and funct., cell signaling, cell growth and prolif. |
| ↑stimulation of cells | 2.6 | 0.003 | |||
| Immune cell trafficking | 24 | ↑adhesion of immune cells | 2.5 | 0.000 | |
| ↑binding of granulocytes | 2.2 | 0.001 | |||
| Carbohydrate metabolism | 25 | ↑metabolism of carbohydrate | 2.6 | 0.010 | |
| quantity of carbohydrate | 1.8 | 0.000 | |||
| Connective tissue devel. and funct. | 26 | ↑quantity of adipose tissue | 2.2 | 0.000 | tissue morphology |
| quantity of white adipose | 2.0 | 0.016 | |||
| Organismal survival | 36 | ↓organismal death | −3.6 | 0.004 | |
| survival of organism | 1.5 | 0.000 | |||
| Protein synthesis | 21 | quantity of IgE | 1.1 | 0.004 | humoral immune response |
| quantity of TNF in blood | 0.4 | 0.002 | |||
| Cardiovascular system devel. and funct. | 28 | ↑binding of endothelial cells | 2.2 | 0.006 | |
| cell movement of endothelial cells | 1.7 | 0.019 | |||
| Cellular growth and proliferation | 45 | ↑stimulation of leukocytes | 2.4 | 0.000 | |
| ↑stimulation of phagocytes | 2.2 | 0.001 | |||
| Cell death and survival | 44 | cell death of breast cancer cell line | −1.7 | 0.027 | |
| fragmentation of DNA | −1.6 | 0.009 | |||
| Cellular development | 35 | ↑prolif. of smooth muscle cells | 2.4 | 0.014 | cell morphology |
| prolif. of vascular smooth muscle cells | 2.0 | 0.016 | |||
| Cellular assembly and organization | 19 | formation of filopodia | 1.2 | 0.019 | cellular funct. and maintenance |
| organization of cytoskeleton | 1.5 | 0.027 | |||
| Digestive sys. devel. and funct. | 22 | mass of liver | 1.0 | 0.002 | hepatic sys. devel. and funct., organ devel, organ morphology |
| inflammation of liver | 0.3 | 0.004 | |||
| Organismal development | 29 | development of blood vessels | 1.4 | 0.005 | |
| endothelial cell development | 1.2 | 0.005 | |||
| Cellular compromise | 17 | degranulation of mast cells | −0.3 | 0.009 | |
| degranulation of cells | 0.2 | 0.001 | |||
| Organ morphology | 22 | size of bone | −1.0 | 0.027 | |
| Energy production | 9 | oxidation of fatty acid | 0.7 | 0.006 | |
| Cell cycle | 15 | mitogenesis | 0.7 | 0.027 | |
| Vitamin and mineral metabolism | 12 | mobilization of Ca2+ | −0.7 | 0.007 | |
| flux of Ca2+ | 0.1 | 0.017 | |||
| Renal and urological sys. devel. and funct. | 12 | proliferation of glomerular cells | 0.3 | 0.009 | |
| DNA replication, recombination, and repair | 9 | fragmentation of DNA | −1.6 | 0.009 | |
| metabolism of DNA | 1.0 | 0.014 | |||
| Tumor morphology | 11 | proliferation of tumor cells | 0.6 | 0.010 | |
| Cell-mediated immune response | 6 | cell movement of T lymphocytes | −1.4 | 0.016 | |
| Free radical scavenging | 10 | metabolism of reactive oxygen species | 0.4 | 0.014 | |
| Hair and skin devel. and funct. | 6 | proliferation of endothelial cells | 0.1 | 0.017 | |
| Organismal functions | 10 | thermoregulation | 1.1 | 0.027 |
Biopsies were obtained from cows during the dry period on day −24 and −9 relative to parturition. Microarray analysis was conducted on purified RNA using Affymetrix GeneChip Bovine Genome Arrays. Genes were identified in the effect of time relative to parturition (day −9 − day −24). Differential expression was attributed to genes meeting the criteria of P ≤ 0.05 and fold-change ≥ |1.5|. All biofunctions met the B-H threshold criteria of P ≤ 0.05. The top 2 functional effects as determined by z-score of activation. Biofunctions with identical functional effects are listed as associated functions. z-Score of activation: IPA prediction of the relationship between experimentally observed gene expression that is either activating or inhibiting (as compiled in the Ingenuity Knowledge Base), z-score > 0 are activating, < 0 are inhibiting. IPA required z-scores ≤ −2.0 or > 2.0 to form a prediction (indicated by arrows). Associated biofunctions measure the probability that genes were randomly associated with a biofunction, P ≤ 0.05 was the threshold for significance.
A central theme in the time relative to parturition dataset was altered metabolism, including 45 differentially expressed genes that enriched five biofunctions highly relevant to lactogenesis: lipid metabolism, protein synthesis, carbohydrate metabolism, vitamin and mineral metabolism, and reproductive system development and function (Tables 4 and 5). Of these differentially expressed genes, 41 were more highly expressed at day −9 compared with day −24 relative to parturition, with an average twofold increase in expression. Genes associated with these functions also enriched the canonical pathway supporting lipid metabolism (LXR/RXR Activation) and immune function (Table 2 and Supplemental Table S3). Several of these differentially expressed genes are established markers of mammary function, including AQP1, LALBA, LPL, LPO, NT5E. In addition to the milk synthesis-related biofunctions, seven genes were functionally associated with involution of the mammary gland as part of the reproductive system development and function biofunction (Table 5, Supplemental Table S2). Comparison to the lactation-related dataset identified 23 genes in common with the bovine lactation genome (33) (Table 5).
Table 5.
Genes differentially expressed (day −9 − day −24) in the mammary gland during the dry period enrich biofunctions related to milk production
| IPA Biofunctions |
||||||||
|---|---|---|---|---|---|---|---|---|
| Fold-change | P Value | LM | PS | VM | CM | RS | ||
| ACSM1* | acyl-CoA synthetase medium-chain 1 | 2.0 | 0.001 | X | ||||
| ALOX15 | arachidonate 15-lipoxygenase | 2.4 | < 0.001 | X | ||||
| ANGPTL4* | angiopoietin-like 4 | 2.1 | 0.019 | X | X | |||
| AQP1* | aquaporin 1 (Colton blood group) | 1.6 | 0.003 | X | ||||
| C3* | complement component 3 | 1.8 | 0.038 | X | X | X | X | |
| CAMP* | cathelicidin antimicrobial peptide | −2.8 | 0.003 | X | X | X | ||
| CD14* | CD14 molecule | 1.6 | 0.012 | X | X | X | ||
| CEBPD* | CCAAT/enhancer binding protein (C/EBP), delta | 1.7 | 0.036 | X | X | |||
| CFB | complement factor B | 2.4 | 0.025 | X | ||||
| CHI3L1* | chitinase 3-like 1 (cartilage glycoprotein-39) | 2.5 | 0.040 | X | ||||
| CIDEA* | cell death-inducing DFFA-like effector a | 2.3 | 0.004 | X | X | |||
| CP | ceruloplasmin (ferroxidase) | 2.7 | 0.002 | X | ||||
| CXCL10 | chemokine (C-X-C motif) ligand 10 | −1.7 | 0.008 | X | X | X | X | |
| CXCL13 | chemokine (C-X-C motif) ligand 13 | −2.1 | 0.033 | X | ||||
| CXCL2* | chemokine (C-X-C motif) ligand 2 | 3.0 | 0.018 | X | ||||
| CYP11A1 | cytochrome P450, family 11, subfamily A1 | 3.2 | 0.016 | X | X | X | ||
| DARC | Duffy blood group, chemokine receptor | 1.7 | 0.008 | X | ||||
| DUSP1* | dual specificity phosphatase 1 | 1.6 | 0.002 | X | X | X | ||
| FABP3* | fatty acid binding protein 3 | 3.0 | 0.001 | X | X | |||
| GK* | glycerol kinase | 1.6 | < 0.001 | X | X | |||
| HP | haptoglobin | 4.5 | 0.025 | X | ||||
| IGFBP5 | insulin-like growth factor binding protein 5 | 1.6 | 0.029 | X | X | X | ||
| IL13RA1 | interleukin 13 receptor, alpha 1 | 1.5 | 0.007 | X | ||||
| IL17RB* | interleukin 17 receptor B | −1.5 | 0.022 | X | ||||
| IL33 | interleukin 33 | 2.1 | 0.001 | X | X | |||
| IL6 | interleukin 6 | 1.7 | 0.002 | X | X | X | X | X |
| KLF6* | Kruppel-like factor 6 | 1.5 | 0.011 | X | ||||
| LALBA* | α-lactalbumin | 1.5 | 0.005 | X | X | |||
| LBP* | lipopolysaccharide binding protein | 3.4 | 0.005 | X | X | |||
| LPIN1 | lipin 1 | 1.7 | 0.023 | X | ||||
| LPL* | lipoprotein lipase | 1.9 | < 0.001 | X | X | X | ||
| MSTN | myostatin | 1.6 | < 0.001 | X | X | |||
| MUC1* | mucin 1, cell surface associated | 2.0 | 0.003 | X | ||||
| NT5E* | 5′-nucleotidase, ecto (CD73) | 1.6 | 0.031 | X | ||||
| NTS | neurotensin | 1.7 | 0.002 | X | X | |||
| PDK4* | pyruvate dehydrogenase kinase, isozyme 4 | 1.9 | 0.014 | X | X | X | ||
| PLVAP | plasmalemma vesicle | 1.6 | < 0.001 | X | X | |||
| PTHLH | parathyroid hormone-like hormone | 2.5 | 0.002 | X | X | X | ||
| RAB35* | Rab 35, member ras | 1.5 | 0.005 | X | X | |||
| RBP1 | retinol binding protein 1, cellular | 1.8 | 0.042 | X | X | |||
| S100A9* | S100 calcium binding protein A9 | 2.4 | 0.013 | X | ||||
| SDC2 | syndecan 2 | 1.5 | 0.005 | X | X | |||
| SFN | stratifin | 1.9 | 0.021 | X | X | |||
| SULF2 | sulfatase 2 | 1.6 | 0.027 | X | ||||
| THBD | thrombomodulin | 1.9 | 0.005 | X | X | |||
Biopsies were obtained from cows during the dry period on day −24 and −9 relative to parturition. Microarray analysis was conducted on purified RNA using Affymetrix GeneChip Bovine Genome Arrays. Genes were identified in the effect of time relative to parturition (day −9 − day −24). Differential expression was attributed to genes meeting the criteria of P ≤ 0.05 and fold-change ≥ |1.5|. IPA biofunctions - LM, lipid metabolism; PS, protein synthesis; VM, vitamin and mineral metabolism; CM, carbohydrate metabolism; RS, reproductive system development and function.
Genes in common with the bovine lactation genome (33).
Genes differentially expressed in response to time were associated with 1,373 upstream regulators in IPA, the top five by P value of overlap were: β-estradiol, lipopolysaccharide, TNF, dexamethasone, and TGF-β1 (Table 6). Together, these upstream regulators were predicted to affect 68 differentially expressed genes (Supplemental Table S4).
Table 6.
Upstream regulators predicted to affect genes differentially expressed in response to time relative to parturition in the mammary gland of cows during the dry period
| Upstream Regulator | Molecule Type | Activation Score | P Value of Overlap | Genes, n |
|---|---|---|---|---|
| β-Estradiol | chemical - endogenous mammalian | 0.81 | <0.0001 | 37 |
| Lipopolysaccharide | chemical drug | 2.57 | <0.0001 | 36 |
| TNF | cytokine | 1.73 | <0.0001 | 35 |
| Dexamethasone | chemical drug | 1.16 | <0.0001 | 35 |
| TGF-β1 | growth factor | 1.42 | <0.0001 | 31 |
Biopsies were obtained from cows during the dry period on day −24 and −9 relative to parturition. Microarray analysis was conducted on purified RNA using Affymetrix GeneChip Bovine Genome Arrays. Genes were identified in the effect of time relative to parturition (day −9 − day −24). Differential expression was attributed to genes meeting the criteria of P ≤ 0.05 and fold-change ≥ |1.5|. Upstream regulators were identified using IPA upstream regulator function. The top 5 upstream regulators by lowest P value of overlap. z-Score of activation: IPA prediction of the relationship between experimentally observed gene expression that is either activating or inhibiting (as compiled in the Ingenuity Knowledge Base), z-score > 0 are activating, < 0 are inhibiting. Genes, the number of differentially expressed genes predicted to be affected by upstream regulator.
Interactive effects of photoperiod and time on mammary gene expression.
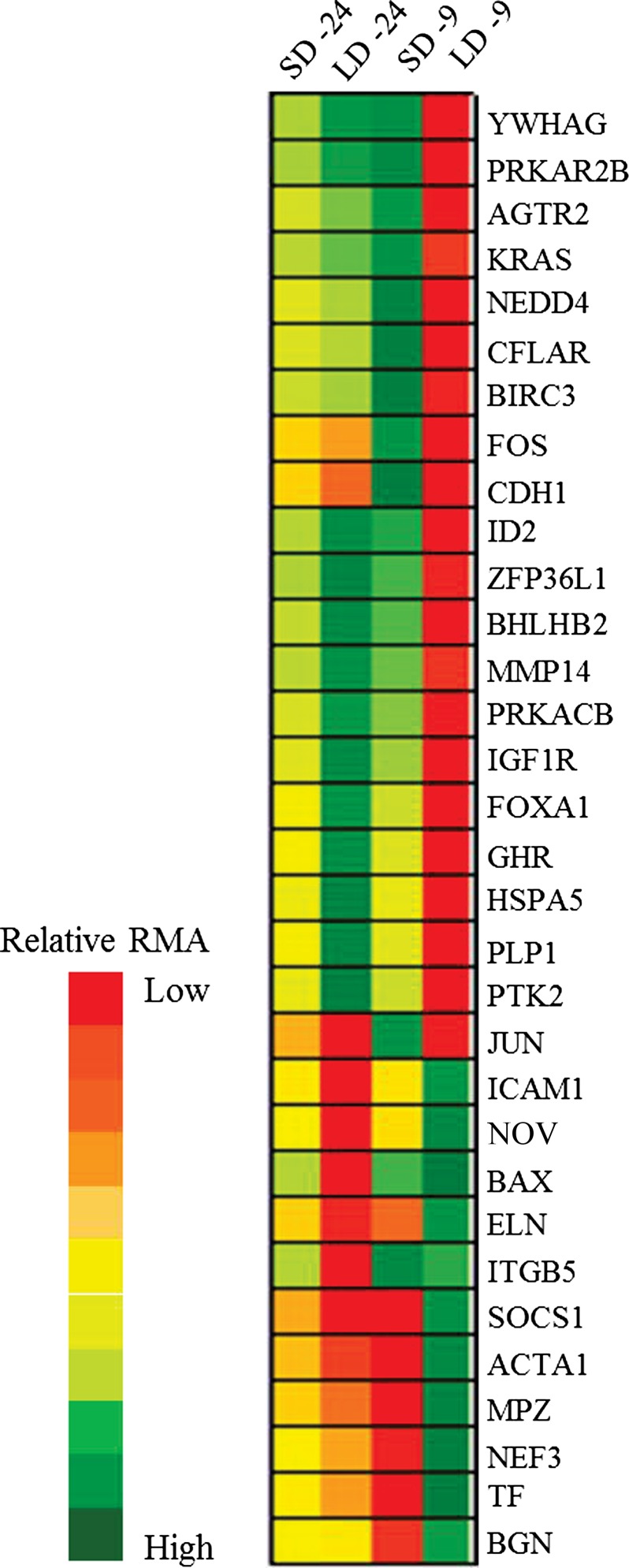
Analysis of the interaction of photoperiod and time relative to parturition identified differential expression of 601 genes (965 probes, Supplemental Table S1). There was overlap among differentially expressed probes in the effects of photoperiod (9 common probes) and time (16 common probes) with those identified in the interaction. Functional analysis identified organismal death (activation score: 3.53) as the most highly enriched functional effect of the differentially expressed genes. The complete list of biofunctions and associated differentially expressed genes is in Supplemental Table S2. One canonical pathway, IGF-I signaling, was enriched by 11 genes (Table 2, Supplemental Table S3). Additionally, IGF-I was a predicted upstream regulator (Supplemental Table S4) of 24 differentially expressed genes. Three genes were present in both lists, resulting in 32 differentially expressed genes associated with IGF-I signaling (Fig. 1). Cows exposed to LD photoperiod had the lowest level of expression of five of these genes on day −24 and 21 genes on day −9 relative to calving. The expression of six genes was lowest in cows exposed to SD on day −9, all of which were most highly expressed in LD-treated cows on the same day (Fig. 1).
Fig. 1.
The relative robust multichip average (RMA) values of genes differentially expressed in bovine mammary gland that are associated with IGF-I signaling. Ingenuity pathway analysis (IPA) associated thirty-two genes, differentially expressed in the interaction of photoperiod and time relative to parturition, with the IGF-I signaling pathway or IGF-I as predicted upstream regulator. Relative RMA values indicate the change in expression in response to long day (LD) and short day (SD) photoperiod on day −9 and −24 and relative to parturition.
DISCUSSION
Effect of photoperiod on mammary gene expression.
Cows exposed to SD photoperiod during the dry period go on to produce more milk than cows on LD photoperiod (4). Comparison of the transcriptome of cows on SD or LD photoperiod, averaged over days −24 and −9 relative to parturition, identified 64 differentially expressed genes. Functional analysis of these genes and their upstream regulators provided insight into their roles as mediators of the effects of photoperiod on mammary cell growth, proliferation, and ultimately function. Among the genes and upstream regulators we identified two lactogenic hormones (prolactin and dexamethasone) and several well-known mammary growth factors (TGF-β1 and TNF) that may mediate the changes in mammary cell proliferation in response to photoperiod (53).
The mammary gland is thought to have evolved as part of the innate immune system, such that milk provided both nourishment and immunological protection for newborns (51). Therefore, lactation and inflammatory responses share many common mechanisms (33, 51). Here, we report photoperiod-responsive expression of genes with known immune regulatory function including antimicrobial and proinflammatory factors: granzyme (gzm)A, secretory leukocyte peptidase inhibitor (SLPI), butyrylcholinesterase (BCHE), and butyrophilin (BTN3A2) (22, 36, 56). The effect of photoperiod on immune function has been well established in mammals (42, 55), including dairy cows during the dry period (5) and specifically the mammary gland defense system (28). Among the differentially expressed genes identified here, members of the BoLA (major histocompatibility complex) gene family have also been associated with mastitis resistance (44), the transition from nonlactating to lactating states (8, 29, 34), and in response to milking frequency in dairy cows (15, 54). Similarly, the immune-related upstream regulator TNF has recently been reported as photoperiod responsive by McFarlane and coworkers (40). In mice, SD photoperiod affects TNF gene expression in the testes, which promotes angiogenesis (48). The expression of IFNG, another immune-related upstream regulator, is responsive to melatonin in human peripheral blood mononuclear cells (27) and is associated with circadian release of hormones from the pituitary gland (11).
Prolactin secretion may further mediate the effects of photoperiod on lactation (4, 16, 17). Calendar cells of the pars tuberalis of the pituitary gland regulate the secretion of prolactin to align with seasonal changes in day length (37); specifically, LD photoperiod increases and SD photoperiod decreases prolactin secretion. In dairy cows, the effects of photoperiod manipulation on immune function are also regulated by prolactin secretion (21, 37). Taken together, genes and immune-related upstream regulators may be part of a mechanism underlying the effects of photoperiod on immune function and lactation in the mammary gland.
The potential for common mechanisms between mammary immune function and lactation provides logical explanations for the effects of photoperiod in the mammary gland. However, we suggest that many of these genes have as-yet undescribed mammary-specific functions, in addition to their immune-related functions listed in the current annotation. Study of these genes and their upstream regulators, in the context of the mammary gland, may clarify their importance in mammary development and lactation as well as the roles they play in the response of the mammary gland to photoperiod manipulation.
Time relative to parturition affects mammary gene expression.
The comparison of gene expression on day −9 to day −24 serves to both validate our method of detecting differentially expressed genes and provide insight into similarities in the mechanisms involved in the mammary response to photoperiod and the initiation of lactation. Of the 125 mapped genes we identified, 45 were associated with lactation promoting biofunctions and were common with the findings of other lactation-related transcriptional studies of the mammary gland (25, 33). These include α-lactalbumin (LALBA), which is the regulatory element of lactose synthase, aquaporin (AQP1), lactoperoxidase (LPO), and 5′-nucleotidase, ecto (NT5E), all of which are established markers of mammary function. Our present dataset also overlaps with our previously reported changes in gene expression in response to milking frequency, which include complement component 3 (C3), mucin 1 (MUC1), chemokine ligand 10 (CXCL10), NT5E, myostatin (MSTN), and chitinase 3-like 1 (CHI3L-1) (54). The commonality between these datasets supports the conclusion that the genes we have identified in the effect of time are indicative of the onset of lactation. Furthermore, the consistent effects of external stimuli on expression of these genes suggest that they play an essential role in modulating mammary gland development and function. These genes may be markers of mammary function and as such could serve as targets for future efforts to maximize milk production.
Genes differentially expressed in response to time were also associated with immune function and inflammation. These genes enriched immune-related biofunctions including migration of granulocytes, adhesion of immune cells, stimulation of leukocytes, as well as immune-related canonical pathways including acute phase response and IL-10 signaling. However, classification of these genes and upstream regulators as “immune” may obscure what are actually mammary-specific functions. The products of several of these genes are found in milk, including acute phase response proteins, complement factors, LALBA, LPO, and mucins (51). As milk components, these proteins serve both nutritional and protective functions for neonates (51) but may function in regulation of colostrogenesis and/or lactogenesis. In goats, changes in the mammary transcriptome during late pregnancy (day 90 vs. day 110), which the authors proposed as essential for functional differentiation of mammary secretory cells (24), correspond to immune response signatures in our dataset. Others have noted the expression of acute phase response proteins in the mammary gland of healthy cows and mice, respectively (10, 49), supporting our supposition that these immune factors may have alternate functions in the mammary gland.
Overall, the genes and upstream regulators identified as responsive to photoperiod were not the same as those identified by time relative to parturition, suggesting these effectors operate through distinct mechanisms (Fig. 2). In addition, photoperiod did not enrich the same lactation-supportive biofunctions that responded to time, which may indicate that LD and SD photoperiod do not directly influence the metabolic pathways associated with the onset of lactation. There is, however, commonality in the immune signature associated with both sets of differentially expressed genes, further suggesting genes annotated as immune related may have mammary-specific functions that support lactation.
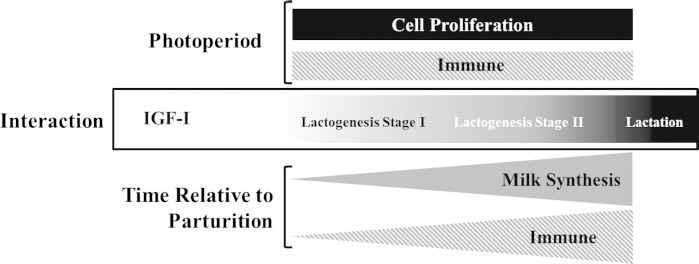
Fig. 2.
The effects of photoperiod, time, and the interaction of photoperiod and time on the mammary transcriptome during the dry period. Photoperiod exposure during the dry period affects genes associated with cell proliferation and immune function during lactogenesis stage I and lactogenesis stage II. Time relative to parturition affected genes associated with milk synthesis and immune function, with expression increasing as lactation is approached. Functional analysis of differentially expressed genes in the interaction of photoperiod and time indicates that the genes associated with the IGF-I signaling pathway may mediate the effects of photoperiod on mammary function.
Interdependent effect of time and photoperiod on mammary gene expression.
Genes identified in the interaction represent those that were differently affected by photoperiod depending on time relative to parturition. This may indicate that photoperiod differentially affects genes important in stage I or stage II of lactogenesis and/or that photoperiod may alter the timing of mammary development. Although this analysis was inherently less robust than that of the main effects (due to lower sample size), it proved useful for identifying functional pathways not significantly enriched by the smaller datasets generated in the main effects of photoperiod and time.
The IGF-I signaling pathway was the only canonical pathway significantly enriched by genes differentially expressed in the interaction of photoperiod treatment and time. IGF-I stimulates growth and proliferation of mammary secretory cells, in response to estrogen stimulation (35), in preparation for milk secretion (3) and has been investigated as a potential mediator of photoperiodic effects on the mammary gland (19, 21, 53). We report that the interaction of photoperiod and time affected expression of 32 genes associated with IGF-I, including growth hormone receptor (GHR), IGF-I receptor (IGFIR), and IGF-I binding proteins. Differential expression of these genes in the interaction indicates the effects of photoperiod on IGF-I signaling differ by time relative to parturition. We interpret this to mean that the physiological state of the gland influences its response to photoperiod. Because Auchtung and coworkers (4) reported an increase in milk production in cows exposed to SD photoperiod during the dry period, we suggest that IGF-I signaling may, in part, mediate the effects of SD photoperiod on enhanced mammary development and function in the ensuing lactation (Fig. 2).
In conclusion, photoperiod manipulation during the dry period induces differential expression of the mammary transcriptome in dairy cows. Photoperiod-responsive genes were associated with mammary development and immune function consistent with the enhancement of milk production in the ensuing lactation. Furthermore, we propose these genes may have mammary-specific functional roles, in addition to their immune-related annotation. Overall, the transcriptomic signatures of photoperiod and time were distinct, suggesting these effectors utilize different mechanisms of action. Molecular signatures identified in the interaction of photoperiod and time implicate IGF-I signaling as a potential mediator of the physiological changes in the mammary gland in response to photoperiod. Further study of gene targets identified here may elucidate mammary-specific gene functions and will ultimately expand our understanding of photoperiodic effects on mammary development and function.
GRANTS
Funding for this research was provided by the National Research Initiative Competitive Grant no. 2007-35206-17983 from the USDA Cooperative State Research, Education, and Extension Service; the Alberta Livestock and Meat Agency; and the Natural Sciences and Engineering Research Council of Canada. Microarray analyses were partially supported by the Vermont Genetics Network through Grant Number P20 RR-16462 from the INBRE Program of the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH). The contents are solely the responsibility of the authors and do not necessarily represent the official views of NCRR or NIH.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
Author contributions: P.A.B., E.H.W., and T.B.M. performed experiments; P.A.B., E.H.W., and T.B.M. analyzed data; P.A.B., E.H.W., and T.B.M. interpreted results of experiments; P.A.B. prepared figures; P.A.B. drafted manuscript; P.A.B., E.H.W., and T.B.M. edited and revised manuscript; P.A.B., E.H.W., G.E.D., and T.B.M. approved final version of manuscript; G.E.D. and T.B.M. conception and design of research.
Supplementary Material
ACKNOWLEDGMENTS
Current affiliation of E. H. Wall: Department of R&D, Pancosma, SA.
The authors thank Scott Tighe and Tim Hunter for assistance with the microarray experiment, Dr. Jeffery Bond for statistical support, Dr. Gina Oliver for discussions of microarray data and analysis, and Dr. Tera Montgomery and the University of Illinois dairy farm staff for contributions to this study.
Footnotes
The online version of this article contains supplemental material.
REFERENCES
- 1.Abribat T, Lapierre H, Dubreuil P, Pelletier G, Gaudreau P, Brazeau P, Petitclerc D. Insulin-like growth factor-I concentration in Holstein female cattle: variations with age, stage of lactation and growth hormone-releasing factor administration. Domest Anim Endocrinol 7: 93–102, 1990. [DOI] [PubMed] [Google Scholar]
- 2.Affymetrix. GeneChip Expression Analysis Technical Manual 2005–2006.
- 3.Akers RM, Ellis SE, Berry SD. Ovarian and IGF-I axis control of mammary development in prepubertal heifers. Domest Anim Endocrinol 29: 259–267, 2005. [DOI] [PubMed] [Google Scholar]
- 4.Auchtung TL, Rius AG, Kendall PE, McFadden TB, Dahl GE. Effects of photoperiod during the dry period on prolactin, prolactin receptor, and milk production of dairy cows. J Dairy Sci 88: 121–127, 2005. [DOI] [PubMed] [Google Scholar]
- 5.Auchtung TL, Salak-Johnson JL, Morin DE, Mallard CC, Dahl GE. Effects of photoperiod during the dry period on cellular immune function of dairy cows. J Dairy Sci 87: 3683–3689, 2004. [DOI] [PubMed] [Google Scholar]
- 6.Benjamini Y, Hochberg Y. Controlling the false discovery rate - a practical and powerful approach to multiple testing. J Roy Stat Soc Ser B 57: 289–300, 1995. [Google Scholar]
- 7.Bionaz M, Loor JJ. Gene networks driving bovine milk fat synthesis during the lactation cycle. BMC Genomics 9: 366, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bionaz M, Periasamy K, Rodriguez-Zas SL, Everts RE, Lewin HA, Hurley WL, Loor JJ. Old and new stories: revelations from functional analysis of the bovine mammary transcriptome during the lactation cycle. PLoS One 7: e33268, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19: 185–193, 2003. [DOI] [PubMed] [Google Scholar]
- 10.Britti D, Peli A, Massimini G, Polci A, Luciani A, Famigli-Bergamini P. Evaluation of TNF-alpha, IL-8 and IL-10 transcriptional activity in milk from healthy dairy cows during lactation period. Vet Res Commun 29, Suppl 2: 281–284, 2005. [DOI] [PubMed] [Google Scholar]
- 11.Cano P, Cardinali DP, Jimenez V, Alvarez MP, Cutrera RA, Esquifino AI. Effect of interferon-gamma treatment on 24-hour variations in plasma ACTH, growth hormone, prolactin, luteinizing hormone and follicle-stimulating hormone of male rats. Neuroimmunomodulation 12: 146–151, 2005. [DOI] [PubMed] [Google Scholar]
- 12.Capuco AV, Wood DL, Baldwin R, McLeod K, Paape MJ. Mammary cell number, proliferation, and apoptosis during a bovine lactation: relation to milk production and effect of bST. J Dairy Sci 84: 2177–2187, 2001. [DOI] [PubMed] [Google Scholar]
- 13.Casey T, Dover H, Liesman J, DeVries L, Kiupel M, Vandehaar M, Plaut K. Transcriptome analysis of epithelial and stromal contributions to mammogenesis in three week prepartum cows. PLoS One 6: e22541, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Collier RJ, Stiening CM, Pollard BC, VanBaale MJ, Baumgard LH, Gentry PC, Coussens PM. Use of gene expression microarrays for evaluating environmental stress tolerance at the cellular level in cattle. J Anim Sci 84, Suppl: E1–E13, 2006. [DOI] [PubMed] [Google Scholar]
- 15.Connor EE, Siferd S, Elsasser TH, Evock-Clover CM, Van Tassell CP, Sonstegard TS, Fernandes VM, Capuco AV. Effects of increased milking frequency on gene expression in the bovine mammary gland. BMC Genomics 9: 362, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crawford HM, Dauderman JL, Morin DE, McFadden TB, Dahl GE. Evidence of a role of prolactin in mediating photoperiodic effects during the dry period. J Dairy Sci 88: 363, 2005. [Google Scholar]
- 17.Dahl GE. Effects of short day photoperiod on prolactin signaling in dry cows: a common mechanism among tissues and environments? J Anim Sci 86: 10–14, 2008. [DOI] [PubMed] [Google Scholar]
- 18.Dahl GE, Buchanan BA, Tucker HA. Photoperiodic effects on dairy cattle: a review. J Dairy Sci 83: 885–893, 2000. [DOI] [PubMed] [Google Scholar]
- 19.Dahl GE, Elsasser TH, Capuco AV, Erdman RA, Peters RR. Effects of a long daily photoperiod on milk yield and circulating concentrations of insulin-like growth factor-1. J Dairy Sci 80: 2784–2789, 1997. [DOI] [PubMed] [Google Scholar]
- 21.Dahl GE, Tao S, Thompson IM. Lactation Biology Symposium: effects of photoperiod on mammary gland development and lactation. J Anim Sci 90: 755–760, 2012. [DOI] [PubMed] [Google Scholar]
- 22.Das UN. Acetylcholinesterase and butyrylcholinesterase as possible markers of low-grade systemic inflammation. Med Sci Monit 13: RA214–RA221, 2007. [PubMed] [Google Scholar]
- 23.Farr VC, Stelwagen K, Cate LR, Molenaar AJ, McFadden TB, Davis SR. An improved method for the routine biopsy of bovine mammary tissue. J Dairy Sci 79: 543–549, 1996. [DOI] [PubMed] [Google Scholar]
- 24.Faucon F, Rebours E, Bevilacqua C, Helbling JC, Aubert J, Makhzami S, Dhorne-Pollet S, Robin S, Martin P. Terminal differentiation of goat mammary tissue during pregnancy requires the expression of genes involved in immune functions. Physiol Genomics 40: 61–82, 2009. [DOI] [PubMed] [Google Scholar]
- 25.Finucane KA, McFadden TB, Bond JP, Kennelly JJ, Zhao FQ. Onset of lactation in the bovine mammary gland: gene expression profiling indicates a strong inhibition of gene expression in cell proliferation. Funct Integr Genomics 8: 251–264, 2008. [DOI] [PubMed] [Google Scholar]
- 26.Gao Y, Lin X, Shi K, Yan Z, Wang Z. Bovine mammary gene expression profiling during the onset of lactation. PLoS ONE 8: e70393, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garcia-Maurino S, Gonzalez-Haba MG, Calvo JR, Rafii-El-Idrissi M, Sanchez-Margalet V, Goberna R, Guerrero JM. Melatonin enhances IL-2, IL-6, and IFN-gamma production by human circulating CD4+ cells: a possible nuclear receptor-mediated mechanism involving T helper type 1 lymphocytes and monocytes. J Immunol 159: 574–581, 1997. [PubMed] [Google Scholar]
- 28.Goldman AS. Evolution of the mammary gland defense system and the ontogeny of the immune system. J Mammary Gland Biol Neoplasia 7: 277–289, 2002. [DOI] [PubMed] [Google Scholar]
- 29.Hou X, Li Q, Huang T. Microarray analysis of gene expression profiles in the bovine mammary gland during lactation. Sci China Life Sci 53: 248–256, 2010. [DOI] [PubMed] [Google Scholar]
- 30.Hurley WL, Loor JJ. Mammary gland: growth, development and involution. In: Encyclopedia of Dairy Sciences (2nd Ed.), edited by Fuquay JW. San Diego: Academic Press, 2011, p. 338–345. [Google Scholar]
- 31.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res 31: e15, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Konishi T. Microarray test results should not be compensated for multiplicity of gene contents. BMC Syst Biol 5, Suppl 2: S6, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lemay DG, Lynn DJ, Martin WF, Neville MC, Casey TM, Rincon G, Kriventseva EV, Barris WC, Hinrichs AS, Molenaar AJ, Pollard KS, Maqbool NJ, Singh K, Murney R, Zdobnov EM, Tellam RL, Medrano JF, German JB, Rijnkels M. The bovine lactation genome: insights into the evolution of mammalian milk. Genome Biol 10: R43, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lemay DG, Neville MC, Rudolph MC, Pollard KS, German JB. Gene regulatory networks in lactation: identification of global principles using bioinformatics. BMC Syst Biol 1: 56, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li RW, Capuco AV. Canonical pathways and networks regulated by estrogen in the bovine mammary gland. Funct Integr Genomics 8: 55–68, 2008. [DOI] [PubMed] [Google Scholar]
- 36.Lieberman J. Granzyme A activates another way to die. Immunol Rev 235: 93–104, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lincoln GA, Andersson H, Loudon A. Clock genes in calendar cells as the basis of annual timekeeping in mammals–a unifying hypothesis. J Endocrinol 179: 1–13, 2003. [DOI] [PubMed] [Google Scholar]
- 38.Littlejohn MD, Walker CG, Ward HE, Lehnert KB, Snell RG, Verkerk GA, Spelman RJ, Clark DA, Davis SR. Effects of reduced frequency of milk removal on gene expression in the bovine mammary gland. Physiol Genomics 41: 21–32, 2010. [DOI] [PubMed] [Google Scholar]
- 39.Marshman E, Streuli CH. Insulin-like growth factors and insulin-like growth factor binding proteins in mammary gland function. Breast Cancer Res 4: 231–239, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McFarlane D, Wolf RF, McDaniel KA, White GL. The effect of season on inflammatory response in captive baboons. J Med Primatol 41: 341–348, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miller AR, Erdman RA, Douglass LW, Dahl GE. Effects of photoperiodic manipulation during the dry period of dairy cows. J Dairy Sci 83: 962–967, 2000. [DOI] [PubMed] [Google Scholar]
- 42.Nelson RJ, Demas GE. Seasonal changes in immune function. Q Rev Biol 71: 511–548, 1996. [DOI] [PubMed] [Google Scholar]
- 43.Norris AW, Kahn CR. Analysis of gene expression in pathophysiological states: balancing false discovery and false negative rates. Proc Natl Acad Sci USA 103: 649–653, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Park YH, Joo YS, Park JY, Moon JS, Kim SH, Kwon NH, Ahn JS, Davis WC, Davies CJ. Characterization of lymphocyte subpopulations and major histocompatibility complex haplotypes of mastitis-resistant and susceptible cows. J Vet Sci 5: 29–39, 2004. [PubMed] [Google Scholar]
- 45.Patterson TA, Lobenhofer EK, Fulmer-Smentek SB, Collins PJ, Chu TM, Bao W, Fang H, Kawasaki ES, Hager J, Tikhonova IR, Walker SJ, Zhang L, Hurban P, de Longueville F, Fuscoe JC, Tong W, Shi L, Wolfinger RD. Performance comparison of one-color and two-color platforms within the MicroArray Quality Control (MAQC) project. Nat Biotechnol 24: 1140–1150, 2006. [DOI] [PubMed] [Google Scholar]
- 46.Pawitan Y, Michiels S, Koscielny S, Gusnanto A, Ploner A. False discovery rate, sensitivity and sample size for microarray studies. Bioinformatics 21: 3017–3024, 2005. [DOI] [PubMed] [Google Scholar]
- 47.Prosser CG, Fleet IR, Corps AN, Froesch ER, Heap RB. Increase in milk secretion and mammary blood flow by intra-arterial infusion of insulin-like growth factor-I into the mammary gland of the goat. J Endocrinol 126: 437–443, 1990. [DOI] [PubMed] [Google Scholar]
- 48.Pyter LM, Hotchkiss AK, Nelson RJ. Photoperiod-induced differential expression of angiogenesis genes in testes of adult Peromyscus leucopus. Reproduction 129: 201–209, 2005. [DOI] [PubMed] [Google Scholar]
- 49.Stein T, Morris JS, Davies CR, Weber-Hall SJ, Duffy MA, Heath VJ, Bell AK, Ferrier RK, Sandilands GP, Gusterson BA. Involution of the mouse mammary gland is associated with an immune cascade and an acute-phase response, involving LBP, CD14 and STAT3. Breast Cancer Res 6: R75–R91, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Swanson KM, Stelwagen K, Dobson J, Henderson HV, Davis SR, Farr VC, Singh K. Transcriptome profiling of Streptococcus uberis-induced mastitis reveals fundamental differences between immune gene expression in the mammary gland and in a primary cell culture model. J Dairy Sci 92: 117–129, 2009. [DOI] [PubMed] [Google Scholar]
- 51.Vorbach C, Capecchi MR, Penninger JM. Evolution of the mammary gland from the innate immune system? Bioessays 28: 606–616, 2006. [DOI] [PubMed] [Google Scholar]
- 52.Wall EH, Auchtung-Montgomery TL, Dahl GE, McFadden TB. Short communication: Short-day photoperiod during the dry period decreases expression of suppressors of cytokine signaling in mammary gland of dairy cows. J Dairy Sci 88: 3145–3148, 2005. [DOI] [PubMed] [Google Scholar]
- 53.Wall EH, Auchtung TL, Dahl GE, Ellis SE, McFadden TB. Exposure to short day photoperiod during the dry period enhances mammary growth in dairy cows. J Dairy Sci 88: 1994–2003, 2005. [DOI] [PubMed] [Google Scholar]
- 54.Wall EH, Bond JP, McFadden TB. Acute milk yield response to frequent milking during early lactation is mediated by genes transiently regulated by milk removal. Physiol Genomics 44: 25–34, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Walton JC, Weil ZM, Nelson RJ. Influence of photoperiod on hormones, behavior, and immune function. Front Neuroendocrinol 32: 303–319, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wilkinson TS, Roghanian A, Simpson AJ, Sallenave JM. WAP domain proteins as modulators of mucosal immunity. Biochem Soc Trans 39: 1409–1415, 2011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.