Abstract
Ghrelin is a multifunctional orexigenic hormone with a unique acyl modification enabled by ghrelin O-acyl transferase (GOAT). Ghrelin is well-characterized in nonmammals, and GOAT sequences of several fishes are available in the GenBank. However, endogenous GOAT in non-mammals remains poorly understood. In this research, GOAT sequence comparison, tissue-specific GOAT expression, and its regulation by nutrient status and exogenous ghrelin were studied. It was found that the bioactive core of zebrafish GOAT amino acid sequence share high identity with that of mammals. GOAT mRNA was most abundant in the gut. GOAT-like immunoreactivity (i.r.) was found colocalized with ghrelin in the gastric mucosa. Food deprivation increased, and feeding decreased GOAT and preproghrelin mRNA expression in the brain and gut. GOAT and ghrelin peptides in the gut and brain showed corresponding decrease in food-deprived state. Intraperitoneal injection of acylated fish ghrelin caused a significant decrease in GOAT mRNA expression, suggesting a feedback mechanism regulating its abundance. Together, these results provide the first in-depth characterization of GOAT in a non-mammal. Our results demonstrate that endogenous GOAT expression is responsive to metabolic status and availability of acylated ghrelin, providing further evidences for GOAT in the regulation of feeding in teleosts.
Introduction
The maintenance of energy homeostasis is critical for the survival of organisms. Hormones play an integral role in energy homeostasis, especially to cope with varying availability of food and changing environmental conditions.1,2 Among hormone-producing tissues, brain (specifically the hypothalamus) plays a critical role to regulate energy homeostasis by secreting appetite-stimulating (orexigenic) and appetite-inhibiting (anorexigenic) endocrine signals.1,3,4 Peripheral organs, including the gastrointestinal tract and adipose tissue also secrete hormones that regulate energy balance. Several central and peripheral neuroendocrine tissues receive input on energy status, and respond to regulate energy intake and expenditure.
Ghrelin is a 28 amino acid acylated peptide hormone5 predominantly produced from the gastric mucosa.6 It is a natural ligand of the growth hormone secretagogue receptor7 and is found in a wide range of cells and tissues, including the brain and digestive tract.8–11 The ghrelinergic system exerts multifunctional regulatory effects in an endocrine, paracrine, and autocrine manner to modulate food intake, energy expenditure, hormone secretion, and reproduction.5,12–14 So far, ghrelin has been identified in various fishes.15–22 As in mammals,23–27 ghrelin increases food intake and promotes body weight gain in fishes.18,28–32 Similar to mammals,23,24 it was demonstrated that endogenous ghrelin levels and ghrelin-induced food intake depend on nutrient/feeding status.20,22,33–35
The unique posttranslational acylation of ghrelin in the third serine is enabled by a membrane-bound O-acyl transferase 4 (MBOAT4), renamed as the ghrelin O-acyl transferase (GOAT).36–38 Octanoylation of ghrelin occurs before proghrelin is transported to the Golgi, where it is cleaved by protein convertase to form mature ghrelin. These findings suggest that GOAT may be located in the membrane of the endoplasmic reticulum compartment and may mediate the translocation of the octanoyl-CoA from the cytosolic side to the ER lumen.37 Studies using genetically modified mice deficient of GOAT (GOAT−/−) showed that GOAT is the only enzyme capable of acylation of ghrelin in vivo.39 GOAT-mediated acylation is critical for most biological activities of ghrelin, especially its orexigenic functions.38,40 The sequence of GOAT is highly conserved across vertebrates, including fish.36,40,41
Expression of GOAT in mammals is tissue specific, and depends on a number of factors, including metabolic status, and appears to vary among species.5,40 In mammals, GOAT is mainly expressed in the stomach, and is colocalized with ghrelin in the gastric oxyntic mucosa.36,37,39,42 Sakata et al.43 observed that GOAT mRNA expression was highly correlated with ghrelin distribution, while another study did not find such a correlation.44 Several studies in mammals also confirmed that acylated ghrelin and/or GOAT mRNA increases following fasting.14,40,45 In contrast, other studies either failed to detect an increase in ghrelin or GOAT following fasting, or observed a decrease.39,46 These discrepancies among studies might be related to the time of food withdrawal, strain of animal used, time of sample collection, type of rodent chow provided, and/or the method of euthanasia.46
Zebrafish GOAT sequence information is available in the GenBank, and was provided in one of the articles reporting its original discovery.36 Further, it was also identified that zebrafish GOAT octanoylates human ghrelin.36 This aside, there are no information on endogenous GOAT in non-mammals. The tissue abundance of GOAT, its cellular localization in the gut, and effects of calorie restriction and exogenous ghrelin on gastric GOAT mRNA are currently unknown. The main objective of this research was to obtain a deeper understanding beyond the sequence information on GOAT in fish.
We investigated the tissue distribution and abundance of GOAT mRNA expression in zebrafish tissues. In addition, we used fluorescence immunohistochemistry to localize GOAT and ghrelin in zebrafish gut. We also examined the periprandial expression of preproghrelin and GOAT mRNAs, and the effects of chronic food deprivation on GOAT mRNA in the brain and gut of zebrafish. Western blot analysis was used to elucidate preproghrelin and GOAT protein abundance in the gut after food deprivation. In addition, the effect of intraperitoneally injected acylated ghrelin on preproghrelin and GOAT mRNA expression in the gut was also determined. The major novel findings of this research indicate that GOAT expression in zebrafish corresponds to preproghrelin profile in the gut, and that food deprivation upregulates, while exogenous acylated ghrelin suppresses both GOAT and preproghrelin.
Materials and Methods
Animals
All in vivo experimental protocols strictly adhered to the national guidelines provided by the Canadian Council for Animal Care, and were approved by the University of Saskatchewan Animal Research Ethics Board. Adult male and female zebrafish (Danio rerio; age: ∼6 months; length: 2–3 cm; and body weight: 1–1.5 g) were purchased from Aquatic Imports (Calgary, Alberta, Canada) and kept on a 14-h light–10-h dark cycle at 28°C. All fish were fed once a day with a commercial flake diet (Nutrafin Max, Rolf C. Hagen, Inc.) at 12:00 p.m.
Structural and phylogenetic analysis of GOAT sequences
GOAT nucleotide and amino acid sequences of organisms assessed in this study were obtained from the NCBI GenBank (www.ncbi.nim.nih.gov). Multiple sequence alignments were conducted using Clustal Omega (www.ebi.ac.uk). Phylogenetic tree based on amino acid sequences was constructed by the Montpellier Laboratory of Informatics, Robotics, and Microelectronics (www.phylogeny.fr/version2_cgi/simple _phylogeny.cgi).
Tissue distribution of GOAT mRNA
To investigate the tissue distribution of GOAT mRNA, brain, gut (J-loop/anterior intestine), liver, heart, eye, muscle, gill, ovary, testes, and skin were sampled from zebrafish. Fish were anesthetized in 0.015% tricaine methanesulfonate (Syndel Laboratories, Vancouver, British Columbia, Canada) before dissection and sampling of tissues. Total RNA was extracted from each tissue using TRIzol™ reagent (Invitrogen Canada, Inc., Toronto, Ontario, Canada) according to the manufacturer's instructions. The concentration of total RNA was estimated from absorbance at 260 nm (A260 nm, Nanodrop2000), and RNA quality was verified by A260 nm/A280 nm ratio (>1.8) and A230/A260 nm ratio (>2). Reverse transcription (RT) was performed using an iScript cDNA Synthesis Kit (Bio-Rad Laboratories, Mississauga, Ontario, Canada) following the manufacturer's instructions.
Real-time quantitative polymerase chain reaction (RT-qPCR) of GOAT was performed on a CFX Connect (Bio-Rad) using iQ SYBR Green Supermix (Bio-Rad). The thermal profile for all reactions was 3 min at 95°C and then 40 cycles of 10 s at 95°C, followed by 30 s at 53°C. The specificity of the amplified product in the RT-qPCR assay was determined by analyzing the melting curve to discriminate target amplicon from primer dimer and other nonspecific products. A single melt curve was observed for each primer set in all RT-qPCR. Each sample was run as duplicates, and the mean threshold cycles (as determined by the linear portion of the fluorescence absorbance curve) were used for the final calculation. Beta (β)-actin (tissue distribution) and eukaryotic elongation factor 2 alpha 1 (eef2a1, for all other mRNA quantification studies) primers served as internal controls to normalize cDNA quantity for each sample. The RT-qPCR data were obtained as CT values, and analyzed using the comparative Ct method (2−ΔΔCt).47 Primers used in this study are provided in Table 1.
Table 1.
Sequences of GOAT, eef2a1, and β-Actin Primers Used for RT and RT-qPCR
| Amplicon | Forward (5′→3′) | Reverse (5′→3′) | Annealing (°C) | Reference |
|---|---|---|---|---|
| GOAT | CACCCTCAGCTGTTTTACCA | GAATCCTCCCATCGCCAAAT | 53 | NM_001122944.1 |
| Ghrelin | TCCTTGTGTCTCGAGTCTGTG | TGGACAGTTCAAATGGAGCG | 53 | NM_001083872.1 |
| eef2a1 | CCTGCATTCCCCTCAAGAAA | TGGCAAGGCACAATTGTTTG | 58 | NM_001045161.2 |
| β-Actin | TTCAAACGAACGACCAACCT | TTCCGCATCCTGAGTCAATG | 58 | NM_131031.1 |
eef2a1, eukaryotic elongation factor 2 alpha 1; GOAT, ghrelin O-acyl transferase; RT, reverse transcription; RT-qPCR, real time, quantitative polymerase chain reaction.
Once the RT-qPCRs were completed, the resulting amplification products underwent electrophoresis on 2% gels prepared using 100 mL TAE buffer, 2 g agarose, and 5 μL ethidium bromide. Gel was run for 1 h at 120 V. A 1 kb ladder (Invitrogen) was run adjacent to samples to determine the size of the resulting band. The gel was then imaged using a Gel Doc™ EZ imager (Bio-Rad).
GOAT and ghrelin-like immunoreactivity in zebrafish J-loop
Zebrafish gut (J-loop) tissues were collected in 4% paraformaldehyde (PFA) for 24 h at 4°C. PFA was replaced and tissues washed thrice with 70% ethanol and tissues were stored in at 4°C. Tissues were processed (dehydrated, embedded in paraffin), at the Pathology Core Facility of the Center for Modeling Human Disease, Toronto Center for Phenogenomics. Paraffin sections of 5 μm thickness were prepared for immunostaining. These sections were deparaffinized with xylene (incubated twice in 100% xylene; 5 min, 25°C) and rehydrated in a graded ethanol series (incubated twice in 100% ethanol, and once each in 95% ethanol, 70% ethanol, 50% ethanol for 2 min at room temperature). The slides were rinsed in distilled water and washed in phosphate-buffered saline (PBS) and Kodak Photo-Flo for 10 min followed by an additional 10 min in PBS and Triton X-100. The sections were then blocked with serum-free protein block reagent (DAKO® Corporation) for 10 min before being incubated overnight in a mixture of rabbit anti-GOAT (targeted against human GOAT amino acids 356–375) (Catalog No. H-032-12; Phoenix Pharmaceuticals) diluted 1:150 and mouse monoclonal to ghrelin (immunogen is the full-length human preproghrelin peptide of 1–118 amino acids, Catalog No. ab57222; Abcam) diluted 1:200 at 4°C. Since heterologous antibodies were used here, it is likely that a certain degree of non-specificity exists in our findings. Therefore, GOAT-like and ghrelin-like, instead of GOAT and ghrelin, were used to refer to immunostaining obtained in this study.
The slides were then washed three times in PBS, and incubated at 37°C for 1 h with the following secondary antibodies: goat pAb to Ms IgG (1:200 dilution, FITC; Vector Laboratories) for the ghrelin primary antibody, and anti-rabbit IgG made in goat (1:100 dilution; Texas Red; Vector Laboratories) for the GOAT primary antibody. All primary and secondary antibodies were diluted in antibody diluent reagent (DakoCytomation®). Slides were rinsed in distilled water and washed in a solution of PBS and Kodak Photo-Flo, Triton X-100 each for 10 min and finally three times in distilled water. Slides were then mounted using VECTASHIELD ® Mounting Medium containing the DAPI nuclear fluorescent dye (Blue; Vector Laboratories). Slides were then imaged using a Nikon Eclipse Ti-Inverted fluorescent microscope (Nikon Canada) and images were captured using a Nikon DS-QI1MC cooled monochrome camera connected to a Dell HP Workstation computer and NIS-Elements Basic Research Imaging Software (Nikon Canada). Fourteen slides (from four fish), each containing eight sections were stained using the above protocol and analyzed. Only representative images of gut staining for GOAT, ghrelin, and colocalization of GOAT and ghrelin are shown. For the quantification of immunopositive cells, first, the total number of cells immunoreactive for GOAT alone (red), ghrelin alone (green), or colocalizing both ghrelin and GOAT (yellow) were counted in all sections assessed. To calculate the percentage distribution, the number of cells under each category (GOAT/ghrelin/GOAT+ghrelin positive) of staining was divided by the total number of immunoreactive cells (GOAT/ghrelin). The result was multiplied by 100 to obtain the percentage population of cells.
Periprandial changes in GOAT and preproghrelin mRNAs in the brain and gut
To examine the pre- and postprandial changes of GOAT and preproghrelin mRNAs in the gut and brain, zebrafish were placed in seven aquaria (n=6 adult zebrafish/tank). Fish were acclimated to the aquaria conditions and scheduled feeding time at 12 p.m. for 2 weeks before the experiments. On the day of the study, gut and brain were collected at 3 h (−3 h, 9 a.m., Aquarium No. 1) and at 1 h (−1 h, 11 a.m., Aquarium No. 2) before the regular feeding time. Zebrafish of Aquarium No. 3 were sampled just after feeding (0 h, 12 p.m.). Zebrafish of Aquaria Nos. 4 and 5 were fed at the regular feeding time, and sampled at 1 (fed +1 h, 1 p.m.) and 3 h (fed +3 h, 3 p.m.) postfeeding schedule, respectively. Two groups of zebrafish remained unfed at the regular feeding time, served as the unfed control and were sampled at 1 h (unfed +1 h, 1 p.m., Aquarium No. 6) and 3 h (unfed +3 h, 3 p.m., Aquarium No. 7). GOAT and preproghrelin mRNA expression was shown as a percentage of expression at −3 h. Total RNA extraction, cDNA synthesis, and RT-qPCRs for GOAT and preproghrelin were conducted employing PCR conditions described earlier in this article. Primers used for preproghrelin mRNA quantification are listed in Table 1. We normalized the data using both β-actin and eef2a1 mRNA expression. Since eef2a1 provided more stable Ct values compared to β-actin, GOAT and preproghrelin mRNA expression normalized to eef2a1 is provided.
Food deprivation and changes in GOAT mRNA in the brain and gut
In this study, we examined the effects of calorie availability/unavailability on GOAT mRNA expression in the brain and anterior intestine of zebrafish during food deprivation over a 7-day period. Adult zebrafish were acclimated as outlined in the periprandial experiment. On the day the study commenced, two groups of fish stopped receiving food (food-deprived group), while two other groups (fed group) continued to have access to food at the regular feeding time (12 p.m.). On days 3 and 7 of the experiment, one fed group (at 2 h postfeeding, 2 p.m.) and one food-deprived group were euthanized. Extraction of total RNA, synthesis of cDNA and RT-qPCR, and normalization of results using internal control genes were performed as explained earlier.
Food deprivation and changes in GOAT and preproghrelin peptides in the brain and gut
In a separate study, distinct from the one discussed above, adult zebrafish were acclimated as outlined in the previous experiment. On the day of the experiment, one group of fish stopped receiving food (food-deprived group), and the other group (fed group) continued to have access to food at the regular feeding time (12 p.m.). On day 3, 2 h post-regular feeding time (2 p.m.), fish were dissected and guts were frozen in liquid nitrogen for western blot analysis. Gut tissues were homogenized in T-PER® Tissue Protein Extraction Reagent (#78510; Thermo Scientific, Mississauga, Ontario, Canada) followed by measurement of protein concentration by Bradford assay. Samples were prepared in 1×Laemmli buffer containing 5% 2-mercaptoethanol (#161-0737 and 161-0710; Bio-Rad), and subsequently boiled at 95°C for 5 min followed by vortexing. The whole sample (20 μL), each containing 50 μg protein was loaded and run on a Mini-PROTEAN® TGX™ 4%–16% gradient gel (#456-1096; Bio-Rad) at 200 V for 30 min. After separation, the proteins were transferred to a 0.2 μm BioTrace™ nitrocellulose membrane (#27377-000; PALL Life Sciences, Mississauga, Ontario, Canada) by Trans-Blot® Turbo™ Transfer Starter System (170-4155; Bio-Rad) with mix molecular weight program. Membrane was blocked in 1×RapidBlock™ solution (#M325; aMReSCO). After blocking, the membranes were incubated in the primary antibody solution: ghrelin antibody diluted 1:1000, rabbit anti-GOAT diluted 1:500 (same antibodies used for immunohistochemistry), and rabbit monoclonal to vinculin (Catalog No. ab129002; Abcam, Toronto, Ontario, Canada) diluted 1:1000. All antibodies were diluted in blocking buffer, and membranes were incubated overnight at 4°C.
Membranes were then washed three times with TBS and incubated with the secondary antibody solution: goat anti-rabbit IgG (H+L) HRP conjugate (#170-6515; Bio-Rad) diluted 1:3000 was used for GOAT and vinculin and goat anti-mouse IgG (H+L) HRP conjugate (#171-1011; Bio-Rad) diluted 1:3000 was used for ghrelin. After 1 h, membranes were washed three times before color development. For protein visualization, the membrane was incubated for 5 min in Clarity™ Western ECL substrate (#170-5061; Bio-Rad) and imaged using ChemiDoc™ MP imaging system (#170-8280; Bio-Rad) with chemiluminescence detection. Membrane stripping in between protein detection was conducted using Restore™ PLUS Western Blot Stripping Buffer (#46430; Thermo Scientific). Precision Plus Protein™ Dual Xtra standards (#161-0377; Bio-Rad) were used as molecular weight markers. The expected size of zebrafish GOAT was approximately 50 kDa, meanwhile zebrafish preproghrelin was expected at 16 kDa.
Effects of intraperitoneal injection of ghrelin on GOAT and preproghrelin mRNA expression in the brain and gut
In this study, we examined the effects of exogenous ghrelin on the expression of GOAT and preproghrelin mRNAs in the brain and anterior intestine of zebrafish 1 h before and after regular feeding time (10 a.m.). Adult zebrafish were acclimated as outlined for the previous experiment. On the day the study commenced, two groups of fish were anesthetized 1 h before regular feeding time (9 a.m.) as described previously and 10 μL of physiological saline or 10 μL of physiological saline containing the 100 ng/g BW synthetic goldfish acylated ghrelin was injected into the peritoneal cavity using a microsyringe (Hamilton). Fish were then immediately placed back into the aquarium and allowed to recover from anesthesia. Normally the fish recovered within 1 min. One hour after injection, fish were euthanized and brain and gut were collected. One hour after feeding at the regular feeding time (11 a.m.), two other groups of fish were injected with saline or ghrelin as described before. An hour post-injection, fish were euthanized and brain and gut were collected. Extraction of total RNA, synthesis of cDNA, and RT-qPCR were performed as explained above.
Statistical analyses
Statistical significance was determined by t-test (when two groups were compared) or ANOVA (when more than two groups were compared) followed by Tukey's test at p<0.05. Homogeneity of variance and normal distribution of data were examined using Levene's test and Kolmogorov–Smirnov test, respectively. Data were log transformed to meet assumptions of normality and homoscedasticity when it was required. All data are expressed as mean+SEM. All statistical analyses were performed using the SPSS 19.0 software package.
Results
Comparison of zebrafish GOAT amino acid sequence to the other fishes and mammals
The objective of this analysis was to identify the sequence similarity of GOAT in vertebrates, especially fish. Zebrafish GOAT sequence exhibited identical amino acid composition to that of other fish, including Mexican tetra (Astyanax mexicanus, Characidae, 54%), rainbow trout (Oncorhynchus mykiss, 47%), bicolor damselfish (Stegastes partitus, 44%), cichlid (Pundamilia nyererei, Cichlidae, 43%), tilapia (Oreochromis niloticus, Cichlidae, 43%), chimaera (Callorhinchus milii, 41%), spotted gar (Lepisosteus oculatus, Lepisosteidae, 47%), and African coelacanth (Latimeria chalumnae, Latimeriidae, 39%) (Fig. 1). Zebrafish GOAT amino acid sequence was also highly similar to that of alligator (Alligator sinensis, 40%), mouse (Mus musculus, 39%), cow (Bos taurus, 39%), and human (Homo sapiens, 38%) (Fig. 1). Compared to human GOAT, the highest similarity was observed in African coelacanth (49%), gar (46%), and zebrafish (38%). In contrast, we found very high similarity between GOAT amino acid sequences across more evolved vertebrates. The cow (B. taurus), mouse (M. musculus), wild pig (Sus scrofa) GOAT amino acid sequences were 80%, 75%, and 75% identical to that of human GOAT amino acid sequence, respectively.
FIG. 1.
Alignment of amino acid sequences in the highly conserved catalytic regions of ghrelin O-acyl transferase (GOAT). Multiple sequence alignments were conducted using Clustal Omega (www.ebi.ac.uk). The proposed catalytic residues (asparagine and histidine) of GOAT are marked by asterisks. Partial sequences of GOAT were obtained from full-length amino acid sequences obtained from the NCBI. The name of the species is given on the left side of the alignment and the number of amino acids is provided above the alignment. The colored amino acids show the conserved regions between species. Species names and GenBank (www.ncbi.nim.nih.gov) accession nos. used in the alignment are as follows: Zebrafish (NP_001116416.1), Mexican tetra (XP_007253942.1), Rainbow trout (CDQ71181.1), Bicolor damselfish (XP_008292386.1), Cichlid (XP_005738327.1) Tilapia (XP_003455315.1), Chimaera (XP_007890232.1), Spotted gar (XP_006627115.1), African coelacanth (XP_006013871.1), Chicken (NP_001186218.1), Alligator (XP_006035341.1), Mouse (NP_001119786.1), Rat (NP_001100787.2), Elephant (XP_003412603.1), Pig (NP_001177352.1), Cow (NP_001179186.1), Sheep (AFV15801.1), Gibbon (XP_003269582.1), Human (NP_001094386.1), Chimpanzee (XP_519692.2), Dolphin (XP_004310679.1), Killer whale (XP_004277196.1), Horse (XP_001494222.2), Cat (XP_003984710.1), Dog (NP_001188260.1), Walrus (XP_004408347.1), and Giant panda (XP_002920871.1). Color images available online at www.liebertpub.com/zeb
Compared to the percentage identity of full-length GOAT amino acid sequences, the bioactive core of GOAT amino acid sequences between fish and mammals exhibited even stronger conservation. The bioactive core of GOAT amino acid sequences in Mexican tetra, spotted gar, rainbow trout, bicolor damselfish, cichlid, and tilapia were 66%, 63%, 59%, 56%, 56%, and 54% identical to zebrafish. Mammalian GOAT bioactive core has high similarity to that of zebrafish (cat 56%, horse 59%, mouse 59%, human 59%). Phylogenetic analyses revealed that zebrafish GOAT was clustered with Mexican tetra within a clade of a larger group containing rainbow trout, bicolor damselfish, tilapia, and cichlid proteins, while sequences from mammals were grouped together as another subclade (Fig. 2). Together, these results indicate that GOAT, especially its bioactive core, is very highly conserved across species.
FIG. 2.
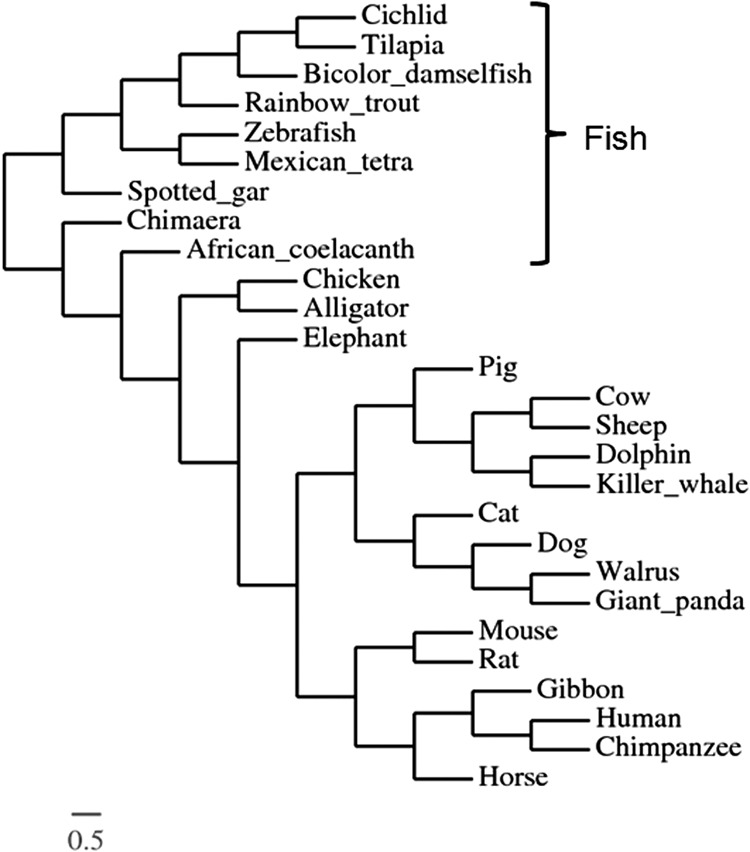
Phylogenetic analysis of GOAT amino acid sequences. A phylogenetic tree based on the amino acid sequences of GOAT was constructed by the Montpellier Laboratory of Informatics, Robotics, and Microelectronics (LIRMM) (www.phylogeny.fr/version2_cgi/simple_phylogeny.cgi). The name of the species is given on the right side of the alignment. The scale bar indicates the average number of substitutions per position (a relative measure of evolutionary distance).
Tissue distribution of GOAT mRNA in zebrafish
While zebrafish GOAT sequence was previously reported, its tissue abundance in vivo remained poorly understood. Analysis of GOAT mRNA expression using RT-PCR showed that it was expressed in the gut, ovary, brain, testis, liver, eye, heart, gill, skin, and muscle (Fig. 3A). Beta-actin served as an internal control to verify the quality and amount of samples (Fig. 3A). The highest expression of GOAT mRNA was observed in the gut, ovary, brain, and testis (Fig. 3B), followed by liver, eye, heart, gills, skin, and muscle with the lowest level of expression observed in the muscle (p<0.05, Fig. 3B). GOAT mRNA is ubiquitously expressed, and is most abundant in zebrafish gut.
FIG. 3.
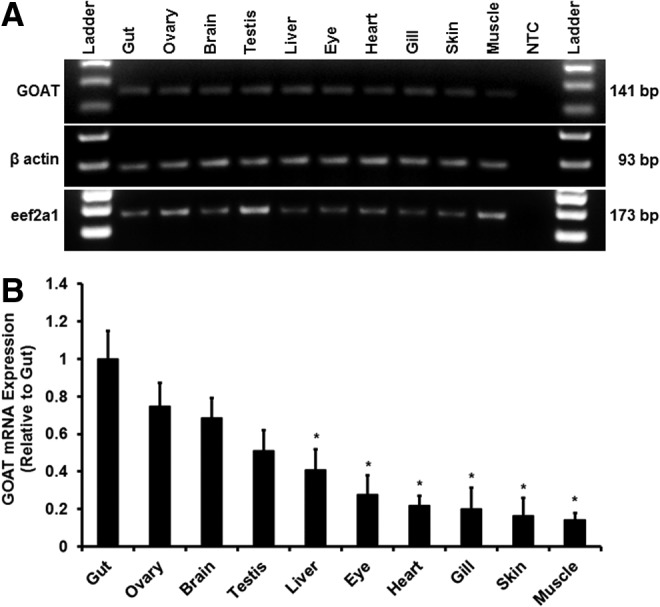
Tissue distribution of GOAT mRNA in zebrafish. (A) Agarose gel showing RT-qPCR amplicons for GOAT mRNA (expected amplicon size: 141 bp), β-actin (expected amplicon size: 93 bp), and eukaryotic elongation factor 2 alpha 1 (eef2a1) (expected amplicon size: 173 bp). Water in place of cDNA was run as a no-template negative control (NTC). Quantitative results for GOAT (B) mRNA expression in various tissues of zebrafish obtained using RT-qPCR. The results were normalized to β-actin, which served as a control to verify the quality and amount of samples. Results are expressed as relative expression levels to the tissue with the highest expression of GOAT. Error bars represent standard error of the mean. Asterisks (*) denote significant differences compared to gut (p<0.05, n=6 zebrafish).
GOAT-like and ghrelin- like immunoreactivity in the gut of zebrafish
Our findings indicate that gut is a major source of GOAT. Are gut endocrine cell sources of GOAT? If so, are they produced in gut ghrelin cells? Cross-sections of the zebrafish gut were stained for ghrelin-like (Fig. 4A, B) and GOAT-like (Fig. 4C, D) immunoreactivity (i.r.) within the cells of the gastric mucosa. GOAT immunopositive cells were scattered deep within the folds of the villi, and was found dispersed along the apical regions of the villi. Ghrelin-like i.r. was also localized on the brush border of the villi of zebrafish (Fig. 4A, B). Colocalization of ghrelin (green) and GOAT (red) was observed in some cells (Fig. 4E, F). No staining was found in negative controls stained with secondary antibody alone (Fig. 4G, H). Quantification of immunoreactive cells demonstrated that ghrelin-like i.r. cells (∼60%) are more abundant than GOAT i.r. cells (∼40%) in zebrafish gut. Among these immunoreactive cells, ∼20% colocalized both ghrelin and GOAT (Fig. 4K). Overall, a small population of gut ghrelin cells were found expressing GOAT in zebrafish.
FIG. 4.
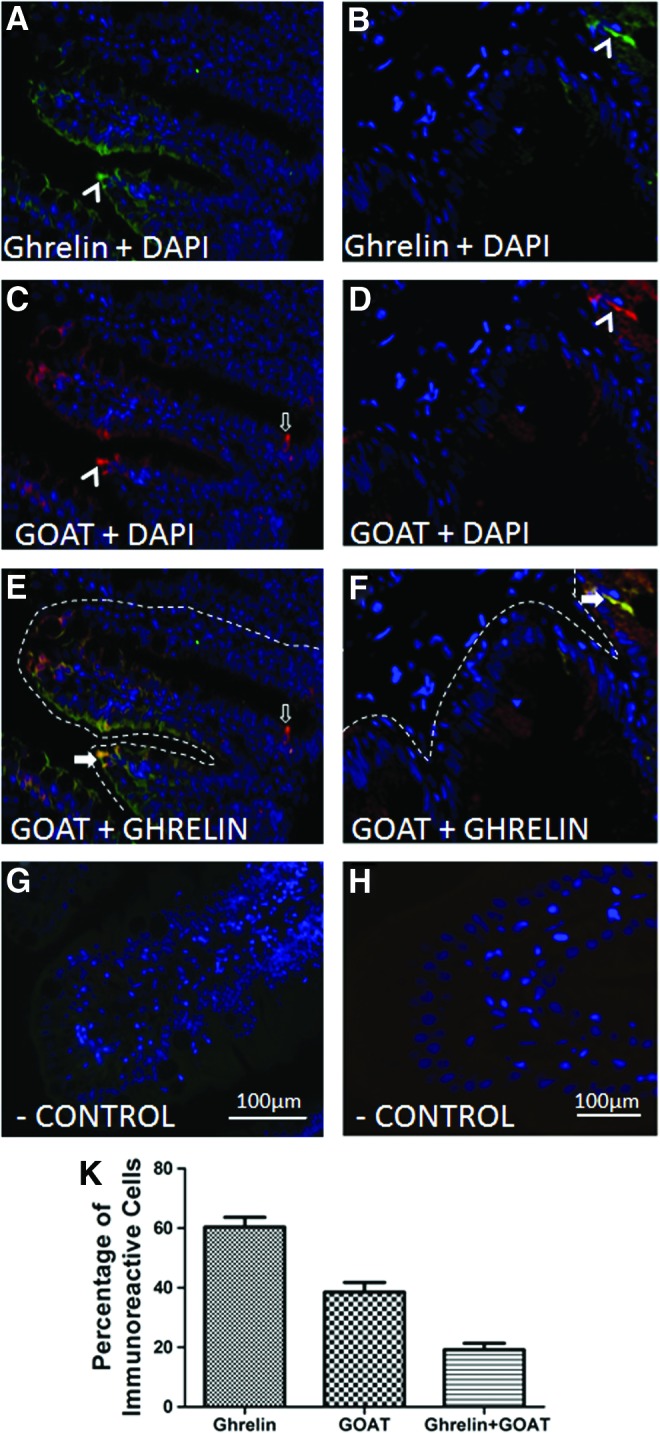
Ghrelin-like and GOAT-like immunoreactivity in zebrafish J-loop. Representative sections showing immunohistochemical staining for ghrelin (A, B, green), GOAT (C, D, red), and merged images of GOAT and ghrelin (E, F, yellow). Arrowheads in (A, D) show cells stained with either GOAT or ghrelin, empty arrow in (C) shows a cell positive for GOAT alone, and solid arrows in (E, F) show cells that colocalize both GOAT and ghrelin. (G, H) A negative control that lacks GOAT and ghrelin immunoreactivity, where slides were labeled only with secondary antibodies. All images are merged with DAPI showing nuclei in blue. Representative images were taken from multiple sections of zebrafish gut, as detailed in the Materials and Methods section. (K) Relative abundance of ghrelin, GOAT, and ghrelin+GOAT immunopositive cells in the gut of zebrafish. For details on methods employed for percentage calculation, please consult the Materials and Methods section. Color images available online at www.liebertpub.com/zeb
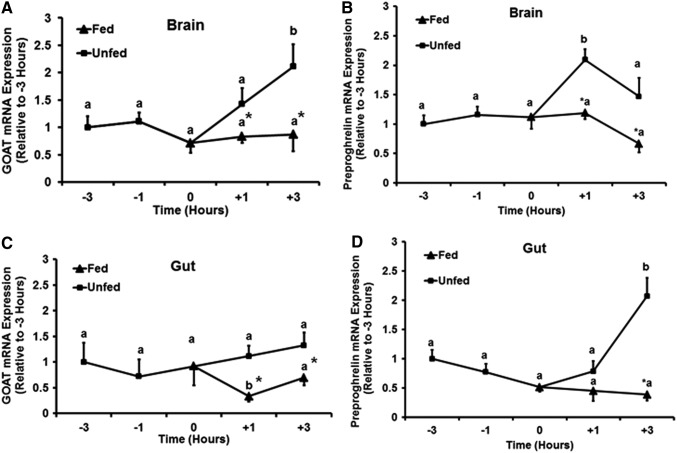
Periprandial changes of GOAT and preproghrelin mRNA in the gut and brain of zebrafish
Ghrelin is a meal regulator in fish. If so, is ghrelin and GOAT expression regulated by meal time and feeding? GOAT mRNA expression remained unchanged in the brain (Fig. 5A) and gut (Fig. 5C) of zebrafish at 1–3 h before the regular feeding time (−1 and −3 h) (p>0.05). It was significantly increased in the brain of unfed zebrafish at 3 h (+3 h) post-regular feeding time, compared to the group fed at the regular feeding time (12 p.m., 0 h) (p<0.05). There were no differences in the brain GOAT mRNA expression in zebrafish fed at the regular feeding time, and the unfed zebrafish at 1 h postfeeding time. In the gut of fed zebrafish, GOAT mRNA expression decreased at +1 and +3 h after the meal (p<0.05; Fig. 5C). Meanwhile, GOAT mRNA expression in the gut of unfed fish remained significantly higher at 1 h (+1 h) and 3 h (+3 h) following the regular feeding time (p<0.05). Preproghrelin mRNA expression remained unchanged in the brain (Fig. 5B) and gut (Fig. 5D) of zebrafish at 1–3 h before the regular feeding time (−1 and −3 h) (p>0.05). It was significantly increased in the brain of unfed zebrafish at 1 h (+1 h), and in the gut at 3 h (+3 h) post-regular feeding time compared to the group fed at the regular feeding time (12 p.m., 0 h) and fed for 1 and 3 h after regular feeding time (+1 and +3 h) (p<0.05). Both ghrelin and GOAT mRNAs were decreased in fed fish, while remained elevated in unfed fish, providing further support to its orexigenic functions.
FIG. 5.
Feeding status affects the periprandial expression of GOAT and preproghrelin mRNAs in the brain and gut of zebrafish. Pre- and postprandial expression of GOAT and preproghrelin mRNAs in the brain (A, B) and gut (C, D) of zebrafish. The expression of GOAT and preproghrelin mRNA was normalized to eef2a1, and represented relative to the expression in the −3 h group. Asterisks represent significant differences between groups at the same time point (p<0.05, n=6 zebrafish). Same letter (a) indicates no differences between time points tested, especially the preprandial sampling points, while changes observed at various times are indicated by different letters (b, c).
Food deprivation effects on GOAT mRNA expression in the gut and brain
Ghrelin was found upregulated during chronic food deprivation. We wanted to test whether GOAT that activates ghrelin also follows the same expression pattern. At day 3 post food deprivation, GOAT mRNA expression was significantly increased in the brain of unfed zebrafish compared to those of fed zebrafish (p<0.05; Fig. 6A). However, the brain GOAT mRNA expression was similar between fed and unfed zebrafish at day 7 post food deprivation (p>0.05; Fig. 6A). In the gut, GOAT mRNA was increased in unfed zebrafish compared to those of fed fish at 3 and 7 days post food deprivation (p<0.05; Fig. 6B). Similar to ghrelin expression changes found during food deprivation,22 GOAT mRNA expression also increased significantly in a tissue-specific manner.
FIG. 6.
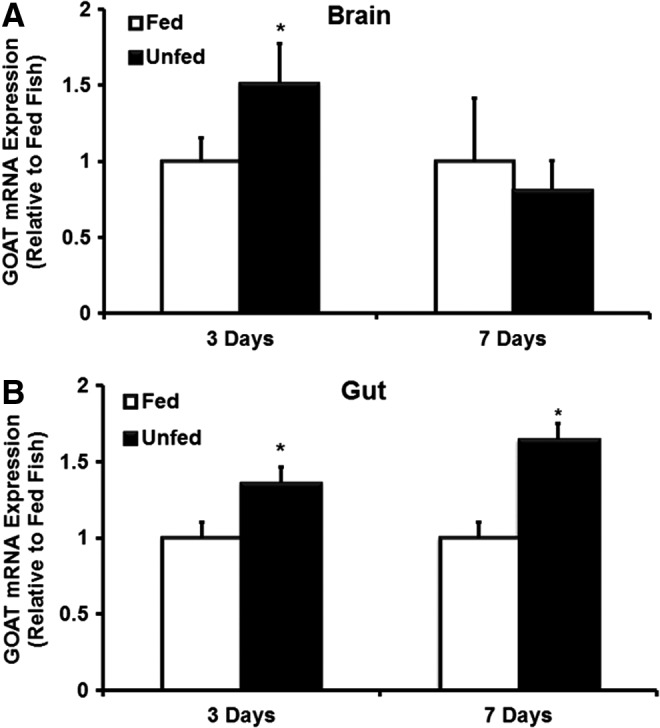
Food deprivation induced changes in GOAT mRNA expression in zebrafish brain and gut at days 3 and 7 postdeprivation. Food deprivation increased the expression of GOAT mRNA in zebrafish brain (A) and gut (B). The GOAT mRNA expression was normalized to eef2a1 and expressed relative to the fed group at each time point. Asterisks represent significant differences between groups at the same time point (p<0.05, n=6 zebrafish).
Western blot analysis of ghrelin and GOAT in the zebrafish gut
We found that ghrelin and GOAT mRNAs increased during fasting. What happens to peptides encoded by these mRNAs? Western blot analysis of zebrafish gut with mouse anti-ghrelin antibody showed full-length preproghrelin protein (16 kDa) (Fig. 7A). Ghrelin protein was significantly decreased in the gut of fed fish compared to unfed fish (Fig. 7A). A western blot using the rabbit anti-GOAT antibody demonstrated 50 kDa GOAT protein in zebrafish gut (Fig. 7B). GOAT was significantly downregulated in the gut of fed fish compared to unfed fish (Fig. 7B). While interindividual variations in band intensities were found, the normalized results clearly indicate that precursors of ghrelin and GOAT are also increased by food deprivation, supporting an orexigenic role for these peptides.
FIG. 7.
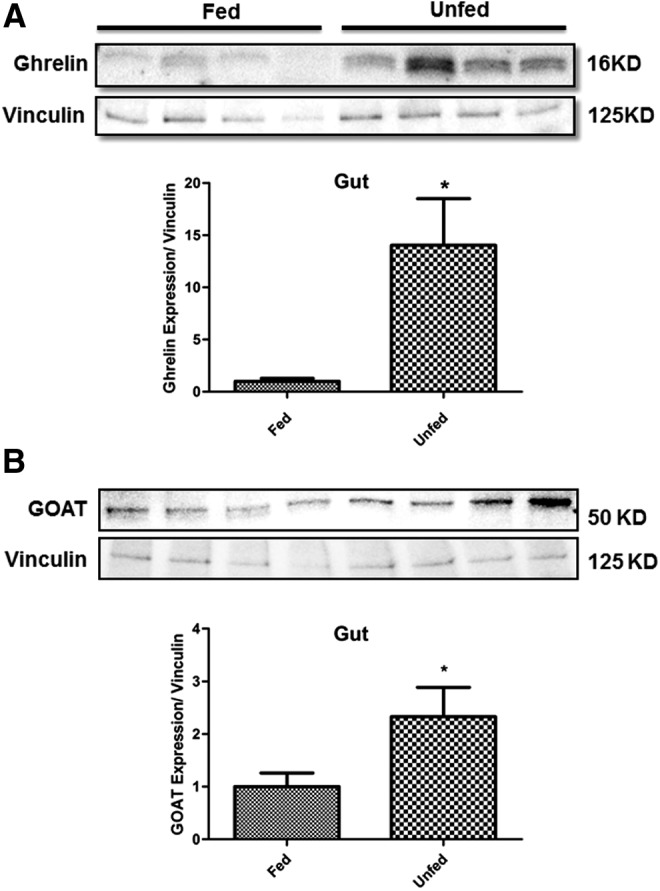
Ghrelin and GOAT precursor proteins are expressed in zebrafish gut and are affected by nutrient status. Fifty micrograms of total protein/well was loaded from the whole tissue lysates. Representative immunoblot shows bands representing preproghrelin corresponding to 16 kDa (A), GOAT corresponding to 50 kDa (B), and vinculin corresponding to 125 kDa. Average relative protein expression levels±SEM from four separate zebrafish gut per treatment group. Asterisk (*) indicates significant difference (p<0.05; n=4 zebrafish).
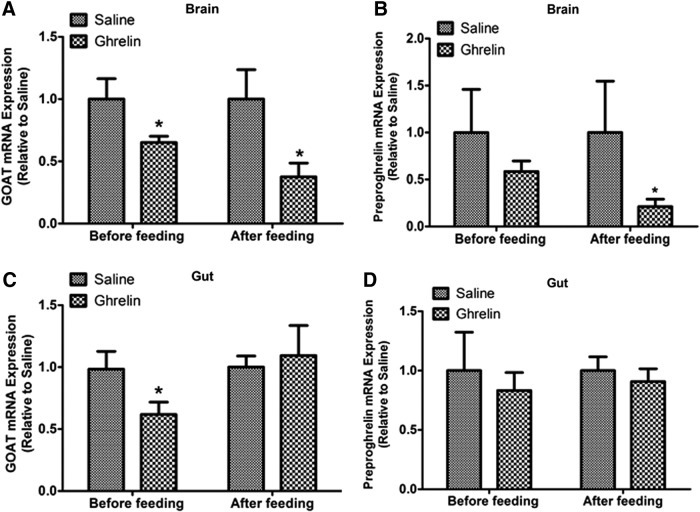
Effects of intraperitoneal injection of ghrelin on GOAT and preproghrelin mRNA expression in the brain and gut
To date, there are no reports on the effects of exogenous acylated ghrelin on endogenous ghrelin and GOAT. In this study, we tested the outcomes of increasing endogenous ghrelin levels by acute administration. Intraperitoneal (IP) injection of 100 ng/g BW of acylated ghrelin before feeding significantly decreased GOAT mRNA expression in the brain (Fig. 8A) and gut (Fig. 8C) compared to the saline-treated controls. Similar suppression of GOAT in the brain was also found in the group of fed zebrafish injected with ghrelin (Fig. 8A), while no such effects were found in the gut of the same fish (Fig. 8C). One hour after regular feeding time, IP administration of 100 ng/g BW of ghrelin significantly decreased brain preproghrelin mRNA expression in ghrelin-injected group compared to the saline-treated controls (Fig. 8B). Gut preproghrelin mRNA expression did not show any significant differences between saline- and ghrelin-treated group (Fig. 8D). An increase in acylated ghrelin causes a reduction in both ghrelin and GOAT, possibly due to a feedback inhibition.
FIG. 8.
Effects of intraperitoneal injection of ghrelin on GOAT and preproghrelin mRNA expression in the brain and gut before and after regular feeding time. Intraperitoneal injection of ghrelin decreased expression of GOAT mRNA in zebrafish brain before and after regular feeding time (A) and zebrafish gut before regular feeding time (C). Intraperitoneal injection of ghrelin decreased expression of preproghrelin mRNA in zebrafish brain after regular feeding time (B). No changes were seen in zebrafish gut either before or after regular feeding time (D). GOAT and preproghrelin mRNA expression was normalized to eef2a1 and presented relative to the saline-injected group. Asterisks (*) represent significant differences between groups at the same time point (p<0.05, n=7 zebrafish).
Discussion
Our results indicate that GOAT amino acid sequence, especially the bioactive core is highly conserved across vertebrates. However, zebrafish GOAT amino acid sequence showed higher identity with other fish GOAT sequences. We observed the most abundant expression of GOAT mRNA in the gut. Identity of GOAT amino acid sequence between zebrafish and other fishes and mammals indicate highly conserved regions in the bioactive core of GOAT, suggesting similar biological functions in vertebrates. Ghrelin is a major orexigenic factor in fish and is expressed in the gut and has been shown to stimulate food intake. The colocalization of GOAT and ghrelin in the gut confirm the potential functional interactions between GOAT and ghrelin in regulating food intake. It was determined that both asparagine in position 307 and histidine in position 338 of mouse GOAT are essential for the catalytic activity of GOAT enzyme.37 The proposed catalytic residues (asparagine and histidine) of GOAT are found conserved in zebrafish bioactive core. It has been shown that zebrafish, rat, and mouse GOAT were able to acylate human ghrelin.36 Zebrafish ghrelin amino acid sequences also exhibited very high identity with other fish species and mammals.22
We observed the expression of GOAT mRNA in various tissues, with the highest expression in the gut and lowest in the muscle of zebrafish. Similarly, wide distribution of GOAT mRNA has been reported in human and mice tissues, where the highest expression was reported in the stomach and intestine.6,9,36,37,42 These suggest that the primary source of GOAT might be gastrointestinal cells. Expression of GOAT in the brain of mammals9,42,48 and zebrafish suggest a potential facilitative role for GOAT in feeding regulation.15,29,31,49 Our data are in general agreement with other reports showing a similar pattern of GOAT mRNA expression in both rodent and human tissues.36,37,48 However, there are some discrepancies in tissue distribution of GOAT in mammals. GOAT mRNA was highest in the stomach and was detectable in the small intestine, colon, and testis and was not detectable in liver, brain, heart, kidney, lung, adipose tissue, and skeletal muscle in mouse.37 Study by Gonzalez et al. (2008)48 did not detect GOAT mRNA expression in testis and liver of rat. These exceptions might be related to species differences or the circadian expression of GOAT.48 GOAT might contribute to the ghrelin-mediated regulation of energy balance in fish and mammals.
In agreement with our PCR results, GOAT immunopositive cells were identified in the gut mucosa of zebrafish. Some GOAT immunoreactive cells were also positive for ghrelin. To the best of our knowledge, this is the first study that shows localization of GOAT i.r. in fish, and its colocalization with ghrelin in a non-mammal. Our result is consistent with the findings in mammals where GOAT and ghrelin are expressed in stomach ghrelin cells.5,50 Ghrelin is mainly produced in the X/A cells located in the gastric mucosa of stomach6 and other mucosal cells in the small intestine.35,51 Similarly, it has been demonstrated that GOAT enzyme expression is highly enriched within the gastric ghrelin-producing cells.43 The coexpression of GOAT in the gut ghrelin cells suggests that acylation is possible within the gastrointestinal tract.40 We found only 20% of GOAT cells immunopositive for ghrelin in zebrafish gut. There appears to be species specificity in the percentage of gut cells expressing both ghrelin and GOAT. For example, about 95% GOAT-positive cells in mice stomach also express ghrelin, meanwhile, only around 56% of GOAT cells colocalize ghrelin in rat stomach.52 While the reason for this species difference is currently unknown, this suggest that GOAT likely has other substrates in the gastrointestinal tract. Future studies employing double-labeled in situ hybridizations and immunohistochemistry using antibodies specific for zebrafish GOAT and ghrelin will help clarify this species difference.
It has been shown that the regulation of food intake by acylated ghrelin is dependent on metabolic status.14 We observed alterations in GOAT mRNA in the brain and the gut of zebrafish under various feeding conditions. Our results indicate that the expression of GOAT mRNA in both brain and gut are influenced by daily feeding period and nutrient status. Periprandial profile of GOAT mRNA expression did not change during 20–24 h (−3 to −1 h) after regular feeding time. In fish that were fed at the regular feeding time, preproghrelin mRNA expression decreased or remained the same as in sampling points before the regular feeding time. However, the most significant result is that in fish that were unfed at the regular feeding time, both preproghrelin and GOAT mRNA expression significantly increased. This increase was tissue specific and time dependent. The circadian pattern of ghrelin and/or GOAT is currently unknown. It is highly likely that ghrelin and GOAT exhibits a circadian pattern in its tissue-specific expression. The fact that there are considerable differences (an increase due to food withholding, and a decrease due to feeding) in the expression profile of GOAT and preproghrelin in fed versus unfed fish suggests that the changes found are likely elicited due to a meal or a lack of food. In other words, diet availability modulates ghrelin and GOAT expression in the gut and brain of zebrafish. Gut GOAT mRNA was increased following fasting of zebrafish at days 3 and 7 in the present study. The brain GOAT mRNA showed an increase following 3 days of fasting and remained unchanged at 7 days fasting. Ghrelin mRNA increased in the brain and gut of zebrafish fasted for 3, 5, and 7 days and refeeding after a 7 day fast caused a significant decrease in preproghrelin mRNA expression in the gut and brain of zebrafish.22 In agreement with this, we found an increase in both preproghrelin and GOAT peptides in the gut of food-deprived fish. Our results are consistent with ghrelin expression level in the gut during short-time fasting (16 and 24 h fasting).33,35 Overall, these meal-related corresponding changes in GOAT and ghrelin in both brain and gut highlight a role for the ghrelinergic system on food intake. It has been shown that 24 h fasting increased stomach GOAT mRNA, but not total ghrelin or stomach ghrelin expression in mice.46,53 Fasting increased stomach GOAT mRNA levels in rats subjected to 21 days of caloric restriction.48 Also fasting (12 and 24 h) increased circulating acylated ghrelin.5,46,54,55 Meanwhile, Kirchner et al. (2009)39 showed that fasting (12, 24, and 36 h) suppressed stomach GOAT mRNA in male mice and increased total ghrelin levels. While species differences in GOAT mRNA expression exist, it is clear that feed availability modulated endogenous GOAT and ghrelin.
Another novel finding of this study is that GOAT mRNA expression in the gut and brain of unfed zebrafish was suppressed after an injection of acylated ghrelin. This points toward a possible feedback inhibition of GOAT, thereby preventing more GOAT to acylated ghrelin, since excess of acylated ghrelin is present in the system. While the same results were also found in the brain of fed fish, this inhibition was not found in the gut of zebrafish. This is likely due to differences in the abundance of GOAT locally produced. Preproghrelin mRNA expression in the brain of fed fish was also attenuated by acyl ghrelin injection, but no such changes were found in the gut of fed and unfed fish and the brain of unfed fish. While the reasons for this tissue-specific expression of ghrelin and GOAT require additional studies, it is clear that the abundance of acylated ghrelin in the system induced by IP injection, in general, has a suppressive effect on the ghrelinergic system in zebrafish. This study only considered mRNA expression, not proteins, but the changes found are suggestive of a negative feedback inhibition of GOAT by active ghrelin.
In conclusion, this study furthers our current understanding of GOAT as a meal responsive orexigenic and metabolic peptide with an evolutionarily conserved structure and function. Presence of GOAT in tissues that are important in regulating metabolism suggests a possible role for GOAT in regulating energy balance in zebrafish. A fasting-induced increase in GOAT mRNA expression suggests a possible orexigenic role for GOAT in zebrafish. Together with our previous findings on zebrafish ghrelin,22 these results indicate that ghrelin/GOAT system are endogenous biologically active peptides that regulate food intake in zebrafish.
Acknowledgments
This research is supported by a Discovery grant and a Discovery Accelerator Supplement award from the Natural Sciences and Engineering Sciences (NSERC) of Canada to S.U. Research facilities used were funded by the Canada Foundation for Innovation—John R. Evans Leaders Fund, and an Establishment Grant from the Saskatchewan Health Research Foundation (SHRF) to S.U. S.U. is a Canadian Institutes of Health Research (CIHR) New Investigator Salary Award. A.H. is a recipient of postdoctoral fellowships from the SHRF and the CIHR.
Disclosure Statement
No competing financial interests exist.
References
- 1.Volkoff H, Canosa LF, Unniappan S, Cerda-Reverter JM, Bernier NJ, Kelly SP, et al. Neuropeptides and the control of food intake in fish. Gen Comp Endocrinol 2005;142:3–19 [DOI] [PubMed] [Google Scholar]
- 2.Valassi E, Scacchi M, Cavagnini F. Neuroendocrine control of food intake. Nutr Metab Cardiovasc Dis 2008;18:158–168 [DOI] [PubMed] [Google Scholar]
- 3.Volkoff H, Xu MY, MacDonald E, Hoskins L. Aspects of the hormonal regulation of appetite in fish with emphasis on goldfish, Atlantic cod and winter flounder: notes on actions and responses to nutritional, environmental and reproductive changes. Comp Biochem Physiol Mol Integr Physiol 2009;153:8–12 [DOI] [PubMed] [Google Scholar]
- 4.Olszewski PK, Schioth HB, Levine AS. Ghrelin in the CNS: from hunger to a rewarding and memorable meal? Brain Res Rev 2008;58:160–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kojima M, Kangawa K. Ghrelin: structure and function. Physiol Rev 2005;85:495–522 [DOI] [PubMed] [Google Scholar]
- 6.Kojima M, Hosoda H, Date Y, Nakazato M, Matsuo H, Kangawa K. Ghrelin is a growth-hormone-releasing acylated peptide from stomach. Nature 1999;402:656–660 [DOI] [PubMed] [Google Scholar]
- 7.Kaiya H, Kangawa K, Miyazato M. Molecular revolution of GPCRS: ghrelin/ghrelin receptors. J Mol Endocrinol 2014;52:T87–T100 [DOI] [PubMed] [Google Scholar]
- 8.Korbonits M, Bustin SA, Kojima M, Jordan S, Adams EF, Lowe DG, et al. The expression of the growth hormone secretagogue receptor ligand ghrelin in normal and abnormal human pituitary and other neuroendocrine tumors. J Clin Endocrinol Metab 2001;86:881–887 [DOI] [PubMed] [Google Scholar]
- 9.Gnanapavan S, Kola B, Bustin SA, Morris DG, McGee P, Fair clough P, et al. The tissue distribution of the mRNA of ghrelin and subtypes of its receptor, GHS-R, in humans. J Clin Endocrinol Metab 2002;87:2988–2991 [DOI] [PubMed] [Google Scholar]
- 10.Unniappan S, Cerda Reverter JM, Peter RE. In situ localization of prepro-galanin mRNA in the goldfish brain and changes in its expression during feeding and starvation. Gen Comp Endocrinol 2004;136:200–207 [DOI] [PubMed] [Google Scholar]
- 11.Peddu SC, Breves JP, Kaiya H, Gordon Grau E, Riley LG., Jr. Pre- and postprandial effects on ghrelin signaling in the brain and on the GH/IGF-I axis in the mozambique tilapia (Oreochromis mossambicus). Gen Comp Endocrinol 2009;161:412–418 [DOI] [PubMed] [Google Scholar]
- 12.Unniappan S. Ghrelin: an emerging player in the regulation of reproduction in non-mammalian vertebrates. Gen Comp Endocrinol 2010;167:340–343 [DOI] [PubMed] [Google Scholar]
- 13.Nishi Y, Yoh J, Hiejima H, Kojima M. Structures and molecular forms of the ghrelin-family peptides. Peptides 2011;32:2175–2182 [DOI] [PubMed] [Google Scholar]
- 14.Gahete MD, Rincon-Fernandez D, Villa-Osaba A, Hormaechea-Agulla D, Ibaez-Costa A, Martinez-Fuentes AJ. Ghrelin gene products, receptors, and GOAT enzyme: biological and pathophysiological insight. J Endocrinol 2014;220:R1–R24 [DOI] [PubMed] [Google Scholar]
- 15.Unniappan S, Lin X, Cervini L, Rivier J, Kaiya H, Kangawa K, et al. Goldfish ghrelin: molecular characterization of the complementary deoxyribonucleic acid, partial gene structure and evidence for its stimulatory role in food intake. Endocrinology 2002;143:4143–4146 [DOI] [PubMed] [Google Scholar]
- 16.Kaiya H, Miyazato M, Kangawa K, Peter RE, Unniappan S. Ghrelin: a multifunctional hormone in non-mammalian vertebrates. Comp Biochem Physiol A 2008;149:109–128 [DOI] [PubMed] [Google Scholar]
- 17.Manning AJ, Murray HM, Gallant JW, Matsuoka MP, Radford E, Douglas SE. Ontogenetic and tissue-specific expression of preproghrelin in the Atlantic halibut, Hippoglossus hippoglossus L. J Endocrinol 2008;196:181–192 [DOI] [PubMed] [Google Scholar]
- 18.Miura T, Maruyama K, Kaiya H, Miyazato M, Kangawa K, Uchiyama M, et al. Purification and properties of ghrelin from the intestine of the goldfish, Carassius auratus. Peptides 2008;30:758–765 [DOI] [PubMed] [Google Scholar]
- 19.Olsson C, Holbrook JD, Bompadre G, Jonsson E, Hoyle CH, Sanger GJ, et al. Identification of genes for the ghrelin and motilin receptors and a novel related gene in fish, and stimulation of intestinal motility in zebrafish (Danio rerio) by ghrelin and motilin. Gen Comp Endocrinol 2008;155:217–226 [DOI] [PubMed] [Google Scholar]
- 20.Terova G, Rimoldi S, Bernardini G, Gornati R, Saroglia M. Sea bass ghrelin: molecular cloning and mRNA quantification during fasting and refeeding. Gen Comp Endocrinol 2008;155:341–351 [DOI] [PubMed] [Google Scholar]
- 21.Xu M, Volkoff H. Molecular characterization of ghrelin and gastrin releasing peptide in Atlantic cod (Gadus morhua): cloning, localization, developmental profile and role in food intake regulation. Gen Comp Endocrinol 2009;160:250–258 [DOI] [PubMed] [Google Scholar]
- 22.Amole N, Unniappan S. Fasting induces preproghrelin mRNA expression in the brain and gut of zebrafish, Danio rerio. Gen Comp Endocrinol 2009;161:133–137 [DOI] [PubMed] [Google Scholar]
- 23.Tschop M, Smiley DL, Heiman ML. Ghrelin induces adiposity in rodents. Nature 2000;407:908–913 [DOI] [PubMed] [Google Scholar]
- 24.Asakawa A, Inui A, Kaga T, Yuzuriha H, Nagata T, Ueno N, et al. Ghrelin is an appetite-stimulatory signal from the stomach with structural resemblance to motilin. Gastroenterology 2001;120:337–345 [DOI] [PubMed] [Google Scholar]
- 25.Wren AM, Seal LJ, Cohen MA, Brynes AE, Frost GS, Murphy KG, et al. Ghrelin enhances appetite and increases food intake in humans. J Clin Endocrinol Metab 2001;86:5992. [DOI] [PubMed] [Google Scholar]
- 26.Nakazato M, Murakami N, Date Y, Kojima M, Matsuo H, Kangawa K, et al. A role for ghrelin in the central regulation of feeding. Nature 2001;409:194–198 [DOI] [PubMed] [Google Scholar]
- 27.Kirchner H, Hofmann SM, Fischer-Rosinský A, Hembree J, Abplanalp W, Ottaway N, et al. Caloric restriction chronically impairs metabolic programming in mice. Diabetes 2012;61:2734–2742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Unniappan S, Peter RE. Structure, distribution and physiological functions of ghrelin in fish. Comp Biochem Physiol A 2005;140:396–408 [DOI] [PubMed] [Google Scholar]
- 29.Matsuda K, Miura T, Kaiya H, Maruyama K, Shimakura S, Uchiyama M, et al. Regulation of food intake by acyl and des-acyl ghrelins in the goldfish. Peptides 2006;27:2321–2325 [DOI] [PubMed] [Google Scholar]
- 30.Miura T, Maruyama K, Shimakura S, Kaiya H, Uchiyama M, Kangawa K, et al. Regulation of food intake in the goldfish by interaction between ghrelin and orexin. Peptides 2007;28:1207–1213 [DOI] [PubMed] [Google Scholar]
- 31.Riley LG, Fox BK, Kaiya H, Hirano T, Grau EG. Long-term treatment of ghrelin stimulates feeding, fat deposition, and alters the GH/IGF-I axis in the tilapia, Oreochromis mossambicus. Gen Comp Endocrinol 2005;142:234–240 [DOI] [PubMed] [Google Scholar]
- 32.Shepherd BS, Johnson K, Silverstein JT, Parhar IS, Vijayan MM, McGuire A, et al. Endocrine and orexigenic actions of growth hormone secretagogues in rainbow trout (Oncorhynchus mykiss). Comp Biochem Physiol A 2007;146:390–399 [DOI] [PubMed] [Google Scholar]
- 33.Unniappan S, Canosa LF, Peter RE. Orexigenic actions of ghrelin in goldfish: feeding induced changes in brain and gut mRNA expression and serum levels, and responses to central and peripheral injections. Neuroendocrinology 2004;79:100–108 [DOI] [PubMed] [Google Scholar]
- 34.Jönsson E, Forsman A, Einarsdottir IE, Kaiya H, Ruohonen K, Björnsson BT. Plasma ghrelin levels in rainbow trout in response to fasting, feeding and food composition, and effects of ghrelin on voluntary food intake. Comp Biochem Physiol 2007;147:1116–1124 [DOI] [PubMed] [Google Scholar]
- 35.Eom J, Hong M, Cone RD, Song Y. Zebrafish ghrelin is expressed in pancreatic endocrine cells and regulated by metabolic state. Biochem Biophys Res Commun 2013;439:115–120 [DOI] [PubMed] [Google Scholar]
- 36.Gutierrez JA, Solenberg PJ, Perkins DR, Willency JA, Knierman MD, Jin Z, et al. Ghrelin octanoylation mediated by an orphan lipid transferase. Proc Natl Acad Sci U S A 2008;105:6320–6325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang J, Brown MS, Liang G, Grishin NV, Goldstein JL. Identification of the acyltransferase that octanoylates ghrelin, an appetite-stimulating peptide hormone. Cell 2008;132:387–396 [DOI] [PubMed] [Google Scholar]
- 38.Mohan H, Unniappan S. Discovery of ghrelin o-acyltransferase. Endocr Dev 2013;25:16–24 [DOI] [PubMed] [Google Scholar]
- 39.Kirchner H, Gutierrez JA, Solenberg PJ, Pfluger PT, Czyzyk TA, Willency JA, et al. GOAT links dietary lipids with the endocrine control of energy balance. Nat Med 2009;15:741–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shlimun A, Unniappan S. Ghrelin O-acyl transferase: bridging ghrelin and energy homeostasis. Int J Pept 2011;217957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Muller TD, Tschop MH, Jarick I, Ehrlich S, Scherag S, Herpertz-Dahlmann B, et al. Genetic variation of the ghrelin activator gene ghrelin O-acyltransferase (GOAT) is associated with anorexia nervosa. J Psychiatr Res 2011;45:706–711 [DOI] [PubMed] [Google Scholar]
- 42.Lim CT, Kola B, Grossman A, Korbonits M. The expression of ghrelin O-acyltransferase (GOAT) in human tissues. Endocr J 2011;58:707–710 [DOI] [PubMed] [Google Scholar]
- 43.Sakata I, Yang J, Lee C, Osborne-Lawrence S, Rovinsky S, Elmquist JK, et al. Co-localization of ghrelin O-acyltransferase (GOAT) and ghrelin in gastric mucosal cells. Am J Physiol Endocrinol Metab 2009;297:E134–E141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gahete MD, Cordoba-Chacon J, Hergueta-Redondo M, Martinez-Fuentes AJ, Kineman RD, Moreno-Bueno G, et al. A novel human ghrelin variant (In1-ghrelin) and ghrelin-O-acyltransferase are overexpressed in breast cancer: potential pathophysiological relevance. PLoS One 2011;6:e23302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu B, Garcia EA, Korbonits M. Genetic studies on the ghrelin, growth hormone secretagogue receptor (GHSR) and ghrelin O-acyl transferase (GOAT) genes. Peptides 2011;32:2191–2207 [DOI] [PubMed] [Google Scholar]
- 46.Gahete MD, Cordoba-Chacon J, Salvatori R, Castano JP, Kineman RD, Luque RM. Metabolic regulation of ghrelin O-acyl transferase (GOAT) expression in the mouse hypothalamus, pituitary, and stomach. Mol Cell Endocrinol 2010;317:154–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real time quantitative PCR and the 2^-ΔΔCT method. Methods 2001;25:402–408 [DOI] [PubMed] [Google Scholar]
- 48.Gonzalez CR, Vazquez MJ, Lopez M, Dieguez C. Influence of chronic under nutrition and leptin on GOAT mRNA levels in rat stomach mucosa. J Mol Endocrinol 2008;41:415–421 [DOI] [PubMed] [Google Scholar]
- 49.Matsuda K, Miura T, Kaiya H, Maruyama K, Uchiyama M, Kangawa K, et al. Stimulatory effect of n-octanoylated ghrelin on locomotor activity in the goldfish, Carassius auratus. Peptides 2006;27:1335–1340 [DOI] [PubMed] [Google Scholar]
- 50.Date Y, Murakami N, Kojima M, Kuroiwa T, Matsukura S, Kangawa K, et al. Central effects of a novel acylated peptide, ghrelin, on growth hormone release in rats. Biochem Biophys Res Commun 2000;275:477–480 [DOI] [PubMed] [Google Scholar]
- 51.Ghelardoni S, Carnicelli V, Frascarelli S, Ronca-Testoni S, Zucchi R. Ghrelin tissue distribution: comparison between gene and protein expression. J Endocrinol Invest 2006;29:115–121 [DOI] [PubMed] [Google Scholar]
- 52.Stengel A, Goebel M, Wang L, Taché Y, Sachs G, Lambrecht NW. Differential distribution of ghrelin-O-acyltransferase (GOAT) immunoreactive cells in the mouse and rat gastric oxyntic mucosa. Biochem Biophys Res Commun 2010;392:67–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kineman RD, Gahete MD, Luque RM. Identification of a mouse ghrelin gene transcript that contains intron 2 and is regulated in the pituitary and hypothalamus in response to metabolic stress. J Mol Endocrinol 2007;38:511–521 [DOI] [PubMed] [Google Scholar]
- 54.Casanueva FF, Dieguez C. Ghrelin: the link connecting growth with metabolism and energy homeostasis. Rev Endocr Metab Disord 2002;3:325–338 [DOI] [PubMed] [Google Scholar]
- 55.Cummings DE. Ghrelin and the short- and long-term regulation of appetite and body weight. Physiol Behav 2006;89:71–84 [DOI] [PubMed] [Google Scholar]