Abstract
Mechanical methods for inducing differentiation and directing lineage specification will be instrumental in the application of pluripotent stem cells. Here, we demonstrate that minimization of cell-substrate adhesion can initiate and direct the differentiation of human pluripotent stem cells (hiPSCs) into cyst-forming trophoblast lineage cells (TLCs) without stimulation with cytokines or small molecules. To precisely control cell-substrate adhesion area, we developed a novel culture method where cells are cultured on microstructured mesh sheets suspended in a culture medium such that cells on mesh are completely out of contact with the culture dish. We used microfabricated mesh sheets that consisted of open meshes (100∼200 μm in pitch) with narrow mesh strands (3–5 μm in width) to provide support for initial cell attachment and growth. We demonstrate that minimization of cell adhesion area achieved by this culture method can trigger a sequence of morphogenetic transformations that begin with individual hiPSCs attached on the mesh strands proliferating to form cell sheets by self-assembly organization and ultimately differentiating after 10–15 days of mesh culture to generate spherical cysts that secreted human chorionic gonadotropin (hCG) hormone and expressed caudal-related homeobox 2 factor (CDX2), a specific marker of trophoblast lineage. Thus, this study demonstrates a simple and direct mechanical approach to induce trophoblast differentiation and generate cysts for application in the study of early human embryogenesis and drug development and screening.
Introduction
Pluripotent stem cells, including human pluripotent stem cells (hiPSCs) and human embryonic stem cells (hESCs) can proliferate rapidly and indefinitely and they can differentiate to form virtually any cell type of the adult organism, making them increasingly attractive for use in regenerative medicine and drug discovery.1,2 In the field of reproductive biology and medicine, a major interest is in developing alternative models for the study of early trophoblast differentiation that would replace the traditional mouse models whose embryogenesis is known to be phenomenologically different from the human case. Indeed, over reliance on animal models due to existing ethical issues associated with the use of human embryos and practical limitations arising from the use of placentas that are less than 6 weeks old have impeded the study of the molecular mechanisms underlying proliferation and differentiation of human trophoblast lineage cells (TLCs).3 Thus, focus is increasingly shifting toward the use of trophoblasts derived from pluripotent cells as models to study early embryogenesis in humans.
A popular model for establishing trophoblast cell lineage in vitro involves treating pluripotent stem cells, mainly hESCs, with bone morphogenetic protein 4 (BMP4).4–8 BMP4 is a member of the transforming growth factor beta superfamily, which controls numerous events of embryonic, fetal, and even adult development in all vertebrates.9 However, although widely used to derive TLCs from hESCs, this model has not been successful with hiPSCs, as illustrated by the fact that only very few studies have succeeded in converting hiPSCs into TLCs by BMP4 treatment.10 It is not clear whether this is a result of underlying differences in genomic and epigenetic signatures between the two,11,12 but considering the ethical concerns associated with embryo-derived hESCs, it would be more desirable to derive TLCs from hiPSCs rather than from hESCs. Thus, there is need to develop methods for generating TLCs from hiPSs more efficiently and reproducibly. Such methods will enable us to establish patient specific in vitro models of the placenta for drug testing and also for studying the mechanisms of placenta-associated diseases.10
It is increasingly becoming clear that cell-substrate interactions and the mechanical cues emanating from them play fundamental roles in the regulation of the signaling pathways that determine cell differentiation and lineage commitment.13 In fact, mounting evidences suggest that the interaction between a stem cell and its extracellular cell matrix via integrin and cadherin-mediated cell–cell interactions determine the pattern of gene expression and the resultant differentiated phenotype despite the blueprint of the genome being the same.14 For instance, physical characteristics of a substrate such as elasticity, topology, and geometry have been implicated in the determination of cell fate and lineage specification,15–18 and it has been demonstrated that stem cells can be tuned to differentiate into a desired cell type by modulating cell adhesion alone.19 An exemplary example of this mechanical approach of differentiation specification has been demonstrated in mesenchymal stem cells (MSCs) where patterns embodied on a substrate could be used to control osteogenesis of MSCs with efficiency similar to chemical stimulation.13,20
This study highlights the possibility to mechanically stimulate hiPSCs to differentiate directly to TLCs by modulating the microscale physical environment of the cells. We demonstrate that minimization of cell-substrate adhesion area achieved by a novel culture method whereby hiPSCs are cultured on microstructured mesh sheets consisting of large-aperture meshes (∼200 μm in pitch) and fine mesh stands (3–5 μm in width) that are suspended in a culture medium can trigger morphogenesis of cell sheets and mechanically induce hiPSCs to differentiate into cyst-forming TLCs without chemical induction.
Materials and Methods
Fabrication of microstructured thin mesh sheets
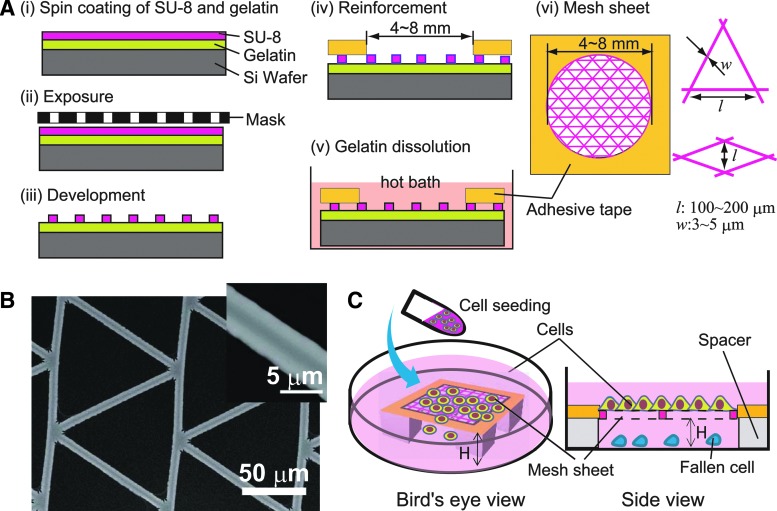
Microstructured mesh sheets used in this study were fabricated by standard photolithography using epoxy-based negative photo resist (SU-8-2; Microchem). The processes of fabrication are shown schematically in Figure 1A (i–v). Briefly, SU-8-2 was spin-coated directly on a silicon wafer precoated with a thin sacrificial layer of gelatin (2% w/v). The thicknesses of spin-coated SU-8 and gelatin were ∼2 μm each. After UV exposure for 5 s at 22 mW power, SU-8 was developed according to the manufacturer's instructions. The developed features were then laminated with Kapton tapes having a hole measuring 4–8 mm at the center to act as a frame reinforcing the ultrathin sheets (2 μm-thick) upon detachment by immersion in a hot water bath (80°C) to dissolve the sacrificial gelatin. Mesh sheets obtained were then sterilized by immersion in 70% ethanol and thereafter kept in an oven at 80°C until used. The resulting mesh sheet is shown schematically in Figure 1A (vi). We fabricated and tested different kinds of mesh shapes, but we focused more on meshes with triangular and rhombus meshes because they gave better performance in terms of ease of cell attachment and proliferation compared with square meshes. The dimension of the meshes were such that the length of the short axis of a rhombus mesh or the base of a triangular mesh was 100∼200 μm [Fig. 1A (vi)]. Regardless of mesh type, the width of the mesh strand was between 3 and 5 μm and the thickness of the sheet was about 2–3 μm. Figure 1B shows an image of a fabricated SU-8 mesh sheet with triangular mesh obtained by a desk-top type scanning electron microscope (Hitachi Tabletop Microscope TM-1000; Hitachi, Inc.).
FIG. 1.
Fabrication of mesh sheet and cell culture. (A) Processes of mesh sheet fabrication by photolithography (i–iii), mesh sheet reinforcement and harvesting (iv–v), and a schematic of prepared mesh sheet showing the dimensions of cell culture area (vi, left) and mesh shapes and dimensions (vi, right). Mesh sheets with rhombus (short axis length, l=100∼200 μm) and triangular shapes (side length, l=100∼200 μm) were used. The width of mesh strand was w=3–5 μm. (B) Scanning electron microscope image of a fabricated triangular mesh. (C) A schematic showing how cell seeding and culture was performed on a mesh sheet suspended in a culture medium. (Left) Orthographic representation of mesh sheet setup during cell seeding, (right) a cross-sectional view of setup during cell culture. Spacers of height H=0.5–1 mm were used to suspend mesh sheets in the medium so as to prevent cells attached on mesh from interacting with the dish bottom.
Cell seeding and culture on mesh sheets
Before cell seeding, mesh sheets were incubated overnight at 4°C with Matrigel (Matrigel Basement Membrane Matrix; Corning, Inc.) diluted at a ratio of 1:20 in DMEM/F12 medium or laminin (iMatrix-511; Wako, Inc.) at a concentration of 10–20 μg/mL in 1× phosphate buffered saline (PBS). For cell seeding, the reinforced mesh sheets were suspended in a culture medium using silicon rubber spacers of thickness, H=0.5–1 mm (Fig. 1C) such that cells seeded on the meshes were completely out of contact with the mesh bottom. In each case, the coating material was briefly rinsed off with 1× PBS buffer just before cell seeding.
Human iPS cells (clone TIG1-4F hiPS cells #1 at ∼40 passages) from lung fibroblasts used in this study were a kind gift from Dr. Takashi Tada of Kyoto University, Japan. Before seeding on a mesh sheet, hiPSCs were cultured on typical culture dishes coated with either Matrigel or laminin to 85% confluence in a humidified incubator at 37°C and 5% CO2 using essential 8 medium (E8 medium)21 (Life Technologies). These cells were then dissociated with 2 mL of a modified trypsin solution (TrypLE; Life Technologies) and transferred to a fresh E8 medium and centrifuged at 100 g for 5 min followed by resuspension in a fresh 2 mL of E8 medium supplemented with a ROCK inhibitor (Y-27632; Wako) at a concentration of 10 μg/mL to prevent apoptosis immediately after seeding. After pipetting briefly to disperse cell aggregates, cells were directly seeded on the suspended mesh sheets at a cell density of ∼106 cells/mL. Since most cells would fall off during seeding, the number of cells that remained attached on the mesh sheet was much less (about 10%) but this depended on mesh size and shape. After seeding, the mesh sheets were incubated in E8 medium for another 12 h in a humidified incubator at 37°C and 5% CO2. Microscopic imaging was periodically performed to monitor cell growth on the mesh sheets. Mesh sheets loaded with cells were transferred to new dishes occasionally to prevent dead or fallen cells that would accumulate at the dish bottom from interfering with microscopic imaging.
Generation of embryonic bodies by suspension culture
To generate embryonic bodies (EBs), we harvested hiPSCs (same clone as above) cultured on laminin-coated dishes to 85% confluence as described above and seeded them as aggregates (without dispersion) on uncoated culture dishes filled with E8 medium. Extended culture was performed in an incubator under the same conditions as described above. Cell aggregation and size growth was monitored and EBs that had grown to over 400 μm in size were harvested for enzyme-linked immunosorbent assay (ELISA) and real-time polymerase chain reaction (RT-PCR) analyses at day 15 and 21.
Live cell staining and immunofluorescence microscopy
For live cell imaging, cells were labeled with 5 μM Calcein AM (Invitrogen) in 1× PBS for 20 min, followed by nucleic staining with 1 μM Hochest33342 (Sigma) for 10 min. Time lapse imaging of unstained live cells was performed using a fluorescence microscope (BZ 9000; Keyence) fitted with a culture chamber. Phase contrast images were acquired using Olympus IX71 fitted with a WAT-221s camera (Watec). Images were slightly processed using Image J (NIH) to improve contrast.
For immunofluorescence microscopy, cells were fixed in 4% formaldehyde (Wako) for 10 min, permeabilized with 0.3% triton X-100 in PBS for 15 min, and blocked with a buffer containing 3% bovine serum albumin (BSA) and 0.3% triton X-100 in 1× PBS for 60 min. The samples were then separately incubated overnight at 4°C with primary antibodies against OCT4, Nanog, CDX2, human chorionic gonadotropin (hCG)-β,E-cadherin, and vinculin, followed by incubation for 60 min with the corresponding secondary antibodies tagged with Alexa Flour dyes (Invitrogen) at 1:500 dilution. Primary antibodies used in this study were mouse anti human OCT4 (Santa Cruz) at 1:50 dilution, goat anti human CDX2 (R&D Systems) at 1:200 dilution, rabbit anti human NANOG (Cell Signaling Technology) at 1:100 dilution, polyclonal rabbit anti human CG (Abcam) at 1:200 dilution, monoclonal Anti-vinculin antibody produced in mouse (clone hVIN-1; Sigma Aldrich), and mouse monoclonal anti-human E-cadherin (Takara Bio) at a dilution of 1:200 dilution. All antibodies were diluted in a buffer containing 1% BSA and 0.3% triton X-100. Immunofluorescence microscopy for CDX2 and hCG-β was performed using confocal microscopes (LSM 700 and LSM 510 microscopes; Zeiss) with a 10× and a 25× objective lenses, while that for E-cadherin and vinculin was performed using Olympus Microscope IX83 fitted with a confocal scanning unit (CSU-W1; Yokogawa Electronics) and Orca flash 4.0 camera (Hamamatsu Photonics). 3D rendering of confocal images of cysts was performed using the maximum intensity projection modethe ZEN Module 3D VisArt software (Zeiss).
Analysis of RNA expression by quantitative RT-PCR analysis
Total RNA was isolated using TRIZOL LS solution (Invitrogen) and reverse-transcribed by the ThermoScript RT-PCR system (Invitrogen) with an oligo-dT primer. Real-time quantitative PCR was performed with an ABI Prism 7300 Thermal Cycler (Applied Biosystems) using FastStart Universal Probe Master (Roche Applied Science) and the indicated combinations of Universal Probe Library (Roche Applied Science). The combination of primer sequences used are shown in Table 1.
Table 1.
A List of Markers and Primer Sequences Used for Real-Time Polymerase Chain Reaction Analysis
| Marker | Left (forward) primer | Right (reverse) primer |
|---|---|---|
| GAPDH | AGCCACATCGCTCAGACAC | GCCCAATACGACCAAATCC |
| OCT4 | GTTGGAGAAGGTGGAACCAA | CCAAGGTGATCCTCTTCTGC |
| TERT (transcript variant 2) | GCCTTCAAGAGCCACGTC | CCACGAACTGTCGCATGT |
| MET | AAATGTGCATGAAGCAGGAA | CCACGAACTGTCGCATGT |
| CDX2 | ATCACCATCCGGAGGAAAG | TGCGGTTCTGAAACCAGATT |
Data were obtained in triplicates and statistical significance was determined by the Student's t-test (unpaired), assuming a normal distribution since the number of the respective features (TLC cysts or EBs) per analyzed sample was sufficiently large (>10 features per analysis).
Analysis of hCG hormone secretion by ELISA
Media samples were collected for analysis from both mesh culture with hiPSC-derived cysts and suspension culture with EBs. Medium was left unchanged the day before to allow for hormone accumulation. ELISA analysis was performed using hCG ELISA kit (BQ Kits) according to the manufacturer's instructions. The results were quantified using an EnSight™ multimode plate reader (PerkinElmer) running a Wallac 1420 software. Data are acquired in quadruplicates and was fitted to the standard curve using a free online software available at <http://elisaanalysis.com/>. The results were imported to a Microsoft excel for further analysis and graph representation. Statistical comparison between TLCs and EBs was done using unpaired Student's t-test.
Results
Adhesion area minimization by mesh culture and induction of hiPSCs differentiation
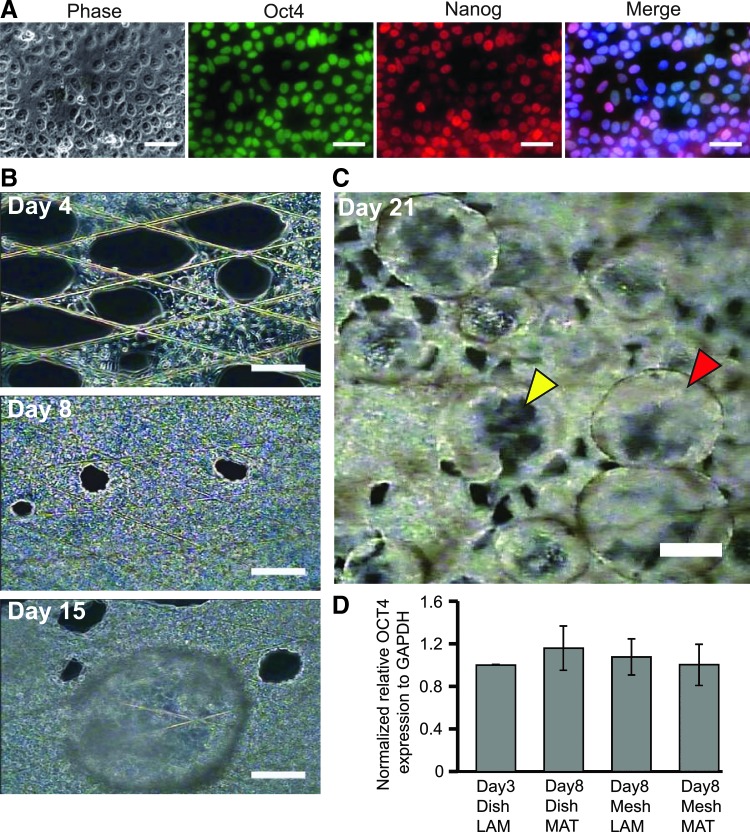
To achieve precise control of cell adhesion area, we developed a novel culture method that involves culturing cells on microstructured thin mesh sheets (Fig. 1A, B) suspended in a culture medium such that cells on mesh are completely out of contact with the culture dish. This method ensures that all cellular activities including attachment, elongation, and proliferation are confined to the narrow mesh strands. To investigate the influence of minimizing cell-substrate adhesion on stem cell differentiation, we monitored the behavior of hiPSCs seeded on the suspended mesh sheets. Before seeding on mesh, we determined by immunofluorescence microscopy that the cells expressed OCT 4 and NANOG and were therefore pluripotent (Fig. 2A).
FIG. 2.
Time lapse monitoring of hiPSCs cultured on a suspended mesh sheet. (A) Results of immunostaining performed to check for pluripotency of dish cultured hiPSCs before seeding on mesh. Merged image also shows blue pigment from Hoechst staining of cell nuclei. (B) Phase contrast images showing growth and proliferation of hiPSCs on mesh. Last image of the column (Day 15) shows a bubble-like cyst formed as a result of cell differentiation. (C) Phase contrast image showing numerous cysts on a mesh sheet at day 21 of culture. Yellow and red arrowheads indicate cysts formed on the upper and lower surfaces of a mesh sheet, respectively. (D) Result of RT-PCR analysis performed to compare pluripotency in hiPSCs cultured on mesh and on typical culture dishes for 8 days. Results were normalized by OCT4 expression in hiPSCs cultured on laminin-coated culture dish for 3 days. MAT, Matrigel; LAM, laminin. Scale bar represent 50 μm in (A) 100 μm in (B), and 200 μm in (C). hiPSCs, human pluripotent stem cells; RT-PCR, real-time polymerase chain reaction.
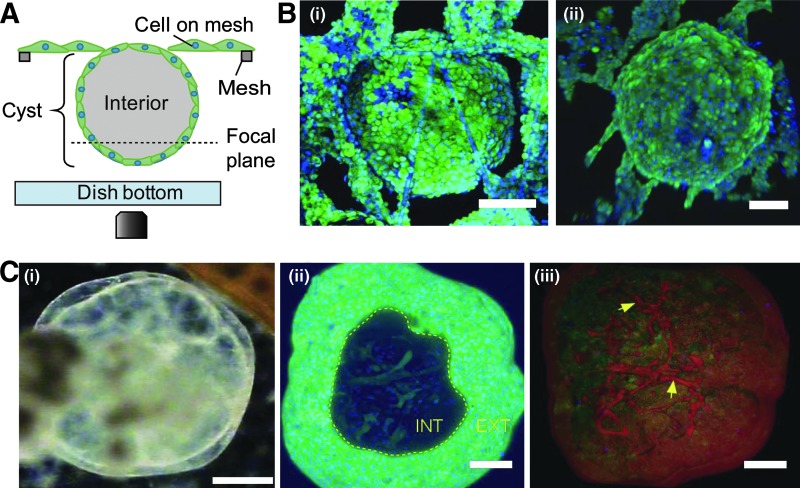
Although most cells would fall off to the dish bottom during seeding, ∼10% of cells adhered successfully on the mesh strands, mainly at the intersection points. These cells could spread along the narrow mesh strands and proliferate normally such that by day 4, cells had begun to form web-like incursions toward the interior of the open meshes, starting from the strand intersections points (Fig. 2B, day 4). By day 8, cells had proliferated, spread, and occupied almost all the initially open meshes, resulting in the formation of a monolayer cell sheet (Fig. 2B, day 8). However, between day 10 and 15, we noticed the appearance of uniquely transparent, bubble-like cysts primarily on the regions of the mesh where cells had formed a uniform cell sheet (Fig. 2B, day 15). Indeed, the features were so transparent that the cells beneath them could be seen clearly under a microscope, hence we could determine that their formation did not result in the disruption of the cell sheet beneath (Fig. 2B, day 15, also Supplementary Fig. S1; Supplementary Data are available online at www.liebertpub.com/tec). Initially, a few cysts would appear but they grew in number and size with time. They had common characteristics of a round shape, a clear interior cavity, and a thin enclosing wall (Fig. 2C). Remarkably, cysts could form on the upper surface (Fig. 2C, red arrow head) and on the lower surface (Fig. 2C, yellow arrow head) of the mesh sheet, with those on the lower surface hanging downward to a height of ∼400 μm. With continuous culture, cysts on the upper surface could grow to a height of more than 800 μm [Fig. 3C (i)].
FIG. 3.
Morphological characterization by confocal microscopy of cysts formed by differentiated hiPSCs on a suspended mesh sheet. (A) A schematic of setup during confocal microscopy. Focus was shifted up and down to capture images of the cysts for 3D rendering. (B) A 3D reconstruction showing both the apical (i) and the basal (ii) surfaces of two different spherical cysts hanging downward from a mesh sheet. Both images illustrate that the cysts were closed spheres. To obtain the images, mesh sheet was turned upside down and imaged as shown in (A). (C) High resolution imaging of a representative cyst cultured continuously on mesh and showing vascularization. (i) A phase contrast image of a mesh generated cyst at day 40. (ii) A 3D reconstruction of a cyst stained with Calcein AM with a view window showing vascular-like cellular networks in the interior. “INT” denotes interior, “EXT” denotes exterior of a cyst. (iii) A 3D reconstruction based on maximum intensity projection showing vascularization (yellow arrows) in the same cyst as in the left image. Pseudo coloring was applied for easy visualization. Scale bar represent 200 μm in (i) and 100 μm in (ii) and (iii).
Since cyst formation occurred mostly after more than 10 days of mesh culture, we sought to examine the pluripotency status of the cells just before this time point. Remarkably, quantitative analysis of RNA expression by RT-PCR revealed that day 8 old hiPSCs on mesh expressed OCT4 at a level comparable with dish cultured cells, regardless of the mesh coating matrix used (Fig. 2D). Notably, we did not observe cyst formation in a separate control experiment in which we cultured the same clone of hiPSCs on a thin nonmesh membrane of SU-8 that was similarly suspended in E8 medium (Supplementary Fig. S2). This suggests that the differentiation pattern shown by hiPSCs on mesh was due to adhesion area minimization rather than physical and chemical factors inherent in the mesh material.
Morphological characterization reveal vascularization of cysts after prolonged culture
Next, we examined the morphology of mesh generated cysts using confocal microscopy as shown schematically in Figure 3A. We could determine that the cysts were indeed spherical and completely enveloped by an epithelial-like cell wall, as shown in the reconstructed 3D images in Figure 3B (i) and (ii). Importantly, reconstructed 3D images of a 40 day-old cyst in Figure 3C (i) revealed the existence of vascular-like cellular network traversing the interior cavity that also stained positive for Calcein AM, suggesting that the network comprised of living cells [Fig. 3C (ii) and Fig. 3C (iii), yellow arrows; Supplementary Movie S1]. Although the exact identity of the interior cellular network remains to be elucidated, we postulate that they may be conduits for nutrient transport that support cell survival in a region where necrosis or apoptosis may not permit cell survival.22 Coincidentally, this observation concurs with other reports suggesting the presence of blood islands in an embryo-like structure formed by in vivo reprogrammed mouse stem cells.23,24
Characterization of trophoblast markers by immunofluorescence microscopy and ELISA
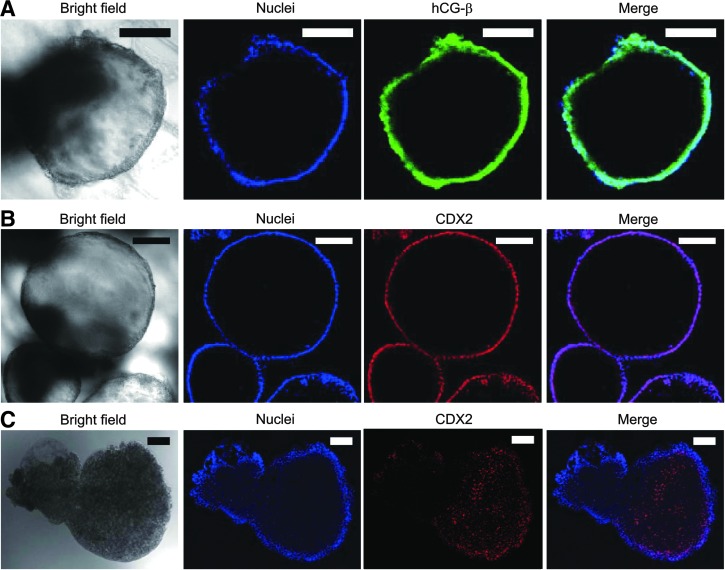
Suspecting that the observed cyst formation could be an indication of trophoblast differentiation, we performed immunofluorescence microscopy to examine the presence of hCG hormone, a key pregnancy hormone secreted by placental trophoblasts and CDX2, a transcription factor strongly associated with trophoblast. Despite the rigors of fixation and permeabilization, cysts still retained their spherical morphology (Fig. 4A, B, bright field), demonstrating that these were stable cellular structures. Importantly, immunofluorescence results revealed the presence of hCG-β (a subunit of hCG hormone) in the cytoplasm (Fig. 4A) and CDX2 in the nuclei (Fig. 4B) of cyst-forming cells, providing evidence for the presence of TLCs as suggested by previous studies.4,5,25,26 A comparative immunostaining of EB for CDX2 turned out negative for the trophoblast marker (Fig. 4C), suggesting that the cysts were distinctively dissimilar to EBs in both structure and expression (compare Fig. 4B, C).
FIG. 4.
Results of immunofluorescence microscopy performed to characterize cysts formed as a result of hiPSC differentiation on mesh. (A) Bright field (first image of row) and immunofluorescence images of a cyst stained for hCG-β subunit of hCG hormone (third image). Hoechst-labeled nuclei is shown in the second image, and merged image in the last image of the row. (B) Immunofluorescence images of a cyst showing the expression of CDX2 (third image). (C) Comparative immunofluorescence images of an EB stained for CDX2. Scale bar represent 100 μm. EB, embryonic body; hCG, human chorionic gonadotropin.
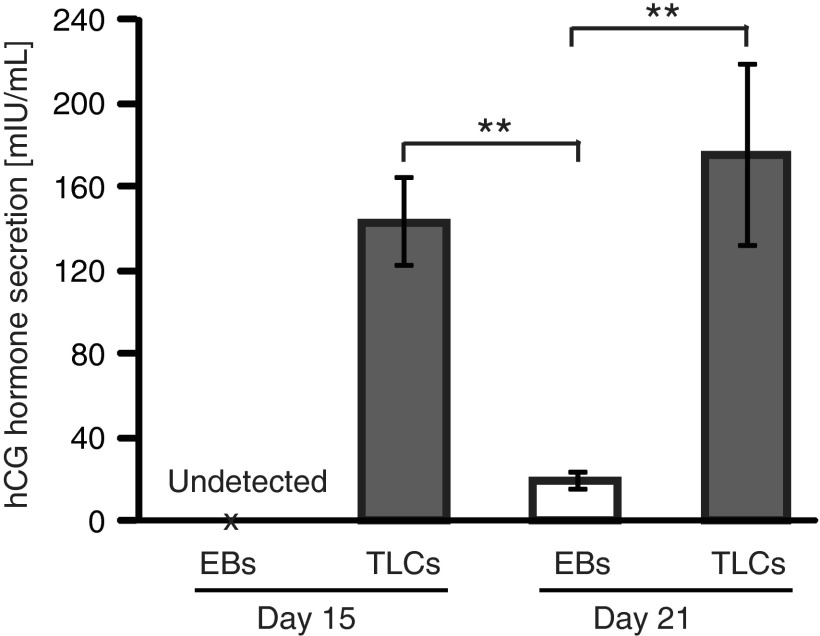
Prompted by the above results, we next sought to determine hCG hormone secretion quantitatively by ELISA analysis. We found significantly high levels of the hormone in the spent culture media from cyst-containing mesh samples at day 15 and 21 (Fig. 5). In contrast, the hormone was undetectably low (Fig. 5, day 15) in the culture medium containing 15 day-old EBs (Supplementary Fig. S3), and extremely low (Fig. 5, day 21) in the culture medium containing 21 day-old EBs. In fact, the level of hCG hormone in the media from mesh samples with cysts was sufficiently high that the hormone could be detected even by an over-the-counter pregnancy testers (Supplementary Fig. S4), and as early as 2 days after spotting the first cysts. Overall, the secretion of hCG hormone by mesh generated cysts, as opposed to EBs, supports the argument for mesh culture-triggered differentiation of hiPSCs to TLCs.
FIG. 5.
Results of quantitative analysis of hCG hormone secretion by ELISA. Assayed culture media were collected from EB culture samples (marked as “EBs”) and mesh culture samples with cysts (marked as “TLCs”) at day 15 and 21. Culture medium was not changed the day before media collection to allow for adequate accumulation of the hormone. Data are presented as mean±SD of four replicates (n=4) and double asterisks (**) denote statistical significance, p<0.05. hCG hormone was undetectably low for day 15 EBs and therefore is marked as “undetected.” ELISA, enzyme-linked immunosorbent assay; TLCs, trophoblast lineage cells.
Characterization by RT-PCR analysis confirmed the presence of trophoblast lineage markers
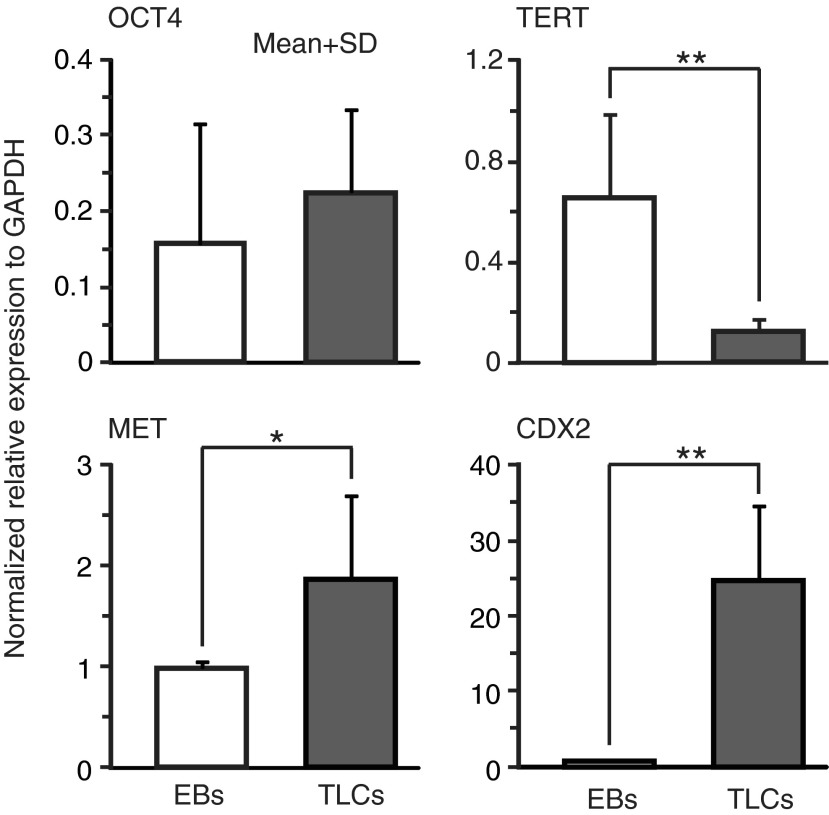
To conclusively determine that hiPSCs on mesh differentiated to cyst-forming TLCs, we performed RT-PCR to examine the expression of OCT4, telomere reverse transcriptase marker (TERT), mesenchymal-epithelial transition marker (MET), and CDX2. These were analyzed in 21 day-old, cyst-containing mesh samples, and the results were compared with those obtained with EBs cultured in a suspension for the same duration. We found considerably low level expression of OCT4 in both TLCs and EBs (Fig. 6, top left), suggesting that both had nearly lost pluripotency over the 21 day period. Conversely, TERT expression was significantly higher (*p<0.1) in EBs than in TLCs (Fig. 6, top right). Since TERT is a marker of cell proliferation,27 this result indicates that EBs maintained proliferation potential relatively much longer than TLCs, although the exact reason for this was not clear to us. Additionally, we examined the expression MET; a transcription marker expressed by cells of epithelial origin and mediates epithelial-mesenchymal transition. The expression of this marker was found to be much higher in TLCs than in EBs (Fig. 6, bottom left), suggesting that cyst-forming cells had to some extent undergone epithelialization, a process known to occur during trophoblast differentiation in vivo.4 Importantly, we found that CDX2 expression was nearly 20-fold higher in TLCs than in EBs (Fig. 6, bottom right). Since CDX2 is the standard marker of trophoblast differentiation, the high level expression provides a compelling evidence for iPSCs differentiation to cyst-forming TLCs under the mesh culture method. Furthermore, these results suggest that the differentiation was not driven by factors inherent in the culture medium, otherwise EBs would have also shown the same expression of markers as TLCs. Further results comparing expression in TLCs and hiPSCs are shown in Supplementary Figure S5.
FIG. 6.
Analysis of RNA expression of selected transcription markers by RT-PCR in differentiated mesh cultured hiPSCs and also in EBs generated by suspension culture at day 21. Top left: OCT4 expression, top right: TERT expression (*p<0.05), bottom left: MET expression (*p<0.1), bottom right: CDX2 expression (**p<0.05). Expression values were obtained relative to GAPDH and normalized by those of control hiPSCs grown on laminin-coated dishes for 3 days. Data are presented as mean±SD of triplicates (n=3). Statistical significance (p-values) was determined by unpaired Student's t-test. TERT, telomere reverse transcriptase marker.
Expression of E-cadherin and vinculin by hiPS cells under mesh culture condition
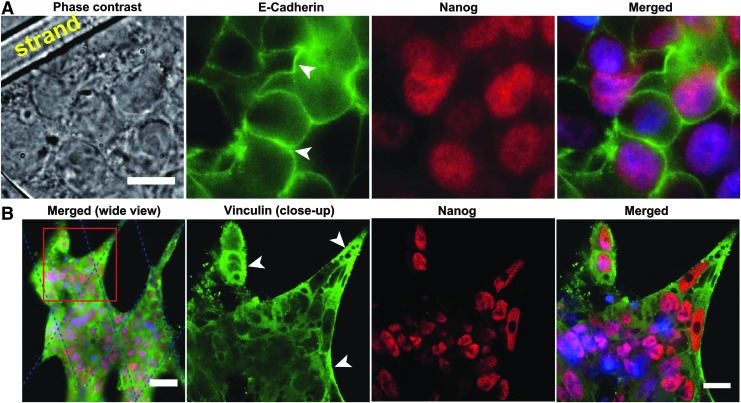
Under mesh culture, cells in close proximity with mesh strands are able to adhere both to the substrate and onto one another, while those in the mesh interior rely solely on cell–cell adhesion. We hypothesized that this differential adhesion may induce the observed cyst formation. To elucidate this, we performed additional immunostaining experiments for E-cadherin and vinculin and examined the expression of these adhesion proteins in hiPSC cultured for 5 days on mesh, just before the onset of differentiation. As expected, markedly stronger expression of E-cadherin was observed at the boundary of cells in the mesh interior compared with cells in close proximity to or in direct contact with the mesh strands (white arrow heads, Fig. 7A). Staining of cytosolic E-cadherin was weak in most cells, suggesting that the adhesion protein was primarily localized at the cell periphery where it functions to maintain cell–cell contact that keep the cells intact in a suspended culture condition. In contrast, vinculin staining was weak particularly in cells in the mesh interior, and appeared to be uniformly distributed in the cytoplasm of these cells. However, we did observe slightly stronger expression of vinculin in cells in direct contact with or close to the mesh strands (white arrow heads, Fig. 7B). As a sign of pluripotency, Nanog was visibly well expressed in all cells, consistent with the results of Figure 1D, which shows that differentiation does not occur until past 8 days of culture on mesh. Although qualitative, the pattern of staining of E-cadherin versus vinculin suggests that the former plays a more pronounced role in cell adhesion under mesh culture. In a nutshell, these results suggest the existence of spatial differences in E-cadherin expression, which may act as a trigger of trophoblast differentiation, as earlier suggested that Ca2+-dependent membrane proteins involved in cell adhesion are required for normal trophoblast differentiation to occur.28
FIG. 7.
Results of immunostaining for E-cadherin and vinculin in hiPSC cultured for 5 days on mesh. Immunofluorescence images showing E-cadherin and Nanog expression (A), and vinculin and Nanog expression (B) by hiPSC on a partially filled mesh. Merged images comprised Hoechst staining of cell nuclei. Comparatively, cells in the mesh interior showed visibly higher expression of E-cadherin stain (white arrow heads in A) compared with cells in contact with or in close proximity to the mesh strands. Contrastingly, the expression of vinculin stain was relatively higher in cells in contact with mesh strands (white arrow heads in B) compared with cells in the mesh interior. Scale bars represent 10 μm in (A), 50 μm in the far left image, and 20 μm in the second to last image of (B). Second to last images are close-up views of the boxed region in the far left image of (B). Blue dotted lines in the far left image of (B) indicate the positions of mesh strands.
Discussion
We have shown that minimization of cell-substrate adhesion using suspended mesh sheets can uniquely initiate the differentiation of hiPSCs to TLCs through a sequence of morphogenetic transformations that begin with individual hiPSCs attached on the strands self-assembling into monolayer cell sheets and ultimately differentiating to form 3D cysts. This demonstrates that the cell-culture milieu presented in this study influences the differentiation, lineage specification, and developmental potential of pluripotent stem cells, in accordance with earlier reports.29
One may wonder as to whether the meshwork imposes some sort of cell selection that may influence the ultimately observed differentiation pattern. At the initial stage of seeding, the adhesion of cells to the meshwork is solely determined by the meshwork architecture, which can be tuned by designing, for instance, meshwork shape and size. We believe that at this stage, given that cells are in a suspension, their interaction with the meshwork is a stochastic process that depend on cell density and the geometry of the interaction surface (in this case, the width of mesh frame and mesh shape). However, although the full mechanism is unclear at this stage, the ability of cells to rapidly mobilize attachment machinery and adhere to the narrow mesh strands can also to some extent determine whether they survive on the mesh or fall off to the dish bottom. In this sense, the meshwork can be regarded as imposing a selection process that may have downstream consequences on differentiation. Remarkably, the process of differentiation observed here is distinctively different from the usual chemical-based derivation model where 3D stem cell aggregates are treated with stimulants such as BMP4 to initiate differentiation to TLCs.30,31
As evidence for trophoblast differentiation, we have examined and found the expression of CDX2, which was significantly highly expressed in mesh generated TLCs compared with EBs generated by suspension culture. We have also determined the secretion of hCG hormone in samples containing cysts formed by differentiated hiPSCs on mesh, which, in combination with CDX2 expression, provides strong evidence for differentiation to TLCs. Although different other factors have been associated with trophoblasts, for simplicity we settled on CDX2, which is the key transcription factor crucial to differentiation and cell fate specification to trophoblast lineage in vivo.4,7,32,33 Indeed, CDX2 is known to drive trophoblast differentiation by downregulating OCT4 and other pluripotency maintaining factors,34–36 and knock out of CDX2 has been shown to interfere with the formation of trophectoderm and even implantation of blastocyst.33,37 Downregulation of OCT4 by CDX2 is consistent with our results that show low level expression of OCT4 versus high level expression of CDX2 (Fig. 6). It should be noted that some studies have associated CDX2 with mesodermal cells in an early stage embryo, but evidences including hCG hormone secretion, high MET expression, and low TERT expression collectively point to a higher likelihood of extraembryonic differentiation in our system as opposed to mesodermal differentiation, otherwise, we would expect a higher expression of pluripotency markers such as OCT4 and TERT instead. Thus, we suggest that the suspended mesh culture method outlined in this study drives the differentiation hiPSCs to TLCs.
Cells interact with the substrate by forming focal adhesions (FAs), which is mediated by the binding of integrin receptors to the substrate (reviewed by Refs.38,39). In the case of mesh culture method described in this study, cell–substrate interaction via FA formation is confined to the narrow mesh strands (<5 μm in width), and considering that the average lateral spacing between adjacent mature FAs is ∼2 μm,40 only a few FA spots (<3 spots) can be accommodated per unit length of the mesh strands. Consequently, cells are stimulated to increase cell–cell adhesion as a compensation for the inadequate cell-substrate adhesion, as supported by relatively high expression of E-cadherin staining along the periphery of cells in the mesh interior. Increased cell–cell interconnectivity results in the formation of a cell sheet by self-assembly organization. The process begins with cells attached on the mesh strands providing support to newly formed cells to spread toward the center of the meshes and continues as more cells divide until the initially open meshes become fully occupied, ultimately resulting in the formation of a cell sheet coverage over the mesh sheet. Thus, E-cadherin-mediated, self-assembly driven cell sheet formation can be regarded as a vital step in the process leading to hiPSC differentiation on mesh, as supported by our observation that cysts could only form on cell sheet covered regions of the mesh sheet.
Based on the results of E-cadherin and vinculin immunostaining, we can suggest that the mesh culture system induces a spatial variation in the expression of E-cadherin, a vital cell–cell adhesion protein with numerous roles ranging from the maintenance of stem cell pluripotency state to induction of trophectoderm formation in early embryos.28,41,42 We postulate that this differential adhesion may play a fundamental role in the differentiation specification to trophoblast. One possible mechanism through which this can occur is via E-cadherin-mediated cell differentiation, sorting, and aggregation in response to differential adhesion, as suggested by the differential adhesive affinity hypothesis proposed by Malcolm Steinberg.43,44 In fact, the essential role of E-cadherin in early embryogenesis has been known for a long time because E-cadherin knockout embryos fail to undergo compaction and do not form the trophectoderm epithelium.42,45 In accordance with this hypothesis, we postulate that cadherin-rich cells may undergo E-cadherin induced mesenchymal-epithelial transition (MET)45 to form an enclosing epithelial envelope around the yet to be differentiated cells. Slowly, differentiation proceeds toward the inside, followed by growth by cell division and swelling due to secretion and inward liquid pumping. In this respect, trophoblast formation in our system may recapitulate the events occurring in early blastocysts, which may not be readily captured by existing embryoid body models. However, elucidating the exact mechanism by which E-cadherin-dependent differential adhesion influences the genetic and epigenetic transformations that lead to differentiation and trophoblast lineage specification in this culture system remains the subject of our future study.
In a nut shell, we show for the first time that adhesion area minimization can trigger direct differentiation of hiPSCs to trophoblast-like cell lineage, providing a novel approach toward the establishment of in vitro human embryo-like models.
Conclusion
This study has investigated the influence of adhesion area minimization on pluripotent stem cell differentiation. We examined the differentiation of hiPSCs cultured on mesh sheets consisting of large size meshes (∼200 μm in pitch) and narrow strands (3–5 μm in width) that are suspended in a culture medium such that cells have to rely on the narrow strands for attachment and growth. Results show that minimization of adhesion area achieved by this novel culture method can initiate the differentiation of hiPSCs to trophoblast-like lineage, resulting in the formation of large spherical cysts that secrete hCG hormone and express trophoblast specific marker such as CDX2. Thus, this study provides a simple mechanical approach to directly induce differentiation of hiPSCs to trophoblasts without necessarily forming EBs and without chemical stimulation. In addition, the cysts can be harvested and utilized as embryo models in in vitro studies of early embryogenesis in humans and also in the development and screening of drugs targeting infertility and placental diseases. Future studies will examine the molecular mechanisms underlying the described differentiation and lineage specification, including detailed analysis of genetic and epigenetic changes in stem cells during culture on mesh.
Supplementary Material
Acknowledgments
We are particularly grateful to Dr. Takashi Tada of Kyoto University for providing the hiPSCs used in this study and for helpful discussions. We thank Dr. Satoshi Tanaka of the University of Tokyo for insightful discussions on characterization of trophoblasts. We would also like to acknowledge the help of Dr. Keiichiro Kushiro of The University of Tokyo in performing ELISA analyses. Finally, we thank Dr. Yuji Teramura of the University of Tokyo and Dr. Yasuhiko Sato of Carl Zeiss Microscopy Co. Ltd for assistance with confocal imaging of trophoblast cysts, and TOBIC (The University of Tokyo IMCB—Olympus Bioimaging Center) for support with immunofluorescence imaging of E-cadherin and vinculin. This work was supported by Japan MEXT KAKENHI grant nos. 25790034 and 23246045, and the Grant-in-Aid for Scientific Research on Innovative Areas; Hyper Bio Assembler for 3D Cellular Innovations no. 26106710.
Author Contributions
K.O.O. conceived and performed experiments, analyzed data, and wrote the article. O.K. performed immunofluorescence and ELISA analyses together with K.O.O. S.Y. performed RT-PCR analyses. H.O., H.K., H.N., and M.W. supervised the work and gave overall guidance during article preparation.
Disclosure Statement
No competing financial interests exist.
References
- 1.Keller G. Embryonic stem cell differentiation: emergence of a new era in biology and medicine. Genes Dev 19, 1129, 2005 [DOI] [PubMed] [Google Scholar]
- 2.Jiang Z., Han Y., and Cao X. Induced pluripotent stem cell (iPSCs) and their application in immunotherapy. Cell Mol Immunol 11, 17, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen Y., Wang K., Gong Y.G., Khoo S.K., and Leach R. Roles of CDX2 and EOMES in human induced trophoblast progenitor cells. Biochem Biophys Res Commun 431, 197, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amita M., Adachi K., Alexenko A.P., Sinha S., Schust D.J., Schulz L.C., Roberts R.M., and Ezashi T. Complete and unidirectional conversion of human embryonic stem cells to trophoblast by BMP4. Proc Natl Acad Sci U S A 110, E1212, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tamm C., Pijuan Galito S., and Anneren C. A comparative study of protocols for mouse embryonic stem cell culturing. PLoS One 8, e81156, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu R., Chen X., Li D., Li R., Addicks G., Glennon C., Zwaka T., and Thomson J. BMP4 initiates human embryonic stem cell differentiation to trophoblast. Nat Biotechnol 284, 143 2002 [DOI] [PubMed] [Google Scholar]
- 7.Bernardo A.S., Faial T., Gardner L., Niakan K.K., Ortmann D., Senner C.E., Callery E.M., Trotter M.W., Hemberger M., Smith J.C., Bardwell L., Moffett A., and Pedersen R.A. BRACHYURY and CDX2 mediate BMP-induced differentiation of human and mouse pluripotent stem cells into embryonic and extraembryonic lineages. Cell Stem Cell 9, 144, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schulz L.C., Ezashi T., Das P., Westfall S.D., Livingston K.A., and Roberts R.M. Human embryonic stem cells as models for trophoblast differentiation. Placenta 29 Supplement, 10, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen D., Zhao M., and Mundy G.R. Bone morphogenetic proteins. Growth Factors 22, 233, 2004 [DOI] [PubMed] [Google Scholar]
- 10.Chen Y., Wang K., Chandramouli G.V., Knott J.G., and Leach R. Trophoblast lineage cells derived from human induced pluripotent stem cells. Biochem Biophys Res Commun 436, 677, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doi A., Park I.H., Wen B., Murakami P., Aryee M.J., Irizarry R., Herb B., Ladd-Acosta C., Rho J., Loewer S., Miller J., Schlaeger T., Daley G.Q., and Feinberg A.P. Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nat Genet 41, 1350, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim K., Doi A., Wen B., Ng K., Zhao R., Cahan P., Kim J., Aryee M.J., Ji H., Ehrlich L.I., Yabuuchi A., Takeuchi A., Cunniff K.C., Hongguang H., McKinney-Freeman S., Naveiras O., Yoon T.J., Irizarry R.A., Jung N., Seita J., Hanna J., Murakami P., Jaenisch R., Weissleder R., Orkin S.H., Weissman I.L., Feinberg A.P., and Daley G.Q. Epigenetic memory in induced pluripotent stem cells. Nature 467, 285, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dalby M.J., Gadegaard N., and Oreffo R.O. Harnessing nanotopography and integrin-matrix interactions to influence stem cell fate. Nat Mater 13, 558, 2014 [DOI] [PubMed] [Google Scholar]
- 14.Nelson C.M., and Bissell M.J. Of extracellular matrix, scaffolds, and signaling: tissue architecture regulates development, homeostasis, and cancer. Annu Rev Cell Dev Biol 22, 287, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scadden D.T. The stem-cell niche as an entity of action. Nature 441, 1075, 2006 [DOI] [PubMed] [Google Scholar]
- 16.Engler A.J., Sen S., Sweeney H.L., and Discher D.E. Matrix elasticity directs stem cell lineage specification. Cell 126, 677, 2006 [DOI] [PubMed] [Google Scholar]
- 17.Li Z., Gong Y., Sun S., Du Y., Lu D., Liu X., and Long M. Differential regulation of stiffness, topography, and dimension of substrates in rat mesenchymal stem cells. Biomaterials 34, 7616, 2013 [DOI] [PubMed] [Google Scholar]
- 18.Kingham E., White K., Gadegaard N., Dalby M.J., and Oreffo R.O. Nanotopographical cues augment mesenchymal differentiation of human embryonic stem cells. Small 9, 2140, 2013 [DOI] [PubMed] [Google Scholar]
- 19.Hogeweg P. Evolving mechanisms of morphogenesis: on the interplay between differential adhesion and cell differentiation. J Theor Biol 203, 317, 2000 [DOI] [PubMed] [Google Scholar]
- 20.Kilian K.A., Bugarija B., Lahn B.T., and Mrksich M. Geometric cues for directing the differentiation of mesenchymal stem cells. Proc Natl Acad Sci U S A 107, 4872, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen G., Gulbranson D.R., Hou Z., Bolin J.M., Ruotti V., Probasco M.D., Smuga-Otto K., Howden S.E., Diol N.R., Propson N.E., Wagner R., Lee G.O., Antosiewicz-Bourget J., Teng J.M.C., and Thomson J.A. Chemically defined conditions for human iPSC derivation and culture. Nat Methods 8, 424, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Coucouvanis E., and Martin G.R. Signals for death and survival: a two-step mechanism for cavitation in the vertebrate embryo. Cell 83, 279, 1995 [DOI] [PubMed] [Google Scholar]
- 23.Abad M., Mosteiro L., Pantoja C., Canamero M., Rayon T., Ors I., Grana O., Megias D., Dominguez O., Martinez D., Manzanares M., Ortega S., and Serrano M. Reprogramming in vivo produces teratomas and iPS cells with totipotency features. Nature 502, 340, 2013 [DOI] [PubMed] [Google Scholar]
- 24.Risau W., Sariola H., Zerwes H.G., Sasse J., Ekblom P., Kemler R., and Doetschman T. Vasculogenesis and angiogenesis in embryonic-stem-cell-derived embryoid bodies. Development 102, 471, 1988 [DOI] [PubMed] [Google Scholar]
- 25.Huang X., Han X., Uyunbilig B., Zhang M., Duo S., Zuo Y., Zhao Y., Yun T., Tai D., Wang C., Li J., Li X., and Li R. Establishment of bovine trophoblast stem-like cells from in vitro-produced blastocyst-stage embryos using two inhibitors. Stem Cells Dev 23, 1501, 2014 [DOI] [PubMed] [Google Scholar]
- 26.Oda M., Tanaka S., Yamazaki Y., Ohta H., Iwatani M., Suzuki M., Ohgane J., Hattori N., Yanagimachi R., Wakayama T., and Shiota K. Establishment of trophoblast stem cell lines from somatic cell nuclear-transferred embryos. Proc Natl Acad Sci U S A 106, 16293, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bodnar A.G., Ouellette M., Frolkis M., Holt S.E., Chiu C.P., Morin G.B., Harley C.B., Shay J.W., Lichtsteiner S., and Wright W.E. Extension of life-span by introduction of telomerase into normal human cells. Science 279, 349, 1998 [DOI] [PubMed] [Google Scholar]
- 28.Coutifaris C., Kao L.C., Sehdev H.M., Chin U., Babalola G.O., Blaschuk O.W., and Strauss J.F., 3rd. E-cadherin expression during the differentiation of human trophoblasts. Development 113, 767, 1991 [DOI] [PubMed] [Google Scholar]
- 29.De Los Angeles A., and Daley G.Q. Stem cells: reprogramming in situ. Nature 502, 309, 2013 [DOI] [PubMed] [Google Scholar]
- 30.Gerami-Naini B., Dovzhenko O.V., Durning M., Wegner F.H., Thomson J.A., and Golos T.G. Trophoblast differentiation in embryoid bodies derived from human embryonic stem cells. Endocrinology 145, 1517, 2004 [DOI] [PubMed] [Google Scholar]
- 31.Lu D., Luo C., Zhang C., Li Z., and Long M. Differential regulation of morphology and stemness of mouse embryonic stem cells by substrate stiffness and topography. Biomaterials 35, 3945, 2014 [DOI] [PubMed] [Google Scholar]
- 32.Roper S., and Hemberger M. Defining pathways that enforce cell lineage specification in early development and stem cells. Cell Cycle 8, 1515, 2009 [DOI] [PubMed] [Google Scholar]
- 33.Strumpf D., Mao C.A., Yamanaka Y., Ralston A., Chawengsaksophak K., Beck F., and Rossant J. Cdx2 is required for correct cell fate specification and differentiation of trophectoderm in the mouse blastocyst. Development 132, 2093, 2005 [DOI] [PubMed] [Google Scholar]
- 34.Roberts R.M., and Fisher S.J. Trophoblast stem cells. Biol Reprod 84, 412, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schiffmacher A.T., and Keefer C.L. CDX2 regulates multiple trophoblast genes in bovine trophectoderm CT-1 cells. Mol Reprod Dev 80, 826, 2013 [DOI] [PubMed] [Google Scholar]
- 36.Niwa H., Toyooka Y., Shimosato D., Strumpf D., Takahashi K., Yagi R., and Rossant J. Interaction between Oct3/4 and Cdx2 determines trophectoderm differentiation. Cell 123, 917, 2005 [DOI] [PubMed] [Google Scholar]
- 37.Hemberger M. Epigenetic landscape required for placental development. Cell Mol Life Sci 64, 2422, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miyoshi H., and Adachi T. Topography design concept of a tissue engineering scaffold for controlling cell function and fate through actin cytoskeletal modulation. Tissue Eng Part B Rev 20, 609, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boettiger D. Mechanical control of integrin-mediated adhesion and signaling. Curr Opin Cell Biol 24, 592, 2012 [DOI] [PubMed] [Google Scholar]
- 40.Kanchanawong P., Shtengel G., Pasapera A.M., Ramko E.B., Davidson M.W., Hess H.F., and Waterman C.M. Nanoscale architecture of integrin-based cell adhesions. Nature 468, 580, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Halbleib J.M., and Nelson W.J. Cadherins in development: cell adhesion, sorting, and tissue morphogenesis. Genes Dev 20, 3199, 2006 [DOI] [PubMed] [Google Scholar]
- 42.Larue L., Ohsugi M., Hirchenhain J., and Kemler R. E-cadherin null mutant embryos fail to form a trophectoderm epithelium. Proc Natl Acad Sci U S A 91, 8263, 1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Steinberg M.S. Reconstruction of tissues by dissociated cells. Some morphogenetic tissue movements and the sorting out of embryonic cells may have a common explanation. Science 141, 401, 1963 [DOI] [PubMed] [Google Scholar]
- 44.Steinberg M.S., and Takeichi M. Experimental specification of cell sorting, tissue spreading, and specific spatial patterning by quantitative differences in cadherin expression. Proc Natl Acad Sci U S A 91, 206, 1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pieters T., and van Roy F. Role of cell-cell adhesion complexes in embryonic stem cell biology. J Cell Sci 127, 2603, 2014 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.