Abstract
Vascularization is a key challenge in tissue engineering. Three-dimensional structure and microcirculation are two fundamental parameters for evaluating vascularization. Microscopic techniques with cellular level resolution, fast continuous observation, and robust 3D postimage processing are essential for evaluation, but have not been applied previously because of technical difficulties. In this study, we report novel video-rate confocal microscopy and 3D postimage processing techniques to accomplish this goal. In an immune-deficient mouse model, vascularized bone tissue was successfully engineered using human bone marrow mesenchymal stem cells (hMSCs) and human umbilical vein endothelial cells (HUVECs) in a poly (d,l-lactide-co-glycolide) (PLGA) scaffold. Video-rate (30 FPS) intravital confocal microscopy was applied in vitro and in vivo to visualize the vascular structure in the engineered bone and the microcirculation of the blood cells. Postimage processing was applied to perform 3D image reconstruction, by analyzing microvascular networks and calculating blood cell viscosity. The 3D volume reconstructed images show that the hMSCs served as pericytes stabilizing the microvascular network formed by HUVECs. Using orthogonal imaging reconstruction and transparency adjustment, both the vessel structure and blood cells within the vessel lumen were visualized. Network length, network intersections, and intersection densities were successfully computed using our custom-developed software. Viscosity analysis of the blood cells provided functional evaluation of the microcirculation. These results show that by 8 weeks, the blood vessels in peripheral areas function quite similarly to the host vessels. However, the viscosity drops about fourfold where it is only 0.8 mm away from the host. In summary, we developed novel techniques combining intravital microscopy and 3D image processing to analyze the vascularization in engineered bone. These techniques have broad applicability for evaluating vascularization in other engineered tissues as well.
Introduction
Vascularization is a major challenge in tissue engineering because nearly all of the tissues and organs in the body receive nourishment and discard waste through vascular systems.1,2 Diffusion can only supply nutrition and oxygen, and remove metabolic waste and carbon dioxide across a distance of 200–250 μm from the end of a microvessel.3 Without adequate vascularization, apoptosis or necrosis will occur, beginning at the center of the engineered tissue construct.
Evaluating vascularization is a crucial step in predicting the fate of engineered implants. The development of new strategies for tissue vascularization is needed to realize the full potential of tissue engineering.4,5 Two critical components of vascularization, morphology, and microcirculation must be evaluated. These reflect the anatomic and functional components of vascularization, respectively. Histology has conventionally been used to evaluate the morphologic aspect of tissue vascularization,6 while blood cell velocity can be used to assess the functional side of microcirculation.7 Unfortunately, histology is an invasive endpoint technique, in which sacrificing the animal is required.8 Furthermore, it precludes analysis of the tissue microcirculation. Some noninvasive methods, such as radiological imaging techniques, have been employed to evaluate the morphologic features of vascularization in engineered tissues.9 Micro-CT provides a good rendering of the 3D structure of the vessels,10 and magnetic resonance imaging can be used to track groups of cells as a stream when cells are labeled with a contrast agent or reporter probe.11 However, each of the current radiologic imaging techniques lack the cellular-level resolution required to distinguish one cell from another. Furthermore, radiology-based techniques cannot differentiate between types of cells and tissues. Since vascular systems are composed of multiple types of cells, radiologic techniques are not sufficient for complete evaluation.
At present, optical microscopy is the method of choice for imaging live tissues at the cellular and subcellular levels.12 With advances in optical imaging technology, the in vivo imaging of live cells and tissues has become possible. Confocal microscopy and multiphoton microscopy have been applied to in vivo imaging and provide 3D morphological information.13 However, standard confocal or multiphoton microscopic techniques do not have a rapid enough scanning rate to capture the motion of fast moving blood cells within the vessels. As a result, they can only be utilized to evaluate the structure of the neovascularization, but not the microcirculation.
Video-rate microscopy is an emerging imaging technique that employs a 30 frames per second (FPS) full frame scanning speed, utilized to track the motion of a single blood cell.14,15 While this advancement in microscopic technology is encouraging, it presents challenges in evaluating tissue vascularization. First, a microvessel system has multiple cell types and a complex structure, especially when blood cells are traveling within the vessel lumen. Visualization of blood cells within a sealed vessel was accomplished previously in our laboratory. Second, video-rate microscopy is still not fast enough to perform continuous z-stack scanning of both the blood vessel and moving blood cells, which means it can only perform one time point z-stack scanning or continuous scanning in one single z-plane.
In this study, we report a novel approach for analyzing the vascularization of tissue-engineered bone, both morphology and functionally, by evaluating the microcirculation within the neovascularization. Our technique enables the simultaneous visualization of both the 3D vascular morphology and the fast moving blood cells. In addition, we performed quantitative postimage analysis on the vascular network and blood cell viscosity.
Materials and Methods
Cell culture and labeling
Human bone marrow samples were obtained according to guidelines approved by our Hospital Institutional Review Board from patients undergoing hip replacement surgery. Human bone marrow mesenchymal stem cell (hMSC) cultures were established as previously described16 and maintained in a mesenchymal stem cell growth medium (MSCGM) (Cambrex). All experiments were performed with hMSCs having less than six cell passages. Human umbilical vein endothelial cells (HUVECs) were maintained in 0.1% gelatin-coated plates in the EGM-2 medium (Cambrex). hMSCs and HUVECs were transduced with lentivirus encoding eGFP and dTomato, respectively, as reported previously.17 10× diluted whole blood from a Balb/C mouse (Jackson Laboratory) was labeled with 100 μM Vibrant DiD before in vivo imaging.
Engineered vascularized bone tissues
Using the protocol established previously in our laboratory,17 porous (d,l-lactide-co-glycolide) (PLGA) scaffolds, 5 mm in diameter, were fabricated using the sucrose leaching technique. 0.1 mL of collagen and fibrin hydrogel containing 1×105 EC and 2.5×104 MSCs were seeded into the scaffolds and cultured in the osteoinductive medium (MSCGM with 50 μg/mL l-ascorbic acid 2-phosphate, 10 mM β-glycerol phosphate, and 10−8 M dexamethasone). Constructs were cultured in the EGM-2 medium during the in vitro experiments or overnight before implanting.
In vivo imaging
All animal procedures were approved by the Institutional Animal Care and Use Committee of the Massachusetts General Hospital and performed according to the National Institutes of Health Guidelines for the Care and Use of Laboratory Animals. Engineered bone constructs were implanted subcutaneously in the backs of severe combined immunodeficient mice (NOD.CB17; Jackson Laboratory) and imaged up to 8 weeks after implantation. Immediately before imaging, mice were anesthetized with ketamine/xylazine and retro-orbitally injected with up to 1×108 DiD-labeled whole blood cells isolated from a Balb/C mouse (Jackson Laboratory). An incision was made to expose the engineered bone grafts for imaging, and the mice were mounted on the heated stage of a custom-built video-rate laser-scanning confocal microscope.18 The video-rate images of fluorescently labeled hMSCs, HUVECs, and whole blood cells were captured using a 30×0.9 N.A. water immersion objective (LOMO) or a 60×1.2 N.A. water immersion objective (Olympus). EGFP and tdTomato were excited with a 491 nm continuous wave laser (Cobalt) and detected through a 520±20-nm bandpass filter (Semrock) and a 593±20-nm bandpass filter (Semrock) by two separate photomultiplier tubes (Hamamatsu Photonics). Vibrant DiD (Invitrogen) was excited using a 635 nm continuous wavelength laser (Coherent) and detected through a 695±27.5-nm bandpass filter (Omega Optical) by a third photomultiplier tube.
When necessary, mosaic image processing was used to extend the visual field, preserving the optical resolution as documented previously.19
Image processing
Static 3D image reconstruction
Three-dimensional volume and orthogonal image reconstruction was performed as documented previously.19 To visualize both the morphology of the vessel and the blood cells, as well as evaluate the location of the blood cells, vessels were presented in the orthogonal view and cells in the volume view. The reconstructed images were overlaid in 3D. The transparency of the reconstructed images was adjusted as needed.
Dynamic 3D image reconstruction
The rationale for engineering prevascularized bone in vitro is the hypothesis that microvessels persist in vivo after implantation and anastomose with the host microvessels so that the blood cells from the host can circulate within the pre-engineered microvessels improving tissue survival. Therefore, the integrity of the microvessels and the circulating blood cells located within the engineered vessels need to be validated.
When imaging blood cells flow through the vessels, the images consist of a relatively static structure (the vessel) and highly dynamic components (the blood cells). Visualizing both structures is essential to confirm that the lumen of the vessel exists and that the blood cells are circulating within the lumen and not just the host tissue. As mentioned previously, continuously performed stacked images to track the moving blood cells are not feasible even on the video-rate microscope. Since the primary purpose for dynamically imaging the blood cells is to track the cells and analyze their velocity so as to evaluate the function of the microcirculation, 3D morphology rendering of the blood cells is not necessary. We, therefore, carried out a hybrid image processing approach, in which we performed a stack scanning of the blood vessel followed by continuously performed single-plane imaging at 30 FPS. Three-dimensional volume reconstruction was then undertaken, and the dynamic images of the blood cells were then overlaid onto the 3D images at the depth of recording.
Microvascular network analysis
The length of the networks and the network intersections were analyzed using our custom-developed software (unpublished data). Z image projection was first performed in the stacked image, and the binary and skeletonization were then performed on the projected image. The total length of the vascular network in the projected image (Lp) was calculated. A digital slicing in the XZ orientation was performed to calculate the average angle (α) between a microvascular branch and the XY-plane. The total length (Lt) was calculated using the equation L=Lp/Cos (α). For the intersection analysis, first the total number of the intersections (It) was calculated and located in the projected image. The located intersections were then verified using our custom-developed software to exclude overhanging branches without connection. The density of the intersections was then calculated.
The mosaic scanned images in each plane of a z-stack image were stitched, followed by a Z-projection process. Binary and skeletonization were then performed on the projected image. The total network length (Lprojection) junctions and junction densities were calculated using our custom-developed computer program using MATLAB (Mathworks). To calculate the total vascular network length in 3D, a digital reslicing in the Z-section was performed, and the average angle (α) between the vascular network and the XY-plane was measured from the Z-section images. The total vascular network length (Ltotal) was then calculated using the following equation: Ltotal = Lprojection/Cos(α).
Blood cell velocity analysis
The microcirculation of the labeled blood cells was recorded at 30 FPS. Individual blood cells were tracked using ImageJ and a custom-developed program using MATLAB. The speed of the cells in the engineered bone (Veng) in its periphery and center, as well as the nearest adjacent area outside the engineered bone (Vadj), was calculated. Since anesthesia affects the blood cell velocity, we calculated relative speed V in our anesthetized mice using the equation V=Veng/Vadj.
Results
Three-dimensional imaging provides morphological evidence that hMSCs are pericytes stabilizing the microvascular network both in vitro and in vivo
As shown in Figure 1 and Supplementary Video S1 (Supplementary materials are available online at http://www.liebertpub.com/tec), using 3D volume reconstruction, the morphology and localization of hMSCs and HUVECs are clearly visualized. The hMSCs attached tightly around the vascular luminal structure formed by HUVECs, thus stabilizing the vascular network for a longer period of time. In contrast, with the HVECs alone, the microvascular structure degraded.
FIG. 1.
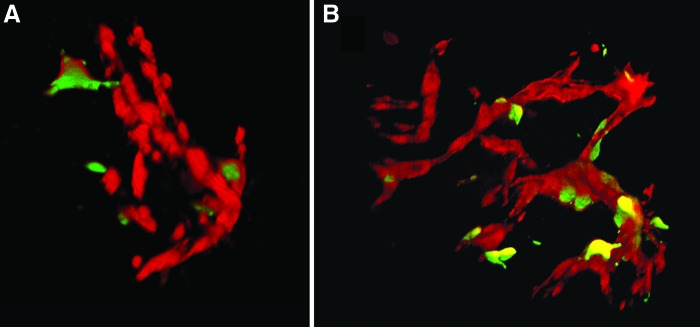
The human bone marrow mesenchymal stem cells (hMSCs) (green) serve as pericytes stabilizing the microvascular network (red) as visualized in vitro (A) and in vivo (B). Yellow is overlay of red and green colors. Color images available online at www.liebertpub.com/tec
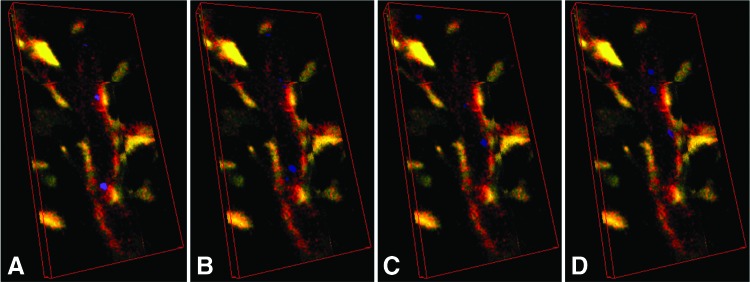
Three-dimensional imaging better elucidates the integrity and architecture of the engineered microvessel and the circulating blood cells are localized within the vessel
A successfully engineered microvascular vessel fulfills two requirements: an intact luminal structure and a conduit for circulating blood cells. As documented above, using in vivo confocal Z axis scanning and 3D image reconstruction, the luminal structure was successfully visualized. Since the blood cells are embedded in the lumen of the vessel, visualization of the vessel and the blood cells is challenging. We successfully accomplished this by (i) dynamically changing the transparency of the vessel wall confirming the localization of the blood cells within the vessels and (ii) by orthogonal viewing, as shown in Figure 2 and Supplementary Video S2. Blood cells were confirmed to be located within the vessel, and the vessel was confirmed to be intact without leakage. The dynamic nature of the blood cell circulation was also visualized through the hybrid image reconstruction, which demonstrated that the blood cells were contained within a static vessel as shown in Figures 2 and 3 and Supplementary Video S3.
FIG. 2.

The orthogonal view of the 3D reconstructed images of the microvascular network composed of hMSCs (green), human umbilical vein endothelial cells (red), and blood cells (blue). (A–C) Represent a different perspective of views. Yellow is overlay of red and green colors. Color images available online at www.liebertpub.com/tec
FIG. 3.
Dynamic hybrid 3D view of the moving blood cells inside the engineered blood vessels. Two-dimensional and 3D view were used to demonstrate the fast moving cells and static vessel, respectively. (A–D) The hMSCs, human umbilical vein endothelial cells (HUVECs) and blood cells were fluorescently labeled with green, red and blue colors respectively. Show representative images at different time points. Yellow is overlay of red and green colors. Color images available online at www.liebertpub.com/tec
Quantitative evaluation of the microvascular network was accomplished through mosaic imaging and postimage processing
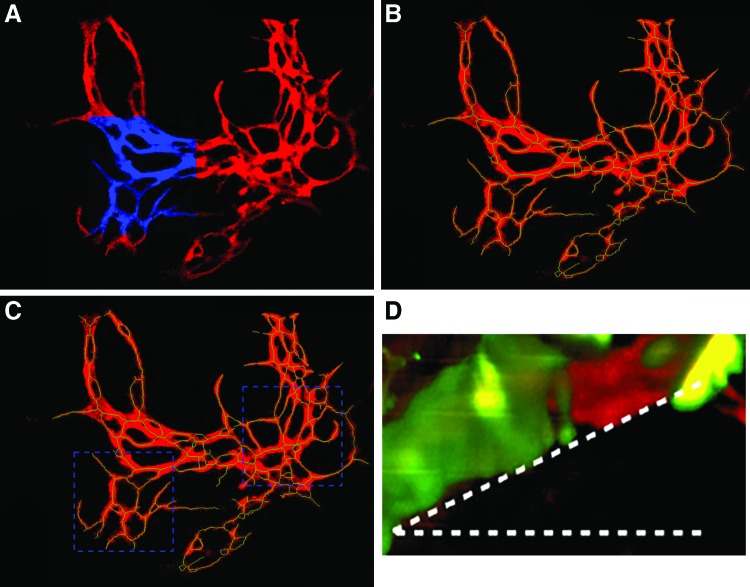
Quantitative analysis on microvascular networks using optical imaging is also challenging. One visual field only provides a few hundred micrometers field of view. This does not well represent the characteristics of the relatively large vascular network. To expand the visual field while preserving the resolution, we used the mosaic imaging technique. As shown in Figure 4A, the high resolution of the optics was preserved, while the visual field was dramatically expanded.
FIG. 4.
The (A) is a representative projected mosaic image where the blue areas represent the regular one visual field. The binary and skeleton images of the (A) are shown in red and green in the (B), respectively. The (C) shows two regions of interest for comparing the degree of the vascularization. The (D) shows a representative angle of 16.5° between a vascular branch and the XY-plane. Color images available online at www.liebertpub.com/tec
Quantitative analysis is also important. It provides the information regarding the degree of angiogenesis so as to optimize the strategies for angiogenesis. Figure 4B shows the skeletonized image overlaying the binary image. The calculated total length of the vascular network is 11,839 μm in the projected image A, and representative angle of 16.5° between a vascular branch and the XY-plane is shown in Figure 4D and the average angle is 17.2°. The total length of the vascular network in 3D is 12,393 μm.
The total identified intersections are 165, as shown in Figure 4B, with a density of 131 intersections/mm2. To compare the degree of angiogenesis between different specific areas, regions of interest (ROIs) were defined as shown in Figure 4C. The two square ROIs are of the same area of 0.11 mm2. The total length and number of intersections of left and right ROI are 1,796 μm and 21 intersections VS 2,824 μm and 47 intersections.
Computer-assisted cell tracking provides functional evaluation of engineered vascular network
The velocity of the blood cells in the microvascular 2network is a crucial parameter that indicates its function. We found that the velocities of the blood cells in the peripheral areas, which connected to the host, showed a similar (p<0.05) velocity compared with the host. However, the velocity drops significantly with increased distance from the interface between the implant and the host to about fourfold at 800 μm away from the junction between the engineered vessel and the host at 8 weeks after implantation. Representative figure and video are shown in Figure 5 and Supplementary Video S4.
FIG. 5.

The (left) (adjacent to host) and (right) (away from the host) are representative images of the blood cells circulating within the engineered bone. The hMSCs, HUVECs and blood cells were fluorescently labeled with green, red and blue colors rspectively. Color images available online at www.liebertpub.com/tec
Discussion
The field of tissue engineering has seen a surge in research and discovery lately. Great progress has been made in many tissue systems that have allowed translational discoveries to be adapted for patient use, including the FDA approval of engineered skin.20 Other tissue systems are currently in clinical trials.21 These translational successes have been in tissues that require less vascularity. The establishment of a competent vascular network remains one of the major challenges in the widespread success of tissue engineering.1,22 Although there are strategies being developed to improve vascularization, we continue to lack effective and efficient tools to observe and evaluate vascularization of engineered tissues in a nondestructive way.
There are currently three strategies for establishing vascularization within engineered tissues.1 (i) Engineering scaffolds with luminal structures.23 (ii) Angiogenic growth factor delivery.24,25 (iii) In vitro engineering a prevascularized tissue before implantation.26,27 It is well accepted that the third strategy is the only viable solution for larger volume tissues and rapid vascularization. Due to this, it is more promising than the others for translational purposes. In addition, the prevascularized tissue has the most complex static and dynamic architecture and its vascular evaluation techniques can be applied to the other two approaches. Therefore, in this project, we utilized a prevascularization model for a broader applicability.
Compared with other techniques, imaging of live animals at microscopic resolution is a powerful tool for addressing how the biological or pathological phenomena happen in the natural environment.15,28 Intravital microscopy (IVM) has been applied to study the vascular networks in tumors29 and evaluate the vascular response in a peripheral arterial disease model. The IVM technology has also been applied in tissue engineering studies previously. Collagen fiber dynamics within the engineered collagenous tissue was studied using intravital nonlinear optical microscopic imaging in an in vitro bioreactor.30 More recently, IVM was performed in a live animal. In a simple cell–hydrogel mixture model, IVM was used to assess blood flow and oxygenation after implantation, but only 2D image processing was applied.
In our current study, we performed IVM within mice and used a more robust engineered bone model with a larger porous scaffold, induced osteoblasts from hMSCs, endothelial cells, and pericyte-like hMSCs. Compared with prior studies, our 3D image reconstruction provided better visualization, and the results were more accurate. We demonstrated that 3D imaging is superior to 2D imaging for evaluating the lumen of the vascular network. It is consistent with our previous finding using 2D imaging that hMSCs serve as pericytes.31 However, in comparison, the 3D imaging provides better evidence for the role of hMSCs in stabilizing the microvessels formed by HUVECs.
We also sought to evaluate whether the blood cells were circulating within intact engineered vessels. We have previously imaged blood cells circulating in a cross-sectional view of the engineered blood vessel. However, there lacked direct evidence that the vessel was intact and the wall of the vessel was composed of only hMSCs and HUVECs rather than host cells. Therefore, we needed to visualize both the 3D vessel wall and the blood cells to confirm that the blood cells were contained and circulating within the engineered vessels. Using a hybrid approach, we successfully accomplished this. To the best of our knowledge, our novel hybrid image reconstruction is the first reported in the literature, in which both the wall of vessel and the blood cells can be visualized using optical sections.
IVM, as with other optical imaging techniques, has a visual field that covers only a small area. Quantitatively, evaluating the vascular network requires a relatively larger area to achieve reliable results. In this study, we used mosaic imaging techniques, which preserved the resolution and expanded the visual field. Through postimage processing, the vascular network length per unit and intersection density were successfully evaluated. Another novel approach we have taken is that by digital reslicing of the stacked confocal images, we were able to calculate the angle between a vascular branch and the XY-plane, thus simplifying the complex 3D length computing.
Functional analysis is another crucial factor for vascular network evaluation. The rapidity with which the blood cells move within a vessel presents a technical challenge. Our video rate microscopy showed the advantage of recording the dynamic circulation of the blood cells in real time. The anastomosis usually occurs 2 days after implantation. We anticipated the velocity of blood cells within the implant to be close to that of the host after 8 weeks. However, using computer-assistant cell tracking, we found that even 8 weeks after implantation, the circulation velocity was up to fourfold lower, only 800 μm away from the interface. Further long observation is planned to elucidate the mechanism.
In summary, we have developed a novel platform to evaluate the vascularization of engineered tissues using IVM and 3D image postprocessing. We have been successfully applying it to engineered bone tissue and it appears to have a broad applicability to other engineered tissues.
Supplementary Material
Acknowledgments
This project was partially found by the Peabody Foundation (B.G. and Y. P.), the Constance and Anthony Franchi Fund (B.G.), and the NVIDIA Academic partnership (Y.P.).
Disclosure Statement
No competing financial interests exist.
References
- 1.Rouwkema J., Rivron N.C., and van Blitterswijk C.A. Vascularization in tissue engineering. Trends Biotechnol 26, 434, 2008 [DOI] [PubMed] [Google Scholar]
- 2.Auger F.A., Gibot L., and Lacroix D. The pivotal role of vascularization in tissue engineering. Annu Rev Biomed Eng 15, 177, 2013 [DOI] [PubMed] [Google Scholar]
- 3.Grimes D.R., Kelly C., Bloch K., and Partridge M. A method for estimating the oxygen consumption rate in multicellular tumour spheroids. J R Soc, Interface 11, 20131124, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Das A., and Botchwey E. Evaluation of angiogenesis and osteogenesis. Tissue Eng Part B Rev 17, 403, 2011 [DOI] [PubMed] [Google Scholar]
- 5.Krishnan L., Willett N.J., and Guldberg R.E. Vascularization strategies for bone regeneration. Ann Biomed Eng 42, 432, 2014 [DOI] [PubMed] [Google Scholar]
- 6.Wang D., Stockard C.R., Harkins L., Lott P., Salih C., Yuan K., Buchsbaum D., Hashim A., Zayzafoon M., Hardy R.W., Hameed O., Grizzle W., and Siegal G.P. Immunohistochemistry in the evaluation of neovascularization in tumor xenografts. Biotech Histochem 83, 179, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rauchfuss F., Scheuerlein H., Ludewig S., Uberruck T., Heise M., Zanow J., and Settmacher U. In vivo assessment of the hepatic microcirculation after mesenterico-portal bypass (REX-shunt) using orthogonal polarization spectral imaging. Liver int 30, 1339, 2010 [DOI] [PubMed] [Google Scholar]
- 8.Seebach C., Henrich D., Kahling C., Wilhelm K., Tami A.E., Alini M., and Marzi I. Endothelial progenitor cells and mesenchymal stem cells seeded onto beta-TCP granules enhance early vascularization and bone healing in a critical-sized bone defect in rats. Tissue Eng Part A 16, 1961, 2010 [DOI] [PubMed] [Google Scholar]
- 9.Civelek A.C., Pacheco E.M., Natarajan T.K., Wagner H.N., Jr., and Iliff N.T. Quantitative measurement of vascularization and vascular ingrowth rate of coralline hydroxyapatite ocular implant by Tc-99m MDP bone imaging. Clin Nucl Med 20, 779, 1995 [DOI] [PubMed] [Google Scholar]
- 10.Munce N.R., Strauss B.H., Qi X., Weisbrod M.J., Anderson K.J., Leung G., Sparkes J.D., Lockwood J., Jaffe R., Butany J., Teitelbaum A.A., Qiang B., Dick A.J., and Wright G.A. Intravascular and extravascular microvessel formation in chronic total occlusions a micro-CT imaging study. JACC Cardiovasc Imaging 3, 797, 2010 [DOI] [PubMed] [Google Scholar]
- 11.Rogers W.J., Meyer C.H., and Kramer C.M. Technology insight: in vivo cell tracking by use of MRI. Nat Clin Pract Cardiovasc Med 3, 554, 2006 [DOI] [PubMed] [Google Scholar]
- 12.Lucotte B., and Balaban R.S. Motion compensation for in vivo subcellular optical microscopy. J Microsc 254, 9, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dickie R., Bachoo R.M., Rupnick M.A., Dallabrida S.M., Deloid G.M., Lai J., Depinho R.A., and Rogers R.A. Three-dimensional visualization of microvessel architecture of whole-mount tissue by confocal microscopy. Microvasc Res 72, 20, 2006 [DOI] [PubMed] [Google Scholar]
- 14.Li C., Pastila R.K., Pitsillides C., Runnels J.M., Puoris'haag M., Cote D., and Lin C.P. Imaging leukocyte trafficking in vivo with two-photon-excited endogenous tryptophan fluorescence. Opt Express 18, 988, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Spencer J.A., Ferraro F., Roussakis E., Klein A., Wu J., Runnels J.M., Zaher W., Mortensen L.J., Alt C., Turcotte R., Yusuf R., Cote D., Vinogradov S.A., Scadden D.T., and Lin C.P. Direct measurement of local oxygen concentration in the bone marrow of live animals. Nature 508, 269, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pang Y., Cui P., Chen W., Gao P., and Zhang H. Quantitative study of tissue-engineered cartilage with human bone marrow mesenchymal stem cells. Arch Facial Plast Surg 7, 7, 2005 [DOI] [PubMed] [Google Scholar]
- 17.Tsigkou O., Pomerantseva I., Spencer J.A., Redondo P.A., Hart A.R., O'Doherty E., Lin Y., Friedrich C.C., Daheron L., Lin C.P., Sundback C.A., Vacanti J.P., and Neville C. Engineered vascularized bone grafts. Proc Natl Acad Sci U S A 107, 3311, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Veilleux I., Spencer J.A., Biss D.P., Côté D., and Lin C.P. In vivo cell tracking with video rate multimodality laser scanning microscopy. IEEE J Sel Top Quantum Electron 14, 10, 2008 [Google Scholar]
- 19.Pang Y., Ucuzian A.A., Matsumura A., Brey E.M., Gassman A.A., Husak V.A., and Greisler H.P. The temporal and spatial dynamics of microscale collagen scaffold remodeling by smooth muscle cells. Biomaterials 30, 2023, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Purdue G.F., Hunt J.L., Still J.M., Jr., Law E.J., Herndon D.N., Goldfarb I.W., Schiller W.R., Hansbrough J.F., Hickerson W.L., Himel H.N., Kealey G.P., Twomey J., Missavage A.E., Solem L.D., Davis M., Totoritis M., and Gentzkow G.D. A multicenter clinical trial of a biosynthetic skin replacement, Dermagraft-TC, compared with cryopreserved human cadaver skin for temporary coverage of excised burn wounds. J Burn Care Rehabil 18, 52, 1997 [DOI] [PubMed] [Google Scholar]
- 21.https://clinicaltrials.gov/
- 22.Post M.J., Rahimi N., and Caolo V. Update on vascularization in tissue engineering. Regen Med 8, 759, 2013 [DOI] [PubMed] [Google Scholar]
- 23.Lee V.K., Lanzi A.M., Haygan N., Yoo S.S., Vincent P.A., and Dai G. Generation of multi-scale vascular network system within 3D hydrogel using 3D bio-printing technology. Cell Mol Bioeng 7, 460, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jay S.M., Shepherd B.R., Andrejecsk J.W., Kyriakides T.R., Pober J.S., and Saltzman W.M. Dual delivery of VEGF and MCP-1 to support endothelial cell transplantation for therapeutic vascularization. Biomaterials 31, 3054, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Freeman I., and Cohen S. The influence of the sequential delivery of angiogenic factors from affinity-binding alginate scaffolds on vascularization. Biomaterials 30, 2122, 2009 [DOI] [PubMed] [Google Scholar]
- 26.Laschke M.W., Mussawy H., Schuler S., Kazakov A., Rucker M., Eglin D., Alini M., and Menger M.D. Short-term cultivation of in situ prevascularized tissue constructs accelerates inosculation of their preformed microvascular networks after implantation into the host tissue. Tissue Eng Part A 17, 841, 2011 [DOI] [PubMed] [Google Scholar]
- 27.Thomson K.S., Korte F.S., Giachelli C.M., Ratner B.D., Regnier M., and Scatena M. Prevascularized microtemplated fibrin scaffolds for cardiac tissue engineering applications. Tissue Eng Part A 19, 967, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pittet M.J., and Weissleder R. Intravital imaging. Cell 147, 983, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carmeliet P., and Jain R.K. Molecular mechanisms and clinical applications of angiogenesis. Nature 473, 298, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu J.J., Humphrey J.D., and Yeh A.T. Characterization of engineered tissue development under biaxial stretch using nonlinear optical microscopy. Tissue Eng Part A 15, 1553, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cai X., Lin Y., Friedrich C.C., Neville C., Pomerantseva I., Sundback C.A., Zhang Z., Vacanti J.P., Hauschka P.V., and Grottkau B.E. Bone marrow derived pluripotent cells are pericytes which contribute to vascularization. Stem Cell Rev 5, 437, 2009 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.