SUMMARY
Norovirus (NV) is an important cause of acute gastroenteritis in children, but is also frequently detected in asymptomatic children, which complicates the interpretation of NV detection results in both the clinical setting and population prevalence studies. A total of 807 faecal samples from children aged <5 years hospitalized for acute gastroenteritis were collected in Thai Binh, Vietnam, from January 2011 to September 2012. Real-time RT–PCR was used to detect and quantify NV-RNA in clinical samples. A bimodal distribution of cycle threshold (Ct) values was observed in which the lower peak was assumed to represent cases for which NV was the causal agent of diarrhoea, whereas the higher peak was assumed to represent cases involving an alternative pathogen other than NV. Under these assumptions, we applied finite-mixture modelling to estimate a threshold of Ct <21·36 (95% confidence interval 20·29–22·46) to distinguish NV-positive patients for which NV was the likely cause of diarrhoea. We evaluated the validity of the threshold through comparisons with NV antigen ELISA results, and comparisons of Ct values in patients co-infected with rotavirus. We conclude that the use of an appropriate cut-off value in the interpretation of NV real-time RT–PCR results may improve differential diagnosis of enteric infections, and could contribute to improved estimates of the burden of NV disease.
Key words: Caliciviruses, diarrhoea, estimating disease prevalence, gastrointestinal infections, modelling
INTRODUCTION
Norovirus (NV) (family Caliciviridae, genus Norovirus) is a major cause of gastrointestinal disease worldwide, and the cause of an estimated 200 000 deaths and 1·1 million hospitalizations in children <3 years of age annually [1, 2]. The NV detection rate in diarrhoeal patients varies from as high as 31–48% [3–5] to as low as 3–5% [6], suggesting that the burden of NV disease may be highly variable geographically. Similarly, a wide range of NV prevalence (5–48%) has been observed in different regions of Vietnam [7–10]. While transmission dynamics may indeed vary across regions, differences between studies may also reflect differences in diagnostic methodology. Detection sensitivities depend on many factors such as sample preparation methods, RNA extraction, presence of reverse-transcriptase inhibitors, primer design, amplification chemistries (e.g. enzymes/buffers), virus concentrations in the sample (influenced by the time elapsed from the onset of diarrhoea to sampling, and sample storage conditions), as well as viral factors such as genetic variation in circulating strains. Interpreting NV detection results in both the clinical setting and in population prevalence studies is complicated by the fact that NV is also frequently detected by reverse-transcriptase–polymerase chain reaction (RT–PCR) in asymptomatic individuals [10–17]. Only a few studies have examined and compared the detection of NV between diarrhoeal cases and concurrent non-diarrhoeal controls (reviewed in [18]). In some studies, similar NV detection rates have been observed in both cases and controls, or even higher rates in controls vs. diarrhoea cases [12, 19]. NV diarrhoeal patients are known to shed virus for prolonged periods of time following recovery [20].
Shedding of microorganisms without diarrhoeal symptoms is a common phenomenon for many enteric pathogens, such as neonatal rotavirus (RV), bocavirus, NV, Vibrio cholerae O1, enterotoxigenic Escherichia coli, enteropathogenic E. coli, Campylobacter jejuni and Giardia lamblia [21–23]. Asymptomatic shedding complicates patient management, studies of disease burden, and monitoring of vaccine trials. Improved methods of inferring a causative role of NV in clinical diagnostics would be a welcome contribution to transmission studies and modelling efforts, with potential applications to clinical diagnostics. Therefore, the objective of this study was to identify a cut-off value for viral load of NV in diarrhoea samples to infer a causative role in diarrhoea.
We started from the observation that distributions of NV Ct values in diarrhoeal cases are typically bimodal [19]. We hypothesized that the lower Ct value peak corresponds to cases for which the cause of the diarrhoea is NV, and the higher Ct value peak corresponds to cases for which another co-infecting agent is the principal cause of diarrhoea. We modelled the NV Ct value bimodal distribution with a finite-mixture model, which allowed us to identify a Ct threshold value associated with disease risk. We then tested our initial hypothesis using information on RV and NV co-infections as determined by antigen enzyme-linked immunosorbent assay (ELISA).
METHODS
Sample collection
The samples and dataset used in this study were generated from a large prospective, hospital-based diarrhoea study in Thai Binh Paediatric Hospital, Thai Binh, Vietnam. Faecal samples of children aged <5 years hospitalized acute gastroenterititis were collected upon obtaining parental consent from 2011 to 2012. Inclusion criteria included diarrhea episode ⩾3 times per 24 h, and admission to hospital within 7 days from onset. The study was approved by the Medical Research Ethical Committee of the National Institute of Hygiene and Epidemiology, Vietnam, and the Internal Review Board of Nagasaki University.
NV detection and genotyping
For all faecal specimens, 20% suspension in DEPC-treated water was prepared for viral RNA extraction using Qiamp viral RNA extraction kit (Qiagen, Germany). NV genogroups GI and GII were detected as previously described [8, 24, 25], for which the detection limit has been determined as 10–50 RNA copies/reaction. The assay can detect the following different genotypes: GI.1, GI.2, GI.3, GI.4, GI.6, GI.8 and GII.2, GII.3, GII.4, GII.6, GII.7, GII.10, GII.12, GII.13. NV genotyping was performed by amplification of the VP1 gene as described previously [26], followed by sequencing of PCR products or pGEM-T cloned vector products on a 3130xl Genetic Analyzer (Applied Biosystems, USA).
Sequences were manually aligned with BioEdit v. 7.0.5 and submitted to the online Norovirus Genotyping Tool v. 1.0 (www.rivm.nl/mpf/norovirus/typingtool) for genotype determination [27] .
Evaluation of cut-off cycle threshold for NV
The cycle threshold (Ct) value from the real time RT–PCR was used as a proxy measure of faecal viral load; Ct <40 was considered positive. Ct values are inversely proportional (on a logarithmic scale) to viral load, hence lower Ct values correspond to higher viral loads. The same positive control was used throughout all experiments, and its Ct value varied within 0·5. Threshold level of the thermocycler (0·03) remained constant throughout the analysis.
Modelling of Ct distributions by finite-mixture models
The typical bimodal distribution of NV Ct values (Figs 1 and 2) was modelled by a finite-mixture model using continuous unimodal distributions from the exponential family [28]. In the absence of prior information on expected distribution for each peak, we evaluated the normal, log-normal, gamma, Weibull distributions, and all possible combinations (4 × 4 = 16). The normal distribution is defined on all the real numbers, whereas the three other distributions (log-normal, Weibull, gamma) are defined on positive reals. These distributions are all characterized by two parameters: a location parameter μ and a scale parameter σ. The first parameter accounts for most of the data and corresponds to the mean of the normal distribution, the mean on a log-scale for the log-normal distribution, and the shape parameter for the gamma and Weibull distributions. The second parameter accounts for the spread of the data around the location parameter and corresponds to the standard deviation in the case of the normal distribution, the standard deviation on a log-scale for the log-normal distribution, the rate parameter (1/scale) for the gamma distribution and the scale parameter for the Weibull distribution. We refer to μ1 and σ1 for the location and scale parameters of the first peak (i.e. lowest Ct values) and μ2 and σ2 for the location and scale parameters of the second peak (i.e. highest Ct values). The density of the bimodal distribution of Ct values thus reads
where D1 and D2 are the distributions accounting for the first (i.e. lowest Ct values) and second (i.e. the highest Ct values) peaks, respectively, and λ and (1 – λ) are the weights for the D1 and D2 distributions, respectively.
Fig. 1.
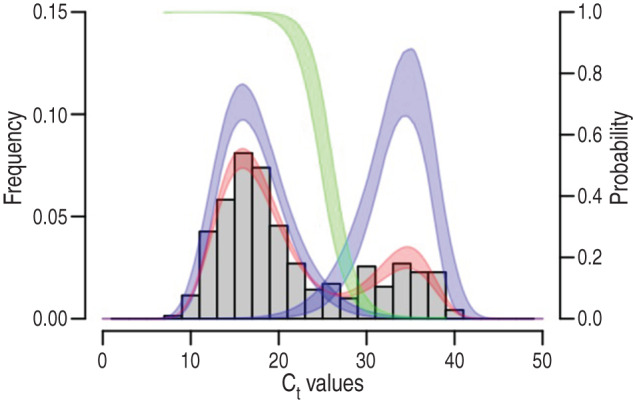
Finite-mixture modelling of the Ct value distribution and identification of the cut-off values. The number of samples with Ct <40 was 346 and is represented by the grey histograms. The best finite-mixture model was D1 with a log-normal distribution (left blue curve), and D2 with a Weibull distribution (right blue curve). This model is shown in red. From these latter two, the probability P of belonging to the left-most peak of Ct values is computed as D1/(D1 + D2) and this is shown by the green curve (see the right vertical scale). The widths of the curves indicate the 95% confidence intervals.
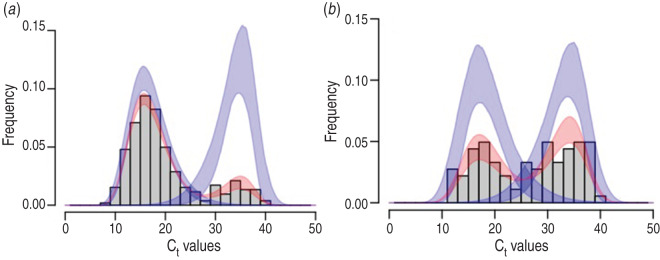
Fig. 2.
Log-normal and Weibull mixture model of norovirus Ct values (same as Fig. 1) applied separately to (a) rotavirus (RV)-negative samples (n = 259) and (b) RV-positive samples (n = 87). Distributions D1 and D2 were log-normal and Weibull, respectively (Supplementary Tables S2 and S3).
For each of the 16 combinations of distributions, parameters λ, μ1, σ1, μ2, σ2 were estimated by maximum likelihood (ML) using the expectation-maximization (EM) algorithm [29], the confidence intervals were calculated as proposed by Oakes [30] and all calculations were done in R [31]. Given the equal numbers of parameters (n = 5) of the 16 models, the best combination of distributions for the two peaks was chosen as the one minimizing the minus log-likelihood. From parameterized distribution D1 and D2 of the best model, we computed the probability P to belong to the low Ct value peak as P = D1/(D1 + D2) and this probability was used to derive a cut-off value separating the two peaks.
Antigen detection of NV and RV by ELISA
RV antigens were tested on all samples whereas NV antigens were tested in a subset of samples (n = 182). Of these 182 samples, 47 were randomly selected from the NV-negative samples (Ct ⩾40) and 135 NV-positive samples were randomly selected to represent a uniform distribution of Ct values <40. NV and RV antigen detection was performed by NV-AD-III kit (Denka Seika, Japan) and Rotaclone™ enzyme immunoassay (Meridian, USA) according to the manufacturers' instructions, respectively. The cut-off for NV and RV positivity was OD450nm = 0·15; samples within 0·01 OD of the cut-off were repeated to confirm results.
RESULTS
Characteristics of children admitted to Thai Binh paediatric hospital
From January 2011 to September 2012, a total of 807 faecal samples were collected and examined for the presence of NV and RV. In Thai Binh Paediatric hospital, 89% and 97% of children hospitalized for diarrhoea were aged less than 24 and 36 months, respectively (Table 1). NV and RV were identified in 43% and 40% of children with diarrhoea, respectively; co-infections with both viruses occurred in 87 (11%) patients, and 227 (28%) were negative for both NV and RV.
Table 1.
Age distribution and molecular screening results for rotavirus (RV) and norovirus (NV) in patients hospitalized with diarrhoea in Thai Binh, Vietnam, January 2011–September 2012
| Age group (months) | Total | NV* Ct cut-off = 40 |
NV* Ct cut-off = 21·36 |
RV* | RV-NV co-infection |
|---|---|---|---|---|---|
| N (%) | n (%) | n (%) | n (%) | n (%) | |
| 0–2 | 26 (3·2) | 4 (1·2) | 2 (0·9) | 5 (1·6) | 0 |
| 3–5 | 126 (15·6) | 53 (15) | 31 (14) | 36 (11) | 13 (15) |
| 6–11 | 327 (40·5) | 148 (43) | 105 (47) | 126 (39) | 33 (38) |
| 12–23 | 243 (30) | 111 (32) | 67 (30) | 118 (37) | 34 (39) |
| 24–35 | 58 (7·2) | 22 (6·4) | 13 (5·8) | 26 (8·1) | 6 (7) |
| 36–47 | 18 (2·2) | 6 (1·7) | 4 (1·8) | 7 (2·2) | 1 (1·1) |
| 48–60 | 9 (1·1) | 2 (0·6) | 1 (0·4) | 3 (0·9) | 0 |
| Total | 807 (100) | 346 (100) | 223 (100) | 321 (100) | 87 |
Including both cases of single infection and co-infection.
A total of 285 NV samples (92% of total samples with Ct <35) were genotyped. NV-GII was the dominant genogroup (91%); GII.3 and GII.4 genotypes constituted 27% and 59%, respectively. Other genotypes of NV-GII included GII.2 (0·6%), GII.7 (0·3%), GII.12 (0·3%), GII.13 (2·6%) and GII.16 (0·6%). Only three (0·9%) cases of GI.8 were identified, one of which was a co-infection with GII.3.
Cut-off Ct value for NV-associated diarrhoea
The 16 finite-mixture models were fitted to the 346 Ct values <40 (represented by the grey histogram in Fig. 1). According to the log-likelihood, the best fit model comprised a log-normal distribution for D1 and Weibull distribution for D2, with λ = 74% [95% confidence interval (CI), 70–79] for the values belonging to the lower peak of Ct values (Supplementary Table S1, Fig. 1). The constitutive distributions D1 and D2 are the left-most and right-most curves, respectively, from which the probability P of belonging to the lower peak Ct values peak is represented by the green curve. From this latter we infer that a probability P of 50% belongs to the lower peak of Ct values, which corresponds to a cut-off of 25·45 (95% CI 25·45–25·45); a probability P of 95% belongs to the lower peak of Ct values corresponding to a cut-off Ct of 21·36 (95% CI 20·29–22·46); a probability P of 99% belongs to the higher peak of Ct values corresponding to a cut-off Ct of 19·01 (95% CI 17·59–20·45). Using the 95% P Ct cut-off (21·36), the adjusted NV detection rates reduced significantly from 43% (95% CI 41–51) to 28% (95% CI 29–38) (Table 1). Thus, using these new criteria, RV and NV were causative agents in 40% and 28% of diarrhoea cases, respectively.
Co-infection with RV
Of the 346 patients positive for NV by real time RT-PCR, 87 were ELISA-positive for RV, and 259 were ELISA-negative for RV. When comparing RV positive and negative cases, we found no significant differences in age (Welch's two-sample t test: t = −0·4389, d.f. = 104·20, P = 0·66, Mann–Whitney two-sample test: W = 6424·50, P = 0·82), gender (Fisher's exact test: P = 0·64), NV genotype composition (Fisher's exact test: P = 0·85) or duration between diarrhoea onset and sampling (Welch's two-sample t test: t = 0·62, d.f. = 76·22, P = 0·54; Mann–Whitney two-sample test: W = 6468, P = 0·75). When fitting the same finite-mixture models separately to the 87 ELISA-RV-positive and the 259 ELISA-RV-negative patients, the best-fit model had a log-normal distribution for D1 and a Weibull distribution for D2 (Fig. 2, Supplementary Tables S2, S3). As expected, the proportion λ of Ct values belonging to the lower peak of Ct values was significantly higher in RV-negative patients (84%, 95% CI 80–89) than in RV-positive patients (44%, 95% CI 35–55).
Effects of host, virus genotypes and time elapsed between diarrhoea onset and sampling
Patients' age did not differ significantly between the two peaks of Ct values (Welch's two-sample t test: t = −0·80, d.f. = 142·80, P = 0·43; Mann–Whitney two-sample test: W = 11 953·5, P = 0·96, see Supplementary Fig. S1A). The proportion of genotype GII.4 (Supplementary Fig. S1D; χ2 = 0·32, d.f. = 279, P = 0·57), and the proportion of males and of genotype GII.3 marginally significantly increased as the Ct values decreased: χ2 = 3·34, d.f. = 279, P = 0·07 and χ2 = 3·17, d.f. = 344, P = 0·08 (see Supplementary Fig. S1C), respectively. By contrast, the time-lag between diarrhoea onset and sampling marginally significantly increased with Ct values (Welch's two-sample t test: t = −1·78, d.f. = 153·69, P = 0·08; Mann–Whitney two-sample test: W = 10 383·50, P = 0·05) (see Supplementary Fig. S1B).
Comparison of NV antigen ELISA and RT–PCR cut-off values
All 47 samples with NV Ct ⩾40 were negative by NV ELISA. Of the 135 samples with NV Ct <40, 72 samples were positive by NV ELISA; 70/72 (97%) antigen-positives had Ct <21·36; and only 2/53 (4%) samples with Ct >21·36 were NV antigen-positive (Supplementary Fig. S2). Of the samples with Ct <21·36, only 70/82 (85%) were antigen positive. The genotypes of samples with high viral load (Ct <21·36) that were negative by antigen ELISA were GII.3 (5/28), GII.4 New Orleans 2009 (n = 3/3), GII.4 2006b (2/42) and GII.13 (1/1). Determination of specificity and sensitivity of the NV antigen ELISA test (Supplementary Table S4) showed 98% specificity (96% if considering only Ct <40) and 85% sensitivity for samples belonging to the lower peak of Ct values (P = 0·95).
DISCUSSION
Several previous studies have evaluated NV viral loads in paediatric populations in order to better understand dynamics of NV shedding in diarrhoeal cases vs. healthy controls [19, 32]. Barreira et al. [19] reported tenfold higher NV RNA copy numbers in diarrhoeal cases compared to controls, and Phillips et al. [32] observed the range of Ct values in children aged <5 years with diarrhoea [interquartile range (IQR) 32–37, median 35] and in age-matched healthy/non-diarrhoeal controls (IQR 34–38, median 37), although these groups overlapped substantially. Phillips et al. used the Youden index and ROC curve analysis of diarrhoeal and healthy control groups to propose a Ct cut-off value for NV gastroenteritis; they suggested a Ct <30 for children aged <5 years, and Ct = 33 for older children and adults. Recently, Elfving et al. presented a study to determine threshold cycle cut-offs for multiple pathogens causing acute diarrhoea [33]. The study proposed cut-off values for Cryptosporidium, Shigella, and ETEC-estA (35, 30, 31, respectively); however, no value could be identified for RV and NV [33].
Here we propose an analytical approach to distinguish diarrhoeal cases in which NV is likely to have played a causative role in disease presentation vs. cases in which an alternative pathogen may be involved. Our method was based on the distribution curves of NV Ct values, and did not require comparison to healthy controls. It was founded on the observations that (i) the distribution of Ct values for NV is generally bimodal, (ii) NV RNA in healthy children are detected at similar or even higher rates than in diarrhoea cases [12, 19]; however, (iii) diarrhoea cases shed larger amount of NV than healthy children [19]. We modelled the bimodal distribution of Ct values with a finite-mixture model, and tested the hypothesis that the lower peak of Ct values corresponded to samples for which the cause of diarrhoea was NV and the higher peak corresponded to samples where other agents were involved. Our working hypothesis was tested by applying our method to a subset of samples with RV co-infections, and by evaluating confirmatory NV diagnoses using NV antigen ELISA. As expected, the proportion of samples with low NV viral load had significantly more RV co-infections, whereas those with high NV viral loads were more likely to be RV negative. By contrast, the interval between onset of diarrhoea and sampling, patient gender, and virus genotypes were not significant in explaining NV viral load.
After applying the cut-off value obtained in this study to our own dataset, the proportion of NV cases decreased from 43% (Ct <40) to 28% (Ct <21·36). However, there was no change in the age distribution of cases. Similar NV prevalence (20·6%) was found in children with diarrhoea in a study conducted in Ho Chi Minh city during 2009–2010 [10]. Due to the complicated nature of NV infections, interpretations of low viral loads are difficult, as these values may still indicate a causative role in symptomatic infections; indeed, the authors suggest that clinical diagnostic laboratories must evaluate appropriate cut-off values for different patient populations.
The probability of positive NV ELISA test increases when the Ct value decreases and this can be explained by the relatively poor sensitivity of antigen detection by ELISA compared to RT-PCR. When comparing our method with ELISA, our proposed Ct <21·36 cut-off agreed well with ELISA, with 98% specificity (96% when considering only the Ct <40). The observation that some NV genotypes and variants were not detected by ELISA may explain the lower sensitivity (85%) of ELISA. Effectively, this means that a positive NV ELISA test can be considered as indicative of a case where NV is the actual cause of the diarrhoea, whereas only 90% (81% if considering only Ct <40) of negative NV ELISA tests can be considered as indicative of a diarrhoea case not caused by NV. It is worth noting that the Ct <21·36 threshold is in the range reported by Costantini et al. (IQR 16·5–22·9, median 19·1) in which most samples were positive by ELISA [34]. Costantini et al. suggested that if samples are collected 48 h after onset of the symptoms, ELISA may yield false negatives.
Due to differences in laboratory practices, it is not feasible to directly compare our suggested cut-off of Ct <21·36 to the value of Ct = 31 proposed by Phillips et al. [32] for distinguishing cases from controls. Of note, our study involved exclusively hospitalized cases of diarrhoea, whereas in the Phillips et al. study cases were recruited from primary care and the community, thus representing a broader spectrum of disease severity. In addition to differences in study design, different sampling techniques, sample quality issues, RNA extraction methods and sample preparation procedures are likely to affect virus genome concentrations, while different RT–PCR assays may vary in sensitivity. We suggest that each laboratory should conduct its own analysis to evaluate distribution curves and determine an appropriate cut-off value for NV-associated disease. The advantage of our approach is that such determinations of a Ct threshold for NV-associated disease may be generated in the absence of data from controls, and thus, could be applied to any laboratory receiving clinical samples on a regular basis.
In conclusion, we propose a threshold Ct value for real-time RT–PCR to assess the aetiological role of NV in children with diarrhoea. Our statistical method allowed determination of a cut-off value without reference to any controls, which is essential for the feasibility of extending this analysis to other laboratories conducting routine epidemiological surveillance and clinical diagnostics. We suggest that cut-off values should be determined individually in each laboratory based on its own assay performance, and that accumulation of these types of data across multiple laboratories would contribute to improved understanding of the burden of NV disease.
ACKNOWLEDGEMENTS
We are grateful for the support from the US Centers for Disease Control and Prevention (Dr Jan Vinje), the National Microbiology Laboratory – Canada and the CAREID-Canada program (Dr Tim Booth and Dr Lai King) for the external quality control of the NV detection and genotyping.
The work was supported by the National Foundation for Science and Technology Development (N. V. Trang, grant no. 106·03-2010·56) and the Japan Initiative for Global Research Network on Infectious Diseases (O. Nakagomi, T. Yamashiro and D. D. Anh).
Marc Choisy was sponsored by the ‘Biodiversity and Infectious Diseases in Southeast Asia’ CNRS-funded GDRI network.
R code of the method as well as tutorial are available from marcchoisy.free.fr/fmm.
DECLARATION OF INTEREST
None.
Supplementary material
For supplementary material accompanying this paper visit https://doi.org/10.1017/S095026881500059X.
click here to view supplementary material
REFERENCES
- 1.Patel MM, et al. Systematic literature review of role of noroviruses in sporadic gastroenteritis. Emerging Infectious Diseases 2008; 14: 1224–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Black RE, et al. Global, regional, and national causes of child mortality in 2008: a systematic analysis. Lancet 2010; 375: 1969–1987. [DOI] [PubMed] [Google Scholar]
- 3.Oh DY, Gaedicke G, Schreier E. Viral agents of acute gastroenteritis in German children: prevalence and molecular diversity. Journal of Medical Virology 2003; 71: 82–93. [DOI] [PubMed] [Google Scholar]
- 4.Amar CF, et al. Detection by PCR of eight groups of enteric pathogens in 4,627 faecal samples: re-examination of the English case-control Infectious Intestinal Disease Study (1993–1996). European Journal of Clinical Microbiology and Infectious Diseases 2007; 26: 311–323. [DOI] [PubMed] [Google Scholar]
- 5.Colomba C, et al. Norovirus and gastroenteritis in hospitalized children, Italy. Emerging Infectious Diseases 2007; 13: 1389–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schnagl RD, et al. Prevalence and genomic variation of Norwalk-like viruses in central Australia in 1995–1997. Acta Virologica 2000; 44: 265–271. [PubMed] [Google Scholar]
- 7.Nguyen TA, et al. Norovirus and sapovirus infections among children with acute gastroenteritis in Ho Chi Minh City during 2005–2006. Journal of Tropical Pediatrics 2008; 54: 102–113. [DOI] [PubMed] [Google Scholar]
- 8.Trang NV, et al. Detection and molecular characterization of noroviruses and sapoviruses in children admitted to hospital with acute gastroenteritis in Vietnam. Journal of Medical Virology 2012; 84: 290–297. [DOI] [PubMed] [Google Scholar]
- 9.Tamura T, et al. Molecular epidemiological study of rotavirus and norovirus infections among children with acute gastroenteritis in Nha Trang, Vietnam, December 2005-June 2006. Japanese Journal of Infectious Diseases 2010; 63: 405–411. [PubMed] [Google Scholar]
- 10.My PV, et al. Endemic norovirus infections in children, Ho Chi Minh City, Vietnam, 2009–2010. Emerging Infectious Diseases 2013; 19: 977–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O'Ryan ML, et al. Symptomatic and asymptomatic rotavirus and norovirus infections during infancy in a Chilean birth cohort. Pediatric Infectious Disease Journal 2009; 28: 879–884. [DOI] [PubMed] [Google Scholar]
- 12.Zhang S, et al. Symptomatic and asymptomatic infections of rotavirus, norovirus, and adenovirus among hospitalized children in Xi'an, China. Journal of Medical Virolology 2011. [DOI] [PubMed] [Google Scholar]
- 13.Garcia C, et al. Asymptomatic norovirus infection in Mexican children. Journal of Clinical Microbiology 2006; 44: 2997–3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ayukekbong J, et al. Enteric viruses in healthy children in Cameroon: viral load and genotyping of norovirus strains. Journal of Medical Virology 2011; 83: 2135–2142. [DOI] [PubMed] [Google Scholar]
- 15.Cheon DS, et al. Seasonal prevalence of asymptomatic norovirus infection in Korean children. Foodborne Pathogens and Diseases 2010; 7: 1427–1430. [DOI] [PubMed] [Google Scholar]
- 16.Bucardo F, et al. Asymptomatic norovirus infections in Nicaraguan children and its association with viral properties and histo-blood group antigens. Pediatric Infectious Disease Journal 2010; 29: 934–939. [DOI] [PubMed] [Google Scholar]
- 17.Kotloff KL, et al. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet 2013; 382: 209–222. [DOI] [PubMed] [Google Scholar]
- 18.Patel MM, et al. Noroviruses: a comprehensive review. Journal of Clinical Virology 2009; 44: 1–8. [DOI] [PubMed] [Google Scholar]
- 19.Barreira DM, et al. Viral load and genotypes of noroviruses in symptomatic and asymptomatic children in Southeastern Brazil. Journal of Clinical Virology 2010; 47: 60–64. [DOI] [PubMed] [Google Scholar]
- 20.Marshall JA, et al. High level excretion of Norwalk-like virus following resolution of clinical illness. Pathology 2001; 33: 50–52. [PubMed] [Google Scholar]
- 21.Levine MM, Robins-Browne RM. Factors that explain excretion of enteric pathogens by persons without diarrhea. Clinical Infectious Diseases 2012; 55 (Suppl. 4): S303–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheng WX, et al. Human bocavirus in children hospitalized for acute gastroenteritis: a case-control study. Clinical Infectious Diseases 2008; 47: 161–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramani S, et al. Comparison of viral load and duration of virus shedding in symptomatic and asymptomatic neonatal rotavirus infections. Journal of Medical Virology 2010; 82: 1803–1807. [DOI] [PubMed] [Google Scholar]
- 24.Kageyama T, et al. Broadly reactive and highly sensitive assay for Norwalk-like viruses based on real-time quantitative reverse transcription PCR. Journal of Clinical Microbiology 2003; 41: 1548–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Neesanant P, et al. Optimization of one-step real-time reverse transcription-polymerase chain reaction assays for norovirus detection and molecular epidemiology of noroviruses in Thailand. Journal of Virological Methods 2013; 194: 317–325. [DOI] [PubMed] [Google Scholar]
- 26.Vinje J, Hamidjaja RA, Sobsey MD. Development and application of a capsid VP1 (region D) based reverse transcription PCR assay for genotyping of genogroup I and II noroviruses. Journal of Virological Methods 2004; 116: 109–117. [DOI] [PubMed] [Google Scholar]
- 27.Kroneman A, et al. Proposal for a unified norovirus nomenclature and genotyping. Archives of Virology 2013; 158: 2059–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schlattmann P. Medical Applications of Finite Mixture Models. Heidelberg: Springer Verlag, 2009. [Google Scholar]
- 29.Do CB, Batzoglou S. What is the expectation maximization algorithm? Nature Biotechnology 2008; 26: 897–899. [DOI] [PubMed] [Google Scholar]
- 30.Oakes D. Direct calculation of the information matrix via the EM algorithm. Journal of Royal Statistic Society 1999: 479–482. [Google Scholar]
- 31.RCoreTeam. R: A language and environment for statistical computing. R Foundation for Statistical Computing. R Foundation: Vienna, Austria. 2013. [Google Scholar]
- 32.Phillips G, et al. Diagnosing norovirus-associated infectious intestinal disease using viral load. BMC Infectious Diseases 2009; 9: 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Elfving K, et al. Real-time PCR threshold cycle (Ct) cut-offs help to identify agents causing acute childhood diarrhea in Zanzibar. Journal of Clinical Microbiology 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Costantini V, et al. Diagnostic accuracy and analytical sensitivity of IDEIA Norovirus assay for routine screening of human norovirus. Journal of Clinical Microbiology 2010; 48: 2770–2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
For supplementary material accompanying this paper visit https://doi.org/10.1017/S095026881500059X.
click here to view supplementary material