Abstract
The red nucleus is located in the rostral midbrain of the vertebrate brain and controls motor coordination during locomotion. It receives input from the cerebellum and sends its output to the spinal cord. The presence of the red nucleus is well established in tetrapods, and its existence has also been suggested in teleosts but its presence and position has still been under discussion. By using wheat germ agglutinin (WGA) as a genetically encoded anterograde tracer, we recently identified contralateral projections from the cerebellum to a putative red nucleus in the zebrafish midbrain tegmentum. In this report we further revealed red nucleus derived from this contralateral afferent from the cerebellum using WGA and contralateral projections to the hindbrain-spinal cord junction site using DiI-mediated retrograde tracing. Thus the structure that we have identified by anterograde and retrograde tracing fulfills the anatomical demands for the red nucleus: the location in the midbrain tegmentum, contralateral afferent from the cerebellum (cerebello-ruber projection) and contralateral efferent to the spinal cord (rubro-spinal projection).
Keywords: red nucleus, wheat germ agglutinin
The red nucleus (nucleus ruber) is an important neuronal structure in the midbrain tegmentum, and it controls motor coordination regulating in particular the forelimbs during locomotion.1 Neurons of this nucleus receive afferents from the contralateral cerebellum and the ipsilateral motor cortex, and they send efferents to the contralateral spinal cord to motor-neurons innervating the forelimbs.1 So far, a rubro-spinal connection has been detected in reptiles, birds and mammals: these animals have in common that they use limbs for locomotion on land. Fish instead use fins for locomotion and fins are thought to have evolved into limbs as the fossils disclose.2-4 Some groups reported the presence of a red nucleus in teleosts as it is seen in tetrapods,5-7 but others could not confirm it.8 Such controversial evidence might arise from 2 reasons. First, previous work focused only either on the afferent or the efferent connections of the red nucleus respectively using traditional tracers but did not confirm both projections within the same study. Second, the red nucleus in tetrapods sends contralateral projection to the spinal cord that governs the motor coordination of forelimbs. In teleosts, pectoral fins are considered to be homologous to the forelimbs but the motor-neuron innervation to the forelimbs and the pectoral fins are anatomically different.9
We have recently disclosed the efferent network of zebrafish cerebellar Purkinje cells - the only output neurons of the cerebellum - by using the genetically encoded anterograde transneuronal tracer, wheat germ agglutinin (WGA).10,11 In that work, WGA staining identified second order Purkinje cell efferents in the thalamus, the preoptic area, the hypothalamus, the optic tectum, the octaval nuclei, the inferior olive, the reticular formation, the nucleus of the medial longitudinal fascicle and others. Among these identified efferents, we detected WGA signals in a part of the midbrain tegmentum as candidate for the red nucleus in zebrafish.
Here, we confirm this location of the zebrafish red nucleus based on both its characteristic afferent and efferent connections. The red nucleus is anatomically characterized by its location in the midbrain tegmentum, and both the afferents and efferents are formed by contralateral projections. To reveal such a contralateral input into midbrain tegmental structures we made use of transient transgenic fish injected with a PC specific construct co-expressing membrane localized FyntagRFP and the transneuronal tracer WGA.
Next, adult transient transgenic fish expressing tagRFP and thus WGA only in one cerebellar hemisphere (Fig. 1A and B) were selected and analyzed for WGA positive neurons. This allowed for discriminating between ipsilateral and contralateral efferent structures. As expected WGA-immunohistochemistry detected WGA signals in both halves of the thalamus (Fig. 1C), in cells of the descending octaval nucleus only on the ipsilateral side (Fig. 1D), and staining of the lateral reticular nucleus in the contralateral brain hemisphere only (Fig. 1E). The WGA signal in the putative red nucleus was clearly found in the contralateral tegmental hemisphere without any contribution to the ipsilateral tegmental side, indicating that this cerebello-rubral projection occurs contralaterally (Fig. 1F).
Figure 1.

Identification of the cerebello-ruber tract in zebrafish WGA immunostaining of zebrafish brains with mosaic expression of WGA only in the right hemisphere of the cerebellum. Transient transgenic Tg(tagRFP-T:PC:WGA) zebrafish with red fluorescence only in the right cerebellar hemisphere were raised to adulthood and processed for WGA expression by immunohistochemistry (A and B). Bilateral WGA expression could be observed in the thalamus (C). Instead, expression in the descending octaval nucleus was ipsilateral to the WGA-expressing hemisphere (D), while expression in the lateral reticular nucleus (E) and red nucleus (F) were only found on the contralateral side of the neuraxis. Solid arrows: WGA signals. Dashed arrows: absence of WGA signals. n = 3.
The efferent projection of the nucleus ruber – the rubro-spinal tract - terminates primarily in the cervical spinal cord in tetrapods, consistent with its function in regulating forelimb movements.12,13 In teleosts, the pectoral fins are considered to be homologous to the forelimbs in tetrapods. The motor-neurons that innervate these pectoral fins originate from further rostral regions compared to tetrapods as these motoneurons are located at the hindbrain-spinal cord junction.9 Injection of the retrograde tracer DiI into the hindbrain-spinal cord junction indeed revealed a contralateral projection from the red nucleus to these spinal motor-neurons (Fig. 2). Instead DiI injection into the spinal cord at more caudal positions did not mark contralateral axonal projections emanating from the tegmental red nucleus (Fig. 2). This suggests that like in mammals, the teleost red nucleus may be important for coordinating movements of pectoral fins but not of the pelvic fins. The difference between tetrapods and teleost in the termination of rubral projections in the spinal cord can explain the controversial results reported so far about a presence of a red nucleus in teleost fish. Thus red nucleus-mediated coordination of the upper extremities may be more conserved than previously anticipated.
Figure 2.
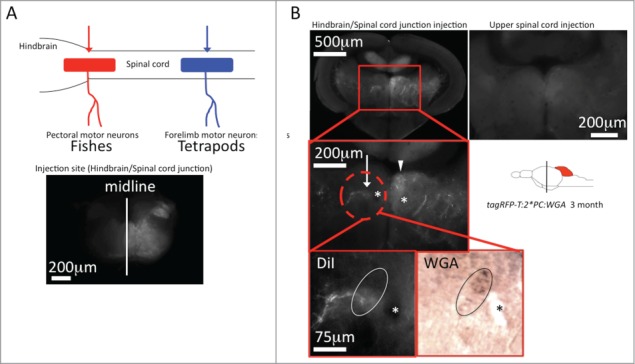
Identification of the rubro-spinal tract in zebrafish DiI tracing from the hindbrain-spinal cord junction. (A) The schematic drawing in the upper left corner illustrates the origin of motor-neurons innervating pectoral fin muscles in fish (red) and forelimb muscles in tetrapods (blue).13 (B) Red and blue arrows indicate the injection site of DiI respectively. Application of DiI into the hindbrain-spinal cord junction of transgenic Tg(tagRFP-T:PC:WGA) zebrafish labeled a contralateral neuronal nucleus containing WGA demonstrating its identity as an efferent structure of Purkinje cells (white arrow). The arrowhead points to the adjacent nucleus of the medial longitudinal fasciculus, asterisks: habenulo-interpeduncular tract. In contrast application of DiI into the upper spinal cord did not label the same WGA-positive structure. n = 5 for each injection site.
Taken together, by using WGA and DiI tracing, we could reveal the presence of a neuronal structure that fulfills the anatomical demands for the red nucleus in zebrafish: a localization in the midbrain tegmentum, the receipt of contralateral cerebello-ruber afferents and the projection of contralateral rubro-spinal efferents. Physiological and functional analysis of this teleost red nucleus is now required to further disclose the role of this important motor coordination structure for which suitable regulatory elements will be needed at first.
Method
Fish maintenance
Fish were raised and maintained under a 14 hours light / 10 hours dark cycle at 28°C according to standard protocols.14,15 To microinject expression constructs and to establish transgenic lines, fish of the brass line brsb2/b2 were used. All animal experiments were approved by the government of Lower Saxonia and were conducted in accordance with legal regulations (EU Directive 2010_63).
Microinjection
Glass capillaries (GC100F-10, Harvard Apparatus) were pulled into microinjection needles by using a vertical needle puller (model 700C, David Kopf Instruments), which were used in a Femtojet Express Microinjector (Eppendorf) equipped with a micromanipulator (Narishige). Plasmid DNA was microinjected into the soma of one-cell stage wild-type embryos (= F0 generation, mosaic) at a concentration of 25 ng/ml (injection volume about 1.5 nl). For the generation of stable transgenic fish, transgene cassettes flanked by recognition sites of the Tol2 transposase were used and the DNA was supplemented with 25 ng/ml mRNA encoding the Tol2 transposase to increase genomic integration of the transgene cassette.16
Immunohistochemistry in the adult zebrafish brain
Immunohistochemistry was performed according to previous studies.11,17
Adult zebrafish were sacrificed by an overdose of Tricaine and brains were explanted and fixed in Bouin fixative (saturated picric acid (Sigma-Aldrich) : formalin (Sigma-Aldrich) : acetic acid (VWR) = 15 : 5 : 1) over night at RT. While paraformaldehyde could also be used the histological perseveration is better by using Bouin fixative. To avoid deformation of the brain, put the sample on a flat place. After a series of ethanol and Xylene washes (70% ethanol 1 hr, 80% ethanol 1 hr, 90% ethanol 1 hr and 3 times 100% ethanol each 1 hr, 3 times Xylen each 1 hr), the brains were embedded in paraffin and sectioned (10 μm) using a microtome (Jung AG). Do not cut the paraffin ribbon at the edge to avoid the loss of important sections.
Paraffin sections were dewaxed in Xylene (2 times each 10 min) and gradually hydrated in an ethanol/PBS series (2 times 100% ethanol each 10 min, 95% ethanol 10 min, 70% ethanol 10 min). After washing in PBS (3 times each 5 min), each section was encircled by PAP PEN Liquid blocker (DAIDO SANGYO) and the sections were incubated for 30 min with 0.1% Pronase (Roche) at RT.
After washing in PBS (3 times each 5 min), the sections were immersed in 0.3% H2O2 in methanol for 30min, washed in PBS (3 times each 5 min) and blocked by 5% skim milk in PBS for 30 min at RT.
Zebrafish larvae at 3 to 5 d post fertilization were fixed in Bouin fixative for 1 hr at RT. Next, these larvae were immersed in cold acetone, and then dried on a filter paper at RT. Dried larvae were crushed intensively and the resulting powder was used for antibody preabsorption.
Goat anti-WGA antibody (Vector Labs) was diluted by 1:200 with 5% skim milk in PBS, and was preabsorbed with acetone powder from zebrafish larvae for 16 hrs at 4°C. Subsequently the antibody-acetone powder mix was centrifuged to collect the supernatant. The pretreated sections were incubated for 16 hrs at 4°C with this antibody supernatant.
After washing in PBS (3 times each 5 min), the sections were incubated with a biotinylated anti-goat IgG (1:500 dilution, Vector Labs) for 1 hr at RT, followed by staining using a Vectastain Elite ABC kit (Vector Labs) for 30 min. Signals were visualized with enhanced diaminobenzidine peroxidase (DAB) (Sigma-Aldrich) reaction; 0.05% DAB, 0.05% Nickel Ammonium Sulfate and 0.015% H2O2 in PBS, pH7.2. The color reaction was stopped before unspecific staining developed in the negative control. The concentrations of DAB and Nickel Ammonium Sulfate were important and high concentration and/or inappropriate storage of these solutions can result in unspecific signals.
After washing in distilled water (5 min), sections were immersed in ethanol (2 times each 5min) and then Xylene (2 times each 5 min) and mounted with Entellan new (Merck Millipore).
Retrograde tracing of neuronal projections
For post mortem DiI-labeling, adult zebrafish brains were fixed with 4% PFA at RT over night. After washing with PBS, DiI crystals (Invitrogen) were applied to the target region with a microneedle pulled from a glass capillary tube (GC100F-10) and a micromanipulator. If necessary, parts of the brain were removed to correctly target the DiI cristals. Samples were incubated in 0.4% PFA for 2 ∼4 weeks at 37°C. Subsequently, specimens were embedded in low-melting agarose and sectioned (200 μm) using a vibratome (Leica VT1000S).
For staining retrogradely labeled tissue for WGA expression, floating slices were washed in PBS/1% TritonX-100 (3 times each 5 min) and then incubated for 30 min with 10 μg/ml proteinase K at RT. After washing in PBS with 1% TritonX-100 (3 times each 10 min), the sections were fixed again in 4% PFA for 30 min followed by washing in PBS/1% TritonX-100 (3 times each 10 min) and blocking in 5% skim milk in PBS/1% TritonX-100 for 6 hours. Afterwards, these samples were incubated for 16 hour at 4°C with a 1:100 dilution of a goat anti-WGA antibody (preabsorbed with acetone powder from zebrafish larvae) in 5% skim milk in PBS/1% TritonX-100. After washing in PBS with 1% TritonX-100 (7 times each 30 min), the sections were incubated in a 1:100 dilution of an anti-goat IgG antibody conjugated to alkaline phosphatase (Vector Labs) for 16 hrs at 4°C. After several washes in PBS/1% TritonX-100 (7 times each 30 min), signals were developed via color reaction with 450 μg/ml NBT, 175 μg/ml BCIP and 5 mM levamisole (Sigma) in AP buffer. The color reaction was stopped by washing with PBS/1% TritonX-100 (3 times each 5 min) and fixation in 4% PFA/PBS for 20 min and then subjected to image recording.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank all members of the lab for continuous discussion, support and helpful advice. We are grateful to Angela Traudt, Eva Saxinger and Timo Fritsch for excellent technical support and zebrafish husbandry. We are thankful to Dr. Mario F. Wullimann for intense discussions and helpful advice about zebrafish neuroanatomy.
Funding
This work was generously supported by an Alexander von Humboldt Fellowship to Dr. Hideaki Matsui and funding from the Deutsche Forschungsgemeinschaft (DFG, KO 1949/7–1).
References
- 1. Carpenter MB. Core text of Neuroanatomy, 4th ed. Philadelphia; Lippincott Williams & Wilkins; 1992. [Google Scholar]
- 2. Holmgren N. On the origin of the tetrapod limb. Acta Zoologica 1933; 14; 185-295; http://dx.doi.org/ 10.1111/j.1463-6395.1933.tb00009.x [DOI] [Google Scholar]
- 3. Holmgren N. Contribution on the question of the origin of the tetrapod limb. Acta Zoologica 1939; 20 89-124; http://dx.doi.org/ 10.1111/j.1463-6395.1939.tb00494.x [DOI] [Google Scholar]
- 4. Boisvert CA, Mark-Kurik E, Ahlberg PE. The pectoral fin of panderichthys and the origin of digits. Nature 2008; 456 636-8; PMID:18806778; http://dx.doi.org/ 10.1038/nature07339 [DOI] [PubMed] [Google Scholar]
- 5. Becker T, Wullimann MF, Becker CG, Bernhardt RR, Schachner M. Axonal regrowth after spinal cord transection in adult zebrafish. J Comp Neurol 1997; 377 577-95; PMID:9007194; http://dx.doi.org/ 10.1002/(SICI)1096-9861(19970127)377:4%3c577::AID-CNE8%3e3.0.CO;2 [DOI] [PubMed] [Google Scholar]
- 6. Prasada Rao PD, Jadhao AG, Sharma SC. Descending projection neurons to the spinal cord of the goldfish, carassius auratus. J Comp Neurol 1987; 265 96-108; PMID:2826554; http://dx.doi.org/ 10.1002/cne.902650107 [DOI] [PubMed] [Google Scholar]
- 7. Wullimann MF, Northcutt RG. Connections of the corpus cerebelli in the green sunfish and the common goldfish: a comparison of perciform and cypriniform teleosts. Brain Behav 1988; 32 293-316; PMID:3233488; http://dx.doi.org/ 10.1159/000116558 [DOI] [PubMed] [Google Scholar]
- 8. Folgueira M, Anadón R, Yáñez J. Afferent and efferent connections of the cerebellum of a salmonid, the rainbow trout (oncorhynchus mykiss): a tract-tracing study. J Comp Neurol 2006; 497 542-565; PMID:16739164; http://dx.doi.org/ 10.1002/cne.20979 [DOI] [PubMed] [Google Scholar]
- 9. Ma LH, Gilland E, Bass AH, Bakera R. Ancestry of motor innervation to pectoral fin and forelimb. Nat Commun 2010; 1 1-8; PMID:20975674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yoshihara Y, Mizuno T, Nakahira M, Kawasaki M, Watanabe Y, Kagamiyama H, Jishage K, Ueda O, Suzuki H, Tabuchi K, Sawamoto K, Okano H, Noda T, Mori K. A genetic approach to visualization of multisynaptic neural pathways using plant lectin transgene. Neuron 1999; 22 33-41; PMID:10027287; http://dx.doi.org/ 10.1016/S0896-6273(00)80676-5 [DOI] [PubMed] [Google Scholar]
- 11. Matsui H, Namikawa K, Babaryka A, Köster RW. Functional regionalization of the teleost cerebellum analyzed in vivo. Proc Natl Acad Sci U S A 2014; 111 11846-51; PMID:25002482; http://dx.doi.org/ 10.1073/pnas.1403105111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ipekchyan NM. Quantitative analysis of the distribution of the motor cortex representations of the fore-and hindlimbs in the red nucleus of the cat. Neurosci Behav Physiol 2008; 38 345-7; PMID:18401723; http://dx.doi.org/ 10.1007/s11055-008-0047-6 [DOI] [PubMed] [Google Scholar]
- 13. Nieoullon A, Rispal-Padel L. Somatotopic localization in cat motor cortex. Brain Res 1976; 105; 405-22; PMID:1260454; http://dx.doi.org/ 10.1016/0006-8993(76)90590-4 [DOI] [PubMed] [Google Scholar]
- 14. Westerfield M. The Zebrafish Book, 4th ed. Eugene; University of Oregon Press; 2000. [Google Scholar]
- 15. Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn 1995; 203 253-310; PMID:8589427; http://dx.doi.org/ 10.1002/aja.1002030302 [DOI] [PubMed] [Google Scholar]
- 16. Kawakami K, Takeda H, Kawakami N, Kobayashi M, Matsuda N, Mishina M. A transposon-mediated gene trap approach identifies developmentally regulated genes in zebrafish. Dev Cell 2004; 7 133-144; PMID:15239961; http://dx.doi.org/ 10.1016/j.devcel.2004.06.005 [DOI] [PubMed] [Google Scholar]
- 17. Matsui H, Taniguchi Y, Inoue H, Uemura K, Takeda S, Takahashi R. A chemical neurotoxin, MPTP induces parkinson's disease like phenotype, movement disorders and persistent loss of dopamine neurons in medaka fish. Neurosci Res 2009; 65 263-71; PMID:19665499; http://dx.doi.org/ 10.1016/j.neures.2009.07.010 [DOI] [PubMed] [Google Scholar]


