Abstract
Soybean is an important crop, with processed soybeans being the second largest source of vegetable oil and the largest source of animal protein feed in the world. Nodules on soybean roots are responsible for symbiotic nitrogen fixation, enabling soybean plants to obtain sufficient nitrogen for growth and seed production. Because nitrogen is an essential, but often limiting, nutrient for plant growth, improvements in nitrogen fixation are highly required in agriculture. We recently reported a comprehensive analysis of rhizosphere bacterial communities during soybean growth in a field in Kyoto prefecture, Japan. The bacterial communities of the rhizosphere changed significantly during growth, with potential plant growth-promoting rhizobacteria, including Bacillus, Bradyrhizobium, and Rhizobium, increasing in a stage-specific manner. In this addendum, we focus on changes in Bradyrhizobium during soybean growth, suggesting that soybean plants select for symbiotic partners.
Keywords: Bradyrhizobium, nitrogen fixation, rhizosphere, soybean, symbiosis
Legumes (Fabaceae) constitute the third largest plant family in the world, with around 700 genera and 20,000 species.1 Most legume plants acquire nitrogen nutrients through symbiosis with soil microbes called rhizobia. This legume-rhizobium symbiosis takes place in specialized organs, called nodules, in which rhizobia effectively convert atmospheric nitrogen into ammonium. In return, plants supply the rhizobia with the products of photosynthesis. Sophisticated signaling cascades to form nodules begin when roots of the host plant secrete flavonoids, which are recognized by the NodD protein in rhizobia, leading to the successive induction of nod genes, which produce the second signaling molecules, lipochitooligosaccharides or Nod-factors.2
Soybean (Glycine max) is the most widely-grown legume crop in the world, with an annual yield of over 200 million tons. Soybeans symbiose with rhizobia such as Bradyrhizobium japonicum and B. elkanii, with about 50 to 60% of nitrogen supplied by atmospheric nitrogen fixation in nodules.3 Maximum symbiotic nitrogen fixation occurs between the R3 and R5 stages of soybean development,4 but is then reduced between the R5 and R7 stages, the seed-filling stages in soybeans. Thus, improvement of nitrogen fixation at these stages is required to optimize agricultural practice.
Genistein and daidzein were identified as signaling flavonoids from soybean roots.5 Using a plasma membrane-enriched vesicles, we detected clear ATP-dependent transport activity of genistein, and demonstrated that an ATP-binding cassette (ABC)-type transporter is involved in the secretion of flavonoids from soybean roots.6,7 The amount and composition of secreted flavonoids vary greatly in soybeans grown axenically in mineral nutrient solution, with secretion dependent on growth stages and conditions (our unpublished results). In Arabidopsis, the contents and compositions of root exudates were shown to correlate with the metabolic activities of bacterial communities in rhizospheres,8,9 suggesting the involvement of plant metabolites in forming rhizosphere microbial communities, which, in turn, affect plant health and growth.10 We recently reported a comprehensive analysis of rhizosphere bacterial communities during soybean growth in a field of Kyoto prefecture, Japan. This analysis showed that the bacterial communities of the rhizosphere changed significantly and in a stage-specific manner during growth, with a higher abundance of potential plant growth promoting rhizobacteria, including Bacillus, Bradyrhizobium, and Rhizobium.11 This addendum focuses on changes in Bradyrhizobium species during soybean growth.
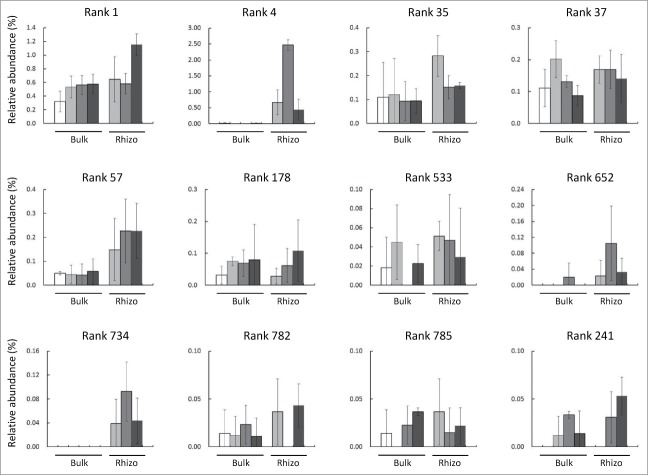
To characterize changes in Bradyrhizobium during soybean growth, the relative abundance of OTUs annotated as Bradyrhizobium were compared. At the genus level, the relative abundance of Bradyrhizobium at all growth stages was higher in rhizosphere than in bulk soils,11 although each OTU annotated as Bradyrhizobium showed different trends during the growth season (Fig. 1). The most abundant Bradyrhizobium (Rank 1) was not more abundant in rhizosphere than in bulk soil, nor did it show a specific pattern during growth. In contrast, Bradyrhizobium in Ranks 4, 57, 652 and 734 were more abundant in rhizosphere than in bulk soils at different stages of growth. These results suggest that soybeans may select specific species, or even strains, of Bradyrhizobium. However, the identities of these OTUs could not be determined at the species or strain level due to the limited resolution of pyrosequencing analysis. In a soybean field, B. japonicum and B. elkanii were the predominant species that formed nodules,12 with both of these species isolated from nodules of soybeans grown in this field (our unpublished results). In addition, Bradyrhizobium with reduced nitrogen fixation activity has been detected in soybean fields.13 It would be interesting to investigate the changes in these Bradyrhizobium at the species or strain level, and to characterize the nitrogen fixation activities of each. Nitrogen fixation activities can be analyzed in correlation with changes of soybean metabolic activities, especially regarding flavonoid biosynthesis and secretion, which play important roles in rhizosphere biological communications.14 Soybean-rhizobacteria interactions offer a good model to test the hypothesis that plants select bacteria from reservoir soils to form rhizosphere bacterial communities.10 This may result in better utilization of rhizosphere microbial communities for agricultural practices.
Figure 1.
Relative abundance of OTUs annotated as Bradyrhizobium. Shown are 12 of the top 1,000 OTUs annotated as Bradyrhizobium. White bars represent bulk soil (bulk) and black bars represent rhizosphere soil (rhizo). Relative abundance was calculated using data from pyrosequencing analysis. The numbers in parentheses are loading values. □; initial soil, ▪; vegetative stage, ▪; flowering stage, ▪; mature stage. Values are mean ± SD (n = 3). The pyrosequencing data have already been published.11
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed
Funding
This study was partly supported by Kyoto University Step-Up Grant for Young Scientists, and Mission Research on Sustainable Humanosphere from RISH, Kyoto University.
References
- 1. Doyle JJ, Luckow MA. The rest of the iceberg. Legume diversity and evolution in a phylogenetic context. Plant Physiol 2003; 131:900-10; PMID:12644643; http://dx.doi.org/ 10.1104/pp.102.018150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wang D, Yang S, Tang F, Zhu H. Symbiosis specificity in the legume: rhizobial mutualism. Cell Microbiol 2012; 14: 334-42; PMID:22168434; http://dx.doi.org/ 10.1111/j.1462-5822.2011.01736.x [DOI] [PubMed] [Google Scholar]
- 3. Salvagiotti F, Cassman KG, Specht JE, Walters DT, Weiss A, Dobermann A. Nitrogen uptake, fixation and response to fertilizer N in soybeans: A review. Field Crop Res 2008; 108: 1-13; http://dx.doi.org/ 10.1016/j.fcr.2008.03.001 [DOI] [Google Scholar]
- 4. Zapata F, Danso SKA, Hardarson G, Fried M. Time course of nitrogen fixation in field grown soybean using nitrogen-15 methodology. Agron J 1987; 79: 172-6; http://dx.doi.org/ 10.2134/agronj1987.00021962007900010035x [DOI] [Google Scholar]
- 5. Kosslak RM, Bookland R, Barkei J, Paaren HE, Appelbaum ER. Induction of Bradyrhizobium japonicum common nod genes by isoflavones isolated from Glycine max. Proc Natl Acad Sci U S A 1987; 84: 7428-32; PMID:16593884; http://dx.doi.org/ 10.1073/pnas.84.21.7428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sugiyama A, Shitan N, Yazaki K. Involvement of a soybean ATP-binding cassette-type transporter in the secretion of genistein, a signal flavonoid in legume-Rhizobium symbiosis. Plant Physiol 2007; 144: 2000-8; PMID:17556512; http://dx.doi.org/ 10.1104/pp.107.096727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sugiyama A, Shitan N, Yazaki K. Signaling from soybean roots to rhizobium: An ATP-binding cassette-type transporter mediates genistein secretion. Plant Signal Behav 2008; 3: 38-40; PMID:19704765; http://dx.doi.org/ 10.4161/psb.3.1.4819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chaparro JM, Badri DV, Bakker MG, Sugiyama A, Manter DK, Vivanco JM. Root exudation of phytochemicals in Arabidopsis follows specific patterns that are developmentally programmed and correlate with soil microbial functions. PLoS One 2013; 8: e55731; PMID:23383346; http://dx.doi.org/ 10.1371/journal.pone.0055731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chaparro JM, Badri DV, Vivanco JM. Rhizosphere microbiome assemblage is affected by plant development. ISME J 2014; 8: 790-803; PMID:24196324; http://dx.doi.org/ 10.1038/ismej.2013.196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Berendsen RL, Pieterse CMJ, Bakker PA. The rhizosphere microbiome and plant health. Trends Plant Sci 2012; 17: 478-86; PMID:22564542; http://dx.doi.org/ 10.1016/j.tplants.2012.04.001 [DOI] [PubMed] [Google Scholar]
- 11. Sugiyama A, Ueda Y, Zushi T, Takase H, Yazaki K. Changes in the bacterial community of soybean rhizospheres during growth in the field. PLoS One 2014; 9: e100709; PMID:24955843; http://dx.doi.org/ 10.1371/journal.pone.0100709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Minamisawa K, Onodera S, Tanimura Y, Kobayashi N, Yuhashi KI, Kubota M. Preferential nodulation of Glycine max, Glycine soja and Macroptilium atropurpureum by two Bradyrhizobium species japonicum and elkanii. FEMS Microbiol Ecol 1997; 24: 49-56; http://dx.doi.org/ 10.1111/j.1574-6941.1997.tb00422.x [DOI] [Google Scholar]
- 13. Itakura M, Saeki K, Omori H, Yokoyama T, Kaneko T, Tabata S, Ohwada T, Tajima S, Uchiumi T, Honnma K, et al. Genomic comparison of Bradyrhizobium japonicum strains with different symbiotic nitrogen-fixing capabilities and other Bradyrhizobiaceae members. ISME J 2009; 3: 326-39; PMID:18971963; http://dx.doi.org/ 10.1038/ismej.2008.88 [DOI] [PubMed] [Google Scholar]
- 14. Sugiyama A, Yazaki K. Flavonoids in plant rhizospheres: secretion, fate and their effects on biological communication. Plant Biotechnol 2014. in press) DOI: 10.5511/plantbiotechnology.14.0917a [DOI] [Google Scholar]