Abstract
SLUG, a member of the SNAIL family of transcriptional repressors, is known to play a diverse number of roles in the cell, and its deregulation has been observed in a variety of cancers including breast. Here, we focus on SLUG's role as a master regulator of mammary epithelial cell (MEC) fate and lineage commitment in the normal mammary gland, and discuss how aberrant SLUG expression can influence breast tumor formation, phenotype, and progression. Specifically, we discuss SLUG's involvement in MEC differentiation, stemness, cellular plasticity, and the epithelial to mesenchymal transition (EMT), and highlight the complex connection between these programs during development and disease progression. Undoubtedly, delineating how molecular factors influence lineage identity and cell-state dynamics in the normal mammary gland will contribute to our understanding of breast tumor heterogeneity.
Keywords: breast cancer, cellular plasticity, differentiation, EMT, mammary stem cells, SLUG
Abbreviations
- MEC
Mammary Epithelial Cell
- EMT
Epithelial to Mesenchymal Transition
- SNAG
Snai.Gfi-1
- E-CAD
E-Cadherin
- ME
Myoepithelial
- BM
Basement Membrane
- CK
Cytokeratin
- SMA
Smooth Muscle Actin
- MaSC
Mammary Stem Cell
- IHC
Immunohistochemical
- HMECs
Human Mammary Epithelial Cells
- WT
Wild type
- CSC
Cancer Stem Cell
- ERα
Estrogen Receptor
- BRCA1
Breast Cancer Associated 1
- BCSC
Breast Cancer Stem Cell
- HDAC
Histone Deacetylasae
- LSD1
Lysine Specific Demethylase 1
SLUG: A SNAIL Family Protein
SLUG (SNAI2) is a member of the SNAIL family of zinc finger transcriptional repressors that mediates sequence-specific interactions with DNA. The most highly studied members of this family include SNAIL (SNAI1) and SLUG (Fig. 1), both of which are conserved among vertebrate species.1 SNAIL family members have been implicated in a variety of developmental and cellular processes, many of which relate to cell motility and induction of the EMT; these include but are not limited to: mesoderm formation, neural crest migration, and determination of left-right asymmetry during embryogenesis, as well as wound re-epithelialization and tissue fibrosis in the adult.2-8 Additionally, SNAIL family members are aberrantly expressed in a variety of cancers where they regulate a diverse number of processes ranging from tumor cell invasion and metastasis to cell survival and proliferation.2, 9-15
Figure 1.
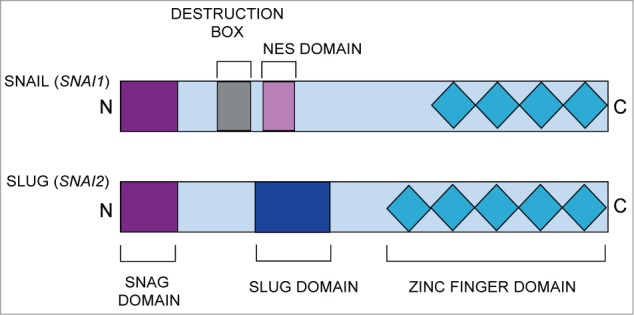
Schematic diagram of the main functional domains of the SNAIL (SNAI1) and SLUG (SNAI2) proteins. The common domains include the N-terminal SNAG domain and C-terminal zinc finger domains. NES, nuclear export sequence.
Proteins of the SNAIL family share a similar structural organization. The carboxy terminus contains 4 to 6 C2H2 zinc finger motifs, which facilitate the protein's direct binding to DNA. The consensus DNA binding site for SNAIL family proteins is the CAGGTG sequence; this motif is a subset of the E-box sequence (CANNTG) to which a number of basic helix-loop-helix transcription factors bind.1,2,16 The DNA-binding domain of SNAIL family transcription factors is necessary, but not sufficient, to mediate transcriptional repression by these proteins.3 At their N-terminus, SNAIL family members contain a highly conserved Snai.Gfi-1 (SNAG) domain; this domain is essential for the protein's nuclear localization and role as a transcriptional repressor.3, 17-19 Despite the similarities at their terminal ends, SNAIL and SLUG proteins differ in their proline-rich central region. While SNAIL contains a destruction box and nuclear export sequence, SLUG possesses a unique 2nine amino acid region known as the SLUG domain.3,20 Therefore, it is likely that the SLUG domain is responsible for many of the functional differences between SLUG and other SNAIL family member proteins. Although recent evidence suggests the SLUG domain interacts with the CtBP1 co-repressor and negatively regulates induction of the EMT, the full functional consequence of this domain remains unknown.21
Despite their considerable homology in protein structure and their involvement in common cellular programs, important functional differences exist between SLUG and SNAIL proteins, thus highlighting the notion that these 2 transcription factors have unique and non-overlapping roles in the cell. For example, while SNAIL is required for early embryogenesis, as SNAIL-null mice die at gastrulation, SLUG-null mice are viable but exhibit a variety of specific tissue and stem cell defects.22,23 Additionally, when over-expressed in MDCK epithelial cells or MCF-7 breast cancer cells, SNAIL and SLUG induce both common and distinct gene expression patterns, suggesting these factors regulate shared as well as specific transcriptional programs in both normal and transformed cells.11,24 Although SNAIL and SLUG can both bind to the E-CADHERIN (E-CAD) promoter and repress transcription, SNAIL does so with a higher efficiency and potency than SLUG.25,26 Furthermore, while SNAIL and SLUG are both aberrantly expressed in a variety of tumors, studies indicate they play different roles related to tumor initiation, progression, and metastasis.27,28 These observations suggest that SNAIL and SLUG have distinct roles in the cell, and thus regulate a unique set of gene targets. However, given their structural similarities, it is likely that one can use SNAIL as a guide to assist in the identification of possible mechanisms of transcriptional regulation by SLUG.
This review will focus on the contribution of the transcription factor, SLUG, to normal mammary gland biology and breast tumor development. Specifically SLUG's influence on mammary epithelial cell lineage commitment and differentiation, cellular plasticity, and the mammary stem cell state will be discussed. In regard to these processes, the consequence of aberrant SLUG expression on breast tumor initiation and breast tumor phenotype will be reviewed. Finally, possible molecular mechanisms by which SLUG may regulate these processes will be explored.
The Evolving Mammary Epithelial Cell Hierarchy
The mammary gland is a complex tissue composed of a branching epithelial ductal tree embedded in a layer of fat and stroma. The mammary ductal tree consists of lobular units interconnected by a network of ducts that ultimately drains into the nipple.29 The mammary epithelium is a bilayered structure composed of 2 types of cells: luminal and basal/myoepithelial (ME) cells (Fig. 2A). Luminal cells, which are polarized and cuboidal in shape, can differentiate into either ductal cells or milk producing alveolar cells. Luminal cells line the inner lumens of ducts and alveoli, and are often separated from the surrounding basement membrane (BM) and stroma by an intervening layer of basal/ME cells.29,30 These cells, which form a single-cell layer sheath separating the luminal cells from the BM, are spindle-shaped and possess contractile abilities that allow them to squeeze milk throughout the ducts during lactation.31 Luminal and basal cells are characterized by distinct markers: luminal cells express cytokeratins (CKs) 8/18 and 19, as well as markers such as MUC1, GATA3, and CD24; basal/ME cells express CKs 5, 14, and 17, as well as smooth muscle actin (SMA) and vimentin.32-34
Figure 2.
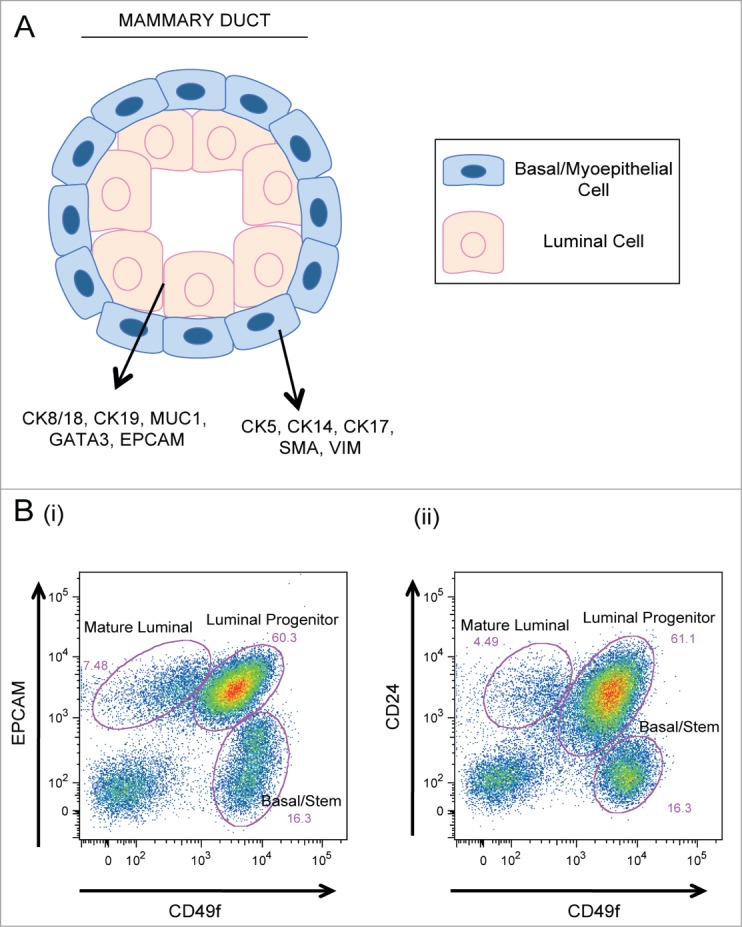
Cellular components of the mammary epithelium. (A) Cross section of a bilayered duct. Luminal cells line the inner duct lumen and are surrounded by an outer layer of contractile basal cells. (B) Flow cytometry plots of primary human mammary epithelial cells analyzed for expression of (i) EPCAM vs. CD49f and (ii) CD24 vs. CD49f expression. Mature luminal, luminal progenitor, and basal/stem populations are indicated.
In addition to mature luminal and basal/ME cells, numerous reports have provided evidence that a mammary epithelial cell hierarchy exists and is responsible for tissue growth and maintenance during periods of development and homeostasis. In a simplistic model, it is thought that bi-potent or multi-potent stem cells give rise to lineage-restricted progenitor cells; these progenitors then divide and differentiate into the committed mature luminal and basal cells of the adult mammary epithelium. Although there have been many attempts to delineate the lineage hierarchy of the mammary epithelium, it is still unclear as to which model, if any, accurately identifies the full spectrum of mammary epithelial cell-types as well as their relationship to one another.35-39
Using cell sorting techniques to isolate subsets of MECs based on their expression of various cell surface markers, researchers have identified subpopulations of luminal progenitor, mature luminal, and basal/stem cells from both human and mouse mammary tissue. While the luminal markers, CD24 and EPCAM, and the basal markers, CD49f and CD29, have consistently been used to segregate luminal and basal MEC populations (Fig. 2B), the identification of intermediate progenitor subsets as well as a pure mammary stem cell (MaSC) population has proven challenging.40-44 Recently, various groups have used different marker combinations to try and define unique MEC progenitor populations.36,40,44,45 While these studies have resulted in the identification of various luminal progenitor subsets, the isolation of a distinct basal progenitor population has been unsuccessful. It is possible, however, that a different combination of markers is necessary to delineate this subset. Similar to the difficulties encountered in defining a basal-progenitor cell subset, attempts to isolate a pure MaSC population have been challenging. Until recently, MaSC-enriched subpopulations were defined predominantly by their ability to form colonies in vitro and to reconstitute a cleared mammary fat pad following transplantation in vivo. Using these techniques, numerous reports concurred that MaSCs reside within the basal epithelial cell subpopulation in both humans and mice; however, markers that can isolate pure MaSCs from within the basal epithelial subset have yet to be discovered.41,42,46,47
Recently, researchers have used a series of lineage tracing studies to further investigate the mammary lineage hierarchy. Lineage tracing allows genetic markers to be tracked in situ, and thus provides a way to trace stem or progenitor cell fate over time under normal, physiological conditions. Using this technique, van Keymeulen and colleagues showed that the mammary gland initially develops from bipotent CK14+ progenitors that persist only during embryogenesis. Following birth, unipotent luminal-restricted and basal/ME-restricted progenitors are responsible for tissue growth and maintenance during puberty, pregnancy, lactation, and involution. The authors did acknowledge, however, that perhaps a rare, adult bipotent mammary stem cell exists that was not targeted by their genetic-labeling system.38
Adding to the complexity of this topic, a study by Rios et al., which also utilized lineage tracing techniques to examine the mammary epithelial cell hierarchy, reported that a bipotent, adult MaSC exists, in addition to long-lived progenitor cells. The results from this study suggested that both MaSCs and progenitor cells contribute to ductal growth, alveolar expansion, and tissue maintenance throughout development of the mammary gland.48 Together, these findings reveal that defining distinct subsets of mammary stem and progenitor cells is challenging, and continued work is necessary to definitively characterize a mammary lineage hierarchy. Undoubtedly, deciphering the mammary epithelial cell hierarchy will provide critical information to aide in our understanding of the cellular and molecular mechanisms that drive breast cancer initiation and progression.
SLUG in mammary epithelial cell differentiation
Recent work has identified a novel role for SLUG as a regulator of mammary epithelial cell differentiation.49,50 In adult human and mouse mammary epithelium, immunohistochemical (IHC) analysis revealed that SLUG localizes to the basal/ME cell layer, suggesting that SLUG may regulate this epithelial cell-state.49,51 Further examination of SLUG expression in different mouse epithelial cell populations, including mature luminal, luminal progenitor, and basal/stem cells, confirmed that SLUG is differentially expressed in the basal/stem subset.42,51,52 Unlike SLUG, SNAIL is expressed at similar levels in luminal and basal cells, but is significantly enriched in the mammary stromal population; this suggests a unique role for SLUG in regulating mammary epithelial cell identity and lineage commitment programs.51,52
Consistent with a role for SLUG in maintaining the basal cell phenotype, stable knockdown of SLUG in immortalized, patient-derived human mammary epithelial cells (HMECs) resulted in increased expression of luminal lineage genes, including EPCAM, E-CAD, CD24, and MUC1; similar results were observed following SLUG inhibition in the spontaneously immortalized MCF10A breast epithelial cell line.50 In both cell lines, the gene signature of SLUG KD cells was significantly enriched in genes characteristic of luminal progenitor cells; this further suggests that SLUG functions to repress luminal lineage differentiation. Additionally, flow cytometry analysis revealed that SLUG inhibition results in a shift in epithelial cell-state proportions, characterized by an expansion of luminal cells and reduction of basal cells.50 Together, these results suggest that SLUG functions to maintain MECs in a basal-like state, and inhibition of SLUG promotes luminal differentiation (Fig. 3A).
Figure 3.
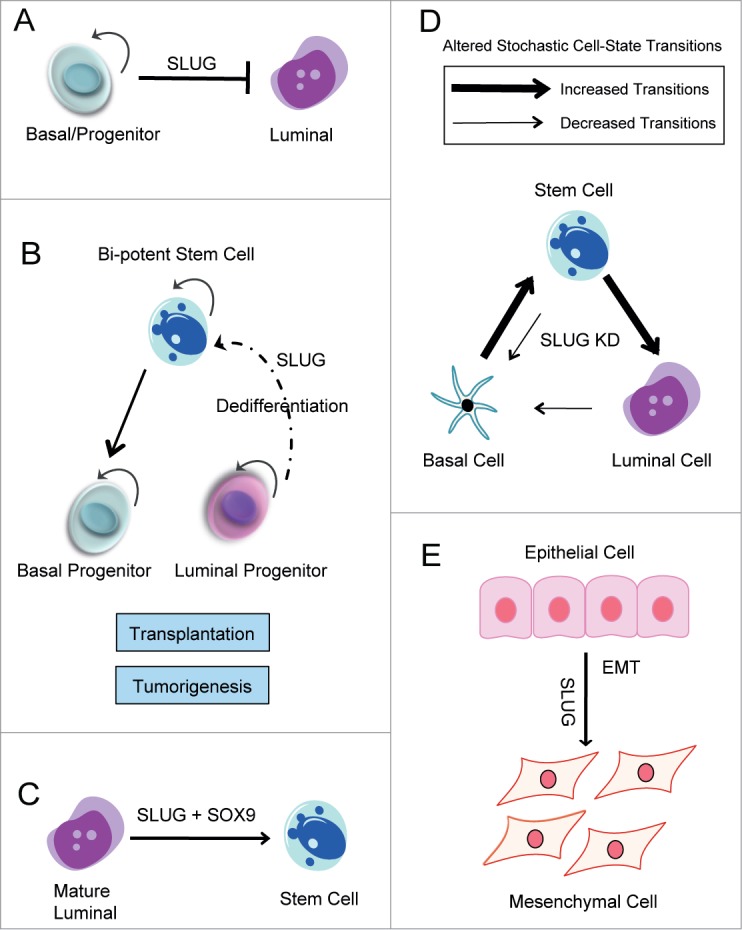
SLUG is a Regulator of Mammary Epithelial Cell-State Dynamics. Schematic diagrams outlining the various functions of SLUG in mammary epithelial cells. (A) SLUG regulates MEC differentiation. As a transcriptional repressor, SLUG represses transcription of luminal lineage genes and maintains cells in a basal/progenitor-like state. (B) During tissue regeneration following transplantation and tumor initiation, SLUG may be necessary for lineage-restricted cells to de-differentiate into a more primitive, stem-like state. (C) SLUG cooperates with SOX9 to promote the mammary stem cell state. (D) SLUG regulates stochastic cell-state transitions between luminal, basal, and stem cells. Following SLUG knockdown, there are decreased transitions into the basal state and increased transitions into the luminal state. SLUG KD also affects transition into and out of the stem cell state. (E) SLUG promotes the EMT.
Confirmation that SLUG regulates MEC differentiation was also shown in vivo using a SLUG-LacZ transgenic mouse model.22,50 Throughout early stages of development and during puberty, strong SLUG expression was observed in the basal/ME layer of mammary ducts.52 Compared to wild type (WT) mice, the mammary epithelium of SLUG-deficient mice displayed increased expression of luminal CKs and luminal-specific genes, including Gata3 and ERα. Histological examination of mammary glands from adult SLUG-LacZ mice revealed severe defects in MEC differentiation characterized by hyperplasia of luminal cells and aberrant expression of luminal markers in the basal/ME cell layer.50 Upon further investigation of this phenotype, a unique population of cells with an EPCAMhi/CD49fhi profile was identified in SLUG-deficient mammary epithelium. This population, termed the luminobasal population, represented basal cells with enhanced features of luminal differentiation. Consistent with this observation, the gene signature of the luminobasal population was significantly enriched in the gene expression patterns of both luminal progenitor and mature luminal cells.50 These data confirm a role for SLUG in regulating MEC identity and differentiation in vivo, and suggest that SLUG is necessary to represses luminal gene expression programs in basal/stem cells. Taken together, in vitro and in vivo models of SLUG-deficiency have highlighted a critical role for SLUG as a regulator of MEC differentiation, whereby SLUG functions to repress luminal gene transcription programs.
The connection between SLUG, cellular plasticity, and the mammary stem cell state
In addition to regulating differentiation, recent work has also shown that SLUG promotes the mammary stem-cell state. This is consistent with SLUG's expression in the basal cell layer of the mammary epithelium, where MaSCs have been reported to reside.46,51,53 Using the mammosphere assay, Nassour and colleagues showed that SLUG-deficient MECs were unable to form secondary and tertiary mammospheres upon serial dissociation and re-plating, suggesting that SLUG may be necessary for stem/progenitor cell self-renewal.52 Additional studies investigating SLUG's role in promoting “stemness” revealed that induction of SLUG correlates with increased proportions of CD44+/CD24− stem-like cells. Cells with this phenotype have in vivo repopulating capabilities, display the ability to self-renew, and exhibit bipotent differentiation potential.54,55 Interestingly, over-expression of SLUG in the MCF10A basal breast epithelial cell line induced formation of CD44+/CD24− stem-like cells; however, the same result was not observed when SLUG was overexpressed in the MCF-7 luminal-type breast cancer cell line.56 This finding suggests that the differentiation state of a cell may affect SLUG's ability to promote stem-like properties. In several studies, SLUG's ability to induce the CD44+/CD24− cancer stem cell (CSC) phenotype has been proposed to result from its induction of the EMT.55-57 However, it is unclear whether a full EMT is necessary to generate these stem cells, or whether certain aspects of the EMT, such as enhanced plasticity, are sufficient to transform cells into bona fide stem cells.
Previously, the mammary transplantation assay had been described as the gold standard for identifying functional stem cells.29 Recently, however, it has been hypothesized that the transplantation assay does not measure a cell's inherent stem cell activity, but instead reflects the potential of lineage-committed cells to de-differentiate and adopt a more primitive, stem-like state.38,58 Recent reports using the mammary transplantation assay to investigate the effect of SLUG loss on a cell's regenerative potential have offered an interesting perspective on this issue.
SLUG-deficient mice do not display stem cell defects during normal growth and development, as they develop full mammary ductal trees. Similarly, when SLUG-deficient tissue fragments were transplanted into the cleared fat pads of WT recipients, they successfully grew and formed outgrowths.52 Together, this suggests that when MECs are maintained in their normal, physiological environment they do not require SLUG for stem cell activity. However, when SLUG-deficient MECs were removed from their natural environment and transplanted as single cell suspensions, they failed to reconstitute the mammary gland.50 This suggests that SLUG may regulate cellular plasticity necessary for differentiated cells to acquire stem-like properties under non-physiological conditions (Fig. 3B).
Consistent with these findings, Guo and colleagues reported that SLUG and the SRY-box transcription factor, SOX9, function together to induce and maintain the mammary stem cell state.51 By ectopically expressing both factors alone or together in primary mouse MECs, this group showed that SLUG and SOX9 are both required for MaSC function. Strikingly, co-expression of both factors in differentiated luminal cells in vitro converted these cells into functional MaSCs that could reconstitute a cleared mammary fat pad following transplantation in vivo (Fig. 3C).51 While this finding provided evidence that SLUG can promote entrance into the stem cell state, it also demonstrated that epithelial cells are plastic, suggesting that certain genetic or epigenetic factors, such as SLUG, may be able to convert differentiated cells into a more primitive stem-like state. In support of a role for SLUG in regulating cell plasticity, we used an in vitro model of cell state dynamics to show that loss of SLUG alters cell-state transitions between normal luminal, basal, and stem cells. Importantly, loss of SLUG affected transitions from and into the stem cell state (Fig. 3D). Together, these findings highlight a novel function of SLUG in regulating the MaSC state and cell-state dynamics.
SLUG and Breast Tumorigenesis
Work from various groups has revealed that disruption of lineage commitment and differentiation programs can alter the epigenetic state of progenitor cells, and thus influence which type of tumor will develop.59-63 Given the important role of SLUG in regulating normal luminal and basal/ME cells in the breast, it should likely play an important role in breast cancer pathogenesis and tumor behavior. In fact, we speculated that breast tumor phenotype would be significantly impacted by SLUG deficiency since the differentiation state of the normal basal precursor target for neoplastic transformation is altered in SLUG-deficient mice. Surprisingly however, we found that SLUG-deficient mice were resistant to Myc-induced mammary tumorigenesis.50 This finding suggests that tumor initiation operates through a mechanism that is similar to tissue regeneration. Indeed, in other contexts, cells must adopt stem-like properties as a pre-requisite for tumor initiation.64,65 These unexpected findings highlight a role for SLUG in regulating mammary epithelial cell plasticity, and suggest that SLUG may be necessary for lineage-committed cells to de-differentiate into a more primitive, stem-like state during tissue regeneration following transplantation and tumor initiation.
SLUG and the EMT
Understanding the full functional consequence of aberrant SLUG expression during malignant transformation is complex, especially given the diverse roles of SLUG in the cell.1 The majority of studies evaluating SLUG's involvement during breast tumorigenesis have focused on SLUG's regulation of the EMT (Fig. 3E), a reversible process in which epithelial cells acquire a mesenchymal phenotype.25, 66-70 In epithelial cells, the EMT is characterized by loss of polarity, cytoskeleton rearrangement, a change in intermediate filaments (from keratin to vimentin), an increase in cell motility, and resistance to apoptosis.68,71 On a molecular level, SLUG's regulation of the EMT is often associated with its ability to transcriptionally repress expression of the epithelial gene, E-CAD.25,72,73 These phenotypic and functional changes have been shown to facilitate cancer cell invasion and metastasis.72,74
Apart from the EMT, transcriptional regulation by SLUG has been shown to affect a diverse number of cellular functions in the mammary epithelium. As previously discussed, recent studies have identified SLUG as a regulator of MEC differentiation; in this context SLUG directly represses the transcription of mammary epithelial luminal lineage genes.49,50,52 SLUG has also been implicated as a regulator of the mammary stem cell state, both in the normal tissue and breast cancer cells.50-52 Consistent with these findings, basal-type breast tumors with elevated SLUG expression were shown to over-express stem-like genes, including CD133 and BMI1.72 Additional studies revealed that breast tumors over-expressing SLUG display increased proportions of CD44+/CD24− CSCs, suggesting that transcriptional programs induced by SLUG promote stemness. However, from these studies it is unclear whether SLUG endows cells with certain properties of stem cells, or fully converts them into bona fide stem cells.55,56,72,75
Interestingly, other studies have recognized that during some developmental and disease processes a full EMT is not always achieved. Termed a partial EMT, cells in this phenotypic state often retain cell-cell junctions but acquire invasive and motile properties.68,76 Physiological examples of a partial EMT include epithelial wound healing and the collective migration of epithelial cells during mammary branching morphogenesis.77 In this sense, the EMT signature appears closely connected to differentiation pathways as well as stem cell signatures.52 The identification of the partial EMT state is critical to our understanding of cell-state identity and cellular plasticity, suggesting that a switch between 2 phenotypic states is not absolute, but exists on a continuum.68 With regards to this, we recently identified SLUG as a regulator of mammary epithelial cell plasticity, and showed that SLUG promotes cell-state transitions into and out of the basal and stem cell states.50 This finding suggests that it may be important to consider the concept of a phenotypic continuum when assessing SLUG's function during development and tumor progression. From the work discussed above, it is clear that SLUG's regulation of cellular differentiation, cellular plasticity, and the mammary stem cell state will have important implications for research focused on understanding breast tumor heterogeneity.
SLUG and the Basal Breast Tumor Phenotype
Breast cancer is a complex disease, comprised of heterogeneous tumors displaying different molecular profiles, metastatic behavior, clinical characteristics, and response to therapeutics. Much research has focused on the 2 major molecular classes of breast cancers: luminal and basal-type tumors. These tumor types were originally classified by their expression of certain markers known to identify with either normal luminal or basal/ME breast cells. Luminal-type tumors are characterized by high expression of genes typically found in normal luminal breast epithelial cells, including CK8/18 and the estrogen receptor (ERα). On the contrary, basal-type tumors consistently express high levels of certain basal/ME-associated genes, including CK5, CK14, and CK17.80-82 Luminal and basal-type tumors differ not only in their molecular profiles, but also in the clinical course of the disease and their response to therapeutics.78,79 Compared to luminal tumors, basal tumors typically are high grade, poorly differentiated, and associated with a poor prognosis.83,84 Interestingly, women with inherited mutations in the Breast Cancer Associated 1 (BRCA1) gene have an increased risk of developing aggressive basal-type breast cancer.85,86
Although known for their high expression of several basal/ME-specific markers, further analysis of the basal tumor subtype revealed that these tumors also express genes characteristic of other cell lineages, including luminal, stem, and progenitor-type cells.80,87,88 Consistent with this, basal breast cancers have been shown to express several stem cell-associated genes, including BMI1.72, 89-91 Recently, a subset of embryonic mammary epithelial genes was also found to be up-regulated in basal-type tumors; several of theses genes, including BCl11a, GRHL3, PROX1, and SOX11, were previously shown to function as regulators of progenitor cell activity or differentiation.92 Furthermore, several studies have reported an enrichment of CD24−/CD44+ breast cancer stem cells (BCSCs) in basal-type tumors.93-96 The discovery that basal-type tumors exhibit stem-like features suggests that properties inherent to stem cells, such as increased plasticity and decreased lineage specification, may contribute to the basal tumor phenotype.
Strikingly, despite their expression of several basal/ME and stem cell markers, recent studies have revealed that the gene signature of basal-type tumors most closely resembles that of normal luminal progenitor cells.42,97 This finding had a major impact in the field and compelled researchers to investigate the connection between basal-type breast cancer and luminal progenitor cells. Recently, studies from several groups have provided evidence that transformed luminal progenitor cells are the cell-of-origin of BRCA1-associated and basal-type breast cancers.49,98,99 This data suggests that factors that regulate cellular differentiation and lineage commitment may play an important role in dictating breast tumor phenotype.
Investigation into possible molecular factors and pathways contributing to the aggressive nature of the basal tumor subtype has often focused on members of the SNAIL family of transcriptional repressors, including SLUG.56,66,68,69,72 In human breast cancer, SLUG is frequently overexpressed in BRCA1-mutated and basal-type tumors.49,69,72 IHC analysis of SLUG expression in primary human breast tumors revealed that elevated SLUG levels correlate with increased metastatic potential and high tumor grade.69 Similarly, SLUG is also over-expressed in basal-type breast cancer cell lines where inhibition of SLUG has been shown to result in increased expression of luminal lineage genes 49,90; this suggests that, similar to the normal MECs, SLUG also represses luminal gene expression in breast cancer cells.49
While these findings clearly establish a connection between SLUG overexpression and the basal tumor subtype, they do not distinguish whether SLUG is the cause or consequence of the basal phenotype. However, some early findings might point to SLUG as the cause of the basal tumor phenotype in at least some breast cancers. In disease-free breast tissue of BRCA1-mutation carriers, SLUG was shown to be overexpressed prior to any evidence of cancer.49 These BRCA1-deficient or mutant MECs were shown to exhibit defects in epithelial cell differentiation as well as progenitor cell function.49, 100-102 In addition, analysis of BRCA1 mutated HMECs prior to transformation revealed that these cells display enhanced features of basal differentiation. BRCA1-mutant HMECs also generated outgrowths in vivo with increased numbers of luminal cells expressing both luminal and basal markers.49 Furthermore, when cells from these women were transformed, they preferentially generated basal-like tumors. This finding suggests that elevated SLUG expression prior to neoplastic transformation might explain, in part, why BRCA1-mutation carriers exhibit a strong predisposition to developing aggressive, basal-type breast tumors.49 In addition, these data suggest that aberrant SLUG expression creates defects in MEC lineage commitment that may ultimately affect tumor phenotype.
Mechanism of transcriptional regulation by SLUG
It is clear that SLUG regulates many critical functions in the cell that are necessary for breast development and tumor formation. Uncovering the molecular mechanisms by which SLUG controls gene expression programs will undoubtedly contribute to our understanding of SLUG's role during breast tumorigenesis and provide possible avenues for new therapies to treat aggressive basal-type tumors. Over the past 20 years, chromatin remodeling and histone modifications have been recognized as major mechanisms controlling gene expression. In particular, histone methylation and acetylation have been frequently implicated in regulating gene transcription.103 The first evidence that SNAIL family members might use epigenetic mechanisms to function as transcriptional repressors was reported in 2000, when treatment of cells with tricostatin A, a histone deactelyase (HDAC) inhibitor, alleviated SNAIL-mediated repression at the E-CAD promoter.3 Since then, SNAIL has been identified in various complexes with epigenetic factors and co-repressors, including HDAC1/2 and the mSIN3A co-repressor,104 the arginine methyltransferase, PRMT5, and the AJUBA co-repressor,105 as well as the methyltransferases SUZ12, G9a, and SUV39H1.106-108 In all cases, the ability of SNAIL to repress the transcription of target genes was linked to its recruitment of chromatin-modifying enzymes and/or co-factors.
More recently, Lysine Specific Demethylase 1 (LSD1) was identified as a novel SNAIL interacting protein and co-regulator of E-CAD transcription.109 LSD1 is an enzyme that removes mono- and dimethyl methyl groups from H3K4 and H3K9, post-translational modifications associated with transcriptional repression.110,111 Lin et al. showed that SNAIL binds LSD1 via its SNAG domain and recruits it to the E-CAD promoter. Once localized, LSD1 removes mono- and di-methyl groups from H3K4, thus promoting gene silencing.109,112
With regards to SLUG, this finding was intriguing since SLUG also contains a SNAG domain and is a known repressor of E-CAD transcription. In fact, it was recently shown that SLUG interacts with LSD1, and this interaction is responsible for epigenetic silencing at SLUG target gene promoters.113 In the MCF10A breast epithelial cell line, we showed that SLUG and LSD1 function together to repress genes associated with luminal differentiation.50 However, whether this complex also functions to regulate mammary stem cell activity is unknown. Interestingly, like SLUG, LSD1 is also over-expressed in aggressive ERα− breast tumors where its expression correlates with aggressive behavior and a poor prognosis.114 Therefore, targeting the SLUG/LSD1 complex may provide a potential avenue for treating aggressive basal-type tumors. However, LSD1 represents only one of potentially many SLUG transcriptional co-regulators, and therefore, further work is necessary to identify other mechanisms by which SLUG may regulate gene transcription programs.
Conclusion
SLUG, a SNAIL family transcription factor, has been highly studied in the context of breast cancer since it is consistently overexpressed in aggressive, basal-type breast tumors. Previous work has predominantly focused on SLUG's regulation of the EMT and its influence on breast tumor invasion and metastasis. More recently, studies have discovered novel roles of SLUG in normal breast epithelial cells, including regulation of cell differentiation, cell-state dynamics, and the mammary stem cell-state. These SLUG-dependent processes have also been shown to influence breast tumor initiation and progression. Understanding the connection between EMT, cellular plasticity, stemness, and cell-state identity during normal development and tumorigenesis is undoubtedly complex; however, SLUG may be an ideal candidate for delineating how these processes relate to one another, and for determining how they collectively contribute to breast cancer initiation and progression.
The studies discussed here offer insight into how targeting SLUG may affect breast tumorigenesis. The role of SLUG in repressing luminal differentiation suggests that inhibiting SLUG in basal-type tumors could alter the differentiation state of these cells. Alternatively, the requirement for SLUG during tissue regeneration following transplantation and Myc-induced tumor formation suggests that targeting SLUG may compromise breast tumor stem cell activity. With regards to EMT during late stages of tumorigenesis, inhibiting SLUG function has already been shown to inhibit cancer cell invasion and metastasis. While it clear that SLUG plays a critical role during multiple stages of breast tumor development, further insight into the cellular components and pathways of the normal mammary hierarchy is necessary to fully comprehend how aberrant SLUG expression influences breast tumor biology.
References
- 1. Hemavathy K, Ashraf SI, Ip YT. Snail/slug family of repressors: Slowly going into the fast lane of development and cancer. Gene 2000; 257:1-12; PMID:11054563; http://dx.doi.org/S0378-1119(00)00371-1 [pii] [DOI] [PubMed] [Google Scholar]
- 2. Nieto MA. The snail superfamily of zinc-finger transcription factors. Nat Rev Mol Cell Biol 2002; 3:155-66; PMID:11994736; http://dx.doi.org/ 10.1038/nrm757 [DOI] [PubMed] [Google Scholar]
- 3. Hemavathy K, Guru SC, Harris J, Chen JD, Ip YT. Human slug is a repressor that localizes to sites of active transcription. Mol Cell Biol 2000; 20:5087-95; PMID:10866665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Isaac A, Sargent MG, Cooke J. Control of vertebrate left-right asymmetry by a snail-related zinc finger gene. Science 1997; 275:1301-4; PMID:9036854 [DOI] [PubMed] [Google Scholar]
- 5. Inukai T, Inoue A, Kurosawa H, Goi K, Shinjyo T, Ozawa K, Mao M, Inaba T, Look AT. SLUG, a ces-1-related zinc finger transcription factor gene with antiapoptotic activity, is a downstream target of the E2A-HLF oncoprotein. Mol Cell 1999; 4:343-52; PMID:10518215 [DOI] [PubMed] [Google Scholar]
- 6. Vega S, Morales AV, Ocana OH, Valdes F, Fabregat I, Nieto MA. Snail blocks the cell cycle and confers resistance to cell death. Genes Dev 2004; 18:1131-43; PMID:15155580; http://dx.doi.org/ 10.1101/gad.294104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Boutet A, De Frutos CA, Maxwell PH, Mayol MJ, Romero J, Nieto MA. Snail activation disrupts tissue homeostasis and induces fibrosis in the adult kidney. EMBO J 2006; 25:5603-13; PMID:17093497; http://dx.doi.org/7601421 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Martinez-Alvarez C, Blanco MJ, Perez R, Rabadan MA, Aparicio M, Resel E, Martinez T, Nieto MA. Snail family members and cell survival in physiological and pathological cleft palates. Dev Biol 2004; 265:207-18; PMID:14697364; http://dx.doi.org/S0012160603005748 [pii] [DOI] [PubMed] [Google Scholar]
- 9. Kajita M, McClinic KN, Wade PA. Aberrant expression of the transcription factors snail and slug alters the response to genotoxic stress. Mol Cell Biol 2004; 24:7559-66; PMID:15314165; http://dx.doi.org/ 10.1128/MCB.24.17.7559-7566.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Barrallo-Gimeno A, Nieto MA. The snail genes as inducers of cell movement and survival: Implications in development and cancer. Development 2005; 132:3151-61; PMID:15983400; http://dx.doi.org/132/14/3151 [pii] [DOI] [PubMed] [Google Scholar]
- 11. Dhasarathy A, Phadke D, Mav D, Shah RR, Wade PA. The transcription factors snail and slug activate the transforming growth factor-beta signaling pathway in breast cancer. PLoS One 2011; 6:e26514; PMID:22028892; http://dx.doi.org/ 10.1371/journal.pone.0026514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kurrey NKKA, Bapat SA. Snail and slug are major determinants of ovarian cancer invasiveness at the transcription level. Gynecol Oncol 2005; 97:155-65; PMID:15790452; http://dx.doi.org/S0090-8258(04)01058-3 [pii] [DOI] [PubMed] [Google Scholar]
- 13. Uygur B, Wu WS. SLUG promotes prostate cancer cell migration and invasion via CXCR4/CXCL12 axis. Mol Cancer 2011; 10:139,4598-10-139; PMID:22074556; http://dx.doi.org/ 10.1186/1476-4598-10-139 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Turner FE, Broad S, Khanim FL, Jeanes A, Talma S, Hughes S, Tselepis C, Hotchin NA. Slug regulates integrin expression and cell proliferation in human epidermal keratinocytes. J Biol Chem 2006; 281:21321-31; PMID:16707493; http://dx.doi.org/M509731200 [pii] [DOI] [PubMed] [Google Scholar]
- 15. Emadi Baygi M, Soheili ZS, Essmann F, Deezagi A, Engers R, Goering W, Schulz WA. Slug/SNAI2 regulates cell proliferation and invasiveness of metastatic prostate cancer cell lines. Tumour Biol 2010; 31:297-307; PMID:20506051; http://dx.doi.org/ 10.1007/s13277-010-0037-5 [DOI] [PubMed] [Google Scholar]
- 16. Cobaleda C, Perez-Caro M, Vicente-Duenas C, Sanchez-Garcia I. Function of the zinc-finger transcription factor SNAI2 in cancer and development. Annu Rev Genet 2007; 41:41-61; PMID:17550342; http://dx.doi.org/ 10.1146/annurev.genet.41.110306.130146 [DOI] [PubMed] [Google Scholar]
- 17. Shirley SH, Hudson LG, He J, Kusewitt DF. The skinny on slug. Mol Carcinog 2010; 49:851-61; PMID:20721976; http://dx.doi.org/ 10.1002/mc.20674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Grimes HL, Chan TO, Zweidler-McKay PA, Tong B, Tsichlis PN. The gfi-1 proto-oncoprotein contains a novel transcriptional repressor domain, SNAG, and inhibits G1 arrest induced by interleukin-2 withdrawal. Mol Cell Biol 1996; 16:6263-72; PMID:8887656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chiang C, Ayyanathan K. Snail/gfi-1 (SNAG) family zinc finger proteins in transcription regulation, chromatin dynamics, cell signaling, development, and disease. Cytokine Growth Factor Rev 2013; 24:123-31; PMID:23102646; http://dx.doi.org/ 10.1016/j.cytogfr.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sefton M, Sanchez S, Nieto MA. Conserved and divergent roles for members of the snail family of transcription factors in the chick and mouse embryo. Development 1998; 125:3111-21; PMID:9671584 [DOI] [PubMed] [Google Scholar]
- 21. Molina-Ortiz P, Villarejo A, MacPherson M, Santos V, Montes A, Souchelnytskyi S, Portillo F, Cano A. Characterization of the SNAG and SLUG domains of Snail2 in the repression of E-cadherin and EMT induction: Modulation by serine 4 phosphorylation. PLoS One 2012; 7:e36132; PMID:22567133; http://dx.doi.org/ 10.1371/journal.pone.0036132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jiang R, Lan Y, Norton CR, Sundberg JP, Gridley T. The slug gene is not essential for mesoderm or neural crest development in mice. Dev Biol 1998; 198:277-85; PMID:9659933 [PubMed] [Google Scholar]
- 23. Carver EA, Jiang R, Lan Y, Oram KF, Gridley T. The mouse snail gene encodes a key regulator of the epithelial-mesenchymal transition. Mol Cell Biol 2001; 21:8184-8; PMID:11689706; http://dx.doi.org/ 10.1128/MCB.21.23.8184-8188.2001 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Moreno-Bueno G, Cubillo E, Sarrio D, Peinado H, Rodriguez-Pinilla SM, Villa S, Bolos V, Jorda M, Fabra A, Portillo F, et al. Genetic profiling of epithelial cells expressing E-cadherin repressors reveals a distinct role for snail, slug, and E47 factors in epithelial-mesenchymal transition. Cancer Res 2006; 66:9543-56; PMID:17018611; http://dx.doi.org/ 10.1158/0008-5472.CAN-06-0479 [DOI] [PubMed] [Google Scholar]
- 25. Bolos V, Peinado H, Perez-Moreno MA, Fraga MF, Esteller M, Cano A. The transcription factor slug represses E-cadherin expression and induces epithelial to mesenchymal transitions: A comparison with snail and E47 repressors. J Cell Sci 2003; 116:499-511; PMID:12508111 [DOI] [PubMed] [Google Scholar]
- 26. Gras B, Jacqueroud L, Wierinckx A, Lamblot C, Fauvet F, Lachuer J, Puisieux A, Ansieau S. Snail family members unequally trigger EMT and thereby differ in their ability to promote the neoplastic transformation of mammary epithelial cells. PLoS One 2014; 9:e92254; PMID:24638100; http://dx.doi.org/ 10.1371/journal.pone.0092254 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Villarejo A, Cortes-Cabrera A, Molina-Ortiz P, Portillo F, Cano A. Differential role of Snail1 and Snail2 zinc fingers in E-cadherin repression and epithelial to mesenchymal transition. J Biol Chem 2014; 289:930-41; PMID:24297167; http://dx.doi.org/ 10.1074/jbc.M113.528026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Come C, Magnino F, Bibeau F, De Santa Barbara P, Becker KF, Theillet C, Savagner P. Snail and slug play distinct roles during breast carcinoma progression. Clin Cancer Res 2006; 12:5395-402; PMID:17000672; http://dx.doi.org/ 10.1158/1078-0432.CCR-06-0478 [DOI] [PubMed] [Google Scholar]
- 29. Visvader JE. Keeping abreast of the mammary epithelial hierarchy and breast tumorigenesis. Genes Dev 2009; 23:2563-77; PMID:19933147; http://dx.doi.org/ 10.1101/gad.1849509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Smalley MJ, Titley J, Paterson H, Perusinghe N, Clarke C, O’Hare MJ. Differentiation of separated mouse mammary luminal epithelial and myoepithelial cells cultured on EHS matrix analyzed by indirect immunofluorescence of cytoskeletal antigens. J Histochem Cytochem 1999; 47:1513-24; PMID:10567435 [DOI] [PubMed] [Google Scholar]
- 31. Warburton MJ, Mitchell D, Ormerod EJ, Rudland P. Distribution of myoepithelial cells and basement membrane proteins in the resting, pregnant, lactating, and involuting rat mammary gland. J Histochem Cytochem 1982; 30:667-76; PMID:6179984 [DOI] [PubMed] [Google Scholar]
- 32. Dairkee S, Heid HW. Cytokeratin profile of immunomagnetically separated epithelial subsets of the human mammary gland. In Vitro Cell Dev Biol Anim 1993; 29A:427-32; PMID:7686143 [DOI] [PubMed] [Google Scholar]
- 33. Gudjonsson T, Adriance MC, Sternlicht MD, Petersen OW, Bissell MJ. Myoepithelial cells: Their origin and function in breast morphogenesis and neoplasia. J Mammary Gland Biol Neoplasia 2005; 10:261-72; PMID:16807805; http://dx.doi.org/ 10.1007/s10911-005-9586-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sopel M. The myoepithelial cell: Its role in normal mammary glands and breast cancer. Folia Morphol (Warsz) 2010; 69:1-14; PMID:20235044 [PubMed] [Google Scholar]
- 35. Jeselsohn R, Brown NE, Arendt L, Klebba I, Hu MG, Kuperwasser C, Hinds PW. Cyclin D1 kinase activity is required for the self-renewal of mammary stem and progenitor cells that are targets of MMTV-ErbB2 tumorigenesis. Cancer Cell 2010; 17:65-76; PMID:20129248; http://dx.doi.org/ 10.1016/j.ccr.2009.11.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shehata M, Teschendorff A, Sharp G, Novcic N, Russell A, Avril S, Prater M, Eirew P, Caldas C, Watson CJ, et al. Phenotypic and functional characterization of the luminal cell hierarchy of the mammary gland. Breast Cancer Res 2012; 14:R134; PMID:23088371; http://dx.doi.org/ 10.1186/bcr/3334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kittrell FS, Carletti MZ, Kerbawy S, Heestand J, Xian W, Zhang M, Lamarca HL, Sonnenberg A, Rosen JM, Medina D, et al. Prospective isolation and characterization of committed and multipotent progenitors from immortalized mouse mammary epithelial cells with morphogenic potential. Breast Cancer Res 2011; 13:R41; PMID:21466693; http://dx.doi.org/ 10.1186/bcr2863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Van Keymeulen A, Rocha AS, Ousset M, Beck B, Bouvencourt G, Rock J, Sharma N, Dekoninck S, Blanpain C. Distinct stem cells contribute to mammary gland development and maintenance. Nature 2011; 479:189-93; PMID:21983963; http://dx.doi.org/ 10.1038/nature10573 [DOI] [PubMed] [Google Scholar]
- 39. dos Santos CO, Rebbeck C, Rozhkova E, Valentine A, Samuels A, Kadiri LR, Osten P, Harris EY, Uren PJ, Smith AD, et al. Molecular hierarchy of mammary differentiation yields refined markers of mammary stem cells. Proc Natl Acad Sci U S A 2013; 110:7123-30; PMID:23580620; http://dx.doi.org/ 10.1073/pnas.1303919110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Asselin-Labat ML, Sutherland KD, Barker H, Thomas R, Shackleton M, Forrest NC, Hartley L, Robb L, Grosveld FG, van der Wees J, et al. Gata-3 is an essential regulator of mammary-gland morphogenesis and luminal-cell differentiation. Nat Cell Biol 2007; 9:201-9; PMID:17187062; http://dx.doi.org/ 10.1038/ncb1530 [DOI] [PubMed] [Google Scholar]
- 41. Eirew P, Stingl J, Raouf A, Turashvili G, Aparicio S, Emerman JT, Eaves CJ. A method for quantifying normal human mammary epithelial stem cells with in vivo regenerative ability. Nat Med 2008; 14:1384-9; PMID:19029987; http://dx.doi.org/ 10.1038/nm.1791 [DOI] [PubMed] [Google Scholar]
- 42. Lim E, Vaillant F, Wu D, Forrest NC, Pal B, Hart AH, Asselin-Labat ML, Gyorki DE, Ward T, Partanen A, et al. Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat Med 2009; 15:907-13; PMID:19648928; http://dx.doi.org/ 10.1038/nm.2000 [DOI] [PubMed] [Google Scholar]
- 43. Keller PJ, Lin AF, Arendt LM, Klebba I, Jones AD, Rudnick JA, DiMeo TA, Gilmore H, Jefferson DM, Graham RA, et al. Mapping the cellular and molecular heterogeneity of normal and malignant breast tissues and cultured cell lines. Breast Cancer Res 2010; 12:R87; PMID:20964822; http://dx.doi.org/ 10.1186/bcr2755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Raouf A, Zhao Y, To K, Stingl J, Delaney A, Barbara M, Iscove N, Jones S, McKinney S, Emerman J, et al. Transcriptome analysis of the normal human mammary cell commitment and differentiation process. Cell Stem Cell 2008; 3:109-18; PMID:18593563; http://dx.doi.org/ 10.1016/j.stem.2008.05.018 [DOI] [PubMed] [Google Scholar]
- 45. Regan JL, Kendrick H, Magnay FA, Vafaizadeh V, Groner B, Smalley MJ. C-kit is required for growth and survival of the cells of origin of Brca1-mutation-associated breast cancer. Oncogene 2012; 31:869-83; PMID:21765473; http://dx.doi.org/ 10.1038/onc.2011.289 [DOI] [PubMed] [Google Scholar]
- 46. Shackleton M, Vaillant F, Simpson KJ, Stingl J, Smyth GK, Asselin-Labat ML, Wu L, Lindeman GJ, Visvader JE. Generation of a functional mammary gland from a single stem cell. Nature 2006; 439:84-8; PMID:16397499; http://dx.doi.org/ 10.1038/nature04372 [DOI] [PubMed] [Google Scholar]
- 47. Stingl J, Eaves CJ, Zandieh I, Emerman JT. Characterization of bipotent mammary epithelial progenitor cells in normal adult human breast tissue. Breast Cancer Res Treat 2001; 67:93-109; PMID:11519870 [DOI] [PubMed] [Google Scholar]
- 48. Rios AC, Fu NY, Lindeman GJ, Visvader JE. In situ identification of bipotent stem cells in the mammary gland. Nature 2014; 506:322-7; PMID:24463516; http://dx.doi.org/ 10.1038/nature12948 [DOI] [PubMed] [Google Scholar]
- 49. Proia TA, Keller PJ, Gupta PB, Klebba I, Jones AD, Sedic M, Gilmore H, Tung N, Naber SP, Schnitt S, et al. Genetic predisposition directs breast cancer phenotype by dictating progenitor cell fate. Cell Stem Cell 2011; 8:149-63; PMID:21295272; http://dx.doi.org/ 10.1016/j.stem.2010.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Phillips S, Prat A, Sedic M, Proia T, Wronski A, Mazumdar S, Skibinski A, Shirley SH, Perou CM, Gill G, et al. Cell-state transitions regulated by SLUG are critical for tissue regeneration and tumor initiation. Stem Cell Reports 2014; In Press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Guo W, Keckesova Z, Donaher JL, Shibue T, Tischler V, Reinhardt F, Itzkovitz S, Noske A, Zurrer-Hardi U, Bell G, et al. Slug and Sox9 cooperatively determine the mammary stem cell state. Cell 2012; 148:1015-28; PMID:22385965; http://dx.doi.org/ 10.1016/j.cell.2012.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Nassour M, Idoux-Gillet Y, Selmi A, Come C, Faraldo ML, Deugnier MA, Savagner P. Slug controls stem/progenitor cell growth dynamics during mammary gland morphogenesis. PLoS One 2012; 7:e53498; PMID:23300933; http://dx.doi.org/ 10.1371/journal.pone.0053498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Stingl J, Eirew P, Ricketson I, Shackleton M, Vaillant F, Choi D, Li HI, Eaves CJ. Purification and unique properties of mammary epithelial stem cells. Nature 2006; 439:993-7; PMID:16395311; http://dx.doi.org/ 10.1038/nature04496 [DOI] [PubMed] [Google Scholar]
- 54. Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A 2003; 100:3983-8; PMID:12629218; http://dx.doi.org/ 10.1073/pnas.0530291100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 2008; 133:704-15; PMID:18485877; http://dx.doi.org/ 10.1016/j.cell.2008.03.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bhat-Nakshatri P, Appaiah H, Ballas C, Pick-Franke P, Goulet R, Jr, Badve S, Srour EF, Nakshatri H. SLUG/SNAI2 and tumor necrosis factor generate breast cells with CD44+/CD24- phenotype. BMC Cancer 2010; 10:411,2407-10-411; PMID:20691079; http://dx.doi.org/ 10.1186/1471-2407-10-411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Morel AP, Lievre M, Thomas C, Hinkal G, Ansieau S, Puisieux A. Generation of breast cancer stem cells through epithelial-mesenchymal transition. PLoS One 2008; 3:e2888; PMID:18682804; http://dx.doi.org/ 10.1371/journal.pone.0002888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. van Amerongen R, Bowman AN, Nusse R. Developmental stage and time dictate the fate of wnt/beta-catenin-responsive stem cells in the mammary gland. Cell Stem Cell 2012; 11:387-400; PMID:22863533; http://dx.doi.org/ 10.1016/j.stem.2012.05.023 [DOI] [PubMed] [Google Scholar]
- 59. Holm K, Hegardt C, Staaf J, Vallon-Christersson J, Jonsson G, Olsson H, Borg A, Ringner M. Molecular subtypes of breast cancer are associated with characteristic DNA methylation patterns. Breast Cancer Res 2010; 12:R36; PMID:20565864; http://dx.doi.org/ 10.1186/bcr2590 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kagara N, Huynh KT, Kuo C, Okano H, Sim MS, Elashoff D, Chong K, Giuliano AE, Hoon DS. Epigenetic regulation of cancer stem cell genes in triple-negative breast cancer. Am J Pathol 2012; 181:257-67; PMID:22626806; http://dx.doi.org/ 10.1016/j.ajpath.2012.03.019 [doi] [DOI] [PubMed] [Google Scholar]
- 61. Chang CJ, Yang JY, Xia W, Chen CT, Xie X, Chao CH, Woodward WA, Hsu JM, Hortobagyi GN, Hung MC. EZH2 promotes expansion of breast tumor initiating cells through activation of RAF1-beta-catenin signaling. Cancer Cell 2011; 19:86-100; PMID:21215703; http://dx.doi.org/ 10.1016/j.ccr.2010.10.035 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Yamamoto M, Ito T, Shimizu T, Ishida T, Semba K, Watanabe S, Yamaguchi N, Inoue J. Epigenetic alteration of the NF-kappaB-inducing kinase (NIK) gene is involved in enhanced NIK expression in basal-like breast cancer. Cancer Sci 2010; 101:2391-7; PMID:20735436; http://dx.doi.org/ 10.1111/j.1349-7006.2010.01685.x [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Byler S, Goldgar S, Heerboth S, Leary M, Housman G, Moulton K, Sarkar S. Genetic and epigenetic aspects of breast cancer progression and therapy. Anticancer Res 2014; 34:1071-7; PMID:24596345; http://dx.doi.org/34/3/1071 [pii] [PubMed] [Google Scholar]
- 64. Schwitalla S, Fingerle AA, Cammareri P, Nebelsiek T, Goktuna SI, Ziegler PK, Canli O, Heijmans J, Huels DJ, Moreaux G, et al. Intestinal tumorigenesis initiated by dedifferentiation and acquisition of stem-cell-like properties. Cell 2013; 152:25-38; PMID:23273993; http://dx.doi.org/ 10.1016/j.cell.2012.12.012 [DOI] [PubMed] [Google Scholar]
- 65. Youssef KK, Lapouge G, Bouvree K, Rorive S, Brohee S, Appelstein O, Larsimont JC, Sukumaran V, Van de Sande B, Pucci D, et al. Adult interfollicular tumour-initiating cells are reprogrammed into an embryonic hair follicle progenitor-like fate during basal cell carcinoma initiation. Nat Cell Biol 2012; 14:1282-94; PMID:23178882; http://dx.doi.org/ 10.1038/ncb2628 [DOI] [PubMed] [Google Scholar]
- 66. Batlle E, Sancho E, Franci C, Dominguez D, Monfar M, Baulida J, Garcia De Herreros A. The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat Cell Biol 2000; 2:84-9; PMID:10655587; http://dx.doi.org/ 10.1038/35000034 [DOI] [PubMed] [Google Scholar]
- 67. Sarrio D, Rodriguez-Pinilla SM, Hardisson D, Cano A, Moreno-Bueno G, Palacios J. Epithelial-mesenchymal transition in breast cancer relates to the basal-like phenotype. Cancer Res 2008; 68:989-97; PMID:18281472; http://dx.doi.org/ 10.1158/0008-5472.CAN-07-2017 [doi] [DOI] [PubMed] [Google Scholar]
- 68. de Herreros AG, Peiro S, Nassour M, Savagner P. Snail family regulation and epithelial mesenchymal transitions in breast cancer progression. J Mammary Gland Biol Neoplasia 2010; 15:135-47; PMID:20455012; http://dx.doi.org/ 10.1007/s10911-010-9179-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Liu T, Zhang X, Shang M, Zhang Y, Xia B, Niu M, Liu Y, Pang D. Dysregulated expression of slug, vimentin, and E-cadherin correlates with poor clinical outcome in patients with basal-like breast cancer. J Surg Oncol 2013; 107:188-94; PMID:22886823; http://dx.doi.org/ 10.1002/jso.23240 [DOI] [PubMed] [Google Scholar]
- 70. Alves CC, Carneiro F, Hoefler H, Becker KF. Role of the epithelial-mesenchymal transition regulator slug in primary human cancers. Front Biosci (Landmark Ed) 2009; 14:3035-50; PMID:19273255 [DOI] [PubMed] [Google Scholar]
- 71. Shook D, Keller R. Mechanisms, mechanics and function of epithelial-mesenchymal transitions in early development. Mech Dev 2003; 120:1351-83; PMID:14623443 [DOI] [PubMed] [Google Scholar]
- 72. Storci G, Sansone P, Trere D, Tavolari S, Taffurelli M, Ceccarelli C, Guarnieri T, Paterini P, Pariali M, Montanaro L, et al. The basal-like breast carcinoma phenotype is regulated by SLUG gene expression. J Pathol 2008; 214:25-37; PMID:17973239; http://dx.doi.org/ 10.1002/path.2254 [DOI] [PubMed] [Google Scholar]
- 73. Hajra KM, Chen DY, Fearon ER. The SLUG zinc-finger protein represses E-cadherin in breast cancer. Cancer Res 2002; 62:1613-8; PMID:11912130 [PubMed] [Google Scholar]
- 74. Martin TA, Goyal A, Watkins G, Jiang WG. Expression of the transcription factors snail, slug, and twist and their clinical significance in human breast cancer. Ann Surg Oncol 2005; 12:488-96; PMID:15864483; http://dx.doi.org/ 10.1245/ASO.2005.04.010 [DOI] [PubMed] [Google Scholar]
- 75. Choi Y, Lee HJ, Jang MH, Gwak JM, Lee KS, Kim EJ, Kim HJ, Lee HE, Park SY. Epithelial-mesenchymal transition increases during the progression of in situ to invasive basal-like breast cancer. Hum Pathol 2013; 44:2581-9; PMID:24055090; http://dx.doi.org/ 10.1016/j.humpath.2013.07.003 [doi] [DOI] [PubMed] [Google Scholar]
- 76. Leroy P, Mostov KE. Slug is required for cell survival during partial epithelial-mesenchymal transition of HGF-induced tubulogenesis. Mol Biol Cell 2007; 18:1943-52; PMID:17344479; http://dx.doi.org/ 10.1091/mbc.E06-09-0823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Ewald AJ, Huebner RJ, Palsdottir H, Lee JK, Perez MJ, Jorgens DM, Tauscher AN, Cheung KJ, Werb Z, Auer M. Mammary collective cell migration involves transient loss of epithelial features and individual cell migration within the epithelium. J Cell Sci 2012; 125:2638-54; PMID:22344263; http://dx.doi.org/ 10.1242/jcs.096875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Sorlie T, Wang Y, Xiao C, Johnsen H, Naume B, Samaha RR, Borresen-Dale AL. Distinct molecular mechanisms underlying clinically relevant subtypes of breast cancer: Gene expression analyses across three different platforms. BMC Genomics 2006; 7:127; PMID:16729877; http://dx.doi.org/1471-2164-7-127 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Rouzier R, Perou CM, Symmans WF, Ibrahim N, Cristofanilli M, Anderson K, Hess KR, Stec J, Ayers M, Wagner P, et al. Breast cancer molecular subtypes respond differently to preoperative chemotherapy. Clin Cancer Res 2005; 11:5678-85; PMID:16115903; http://dx.doi.org/11/16/5678 [pii] [DOI] [PubMed] [Google Scholar]
- 80. Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, et al. Molecular portraits of human breast tumours. Nature 2000; 406:747-52; PMID:10963602; http://dx.doi.org/ 10.1038/35021093 [DOI] [PubMed] [Google Scholar]
- 81. Rakha EA, Ellis IO. Triple-negative/basal-like breast cancer: Review. Pathology 2009; 41:40-7; PMID:19089739; http://dx.doi.org/ 10.1080/00313020802563510 [DOI] [PubMed] [Google Scholar]
- 82. Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A 2001; 98:10869-74; PMID:11553815; http://dx.doi.org/ 10.1073/pnas.191367098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Bertucci F, Finetti P, Birnbaum D. Basal breast cancer: A complex and deadly molecular subtype. Curr Mol Med 2012; 12:96-110; PMID:22082486; http://dx.doi.org/BSP/CMM/E-Pub/00017 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Ishihara A, Tsuda H, Kitagawa K, Yoneda M, Shiraishi T. Morphological characteristics of basal-like subtype of breast carcinoma with special reference to cytopathological features. Breast Cancer 2009; 16:179-85; PMID:19466513; http://dx.doi.org/ 10.1007/s12282-009-0108-x [doi] [DOI] [PubMed] [Google Scholar]
- 85. Arnes JB, Brunet JS, Stefansson I, Begin LR, Wong N, Chappuis PO, Akslen LA, Foulkes WD. Placental cadherin and the basal epithelial phenotype of BRCA1-related breast cancer. Clin Cancer Res 2005; 11:4003-11; PMID:15930334; http://dx.doi.org/11/11/4003 [pii] [DOI] [PubMed] [Google Scholar]
- 86. Foulkes WD, Brunet JS, Stefansson IM, Straume O, Chappuis PO, Begin LR, Hamel N, Goffin JR, Wong N, Trudel M, et al. The prognostic implication of the basal-like (cyclin E high/p27 low/p53+/glomeruloid-microvascular-proliferation+) phenotype of BRCA1-related breast cancer. Cancer Res 2004; 64:830-5; PMID:14871808 [DOI] [PubMed] [Google Scholar]
- 87. Livasy CA, Karaca G, Nanda R, Tretiakova MS, Olopade OI, Moore DT, Perou CM. Phenotypic evaluation of the basal-like subtype of invasive breast carcinoma. Mod Pathol 2006; 19:264-71; PMID:16341146; http://dx.doi.org/3800528 [pii] [DOI] [PubMed] [Google Scholar]
- 88. Gusterson B. Do ‘basal-like’ breast cancers really exist? Nat Rev Cancer 2009; 9:128-34; PMID:19132008; http://dx.doi.org/ 10.1038/nrc2571; 10.1038/nrc2571 [DOI] [PubMed] [Google Scholar]
- 89. Bertucci F, Cervera N, Birnbaum D. A gene signature in breast cancer. N Engl J Med 2007; 356:1887,8; author reply 1887-8; PMID:17476019; http://dx.doi.org/356/18/1887 [pii] [DOI] [PubMed] [Google Scholar]
- 90. Charafe-Jauffret E, Ginestier C, Monville F, Finetti P, Adelaide J, Cervera N, Fekairi S, Xerri L, Jacquemier J, Birnbaum D, et al. Gene expression profiling of breast cell lines identifies potential new basal markers. Oncogene 2006; 25:2273-84; PMID:16288205; http://dx.doi.org/1209254 [pii] [DOI] [PubMed] [Google Scholar]
- 91. Liu R, Wang X, Chen GY, Dalerba P, Gurney A, Hoey T, Sherlock G, Lewicki J, Shedden K, Clarke MF. The prognostic role of a gene signature from tumorigenic breast-cancer cells. N Engl J Med 2007; 356:217-26; PMID:17229949; http://dx.doi.org/356/3/217 [pii] [DOI] [PubMed] [Google Scholar]
- 92. Zvelebil M, Oliemuller E, Gao Q, Wansbury O, Mackay A, Kendrick H, Smalley MJ, Reis-Filho JS, Howard BA. Embryonic mammary signature subsets are activated in Brca1-/- and basal-like breast cancers. Breast Cancer Res 2013; 15:R25; PMID:23506684; http://dx.doi.org/ 10.1186/bcr3403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Fillmore CM, Kuperwasser C. Human breast cancer cell lines contain stem-like cells that self-renew, give rise to phenotypically diverse progeny and survive chemotherapy. Breast Cancer Res 2008; 10:R25; PMID:18366788; http://dx.doi.org/ 10.1186/bcr1982 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Sheridan C, Kishimoto H, Fuchs RK, Mehrotra S, Bhat-Nakshatri P, Turner CH, Goulet R, Jr, Badve S, Nakshatri H. CD44+/CD24- breast cancer cells exhibit enhanced invasive properties: An early step necessary for metastasis. Breast Cancer Res 2006; 8:R59; PMID:17062128; http://dx.doi.org/bcr1610 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Park SY, Lee HE, Li H, Shipitsin M, Gelman R, Polyak K. Heterogeneity for stem cell-related markers according to tumor subtype and histologic stage in breast cancer. Clin Cancer Res 2010; 16:876-87; PMID:20103682; http://dx.doi.org/ 10.1158/1078-0432.CCR-09-1532 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Honeth G, Bendahl PO, Ringner M, Saal LH, Gruvberger-Saal SK, Lovgren K, Grabau D, Ferno M, Borg A, Hegardt C. The CD44+/CD24- phenotype is enriched in basal-like breast tumors. Breast Cancer Res 2008; 10:R53; PMID:18559090; http://dx.doi.org/ 10.1186/bcr2108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Keller PJ, Arendt LM, Kuperwasser C. Stem cell maintenance of the mammary gland: It takes two. Cell Stem Cell 2011; 9:496-7; PMID:22136921; http://dx.doi.org/ 10.1016/j.stem.2011.11.008 [DOI] [PubMed] [Google Scholar]
- 98. Keller PJ, Arendt LM, Skibinski A, Logvinenko T, Klebba I, Dong S, Smith AE, Prat A, Perou CM, Gilmore H, et al. Defining the cellular precursors to human breast cancer. Proc Natl Acad Sci U S A 2012; 109:2772-7; PMID:21940501; http://dx.doi.org/ 10.1073/pnas.1017626108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Molyneux G, Geyer FC, Magnay FA, McCarthy A, Kendrick H, Natrajan R, Mackay A, Grigoriadis A, Tutt A, Ashworth A, et al. BRCA1 basal-like breast cancers originate from luminal epithelial progenitors and not from basal stem cells. Cell Stem Cell 2010; 7:403-17; PMID:20804975; http://dx.doi.org/ 10.1016/j.stem.2010.07.010 [DOI] [PubMed] [Google Scholar]
- 100. Furuta S, Jiang X, Gu B, Cheng E, Chen PL, Lee WH. Depletion of BRCA1 impairs differentiation but enhances proliferation of mammary epithelial cells. Proc Natl Acad Sci U S A 2005; 102:9176-81; PMID:15967981; http://dx.doi.org/0503793102 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Kubista M, Rosner M, Kubista E, Bernaschek G, Hengstschlager M. Brca1 regulates in vitro differentiation of mammary epithelial cells. Oncogene 2002; 21:4747-56; PMID:12101413; http://dx.doi.org/ 10.1038/sj.onc.1205580 [doi] [DOI] [PubMed] [Google Scholar]
- 102. Liu S, Ginestier C, Charafe-Jauffret E, Foco H, Kleer CG, Merajver SD, Dontu G, Wicha MS. BRCA1 regulates human mammary stem/progenitor cell fate. Proc Natl Acad Sci U S A 2008; 105:1680-5; PMID:18230721; http://dx.doi.org/ 10.1073/pnas.0711613105 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Cloos PA, Christensen J, Agger K, Helin K. Erasing the methyl mark: Histone demethylases at the center of cellular differentiation and disease. Genes Dev 2008; 22:1115-40; PMID:18451103; http://dx.doi.org/ 10.1101/gad.1652908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Peinado H, Ballestar E, Esteller M, Cano A. Snail mediates E-cadherin repression by the recruitment of the Sin3A/histone deacetylase 1 (HDAC1)/HDAC2 complex. Mol Cell Biol 2004; 24:306-19; PMID:14673164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Hou Z, Peng H, Ayyanathan K, Yan KP, Langer EM, Longmore GD, Rauscher FJ, 3rd. The LIM protein AJUBA recruits protein arginine methyltransferase 5 to mediate SNAIL-dependent transcriptional repression. Mol Cell Biol 2008; 28:3198-207; PMID:18347060; http://dx.doi.org/ 10.1128/MCB.01435-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Dong C, Wu Y, Wang Y, Wang C, Kang T, Rychahou PG, Chi YI, Evers BM, Zhou BP. Interaction with Suv39H1 is critical for snail-mediated E-cadherin repression in breast cancer. Oncogene 2013; 32:1351-62; PMID:22562246; http://dx.doi.org/ 10.1038/onc.2012.169 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Dong C, Wu Y, Yao J, Wang Y, Yu Y, Rychahou PG, Evers BM, Zhou BP. G9a interacts with snail and is critical for snail-mediated E-cadherin repression in human breast cancer. J Clin Invest 2012; 122:1469-86; PMID:22406531; http://dx.doi.org/ 10.1172/JCI57349 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Herranz N, Pasini D, Diaz VM, Franci C, Gutierrez A, Dave N, Escriva M, Hernandez-Munoz I, Di Croce L, Helin K, et al. Polycomb complex 2 is required for E-cadherin repression by the Snail1 transcription factor. Mol Cell Biol 2008; 28:4772-81; PMID:18519590; http://dx.doi.org/ 10.1128/MCB.00323-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Lin T, Ponn A, Hu X, Law BK, Lu J. Requirement of the histone demethylase LSD1 in Snai1-mediated transcriptional repression during epithelial-mesenchymal transition. Oncogene 2010; 29:4896-904; PMID:20562920; http://dx.doi.org/ 10.1038/onc.2010.234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Shi YJ, Matson C, Lan F, Iwase S, Baba T, Shi Y. Regulation of LSD1 histone demethylase activity by its associated factors. Mol Cell 2005; 19:857-64; PMID:16140033; http://dx.doi.org/ 10.1016/j.molcel.2005.08.027 [DOI] [PubMed] [Google Scholar]
- 111. Saleque S, Kim J, Rooke HM, Orkin SH. Epigenetic regulation of hematopoietic differentiation by gfi-1 and gfi-1b is mediated by the cofactors CoREST and LSD1. Mol Cell 2007; 27:562-72; PMID:17707228; http://dx.doi.org/ 10.1016/j.molcel.2007.06.039 [DOI] [PubMed] [Google Scholar]
- 112. Lin Y, Wu Y, Li J, Dong C, Ye X, Chi YI, Evers BM, Zhou BP. The SNAG domain of Snail1 functions as a molecular hook for recruiting lysine-specific demethylase 1. EMBO J 2010; 29:1803-16; PMID:20389281; http://dx.doi.org/ 10.1038/emboj.2010.63 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Wu ZQ, Li XY, Hu CY, Ford M, Kleer CG, Weiss SJ. Canonical wnt signaling regulates slug activity and links epithelial-mesenchymal transition with epigenetic breast cancer 1, early onset (BRCA1) repression. Proc Natl Acad Sci U S A 2012; 109:16654-9; PMID:23011797; http://dx.doi.org/ 10.1073/pnas.1205822109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Lim S, Janzer A, Becker A, Zimmer A, Schule R, Buettner R, Kirfel J. Lysine-specific demethylase 1 (LSD1) is highly expressed in ER-negative breast cancers and a biomarker predicting aggressive biology. Carcinogenesis 2010; 31:512-20; PMID:20042638; http://dx.doi.org/ 10.1093/carcin/bgp324. [DOI] [PubMed] [Google Scholar]


