Abstract
A fundamental question in biology is how multicellular organisms can arise from their single-celled precursors. The evolution of multicellularity requires the adoption of new traits in unicellular ancestors that allows the generation of form by, for example, increasing the size and developing new cell types. But what are the genetic, cellular and biochemical bases underlying the evolution of multicellularity? Recent advances in evolutionary developmental biology suggest that the regulation of gene expression by cis-regulatory factors, gene duplication and alternative splicing contribute to phenotypic evolution. These mechanisms enable different degrees of phenotypic divergence and complexity with variation in traits from genomes with similar gene contents. In addition, signaling pathways specific to cell types are developed to guarantee the modulation of cellular and developmental processes matched to the cell types as well as the maintenance of multicellularity.
Keywords: alternative splicing, cellular differentiation, cis-regulatory elements, evolution, gene duplication, micrornas, multicellularity
“…one of the most important discoveries in recent developmental genetics has been the context-dependent actions of regulatory genes.” (Scott F. Gilbert, 2005)1
The confusing maze behind the molecular mechanisms underlying phenotypic divergence is one of the most interesting and, at the same time, the most complicated processes in biology. This also includes the transition from unicellular ancestors to multicellular organisms and the generation of new cell types. The main challenge to understanding the molecular mechanisms of evolution is to identify the genetic basis behind developmental processes that lead to shapes. For a long time, it was largely assumed that species-specific proteins contribute, almost single-handedly, to phenotypic divergence. Although changes in protein function can generate new phenotypes, there is growing evidence indicating that many homologous proteins show highly conserved functions among closely related species. Recent advances in evolutionary developmental biology show that the regulation of gene activity is the main source of biodiversity.2 The first evidence of the importance of gene regulatory mechanisms in evolutionary changes came from studies during the 1960s and 1970s that proposed that affecting gene expression by mutations and other regulatory mechanisms probably led to the evolution of organismal diversity.3,4 Knowledge gained in recent decades confirms this assessment and shows that gene regulatory factors play a central role in the evolution of biodiversity and the generation of form (reviewed in5). Collaboration between various sophisticated gene regulatory mechanisms enables evolutionary novelties by fine-tuning gene expression and depends on location, timing and cell type, notably with respect to the developmental history of the cell. In this way, various phenotypes can form from genomes with similar gene contents. Cis-regulatory elements,6 gene duplication,7 alternative splicing8 and potentially micoRNAs9,10 are the key actors in evolutionary developmental biology. Armed with these insights, I will give an overview of the potential impact of these regulatory mechanisms on the evolution of multicellular organisms from unicellular ancestors.
The members of the volvocine algae, a group of chlorophytes including unicellular Chlamydomonas reinhardtii (hereafter Chlamydomonas) and multicellular Volvox carteri (hereafter Volvox), represent a recent case of transition from unicellular to multicellular life. Based on molecular-phylogenetic studies, Volvox and Chlamydomonas probably diverged ∼200 million years ago from a common unicellular ancestor.11 The evolution of multicellular life from Chlamydomonas to Volvox required several developmental traits, including asymmetric cell division and embryonic morphogenesis. However, the most exiting developmental trait was the evolution of germ-soma differentiation.12 Unlike Chlamydomonas, Volvox has 2 cell types, i.e. 2000-4000 biflagellate, motile, terminally differentiated somatic cells and around 16 much larger immotile reproductive cells, with a clear division of labor (Figure 1). The cells are embedded in a transparent sphere of a glycoprotein-rich extracellular matrix (ECM).13 At the molecular level, however, both organisms possess similar gene contents. The nuclear genome of Chlamydomonas contains 118 Mbp, and that of its multicellular relative Volvox contains 138 Mbp. The larger genome of Volvox (∼17%) is attributed to its higher content of transposons and repetitive DNA, because both species have almost identical protein-coding potentials, i.e., 14 516 and 14 520 protein-coding genes in Chlamydomonas and Volvox, respectively.14,15 Only a few gene families, i.e. the pherophorin genes, the VMP genes (Volvox matrix metalloproteases) and the cyclin-D-related genes, have more members in Volvox than in Chlamydomonas.14 The same situation can be observed in the human genome, which contains almost as many genes as that of Caenorhabditis elegans.16 This fact strongly supports the theory of evolutionary developmental biology and suggests that the transition from a unicellular Chlamydomonas-like ancestor to multicellular Volvox did not require major changes in gene content.14,17 Based on this observation together with the fact that Volvox cell types represent differential patterns of gene expression in various functional classes,18-20 the development of species-specific proteins could not account for the development of Volvox from a Chlamydomonas-like ancestor. This is also supported by experimental evidence that showed that 2 important proteins, GlsA and InvA, which are responsible for essential developmental processes behind the evolution of multicellularity in Volvox, namely asymmetric division and embryo inversion, respectively, are conserved in unicellular Chlamydomonas. Interestingly, Chlamydomonas orthologs can rescue Volvox glsA and invA mutants.21-23 Thus, rather than species-specific proteins, the functional divergence of gene regulatory elements could be the main contributor to the development of multicellularity during evolution.
Figure 1.
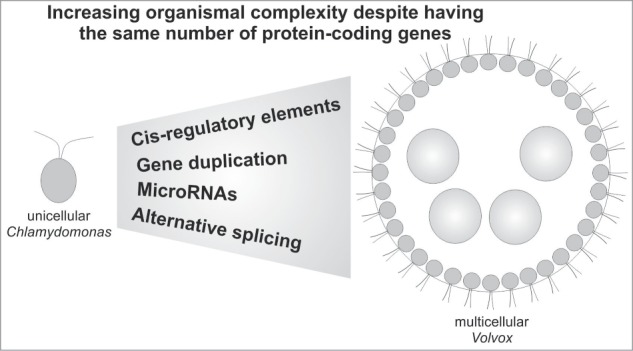
Gene regulatory mechanisms behind the evolution of multicellularity. Model illustrating the role of gene regulatory mechanisms in the evolution of multicellular Volvox from a Chlamydomonas-like ancestor.
We and others have recently shown that alternative splicing could contribute to the appearance of multicellularity by generating multiple transcripts from a single gene.24,25 In many cases, alternative splicing seems to be a part of the molecular mechanisms that allow organisms to decrease the expression of specific genes by generating non-functional or modified variants toward attenuation or alteration of specific cellular and physiological processes. Our analyses show that at least ∼2.9% of the intron-containing genes in Volvox are alternatively spliced. Considering the number of analyzed ESTs, it is very likely that the Volvox genome possesses more favorable conditions, e.g. changes in the length and GC content of introns, for the occurrence of alternative splicing than those of the closely related Chlamydomonas.24,26 On the other hand, an analysis of the alternative-splicing status of homologous genes from the closely related alga Chlamydomonas could show that a large fraction of the genes that are alternatively spliced in Volvox are not alternatively spliced in Chlamydomonas. Concurrently with our study, Urrutia and colleagues examined how alternative splicing was related to organismal complexity by analyzing alternative splicing in 47 eukaryotic species. They found that alternative splicing has steadily increased over eukaryotic evolution and is strongly associated with organismal complexity and cell-type number.25 Therefore, it might be conceivable that alternative splicing acts as a key regulatory factor to facilitate the evolution of multicellularity in volvocine algae. However, more effort should be made to provide more insight into the evolutionary aspects of alternative splicing behind the development of multicellularity, for example by a genome-wide comparative analysis of alternative-splicing events between Chlamydomonas and Volvox (to investigate species-specific alternative-splicing events) as well as by investigating the cell-type-specific regulation of events in multicellular Volvox. In this respect, it is also worth noting that the impact of environmental factors, e.g., light cues, which have a large impact on the growth and development of photosynthetic organisms, on the regulation of alternative splicing should be taken into account. Light-regulated gene expression, mediated by photoreceptors, acts as a multifaceted regulator to control the abundance of functional genes at different levels (reviewed in27). Surprisingly, Volvox photoreceptors are mostly expressed in a cell-type-specific manner,28 enabling the alga to use distinct light-signaling pathways to modulate the expression of genes involved in various cellular and metabolic pathways in a cell-type-specific manner.29 This reflects an early development of cell-type-specific signaling mechanisms during evolution to ensure the development as well as the maintenance of cellular differentiation.30
Another important regulatory mechanism that should be moved increasingly into focus is the role of cis-regulatory elements. Cis-regulatory elements (such as promoters and enhancers) are transcription-factor binding sites and other non-coding DNA that are normally located upstream, downstream or in the introns of genes. These regulatory elements regulate gene expression in a cell-type-, tissue- or developmental-stage-specific fashion.2 Considering the fact that Volvox has almost as many genes as Chlamydomonas, the species-specific regulation of gene expression could be the main source of diversity across volvocine algae. To go further, the first step will be to identify the cis-regulatory elements in those genes (as has been partially done for regA gene)31 that are of particular importance for the evolution of multicellularity. It has been shown that around 50% of the genes from closely related species show differences in cis-regulatory elements.32,33 However, again genome-wide comparative analyses (e.g. prediction of cis-regulatory elements)34,35 and supporting experiments would be of great benefit for identifying the developmentally important elements– as well as their candidate sites–and for studying cis-regulatory divergence and activity during the transition from unicellular to multicellular life.
Gene duplication and microRNAs (small noncoding RNAs that regulate gene expression post-transcriptionally) could also be considered as additional sources of novel evolutionary diversity. In particular, gene duplication could contribute to the creation of species-specific transcription factors and other essential proteins for the evolution of gene regulatory networks.36 An example of species-specific transcription factors is RegA, a Volvox-specific transcription factor involved in cellular differentiation. Interestingly, regA gene seems to be absent in the closely related Chlamydomonas. Phyogenetic analysis suggests that regA gene was present in a common unicellular ancestor of Volvox and Chlamydomonas, but was later lost in Chlamydomonas.37 In Volvox, reproductive activities (and subsequently growth) are suppressed in somatic cells by the transcription factor RegA, which is expressed at very high level in these cells.18,38 Conversely, the dark green reproductive cells, which show a low regA transcript level, possess more photosynthetic activities.39
It is known that an inverse correlation exists between the size of a gene's family and its use of alternatively spliced isoforms in humans, mice and worms.40-44 However, a recent study by Cooper and colleagues demonstrated that the reduction in alternative splicing was independent of the size of the gene family in zebrafish.45 In Volvox, conversely, it even seems that the more gene duplicates there are, the more alternative splicing is observed (Kianianmomeni et al., unpublished data). Thus, coordination between gene duplication and alternative splicing provides resources for functional innovation to expand protein diversity during the evolution of multicellularity. Moreover, microRNAs, which play an important role in diverse developmental processes,9,10,46 could contribute to evolvability during the transition to multicellularity. Many microRNAs have been identified in both Chlamydomonas and Volvox.47-49 Intriguingly, many of the Volvox microRNAs are expressed in a cell-type-specific manner. On the other hand, only one miRNA was found to be conserved between Chlamydomonas and Volvox,49 suggesting a high rate of microRNA de novo emergence in volvocine algae that could contribute to the creation of species-specific gene repertoires.
In conclusion, a comparison of the genomes of 2 closely related organisms, unicellular Chlamydomonas and multicellular Volvox, highlighted that the transition from unicellular to multicellular life does not require large changes in gene content.17 The evolution of gene regulatory mechanisms, rather than the development of species-specific proteins, seems to play a central role in diverse developmental processes during the unicellular-multicellular transition.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
I gratefully acknowledge the support by the Deutsche Forschungsgemeinschaft (DFG grant KI 1779/1-1).
References
- 1. Gilbert SF. Putting evo-devo into focus. An interview with Scott F. Gilbert. Interview by Alexander T. Mikhailov. Int J Dev Biol 2005; 49:9-16; PMID:15744662; http://dx.doi.org/ 10.1387/ijdb.041972am [DOI] [PubMed] [Google Scholar]
- 2. Wittkopp PJ, Kalay G. Cis-regulatory elements: molecular mechanisms and evolutionary processes underlying divergence. Nat Rev Genet 2012; 13:59-69; http://dx.doi.org/ 10.1038/nri3362 [DOI] [PubMed] [Google Scholar]
- 3. Britten RJ, Davidson EH. Gene regulation for higher cells: a theory. Science 1969; 165:349-57; PMID:5789433; http://dx.doi.org/ 10.1126/science.165.3891.349 [DOI] [PubMed] [Google Scholar]
- 4. King MC, Wilson AC. Evolution at two levels in humans and chimpanzees. Science 1975; 188:107-16; PMID:1090005; http://dx.doi.org/ 10.1126/science.1090005 [DOI] [PubMed] [Google Scholar]
- 5. Carroll SB. Evolution at two levels: on genes and form. PLoS Biol 2005; 3:1159-66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Prud'homme B, Gompel N, Carroll SB. Emerging principles of regulatory evolution. Proc Natl Acad Sci U S A 2007; 104 Suppl 1:8605-12; PMID:17494759; http://dx.doi.org/ 10.1073/pnas.0700488104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Amoutzias GD, Robertson DL, Oliver SG, Bornberg-Bauer E. Convergent evolution of gene networks by single-gene duplications in higher eukaryotes. EMBO Rep 2004; 5:274-9; PMID:14968135; http://dx.doi.org/ 10.1038/sj.embor.7400096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Merkin J, Russell C, Chen P, Burge CB. Evolutionary dynamics of gene and isoform regulation in Mammalian tissues. Science 2012; 338:1593-9; PMID:23258891; http://dx.doi.org/ 10.1126/science.1228186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jasinski S, Vialette-Guiraud AC, Scutt CP. The evolutionary-developmental analysis of plant microRNAs. Philos Trans R Soc Lond B Biol Sci 2010; 365:469-76; PMID:20047873; http://dx.doi.org/ 10.1098/rstb.2009.0246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kosik KS. MicroRNAs tell an evo-devo story. Nat Rev Neurosci 2009; 10:754-9; PMID:19738624; http://dx.doi.org/ 10.1038/nrn2713 [DOI] [PubMed] [Google Scholar]
- 11.Herron MD, Hackett JD, Aylward FO, Michod RE. Triassic origin and early radiation of multicellular volvocine algae. Proc Natl Acad Sci U S A 2009; 106:3254-8; PMID:19223580; http://dx.doi.org/ 10.1073/pnas.0811205106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kirk DL. A twelve-step program for evolving multicellularity and a division of labor. BioEssays 2005; 27:299-310; PMID:15714559; http://dx.doi.org/ 10.1002/bies.20197 [DOI] [PubMed] [Google Scholar]
- 13. Kirk D. Volvox: Molecular-genetic Origins of Multicellularity and Cellular Differentiation. Taylor & Francis, Cambridge, UK: 1998 [Google Scholar]
- 14. Prochnik SE, Umen J, Nedelcu AM, Hallmann A, Miller SM, Nishii I, Ferris P, Kuo A, Mitros T, Fritz-Laylin LK, et al. Genomic analysis of organismal complexity in the multicellular green alga Volvox carteri. Science 2010; 329:223-6; PMID:20616280; http://dx.doi.org/ 10.1126/science.1188800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Merchant SS, Prochnik SE, Vallon O, Harris EH, Karpowicz SJ, Witman GB, Terry A, Salamov A, Fritz-Laylin LK, Maréchal-Drouard L, et al. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 2007; 318:245-50; PMID:17932292; http://dx.doi.org/ 10.1126/science.1143609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature 2001; 409:860-921; PMID:11237011; http://dx.doi.org/ 10.1038/35057062 [DOI] [PubMed] [Google Scholar]
- 17. Pennisi E. Genetics. Volvox genome shows it doesn't take much to be multicellular. Science 2010; 329:128-9; PMID:20616240; http://dx.doi.org/ 10.1126/science.329.5988.128-a [DOI] [PubMed] [Google Scholar]
- 18. Nematollahi G, Kianianmomeni A, Hallmann A. Quantitative analysis of cell-type specific gene expression in the green alga Volvox carteri. BMC Genomics 2006; 7:321; PMID:17184518; http://dx.doi.org/ 10.1186/1471-2164-7-321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tam LW, Kirk DL. Identification of cell-type-specific genes of Volvox carteri and characterization of their expression during the asexual life cycle. Devl Biol 1991; 145:51-66; http://dx.doi.org/ 10.1016/0012-1606(91)90212-L [DOI] [PubMed] [Google Scholar]
- 20. Meissner M, Stark K, Cresnar B, Kirk DL, Schmitt R. Volvox germline-specific genes that are putative targets of RegA repression encode chloroplast proteins. Curr Genet 1999; 36:363-70; PMID:10654090; http://dx.doi.org/ 10.1007/s002940050511 [DOI] [PubMed] [Google Scholar]
- 21. Cheng Q, Fowler R, Tam LW, Edwards L, Miller SM. The role of GlsA in the evolution of asymmetric cell division in the green alga Volvox carteri. Dev Genes Evol 2003; 213:328-35; PMID:12743823; http://dx.doi.org/ 10.1007/s00427-003-0332-x [DOI] [PubMed] [Google Scholar]
- 22. Nishii I, Ogihara S, Kirk DL. A kinesin, invA, plays an essential role in Volvox morphogenesis. Cell 2003; 113:743-53; PMID:12809605; http://dx.doi.org/ 10.1016/S0092-8674(03)00431-8 [DOI] [PubMed] [Google Scholar]
- 23. Miller SM. Volvox, Chlamydomonas, and the evolution of multicellularity. Nat Education 2010; 3:65 [Google Scholar]
- 24. Kianianmomeni A, Soon Ong C, Ratsch G, Hallmann A. Genome-wide analysis of alternative splicing in Volvox carteri. BMC Genomics 2014; 15:1117; PMID:25516378; http://dx.doi.org/ 10.1186/1471-2164-15-764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chen L, Bush SJ, Tovar-Corona JM, Castillo-Morales A, Urrutia AO. Correcting for differential transcript coverage reveals a strong relationship between alternative splicing and organism complexity. Mol Biol Evol 2014; 31:1402-13; PMID:24682283; http://dx.doi.org/ 10.1093/molbev/msu083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Labadorf A, Link A, Rogers MF, Thomas J, Reddy AS, Ben-Hur A. Genome-wide analysis of alternative splicing in Chlamydomonas reinhardtii. BMC Genomics 2010; 11:114; PMID:20163725; http://dx.doi.org/ 10.1186/1471-2164-11-114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kianianmomeni A. More light behind gene expression. Trends Plant Sci 2014; 19:488-90; PMID:24928178; http://dx.doi.org/ 10.1016/j.tplants.2014.05.004 [DOI] [PubMed] [Google Scholar]
- 28. Kianianmomeni A, Hallmann A. Transcriptional analysis of Volvox photoreceptors suggests the existence of different cell-type specific light-signaling pathways. Current Genet 2015, 61:3-18; http://dx.doi.org/ 10.1007/s00294-014-0440-3 [DOI] [PubMed] [Google Scholar]
- 29. Kianianmomeni A. Cell-type specific light-mediated transcript regulation in the multicellular alga Volvox carteri. BMC Genomics 2014; 15:764; PMID:25194509; http://dx.doi.org/ 10.1186/1471-2164-15-764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kianianmomeni A. Cell-type specific photoreceptors and light signaling pathways in the multicellular green alga Volvox carteri and their potential role in cellular differentiation. Plant Signal Behav 2015; In Press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Stark K, Kirk DL, Schmitt R. Two enhancers and one silencer located in the introns of regA control somatic cell differentiation in Volvox carteri. Genes Dev 2001; 15:1449-60; PMID:11390364; http://dx.doi.org/ 10.1101/gad.195101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tirosh I, Reikhav S, Levy AA, Barkai N. A yeast hybrid provides insight into the evolution of gene expression regulation. Science 2009; 324:659-62; PMID:19407207; http://dx.doi.org/ 10.1126/science.1169766 [DOI] [PubMed] [Google Scholar]
- 33. McManus CJ, Coolon JD, Duff MO, Eipper-Mains J, Graveley BR, Wittkopp PJ. Regulatory divergence in Drosophila revealed by mRNA-seq. Genome Res 2010; 20:816-25; PMID:20354124; http://dx.doi.org/ 10.1101/gr.102491.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Elati M, Nicolle R, Junier I, Fernandez D, Fekih R, Font J, Képès F. PreCisIon: PREdiction of CIS-regulatory elements improved by gene's positION. Nucleic Acids Res 2013; 41:1406-15; PMID:23241390; http://dx.doi.org/ 10.1093/nar/gks1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. He X, Ling X, Sinha S. Alignment and prediction of cis-regulatory modules based on a probabilistic model of evolution. PLoS Comput Biol 2009; 5:e1000299; PMID:19293946; http://dx.doi.org/ 10.1371/journal.pcbi.1000299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nowick K, Stubbs L. Lineage-specific transcription factors and the evolution of gene regulatory networks. Brief Funct Genomics 2010; 9:65-78; PMID:20081217; http://dx.doi.org/ 10.1093/bfgp/elp056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Duncan L, Nishii I, Harryman A, Buckley S, Howard A, Friedman NR, Miller SM. The VARL gene family and the evolutionary origins of the master cell-type regulatory gene, regA, in Volvox carteri. J Mol Evol 2007; 65:1-11; PMID:17646893; http://dx.doi.org/ 10.1007/s00239-006-0225-5 [DOI] [PubMed] [Google Scholar]
- 38. Kirk MM, Stark K, Miller SM, Muller W, Taillon BE, Gruber H, Schmitt R, Kirk DL. regA, a Volvox gene that plays a central role in germ-soma differentiation, encodes a novel regulatory protein. Development 1999; 126:639-47; PMID:9895312 [DOI] [PubMed] [Google Scholar]
- 39. Choi G, Przybylska M, Straus D. Three abundant germ line-specific transcripts in Volvox carteri encode photosynthetic proteins. Curr Genet 1996; 30:347-55; PMID:8781179; http://dx.doi.org/ 10.1007/s002940050143 [DOI] [PubMed] [Google Scholar]
- 40. Kopelman NM, Lancet D, Yanai I. Alternative splicing and gene duplication are inversely correlated evolutionary mechanisms. Nat Genet 2005; 37:588-9; PMID:15895079; http://dx.doi.org/ 10.1038/ng1575 [DOI] [PubMed] [Google Scholar]
- 41. Su Z, Wang J, Yu J, Huang X, Gu X. Evolution of alternative splicing after gene duplication. Genome Res 2006; 16:182-9; PMID:16365379; http://dx.doi.org/ 10.1101/gr.4197006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Talavera D, Vogel C, Orozco M, Teichmann SA, de la Cruz X. The (in)dependence of alternative splicing and gene duplication. PLoS Comput Biol 2007; 3:e33; PMID:17335345; http://dx.doi.org/ 10.1371/journal.pcbi.0030033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hughes AL, Friedman R. Alternative splicing, gene duplication and connectivity in the genetic interaction network of the nematode worm Caenorhabditis elegans. Genetica 2008; 134:181-6; PMID:18026854; http://dx.doi.org/ 10.1007/s10709-007-9223-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chen L, Tovar-Corona JM, Urrutia AO. Alternative splicing: a potential source of functional innovation in the eukaryotic genome. Int J Evol Biol 2012; 2012:596274; PMID:22811948; http://dx.doi.org/ 10.1155/2012/596274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lambert MJ, Olsen KG, Cooper CD. Gene duplication followed by exon structure divergence substitutes for alternative splicing in zebrafish. Gene 2014; 546:271-6; PMID:24942242; http://dx.doi.org/ 10.1016/j.gene.2014.05.068 [DOI] [PubMed] [Google Scholar]
- 46. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004; 116:281-97; PMID:14744438; http://dx.doi.org/ 10.1016/S0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- 47. Zhao T, Li G, Mi S, Li S, Hannon GJ, Wang XJ, Qi Y. A complex system of small RNAs in the unicellular green alga Chlamydomonas reinhardtii. Genes Dev 2007; 21:1190-203; PMID:17470535; http://dx.doi.org/ 10.1101/gad.1543507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Molnar A, Schwach F, Studholme DJ, Thuenemann EC, Baulcombe DC. miRNAs control gene expression in the single-cell alga Chlamydomonas reinhardtii. Nature 2007; 447:1126-9; PMID:17538623; http://dx.doi.org/ 10.1038/nature05903 [DOI] [PubMed] [Google Scholar]
- 49. Li J, Wu Y, Qi Y. MicroRNAs in a multicellular green alga Volvox carteri. Sci China Life Sci 2014; 57:36-45; PMID:24369344; http://dx.doi.org/ 10.1007/s11427-013-4580-3 [DOI] [PubMed] [Google Scholar]


