Abstract
During the early stages of development, the embryo depends on the placenta as provider of oxygen and calcium, among other essential compounds. Although fetal liver accomplishes a well-known haematopoietic function, its contribution to calcium homeostasis upon development is poorly understood. The homeostasis of cell calcium contributes to diverse signaling pathways across developmental stages of most tissues and the calcium-ATPase located at the plasma membrane (PMCA) helps pumping excess calcium into the extracellular space. To date, the understanding of the equilibrium shift between PMCA isoforms during liver development is still missing. This review focuses on the characterization of the hepatic PMCA along the early stages of development, followed by a description of modern approaches to study calcium homeostasis involving several types of pluripotent cells. The application of interdisciplinary techniques to improve our understanding of liver development and the role calcium homeostasis plays in the definition of pathogenesis is also discussed.
Keywords: Ca2+ homeostasis, embryonic stem cells, fetal liver, induced pluripotent stem cells, plasma membrane calcium ATPase
Introduction
Our current knowledge of the mechanisms that regulate calcium homeostasis of fetal liver during the embryonic development remains incomplete. This review establishes the most important differences between fetal and adult liver physiology in order to highlight how calcium contributes to the specific physiological demands of each developmental stage.
Approximately 80% of liver tissue contains parenchymal cells (i.e., hepatocytes), while the remaining 20% includes nonparenchymal cells such as cholangiocytes, sinusoidal endothelial cells, Kupffer cells, lymphocytes, biliary cells, and hepatic stellate cells (or Ito cells). A fetal liver also contains stromal cells including macrophage, endothelial cells, epithelial cells, and fibroblasts that produce cytokines and synthesize extracellular matrix proteins.1 Although nonparenchymal cells represent a small proportion of the total hepatic tissue, they are key contributors for proper hepatocyte function that is mediated by paracrine mechanisms under normal and pathological conditions. Furthermore, during the fetal stages, nonparenchymal cells are needed to support the differentiation of hepatoblasts into hepatocytes.2,3 In vitro, nonparenchymal cells also importantly contribute to obtain hepatocyte-like cells from embryonic stem cells (ESCs).4
In contrast to the active proliferation observed in fetal hepatoblasts and hepatic progenitor cells during liver organogenesis, the adult liver has been considered as a quiescent tissue since it has been reported that one mitosis is detected for every 10,000 to 20,000 cells.5 However, following exposure to certain chemicals or physical injury, mature hepatocytes reenter the cell cycle in a process known as liver regeneration (or compensatory hyperplasia) showing an extraordinary replicative capacity. In mice, this process involves only hypertrophy, or hypertrophy and hyperplasia after 30% and 70% partial hepatectomy, respectively.6 Also, the nuclei number does not change and ploidy slightly increases in regenerated liver after 30% hepatectomy; nevertheless, nuclei number decreases and ploidy increases after 70% hepatectomy. Interestingly, in fetal and adult stages, even while regenerating, the liver performs complex functions involving the intermediary metabolism of amino acids, lipids, and carbohydrates, as well as the detoxification of xenobiotics and bile secretion. Moreover, functions such as vesicular trafficking, cell growth regulation, glycogen metabolism, and importantly, the synthesis of bile salts among others are related to the homeostasis of calcium in the liver. The canalicular membranes show a specialized composition to perform the secretion of bile including a 2-fold higher content of sphingomyelin and cholesterol than those present in basolateral and sinusoidal domains.7
Although the liver is not an excitable tissue, when several receptors are activated in the plasma membrane of hepatocytes, phosphoinositide signaling cascades are triggered and Ca2+ oscillations originating in the endoplasmic reticulum constitute part of the calcium signaling process.8 In the canalicular zone of hepatocytes there is also a "trigger zone" that is rich in type II receptors (InsP3R2) and shown to be important for the initiation of Ca2+ waves.9
The fetal liver provides temporary functions related to hematopoiesis with the release of factors, primarily cytokines from haematopoietic cells. These factors are needed for the proper growth and differentiation of the liver.10 In mice, the hematopoiesis process is initially observed around 12.5 d post-coitum (dpc). Between 11 dpc and 16 dpc, active erythropoiesis is observed, with the presence of precursors for myeloid and B-cells from 13 dpc up to birth. Lymphoid precursors of p-T type cells also increase 12 dpc, while precursors of p-B type cells are scarce and increase up to 18–19 d of gestation. In mice, at the start of hematopoiesis, the association between macrophages and erythroblasts during erythropoiesis has been clearly observed,1 with macrophages at the center of erythroblastic islands contributing to maturation and enucleation of surrounding erythroblasts.1
While in the developing fetus calcium is transported through the placenta against a blood calcium gradient mediated by parathyroid hormone (PTH) and PTH-related protein (PTHrP), in adult tissue, PTH is the main regulator of blood calcium. In studies of mice deficient for Hoxa3, a transcription factor involved in parathyroid organogenesis, an absence of parathyroid glands and therefore PTH, were observed.11 As a result, low calcium levels have been detected in both blood and amniotic fluid samples, while serum levels of PTHrP and the placental transport of calcium remained unaffected. In combination with studies of Pthrp-/- mutants, where serum PTHrP levels were found to increase concomitant with its overexpression in the liver and placenta, these findings suggest that these organs play active roles in fetal calcium homeostasis mediated by PTHrP instead of PTH.11
On the other hand, several Ca2+ mobilizing systems are located in the plasma membrane and cytosolic organelles of hepatic cells, also working to maintain very low levels of calcium. In the plasma membrane of hepatocytes, the plasma membrane calcium ATPase (PMCA) is primarily responsible for maintaining low levels of calcium, since under normal conditions, the Na+/Ca2+ exchanger appears to be inactive.12,13 This review focuses on the contributions of PMCA to the overall system that controls calcium homeostasis during the embryonic development of the liver.
Plasma Membrane Calcium ATPase (PMCA)
In eukaryotic cells, PMCA is part of a specialized battery of proteins that maintain cytosolic calcium levels by extruding calcium against a concentration gradient. These proteins belong to a family of P-type ATPases, which undergo phosphorylation to form high and low affinity conformations (E1-E2) for Ca2+ in order to mediate a calcium transport cycle. The function of several calcium ATPases has been elucidated from the first studies with erythrocytes conducted over 40 y ago to characterize their canonical features.14 In particular, it has been identified that PMCAs are generally regulated by calmodulin (CaM), require as cofactors, Ca2+ and Mg2+ and exhibit inhibitory sensitivity to vanadate and lanthanum.
Although PMCAs exhibit ubiquitous distribution, tissue-specific combinations of PMCA isoforms have been observed. For example, in excitable cells where signal transduction is strictly regulated, a wide variety of PMCA variants have been described according to their kinetic properties. For our laboratory, the role of PMCA in liver function under normal and pathological conditions, as well as during the normal process of liver regeneration, has been of particular interest,15-17 The PMCA present in the liver tissue seems not to be modulated by CaM since the purification of the enzyme employing CaM-coupled columns has been unsuccessful.18 Therefore, it is hypothesized that alternative mechanisms of regulation are involved, since liver Ca2+-ATPases can be modulated by several hormones, such as PTH via G protein mediated signaling19 or by lipids such as cholesterol as recently studied by us (unpublished results).
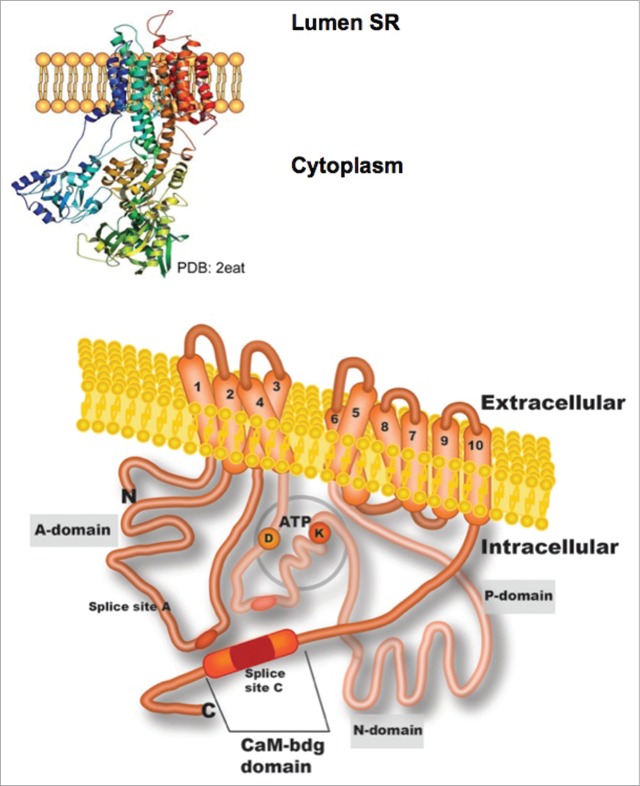
To date, the PMCA crystal structure has not been reported, therefore the accepted topological model is based on the high resolution crystal structure from other member of the P-ATPases family, the sarco/endoplasmic reticulum Ca2+-ATPase (SERCAa1) solved by Toyoshima et al.20 The crystal structure of SERCAa1 expressed in bovine fast-twitch skeletal muscle has also been solved showing slight differences in comparison to that from rabbit.21 At this stage, it is important to note that the topological model of PMCA accepted for many years based only on predicted DNA sequences of SERCA, is still accurate.22 The model includes a single polypeptide (125–140 kDa) corresponding to 10 transmembrane (TM) domains with the N- and C- ends located on the cytosolic side (Fig. 1). In addition, 2 main intracellular loops have been described composed of the A domain (located between TM domains 2 and 3), a nucleotide-binding/catalytic phosphorylation (N/P) domain (between TM domains 4 and 5), and a CaM-binding site in the C-terminus segment that extended from TM domain 10.23
Figure 1.
Illustrated topology of a plasma membrane Ca2+-ATPase (PMCA). The three-dimensional representation of reticulum sarcoplasmic Ca2+-ATPase (SERCA; PDB:2eat) that is shown at the top of the figure was used to construct the PMCA model below. The structure predicted for PMCA includes most of the domains being oriented toward the cytoplasmic face and 10 transmembrane domains (TM). Domain A, which is between TM domains 2 and 3, contains splice site A and a site for phospholipid binding. The intracellular loop located between TM domains 4 and 5 comprises the P- and N-domains where phosphorylation of an aspartate residue (D) and ATP-binding of a lysine residue (K) occur. The C-terminus also contains a calmodulin-binding site in splice site C.
There are 4 genes (Atp2b1-Atp2b4) that encode the isoforms PMCA1–4. These genes undergo complex alternative splicing at sites A and C to produce various combinations of PMCAs that have specific kinetic properties and tissue-specific expression profiles (Fig. 2). Site A is located in the first cytosolic loop close to a phospholipid-binding domain, while Site C is located close to the C-terminus and comprises a CaM-binding domain and internal splicing sites that serve to increase the number of variants that are produced.24-26 The C-end domain also modulates enzyme activity based on an autoinhibitory sequence which mediates interactions with both intracellular loops to potentially decrease PMCA activity at low intracellular calcium concentrations.
Figure 2.
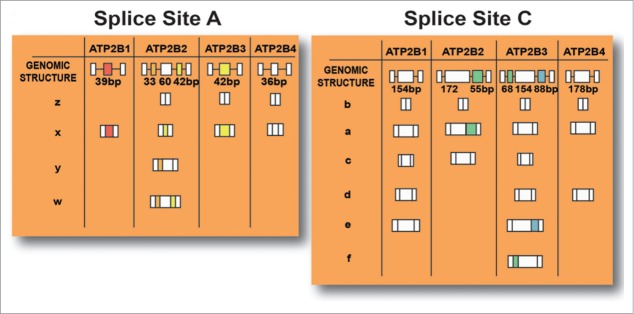
Possible isoforms of PMCA which are produced based on alternative splicing sites that are present at site A (resulting in w-z variants) and site C (resulting in a-f variants) of the 4 genes that encode PMCAs.
In general, it is well-accepted that PMCA1 and PMCA4 have a lower affinity for Ca2+ with respect to PMCA2 and PMCA3, and represent housekeeping isoforms that are ubiquitously expressed. In contrast, isoforms PMCA2 and PMCA3 are expressed in excitable tissues,16,27 that during embryonic development exhibit spatiotemporal expression profiles. Although PMCA has also been shown to play a role in the homeostasis of calcium in the placenta,28 the role of this enzyme in fetal liver remains to be investigated.
PMCA Expression in the Early Stages of Liver Development
In order to study hepatic cells during fetal and adult stages of development, several methodological issues related to the availability of biological material and proper tracing techniques need to be addressed. Correspondingly, there have been diverse efforts to study PMCA expression in different species during the fetal stages of liver differentiation (Table 1). Zacharias and Kappen have reported the ATPases identified throughout the stages of embryonic development in the mouse from 9.5 dpc to 18.5 dpc.29 In situ hybridization images have shown that PMCA1 is expressed as a constitutive variant during all stages studied. In contrast, PMCA4 is abundantly expressed in fetal liver at 12.5 dpc (Table 1). The latter result is interesting since it corresponds with the initiation of hematopoiesis and the observation that PMCA4 is the predominant isoform in erythrocytes.30 Zacharias and Kappen have also demonstrated that PMCA1–4 transcripts are expressed in ESCs that can potentially differentiate into hepatocytes.29 In this study, PMCA1 and PMCA3 were found to be expressed at high levels, while PMCA2 and PMCA4 were expressed at low levels.29 However, in another study only PMCA1 and PMCA4 expression were detected in mouse ESCs.31 These differences may be due to the methodological approaches used in each study.
Table 1.
Comparative expression of PMCA transcripts in fetal and adult liver
| Species | Age | PMCA1 | PMCA2 | PMCA3 | PMCA4 | Reference |
|---|---|---|---|---|---|---|
| Human | Fetal (20–22 w) | 1b | 2 | ND | 4a | 32 |
| Human | Fetal | U | 2x | U | U | 33 |
| Human | Fetal | U | 2w | U | U | 35 |
| Human | Adult | 1b | 2w | ND | 4b | 35 |
| Human | Adult 75 y | 1x (70%) | 2x, 2w (≤1%) | ND | 4x (28%) | 25 |
| 1b (70%) | 2b (≤1%) | 4b (28%) | ||||
| Rat | Adult | 1b | 2w | ND | ND | 35 |
| Rat | 13 d | 1x > 1b | 2w | U | 4b > 4x | 15 |
| Rat | 17 d | 1x > 1b | 2w | U | 4b > 4x > 4z | 15 |
| Rat | Neonatal | 1x > 1b | 2w | U | 4b > 4x > 4z | 15 |
| Rat | Adult | 1x > 1b | 2w | U | 4b > 4a, 4d, 4x | 15 |
| Rat | Adult | H 1x, 1c | H 2A* | H 3A* | H 4a, 4d | 17 |
| KC 1x, 1c | KC 2A, 2a | KC 3A, 3e | KC 4a, 4d | |||
| HSC 1x, 1c | HSC 2A, 2a | HSC 3A, 3e | HSC 4a, 4d | |||
| Mouse | Fetal 12.5 d | Present | ND | ND | Maximal | 29 |
| Mouse | Fetal 18.5 d | Maximal | ND | ND | Minimal | 29 |
Isoforms 2A and 3A correspond to variants detected with primers designed upstream of splicing site A. Isoform 3e: HSC>
KC>H. ND; not detected; U; undetermined; HSC: Hepatic stellate cells, KC: Kupffer cells; H: Hepatocytes.
On the other hand, employing a cDNA library of human fetal liver at an unspecified gestational age, expression of PMCA1b, PMCA4a, and PMCA2 was detected.32 However, Heim et al. only detected expression of isoform 2x when using a cDNA library constructed from human fetal liver.33 Furthermore, in a quantitative report of PMCA expression in human adult liver, splicing of PMCA1 at site A (1x) or at site C (1b) was found to produce 70% of the total PMCA expression detected for each splicing site. This was followed by PMCA4 isoforms (4x and 4b) representing 28% of total PMCA expression.25 In addition, the expression of isoform 2w was detected in fetal human livers between 7.6 and 19.5 weeks of gestation.34
Using the liver from adult rats, Howard et al. identified PMCA1b and PMCA4b as the main isoforms in this tissue, with levels of PMCA1 being greater than those of PMCA4. In contrast, adult human liver samples exhibited higher levels of PMCA4 expression than PMCA1.35 Previously, we have characterized the expression of the housekeeping isoforms of PMCA (i.e., PMCA1 and PMCA4) in both normal liver and liver tissue undergoing regeneration after 70% partial hepatectomy.15 These isoforms have also been studied in several fetal development stages and in a rat hepatocarcinoma model.15 Transcripts for isoforms 1x, 1b, and 4b have consistently been found to be the most abundant, while the pattern of expression for these isoforms during regeneration was found to be more similar to that of normal liver than that of fetal liver (Table 1). This phenomenon can be explained due to the significant role that mature hepatocytes play during liver regeneration.36 In addition, 2w transcripts were detected in all samples analyzed.15 Although a comprehensive analysis of PMCA expression during the fetal stages of rat liver development is still ongoing, 4 PMCA variants have been analyzed in our laboratory using single-cell quantitative reverse transcription PCR techniques. Various cell types derived from hepatic tissue, including hepatocytes, Kupffer cells, and hepatic stellate cells, have been analyzed, and for the first time facilitated the study of the PMCA3 isoform in adult rat liver tissue.17
Taken together, these observations show that available knowledge related to the expression of PMCAs in liver tissue during the embryonic stages of development, is scarce, largely due to challenges that remain to be solved related to the obtention of sufficient and good quality biological material to study.
Stem Cells as a Tool for Studying Embryonic Development
In the early 1980s, the use of pluripotent stem cells such as ESCs, emerged as an alternative to the low availability of hepatocytes for transplantation. However, important ethical issues emerged since the method for obtaining ESCs implied the destruction of an embryo. In 2006, Takahashi and Yamanaka described the use of induced pluripotent stem cells (iPSCs) produced from mouse embryonic and adult fibroblasts that could overcome the immunological rejection associated to ESCs.37
To date, iPSCs have been found to be produced by several species including humans, and shown to differentiate to form distinct cell types.38 Also, iPSCs can be obtained from diverse cell lineages that are "forced" through reprogramming mediated by retroviral or lentiviral induction of 4 essential pluripotency transcription factors, to differentiate into a hepatocyte phenotype.37 Among these factors, apparently c-myc plays a role as a major component promoting the expression of features associated to ESCs and repressing fibroblast-specific genes.39 Subjacent mechanisms involved in the reprogramming are under not only genetic, but under complex epigenetic control addressing the strict transcription of genes.37,40,41 In other studies where reprogramming is performed in the absence of transgenes, no transcriptomic differences were identified in cells that were reprogrammed with or without c-myc.42 Of particular interest is the finding that among more than 2,000 genes that exhibited significant differential expression when the transgene is absent (most of them related to epigenetic regulation and chromatin assembly), the housekeeping PMCA isoforms were expressed. Furthermore, PMCA4 appeared to be overexpressed, while PMCA1 was underexpressed.42 Although the meaning of these results is not readily apparent, they may be understood in terms of the unique features of each isoform associated to the specific demands that the microenvironment might impose.
In this sense, an advantage of reprogramming is that the experiments can be performed under conditions that mimic the natural development of the liver. As a result, hepatocytes can be obtained for transplantation or for studies of developmental biology.43-47 For example, there is a 25-day protocol for obtaining hepatocyte-like cells from ESCs or iPSCs that reproduces the main stages of embryonic development in mice.44,45 Recently, it has been shown that human iPSCs spontaneously form 3-dimensional clusters 48 h after seeding, where after 6 d of culture they develop a bud-like structure which corresponds to 9.5–10.5 d of gestation in mice provided of a functional endothelial network and proliferating hepatoblasts.46
To date, few reports have explored the phenomena of calcium homeostasis in excitable tissues using human iPSC and ESC lines differentiated into cardiomyocytes. It has been demonstrated that iPSCs exhibit an apparent immaturity in the expression of important calcium mobilizing systems involving the ryanodine receptor and SERCA.48 Employing iPSCs derived from patients suffering from hypertrophic cardiomyopathy or Parkinson disease, alterations in calcium handling and an increase in the levels of intracellular Ca2+ have been found in close relationship to the genesis of these pathologies.49,50 Therefore, the role of PMCAs in regulating calcium perturbations in excitable and nonexcitable tissues represents an active area of ongoing research.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors thank Jorge Bravo-Martínez for graphic art support and helpful discussions, and Javier Gallegos for the specialized literature search performed. We also thank Caren Smith for editorial help.
Funding
Studies discussed in this review performed by Mas-Oliva's group have been supported by DGAPA-UNAM and CONACyT grants.
References
- 1. Li D, Wang G-Y, Liu Z-F, Shi Y-X, Zhang H, Bai Z-L. Macrophage-associated erythropoiesis and lymphocytopoiesis in mouse fetal liver: ultrastructural and ASH analysis, Cell Biol Int 2004; 28:457-61; PMID:15223022; http://dx.doi.org/ 10.1016/j.cellbi.2004.03.015 [DOI] [PubMed] [Google Scholar]
- 2. Matsumoto K, Yoshitomi H, Rossant J, Zaret KS. Liver organogenesis promoted by endothelial cells prior to vascular function. Science 2001; 294:559-63; PMID:1577199; http://dx.doi.org/ 10.1126/science.1063889 [DOI] [PubMed] [Google Scholar]
- 3. Nitou M, Sugiyama Y, Ishikawa K, Shiojiri N. Purification of fetal mouse hepatoblasts by magnetic beads coated with monoclonal anti-e-cadherin antibodies and their in vitro culture. Exp Cell Res 2002; 279:330-43; PMID:12243758; http://dx.doi.org/ 10.1006/excr.2002.5615 [DOI] [PubMed] [Google Scholar]
- 4. Soto-Gutiérrez A, Navarro-Alvarez N, Zhao D, Rivas-Carrillo JD, Lebkowski J, Tanaka N, Fox I, Kobayashi N. Differentiation of mouse embryonic stem cells to hepatocyte-like cells by co-culture with human liver nonparenchymal cell lines. Nat Protoc 2007; 2:347-56; PMID:17406596; http://dx.doi.org/ 10.1038/nprot.2007.18 [DOI] [PubMed] [Google Scholar]
- 5. Brues AM, Marble BB. An analysis of mitosis in liver restoration. J Exp Med 1937; 65:15-27; PMID:19870587; http://dx.doi.org/ 10.1084/jem.65.1.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Miyaoka Y, Miyajima A. To divide or not to divide: revisiting liver regeneration. Cell Div 2013; 8:8; PMID:23786799; http://dx.doi.org/ 10.1186/1747-1028-8-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Meier PJ, Sztul ES, Reuben A, Boyer JL. Structural and functional polarity if canalicular and basolateral plasma membrane vesicles isolated in high yield from rat liver. J Cell Biol 1984; 98:991-1000; PMID:6699096; http://dx.doi.org/ 10.1083/jcb.98.3.991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Garcin I, Tordjmann T. Calcium signaling and liver regeneration. Int J Hepatol 2012; 2012:630670; PMID:23119169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kruglov E, Gautam S, Guerra MT, Nathanson MH. Type 2 inositol 1,4,5-trisphosphate receptor modulates bile salt export pump activity in rat hepatocytes. Hepatology 2011; 54:1790-1799; PMID:21748767; http://dx.doi.org/ 10.1002/hep.24548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Navarro-Alvarez N, Soto-Gutierrez A, Kobayashi N. Hepatic stem cells and liver development. Meth Mol Biol 2010; 640:181-236; PMID:20645053; http://dx.doi.org/ 10.1007/978-1-60761-688-7_10 [DOI] [PubMed] [Google Scholar]
- 11. Kovacs CS, Manley NR, Moseley JM, Martin TJ, Kronenberg HM. Fetal parathyroids are not required to maintain placental calcium transport. J Clin Invest 2001; 107:1007-15; PMID:11306604; http://dx.doi.org/ 10.1172/JCI11321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lidofsky SD, Xie MH, Scharschmidt BF. Na+-Ca2+ exchange in cultured rat hepatocytes: evidence against a role in cytosolic Ca2+ regulation or signaling. Am J Physiol 1990; 259:G56-G61; PMID:2372064 [DOI] [PubMed] [Google Scholar]
- 13. Ikari A, Sakai H, Takeguchi N. Protein kinase C-mediated up-regulation of Na+/Ca2+- exchanger in rat hepatocytes determined by a new Na+/Ca2+-exchanger inhibitor, KB-R7943. Eur J Pharmacol 1998; 360:91-8; PMID:9845277; http://dx.doi.org/ 10.1016/S0014-2999(98)00659-1 [DOI] [PubMed] [Google Scholar]
- 14. Schatzmann HJ. Dependence on calcium concentration and stoichiometry of the calcium pump in human red cells. J Physiol 1973; 27:743-5; PMID:4271735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Delgado-Coello B, Santiago-García J, Zarain-Herzberg A, Mas-Oliva J. Plasma membrane Ca2+-ATPase mRNA expression in murine hepatocarcinoma and regenerating liver cells. Mol Cell Biochem 2003; 247:177-84; PMID:12841646; http://dx.doi.org/ 10.1023/A:1024119831983 [DOI] [PubMed] [Google Scholar]
- 16. Delgado-Coello B, Trejo R, Mas-Oliva J. Is there a specific role for the plasma membrane Ca2+-ATPase in the hepatocyte? Mol Cell Biochem 2006; 285:1-15: PMID:16477375; http://dx.doi.org/ 10.1007/s11010-005-9060-z [DOI] [PubMed] [Google Scholar]
- 17. Delgado-Coello B, Bravo-Martínez J, Sosa-Garrocho M, Briones-Orta MA, Macías-Silva M, Mas-Oliva J. Plasma membrane calcium ATPase isoform 3 expression in single cells isolated from rat liver. Mol Cell Biochem 2010; 344:117-24; PMID:20625796; http://dx.doi.org/ 10.1007/s11010-010-0535-1 [DOI] [PubMed] [Google Scholar]
- 18. Kessler F, Bennardini F, Bachs O, Serratosa J, James P, Caride A, Gazzoti P, Penniston JT, Carafoli E. Partial purification and characterization of the Ca2+-pumping ATPase of the liver plasma membrane. J Biol Chem 1990; 265:16012-19; PMID:2144292 [PubMed] [Google Scholar]
- 19. McKenzie RC, Lotersztajn S, Pavoine C, Pecker F, Epand RM, Orlowski RC. Inhibition of the calcium pump by human parathyroid hormone (1-34) and human calcitonin in liver plasma membrane. Biochem J 1990; 266:817-22. PMID:2158300 [PMC free article] [PubMed] [Google Scholar]
- 20. Toyoshima C, Nakasako M, Nomura H, Ogawa H. Crystal structure of the calcium pump of sarcoplasmic reticulum at 2.6Å resolution. Nature 2000; 405:647-55; PMID:10864315; http://dx.doi.org/ 10.1038/35015017 [DOI] [PubMed] [Google Scholar]
- 21. Sacchetto R. Bertipaglia I, Gianetti S, Cendron L, Mascarello F, Damiani E, Carafoli E, Zanotti G. Crystal structure of sarcoplasmic reticulum Ca2+-ATPase (SERCA). J Struct Biol 2012; 178:38-44; PMID:22387132; http://dx.doi.org/ 10.1016/j.jsb.2012.02.008 [DOI] [PubMed] [Google Scholar]
- 22. MacLennan DH, Brandl CJ, Korczak B, Green NM. Amino acid sequence of a Ca2+- Mg2+-dependent ATP from rabbit muscle sarcoplasmic reticulum, deduced from its complementary DNA sequence. Nature 1985; 316:696-700; PMID:2993904; http://dx.doi.org/ 10.1038/316696a0 [DOI] [PubMed] [Google Scholar]
- 23. Strehler EE. Plasma membrane calcium ATPases as novel candidates for therapeutic agent development. J Pharm Pharmaceut Sci 2013; 16:190-206; PMID:23958189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Strehler EE. Strehler–Page MA, Vogel G, Carafoli E. mRNAs for plasma membrane calcium pump isoforms differing in their regulatory domain are generated by alternative splicing that involves two internal donor sites in a single exon. Proc Natl Acad Sci 1989; 86:6908-12; PMID:2528729; http://dx.doi.org/ 10.1073/pnas.86.18.6908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Stauffer TP, Hilfiker H, Carafoli E, Strehler EE. Quantitative analysis of alternative splicing options for human plasma membrane calcium pump genes. J Biol Chem 1993; 68:25993-6003; PMID:8245032 [PubMed] [Google Scholar]
- 26. Keeton TP, Burk SE, Shull GE. Alternative splicing of exons encoding the calmodulin-binding domains and C termini of plasma membrane Ca(2+)-ATPase isoforms 1, 2, 3, and 4. J Biol Chem 1993; 268:2740-48; PMID:8428948 [PubMed] [Google Scholar]
- 27. Bravo-Martínez J, Delgado-Coello B, Mas-Oliva J. Cell calcium extrusion systems and their role in epileptogenesis. Open Neurosci J 2010; 4:1-12; http://dx.doi.org/ 10.2174/1874082001004010001 [DOI] [Google Scholar]
- 28. Lafond J, Simoneau L. Calcium homeostasis in human placenta: role of calcium-handling proteins. Int Rev Cytol 2006; 250:109-74; PMID:16861065; http://dx.doi.org/ 10.1016/S0074-7696(06)50004-X [DOI] [PubMed] [Google Scholar]
- 29. Zacharias DA, Kappen C. Developmental expression of the four plasma membrane calcium ATPase (Pmca) genes in the mouse. Biochim Biophys Acta 1999; 1428:397-405; PMID:10434059; http://dx.doi.org/ 10.1016/S0304-4165(99)00058-6 [DOI] [PubMed] [Google Scholar]
- 30. Stauffer TP, Guerini D, Carafoli E. Tissue distribution of the four gene products of the plasma membrane Ca2+ pump. J Biol Chem 1995; 270:12184-90; PMID:7538133; http://dx.doi.org/ 10.1074/jbc.270.20.12184 [DOI] [PubMed] [Google Scholar]
- 31. Yanagida E, Shoji S, Hirayama Y, Yoshikawa F, Otsu K, Uematsu H, Hiroshi U, Hiraoka M, Furuichi T, Kawano S. Functional expression of Ca2+ signaling pathways in mouse embryonic stem cells, Cell Calcium 2004; 36:135-46; PMID:15193861; http://dx.doi.org/ 10.1016/j.ceca.2004.01.022 [DOI] [PubMed] [Google Scholar]
- 32. Brandt P, Neve RL. Kammesheidt A, Rhoads RE, Vanaman TC. Analysis of the tissue-specific distribution of mRNAs encoding the plasma membrane calcium-pumping ATPases and characterization of an alternately spliced form of PMCA4 at the cDNA and genomic levels. J Biol Chem 1992; 267:4376-85; PMID:1531651 [PubMed] [Google Scholar]
- 33. Heim R., Hug M. Iwata T, Strehler EE, Carafoli E. Microdiversity of human-plasma-membrane calcium-pump isoform 2 generated by alternative splicing in the N-terminal coding region. Eur J Biochem 1992; 205:333-40; PMID:1313367; http://dx.doi.org/ 10.1111/j.1432-1033.1992.tb16784.x [DOI] [PubMed] [Google Scholar]
- 34. Selden C, Jones M, Wade D, Hodgson H. Hepatotropin mRNA expression in human foetal liver development and in liver regeneration. FEBS Lett 1990; 270:81-4; PMID:2146150; http://dx.doi.org/ 10.1016/0014-5793(90)81239-K [DOI] [PubMed] [Google Scholar]
- 35. Howard A, Barley NF, Legon S, Walters JR. Plasma-membrane calcium-pump isoforms in human and rat liver. Biochem J 1994; 303:275-9; PMID:7945253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Michalopoulos GK, DeFrances MC. Liver regeneration. Science 1997; 276:60-6; PMID:9082986; http://dx.doi.org/ 10.1126/science.276.5309.60 [DOI] [PubMed] [Google Scholar]
- 37. Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006; 126:663-76. PMID:16904174; http://dx.doi.org/ 10.1016/j.cell.2006.07.024 [DOI] [PubMed] [Google Scholar]
- 38. Chiang C-H, Huo T-I, Sun C-C, Hsieh J-H, Chien Y, Lu K-H, Lee S-D. Induced pluripotent cells and hepatic differentiation. J Chin Med Assoc 2013; 76:599-605; PMID:23933345; http://dx.doi.org/ 10.1016/j.jcma.2013.07.007 [DOI] [PubMed] [Google Scholar]
- 39. Sridharan R, Tchieu J, Mason MJ, Yachechko R, Kuoy E, Horvath S, Zhou Q, Plath K. Role of murine reprogramming factors in the induction of pluripotency. Cell 2009; 136:364-77; PMID:19167336; http://dx.doi.org/ 10.1016/j.cell.2009.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Snykers S, Henkens T, De Rop E, Vinken M, Fraczek J, De Kock J, De Prins E, Geerts A, Rogiers V, Vanhaecke T. Role of epigenetics in liver-specific gene transcription, hepatocyte differentiation and stem cell reprogramming. J Hepatol 2009; 51:187-211; PMID:19457566; http://dx.doi.org/ 10.1016/j.jhep.2009.03.009 [DOI] [PubMed] [Google Scholar]
- 41. Kim H, Jang M-J, Kang M-J, Han Y-M. Epigenetic signatures and temporal expression of lineage-specific genes in hESCs during differentiation to hepatocytes in vitro. Hum Mol Genet 2011; 20:401-12; PMID:21059703; http://dx.doi.org/ 10.1093/hmg/ddq476 [DOI] [PubMed] [Google Scholar]
- 42. Sommer CA, Christodoulou C, Gianotti-Sommer A, Shen SS, Sailaja BS, Hezroni H, Spira A, Meshorer E, Kotton DN, Mostoslavsky G. Residual expression of reprogramming factors affects the transcriptional program and epigenetic signatures of induced pluripotent stem cells. PLOS One 2012; 7:e51711; PMID:23272148; http://dx.doi.org/ 10.1371/journal.pone.0051711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Touboul T, Hannan NRF, Corbineau S, Martinez A, Martinet C, Branchereau S, Mainot S, Strick-Marchand H, Pedersen R, Di Santo J, et al. Generation of functional hepatocytes from human embryonic stem cells under chemically defined conditions that recapitulate liver development. Hepatology 2010; 5:754-65; PMID:20301097 [DOI] [PubMed] [Google Scholar]
- 44. Zhao R, Duncan SA. Embryonic development of the liver. Hepatology 2005; 41:956-67; PMID:15841465; http://dx.doi.org/ 10.1002/hep.20691 [DOI] [PubMed] [Google Scholar]
- 45. Hannan NRF, Segeritz C-P, Touboul T, Vallier L. Production of hepatocyte-like cells from human pluripotent stem cells. Nat Protoc 2013; 8:430-7; PMID:23424751; http://dx.doi.org/ 10.1038/nprot.2012.153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Takebe T, Sekine K, Enomura M, Koike H, Kimura M, Ogaeri T, Zhang RR, Ueno Y, Zheng YW, Koike N, et al. Vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature 2013; 499:481-4; PMID:23823721; http://dx.doi.org/ 10.1038/nature12271 [DOI] [PubMed] [Google Scholar]
- 47. Zhang R, Takebe T, Sekine K, Koike H, Zheng Y, Taniguchi H. Identification of proliferating human hepatic cells from human induced pluripotent stem cells. Transplant Proc 2014; 46:1201-4; PMID:24815160; http://dx.doi.org/ 10.1016/j.transproceed.2013.12.021 [DOI] [PubMed] [Google Scholar]
- 48. Lee YK, Ng KM, Lai WH, Chan YC, Lau YM. Lian Q, Tse HF, Siu CW. Calcium homeostasis in human induced pluripotent stem cell-derived cardiomyocytes. Stem Cell Rev and Rep 2011; 7:976-86; PMID:21614516; http://dx.doi.org/ 10.1007/s12015-011-9273-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lan F, Lee AS, Liang P, Sanchez-Freire V, Nguyen PK, Wang L, Han L, Yen M, Wang Y, Sun N, et al. Abnormal calcium handling properties underlie familial hypertrophic cardiomyopathy pathology in patient-specific induced pluripotent stem cells. Cell Stem Cell 2013; 12:101-13; PMID:21614516; http://dx.doi.org/ 10.1016/j.stem.2012.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Schöndorf DC, Aurell M, Mcallister FE, Hindley CJ, Mayer F, Schmid B, Sardi SP, Valsecchi M, Hoffmann S, Schwarz LK, et al. iPSC-derived neurons from GBA1-associated Parkinson disease patients show autophagic defects and impaired calcium homeostasis. Nat Commun 2014; 5:4028; PMID:24905578 [DOI] [PubMed] [Google Scholar]