Abstract
In organisms from all kingdoms of life, ammonia and its conjugated ion ammonium are transported across membranes by proteins of the AMT/Rh family. Efficient and successful growth often depends on sufficient ammonium nutrition. The proteins mediating this transport, the so called Ammonium Transporter (AMT) or Rhesus like (Rh) proteins, share a very similar trimeric overall structure and a high sequence similarity even throughout the kingdoms. Even though structural components of the transport mechanism, like an external substrate recruitment site, an essential twin histidine pore motif, a phenylalanine gate and the hydrophobic pore are strongly conserved and have been analyzed in detail by molecular dynamic simulations and mutational studies, the substrate(s), which pass the central pores of the AMT/Rh subunits, NH4+, NH3 + H+, NH4+ + H+ or NH3, are still a matter of debate for most proteins, including the best characterized AmtB protein from Escherichia coli. The lack of a robust expression system for functional analysis has hampered proof of structural and mutational studies, although the NH3 transport function for Rh-like proteins is rarely disputed. In plant transporters belonging to the subfamily AMT1, transport is associated with electrical currents, while some plant transporters, notably of the AMT2 type, were suggested to transport NH3 across the membrane, without associated ionic currents. Here we summarize data in favor of each substrate for the distinct AMT/Rh classes, discuss mutants and how they differ in structure and functionality. A common mechanism with deprotonation and subsequent NH3 transport through the central subunit pore is suggested.
Keywords: ammonia/ammonium, ammonium transport, membrane transport, molecular dynamics simulations, proton transport
Introduction
Long before the molecular identification of transport proteins for ammonia (this term refers here to the sum of ammonia and ammonium, the molecular species are further distinguished by using NH3 and NH4+), the presence of such high affinity, energy-dependent transport systems for ammonia had been shown in all domains of life.1-4 With the help of a bakers yeast mutant, that was deficient in the endogenous high affinity ammonia uptake, in 1994, the molecular basis of ammonia transport was identified by yeast complementation cloning assays, revealing the yeast methylammonia permease MEP15 and the Arabidopsis thaliana AMT1,1 transporter.6 Homology to these allowed the functional identification of further ammonia transporters from organisms of all domains of life. Over the years, yeast remained an important model organism for ammonia transport studies, but the first X-ray crystal structure of an AMT/Rh protein was published for the AmtB transporter from Eschericha coli,7,8 making that homolog the model transporter of choice for structural and molecular studies. Key structural determinants for the transport mechanism that are relevant to plant ammonium transporters are discussed here, but a more detailed overview is given in Lamoureux et al. 2010.9
For plants, the molecular basis of ammonium transport is currently best understood in Arabidopsis thaliana. The genome of this plant comprises 6 AMT genes and transcriptional, post-transcriptional and post-translational regulation of individual AMTs has been identified.10-12 The 6 AMT proteins divide into 2 subfamilies. The AMT1 family has 5 members, AMT1,1–5, and the AMT2 family has a single member. Three vmembers of the AtAMT1 subfamily, AtAMT1,1; AtAMT1,2 and AtAMT1,3 are responsible for 90% of the high affinity ammonium uptake at the roots.13 They mainly reside in the plasma membrane of root (and shoot) cells. In the root, they show a spatial radial arrangement in the order of their affinity.13,14 AMT1,1 and AMT1,3 build heterotrimers in the plasma membrane of the root epidermis.15 With a KmAMT1,1 = 5–34 μM16-18 and KmAMT1,3 = 11 μM17 these 2 transporters mediate the very high affinity uptake of ammonia from the rhizosphere into the root. AMT1,2 with a KmAMT1,2 = 140 μM14 is primarily located in the cortical root cell layers and also mediates uptake of ammonia into the endodermis to facilitate the transfer of ammonia across the impermeable casparian strip.14
Plants seem to possess NH4+ and NH3 transporting proteins,19,20 which may also apply to C. elegans and Drosophila melanogaster, which encode both Amt and Rh homologues in their genomes.21 Plant AMT1 proteins not only from Arabidopsis, but also from tomato,22 bean23 and many other plants were shown to mediate electrogenic, secondary active transport, which might be molecularly as NH4+ ion, NH3 + H+ or even NH4+ + H+ transport (Fig. 1A and B). The latter was suggested for the bean AMT1,1 homolog.23 Until now, functional assays with plant AMT proteins belonging to subfamily 2 suggest these to be electroneutral NH3 transporters, although they also likely recruit NH4+ to the pore entrance.19,20,24 AtAMT2 is co-localized with AtAMT1,1 and AtAMT1,3 in the plasma membrane of the root epidermis cells, but root ammonia uptake was unchanged in a loss-of-function mutant.13,25 This raises the question how plants regulate AMT activity to avoid concurrent activity and futile cycling of ammonium and ammonia, which would lead to the breakdown of the essential proton gradient across the membranes.
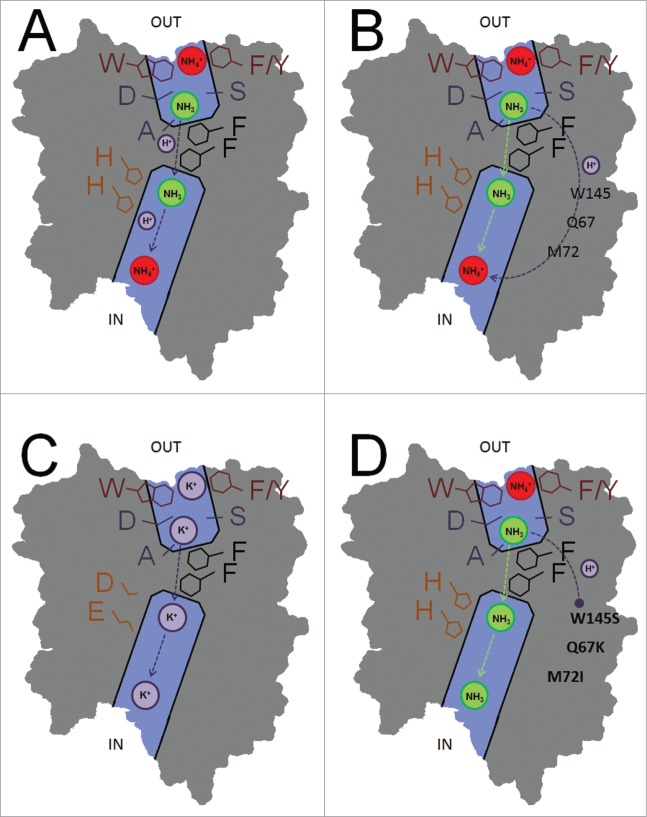
Figure 1.
Schematic transport mechanisms in AMT subunit pores. (A) Electrogenic wild type transport in which the proton is co-transported with the ammonia molecule in the central subunit pore. (B) Wild type electrogenic transport in which the proton is transported through the protein via a specific (unknown) proton pathway. (C) Mutation of the 2 pore-lining histidines in E.c. AmtB results in direct K+ transport. (D) Mutations at the subunit interfaces disrupt proton transport in A.t. AMT1;2, which results in net NH3 transport.
Lessons from the structure
The high-resolution X-ray structures of ammonium transporters from different species were a breakthrough for AMT research, beginning with EcAmtB from E. coli,7,8 followed by AfAMT-1 from A. fulgidus,26 NeRh50 of N. europaea,27 and finally the human RhCG.28 All these proteins share a similar overall structure, especially in the central subunit pore, with at least 3 important conserved molecular arrangements:
Ammonium binding site, deprotonation and mechanistic simulations with computer models
The structure of EcAmtB suggested that a NH4+ recruitment site selects against water and cations at the external pore entrance of each subunit.7,8 While the structure implied that only NH3 crosses the pore center,7 others suggested that NH4+ may be translocated.8 Most computational analyses are in line with the idea that the ion must be deprotonated before NH3 passes the central histidines along the pore, which may even be co-transported with a proton in such simulations.29 Rh proteins, by contrast, for which most functional and structural evidence suggests transport of the uncharged NH3, lack a bona fide external ion recruitment site.27,30,31 Disruption of the proposed ammonium ion recruitment site in EcAmtB lead to hyperactive transporters,32 while its disruption in LeAMT1;1 decreased massively the transport rate and decreased the Km.16 The molecular architecture of an aromatic cation recruitment site in the external pore vestibule is highly similar in the functionally less characterized AfAMT-1, but in contrast to the situation in EcAmtB, the recruitment site was not occupied with electron-dense substrate, when co-crystallized with ammonium or the transport analog methylammonium (MeA).26 Only recently, ionic currents have been detected from AfAMT-1 using a cell-free solid-supported membrane (SSM)-based electrophysiology system, with extremely low maximal transport rates at ∼300 per second and an unusual low affinity in the mM range.33
In an attempt to visualize transport events in an AfAMT-1, we performed molecular dynamics simulations of the highly stable trimeric structure embedded into a POPE lipid membrane and surrounded by water (Fig. 2A and B), a similar setup had been used to study the permeation of solutes in aquaporins.36 The simulations were carried out using the NAMD234 program and classical force field (Amber03). Minimization and equilibration preceded the simulations, as described.36 A constant force vector (44.5 pN) in the z-direction was applied to NH3 and NH4+, to increase the probability that the substrate enters the outer vestibule and eventually crosses the pore. The trajectories of the substrates along the z-axis were calculated relative to the Cα of histidine 157 (Fig. 2D and E). However, the computational data did not identify NH3 or NH4+ transport events, despite the steering force into the AMT pore (Fig. 2). This may be a consequence of the short computational simulation time used, may suggest that none of these molecular species are effectively transported or, most likely, powerfully illustrates the limited value of classical force fields in molecular simulations of this unique transport system. Thermodynamic free energy calculations had previously identified the critical importance of polarizable force fields in cation-π interactions and suggested that the NH4+ is fragmented during transport in EcAmtB.29 Commonly used classical force fields failed to identify essential deprotonation and reprotonation events. NH4+ forms π-interactions with aromatic residues and a H-bridge with γO from Ser 208, in the external pore vestibule of AfAMT-1.26 In our steered molecular dynamics (MD) simulations with NH4+ on AfAMT-1, the ion entered the external vestibule and localized preferentially to that position (Fig. 2C). However, a second distinct position along the z-axis was also recognized, and occasionally hydrogen bonds with other residues of the vestibule were formed (Fig. 2C). H-bonds were frequently formed with the backbone O from Ser 208, Oε1 from Gln 93, and the backbone oxygen from Phe 150 and Ala 151. The existence of 2 neighboring positions for the ion in the recruitment site of AfAMT-1 may explain the lack of a defined substrate electron density peak in the structure.26 Slightly different H-bonding partners in the outer vestibule and a different entry depth were identified when NH4+ was replaced by NH3. Although steering forces were applied to NH4+ and NH3, both molecules never entered the pore beyond the Phe gate. Identical steering forces allowed rapid NH3 permeation events (within a few ns) in similar simulations with (open) water channels.36 When the forces on the substrates were released, NH3 and NH4+ eventually exited from the recruitment vestibule into the extracellular space (Fig. 2D and E). While a closed pore was also encountered in previous simulations with EcAmtB, this was mostly ignored in the interpretations.37 Opening of the phenylalanine gate side chains, which showed more flexibility than other pore residues, as in EcAmtB,38 was sufficient in other simulations to allow substrate to pass, but opening may involve larger conformational changes of AfAMT-1, not captured by short molecular simulations. If deprotonation of NH4+ to NH3 is a prerequisite for the pore opening, only molecular dynamics simulations with polarizable force fields describing quantum mechanical effects are able to mechanistically reveal the true transport events.
Figure 2.
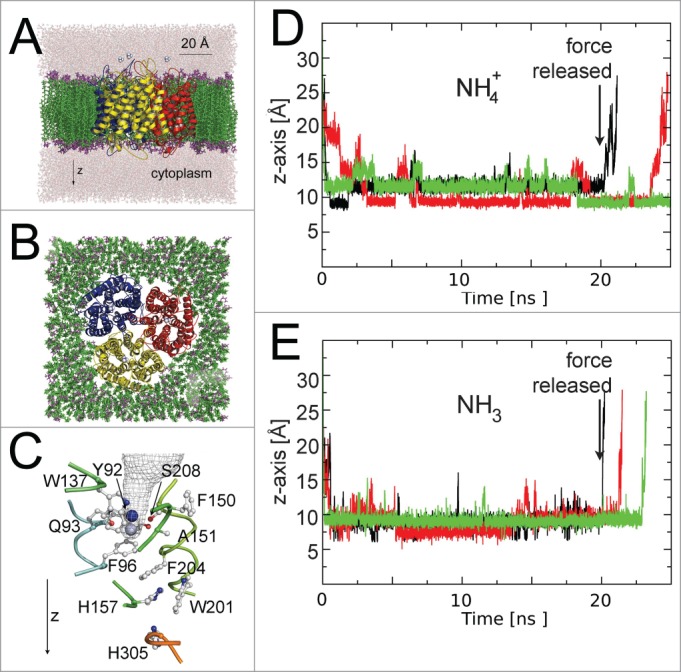
MD simulations with AfAMT-1. (A) Side view (B) top view of the trimer embedded in lipid membranes. (C) Detail of the pore with 2 preferential residency sites of NH4+ along the z-axis shown in blue and gray. (D and E) Z-axis trajectories of NH4+ (D) and NH3 (E) relative to His 157. Individual trajectories are given in different colors. Note that a steering force was applied to increase the probability of the substrate to enter the external vestibule. The force was released after 20 ns.
Because the neighboring AMT subunits of the trimer allosterically regulate activity via the carboxyl-terminus, the flexibility of this region was particularly monitored, but only minimal conformational movements occurred. All simulations, including those above, agree that the overall AMT arrangement represents a relatively stable conformation of the pore. However, studies with a fluorescent sensor coupled to AtAMT1,1 imply that conformational changes occur during transport in AMTs, and that the change between these conformations is ammonia- and activity-dependent.39
Most simulations on EcAmtB were initially in agreement with a NH4+ recruitment, transient deprotonation and NH3 conduction mechanism, although the site of de-protonation and the H+ acceptor are disputed.37,40–43 Some molecular dynamics (MD) simulations suggested that the essential Asp160 stabilizes NH4+ indirectly at the external pore vestibule, where water accepts a proton from NH4+.42,44 Alternatively, Asp160 may play a direct role with Ser219 and Ala162; the corresponding residues in AtAMT1,2 are Asp211, Ser275, Ala213.43,45 In electrophysiological measurements with Xenopus leavis oocytes expressing AtAMT1,2, the Km(NH4+) was highly voltage dependent, suggesting that the site that determines the affinity for NH4+ is deeply buried in the membrane electric field. The fractional electrical distances δ (NH4+) = 0.56 and δ (MeA+) = 0.26 suggest that the “binding” sites for NH4+ and MeA+ are located 56% and 26%, respectively, inside the membrane electric field, measured from the outside. These functional data suggest NH4+ crosses more than half of the membrane electric field to reach the hydrophobic pore environment that prefers the conjugated base, NH3 and thus the catalytic site of deprotonation.14 Clearly, rapid transport excludes tight “binding” of the substrate to the pore and high selectivity against K+ together with high transport rates may thus require fragmenting the substrate, NH4+ into NH3 and H+. The proposed recruitment sites are highly conserved in plant AMT1, AMT2, AfAMT-1 and AmtB proteins.20 However, the substrate affinities among distinct isoforms can differ in more than fold10-, which further implies that the recruitment site does not define the affinity of the transporters. Mutations in the cytoplasmic part of a wheat AMT not only changed the pH dependence of the transport, but also its affinity for NH4+ and MeA+, highlighting the importance of internal protein regions and inner-subunit interactions for the determination of the transport activity.46 Several other mutations in regions distant from the recruitment site massively changed the substrate saturation properties of AMTs by NH4+ and MeA+.39
After the pore transfer, NH3 is quickly re-protonated at the cytoplasmic pore exit, as the equilibrium between NH4+ and NH3 in the cytoplasmic pH strongly favors NH4+. More recent simulations are in favor of a proton-co transport with NH3,29 but molecular simulations could not unambiguously identify whether the NH3 conduction in the pore of EcAmtB leads to net transfer of NH3 (“passive”) or NH4+ (secondary active) across the membrane.
A recent screen with plant AMT1; 2 for functional ammonia-transporting mutants that rescued yeast growth on methylammonia revealed mutants with residual transport activity. Amino acids altered in these mutant proteins were in positions not directly in contact with the hydrophobic substrate pore and rather occurred in amino acids close to the subunit interface. Interestingly, the activity of AMT1;1 had been increased by a corresponding mutation in the first membrane span (Q57H), again not directly facing the hydrophobic pore.18 In the above mentioned screen, the corresponding glutamine was found to be mutated to lysine and the activity of this mutant was reduced, rather than elevated. Three identified mutations showed reduced ammonia and methylammonia transport, but surprisingly, the residual transport was not coupled to any electric currents, as is the ammonia transport in the AMT1;2 wild type protein (Fig. 1D). This loss of flux/charge coupling in these mutants is naturally explained by a NH3/H+ co-transport mode in the AMT1;2 wild type (Fig. 1A and B), but the co-transported charge is lost in the mutant (Fig. 1D). This interpretation takes into account that mutations were not directly associated with the pore and thus, the selectivity of pore to the major substrate (NH3) was unchanged. Because uncoupling mutants were not identified in residues with direct pore contact, but rather at sites between subunits, it opens the possibility that the H+ is co-transported in wild type proteins via a distinct pathway than the hydrophobic pore.47 Clearly, because all structural key elements of the pore are unchanged in these mutants, the loss of charge coupling by mutations strongly implies that NH3 and not NH4+ is generally passing typical AMT pores (although it cannot be excluded that large conformational changes occured in the mutants).
The Phenylalanine Gate
In AMT/Rh crystal structures, the pore is occluded to different levels by a “gate” that is formed by 2 phenylalanines (Phe). Some mutations of these phenylalanines to smaller aliphatic residues did not strongly influence the transport or even improved the transport rate in EcAmtB, while others are not tolerated.32 The “Phe gate” in RhC glycoproteins appears less strictly occluded27 and the simultaneous deletion of both Phe in the gate of the RhC glycoprotein yielded a NH3 conducting channel.48 The outer Phe was absolutely required for the function of EcAmtB, as it may be critical for the deprotonation of NH4+.49 All these observations are in accordance with the notion that such a gate is required for stripping off a proton from the NH4+ substrate in EcAmtB and AfAMT-1.
The Twin His Motif
Each AMT protein is arranged as a trimer with the subunits forming hydrophobic pores that are aligned by 2 pore facing conserved histidines.7,8,26,27 These histidines were proposed to be involved in deprotonation of ammonium prior to transport.7,8 Mutational and functional analysis had experimentally demonstrated an important, but not crucial role of the twin-histidine motif for EcAmtB.50 In initial experiments, the first histidine (His168), but not the second, could be exchanged to glutamate (Glu) and yielded a transporter with residual activity. This is in agreement with the fact that many native AMTs from fungi have a Glu at that position. Further analysis showed that the second histidine could also be exchanged to acidic amino acids, to yield a partially active transporter, but only in combinations with other mutations.32 All double mutants of the 2 histidines in the E.coli AmtB transporter were not functional in ammonia transport.32,50–52 For all wild type AMTs analyzed in detail, other cations with an ionic radius similar to ammonium (like the potassium ion) are strongly excluded from transport. However, the mutation of H318D in EcAmtB and the double mutation of H168D, H318E did not change the overall protein structure, but led to a loss of AmtB transport specificity and allowed the transport of K+ (Fig. 1C), even though ammonia transport was abolished by the mutations.52 Single mutations of the histidines to alanines, which make the pore more hydrophobic, excluded the transport of the ammonium analog MeA, but not that of NH4+ itself. Mutations of the histidines to residues with a stronger polarity might raise the hydrophilicity of the pore, facilitate a deeper entry of K+ and eventually allow passage of this ion. In a plant AMT from bean, the exchange of the first histidine had an effect on vmax, and Km, and the proposed H+/NH4+ co-transport mechanism.23 These findings demonstrate that individual AMT pore architecture principally can accommodate and pass cations, such as K+, but this depends on whether potentially negatively charged residues are positioned in the pore substrate pathway and determines the electrostatics of the pore lumen. While the central histidines play a crucial role in the transport mechanism, their relevance to each individual AMT is not the same. Comparative studies with histidine mutants in functionally distinct AMTs, such as plant AMT1 and AMT2 transporters, might not only help to clarify their role in these proteins, but finally lead to a common consensus requirement for substrate transport.
Deprotonation as a prerequisite for transport
Although initially disputed, electrogenic transport and cellular accumulation of NH4+ was found repeatedly for several AMTs.14,53 The substrate/charge coupling of 1:1 was determined for AMTs from tomato and wheat, respectively, which is compatible with NH4+ uniport and NH3/H+ co-transport.16,46 Because of the high sequence identity of prokaryotic and plant AMTs, their pore is likely very similar in structure, but unfortunately, structural homology models were too unstable to derive mechanistic conclusions for plant AMT function using molecular dynamics simulations.22,46 AMT function can be inactivated by the C-terminus of each monomer, which also contacts neighboring subunits and mutations in several protein regions apparently compensated an inactivating carboxy-terminus mutant of plant AMTs, suggesting that subunit interactions and inter-protein movements are crucial for transport.54
If there is a general NH3 conduction mechanism through the central AMT/Rh pore, the electrogenic ammonium transport in plant AMT1 proteins must be explained by a mechanism, where NH3 transit is coupled to H+ co-transport. Although functional assays with liposomes initially supported a facilitated diffusion mechanism for NH3 in the prototype EcAmtB,7 these results could not be confirmed by others.55 Heterologous expression of EcAmtB in oocytes is in agreement with a net NH3 transport mechanism,56 but NH4+ transport was deduced from the capacity of an EcAmtB mutant to accumulate ammonium in bacterial cells.57 The exact transport mechanism is likely crucial for the cellular physiology, as in the presence of a membrane potential and a proton gradient, this determines whether ammonia is taken up in a secondary active way and accumulated by exploiting the membrane potential and proton gradient, or whether NH3 is passively excreted and lost.58
AMTs are unlikely static “holes” in the membrane, and several findings suggest that they are “gated.” The carboxy-terminus of the AMT is involved in this gating mechanism, but apparently also diverse other membrane integral parts of the protein.14,54 Differential, contrasting gating is therefore expected for functionally opposite AMT1 and AMT2-type transporters in plants, as these are co-localized in the same membranes, but it is still unclear under which conditions net NH3-flux is relevant. Simple facilitated NH3 diffusion may occur in Rh glycoproteins or some aquaporins, but in AMT2 transporters, the transport mechanism is clearly more complex, as NH4+ is typically recruited with high affinity and the deprotonation step is likely essential for transport activity. This may impose one barrier to impede futile cycling of ammonia, in addition to gating and low cytosolic NH4+ levels through effective metabolic trapping by glutamine synthetase. As long as energy (supplied as photosynthates to plant roots) is not limiting, but nitrogen is limiting, the plant may decide to increase the ammonia uptake irrespective of any potential energetic losses. However, when energy gets limiting, strict control of net NH4+ must be in place to avoid any potential losses of nitrogen and energy.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1. Hackette SL, Skye GE, Burton C, Segel IH. Characterization of an ammonium transport system in filamentous fungi with methylammonium-14C as the substrate. J Biol Chem 1970; 45:4241-50; PMID:5498410 [PubMed] [Google Scholar]
- 2. Kleiner D. Energy expenditure for cyclic retention of NH3/NH4 +during N2 fixation by Klebsiella pneumoniae. FEBS Lett 1985; 187:237-239; PMID:3894049; http://dx.doi.org/ 10.1016/0014-5793(85)81249-7 [DOI] [PubMed] [Google Scholar]
- 3. Dubois E, Grenson M. Methylamine/ammonia uptake systems in Saccharomyces cerevisiae: multiplicity and regulation. Mol Gen Genet MGG 1979; 175:67-76; http://dx.doi.org/ 10.1007/BF00267857 [DOI] [PubMed] [Google Scholar]
- 4. Knepper MA. NH4 +transport in the kidney. Kidney Int Suppl 1991; 33; S95-102; PMID:1890804 [PubMed] [Google Scholar]
- 5. Marini AM, Vissers S, Urrestarazu A, André B. Cloning and expression of the MEP1 gene encoding an ammonium transporter in Saccharomyces cerevisiae. EMBO J 1994; 13:3456-63; PMID:8062822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ninnemann O, Jauniaux JC, Frommer WB. Identification of a high affinity NH4+ transporter from plants. EMBO J 1994; 13:3464-71; PMID:8062823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Khademi S, O'Connell J, 3rd, Remis J, Robles-Colmenares Y, Miercke LJ, Stroud RM. Mechanism of ammonia transport by Amt/MEP/Rh: structure of AmtB at 1.35 A. Science 2004; 305:1587-94; PMID:15361618; http://dx.doi.org/ 10.1126/science.1101952 [DOI] [PubMed] [Google Scholar]
- 8. Zheng L, Kostrewa D, Bernèche S, Winkler FK, Li X-D. The mechanism of ammonia transport based on the crystal structure of AmtB of Escherichia coli. Proc Natl Acad Sci U S A 2004; 101:17090-5; PMID:15563598; http://dx.doi.org/ 10.1073/pnas.0406475101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lamoureux G, Javelle A, Baday S, Wang S, Bernèche S. Transport mechanisms in the ammonium transporter family. Transfus Clin Biol 2010; 17:168-75; PMID:20674437; http://dx.doi.org/ 10.1016/j.tracli.2010.06.004 [DOI] [PubMed] [Google Scholar]
- 10. Loqué D, von Wirén N. Regulatory levels for the transport of ammonium in plant roots. J Exp Bot 2004; 55:1293-305; http://dx.doi.org/ 10.1093/jxb/erh147 [DOI] [PubMed] [Google Scholar]
- 11. Ludewig U, Neuhäuser B, Dynowski M. Molecular mechanisms of ammonium transport and accumulation in plants. FEBS Lett 2007; 581:2301-8; PMID:17397837; http://dx.doi.org/ 10.1016/j.febslet.2007.03.034 [DOI] [PubMed] [Google Scholar]
- 12. Lanquar V, Loqué D, Hörmann F, Yuan L, Bohner A, Engelsberger WR, Lalonde S, Schulze WX, von Wirén N, Frommer WB. Feedback inhibition of ammonium uptake by a phospho-dependent allosteric mechanism in Arabidopsis. Plant Cell 2009; 21:3610-22; PMID:19948793; http://dx.doi.org/ 10.1105/tpc.109.068593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yuan L, Loqué D, Kojima S, Rauch S, Ishiyama K, Inoue E, Takahashi H, von Wirén N. The organization of high-affinity ammonium uptake in Arabidopsis roots depends on the spatial arrangement and biochemical properties of AMT1-type transporters. Plant Cell 2007; 19:2636-52; PMID:17693533; http://dx.doi.org/ 10.1105/tpc.107.052134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Neuhäuser B, Dynowski M, Mayer M, Ludewig U. Regulation of NH4 +transport by essential cross talk between AMT monomers through the carboxyl tails. Plant Physiol 2007; 143:1651-9; http://dx.doi.org/ 10.1104/pp.106.094243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yuan L, Gu R, Xuan Y, Smith-Valle E, Loqué D, Frommer WB, von Wirén N. Allosteric regulation of transport activity by heterotrimerization of Arabidopsis ammonium transporter complexes in vivo. Plant Cell 2013; 25:974-84; PMID:23463773; http://dx.doi.org/ 10.1105/tpc.112.108027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mayer M, Ludewig U. Role of AMT1;1 in NH4+ acquisition in Arabidopsis thaliana. Plant Biol (Stuttg) 2006; 8:522-8; PMID:16917981 [DOI] [PubMed] [Google Scholar]
- 17. Gazzarrini S, Lejay L, Gojon A, Ninnemann O, Frommer WB, von Wirén N. Three functional transporters for constitutive, diurnally regulated, and starvation-induced uptake of ammonium into Arabidopsis roots. Plant Cell 1999; 11:937-48; PMID:10330477; http://dx.doi.org/ 10.1105/tpc.11.5.937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Loqué D, Mora SI, Andrade SLA, Pantoja O, Frommer WB. Pore mutations in ammonium transporter AMT1 with increased electrogenic ammonium transport activity. J Biol Chem 2009; 284:24988-95; http://dx.doi.org/ 10.1074/jbc.M109.020842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Guether M, Neuhäuser B, Balestrini R, Dynowski M, Ludewig U, Bonfante P. A mycorrhizal-specific ammonium transporter from Lotus japonicus acquires nitrogen released by arbuscular mycorrhizal fungi. Plant Physiol 2009; 150:73-83; PMID:19329566; http://dx.doi.org/ 10.1104/pp.109.136390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Neuhäuser B, Dynowski M, Ludewig U. Channel-like NH3 flux by ammonium transporter AtAMT2. FEBS Lett 2009; 583:2833-8; http://dx.doi.org/ 10.1016/j.febslet.2009.07.039 [DOI] [PubMed] [Google Scholar]
- 21. Ludewig U, von Wirén N, Rentsch D, Frommer WB. Rhesus factors and ammonium: a function in efflux? Genome Biol 2001; 2; REVIEWS1010; PMID:11276430; http://dx.doi.org/ 10.1186/gb-2001-2-3-reviews1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mayer M, Dynowski M, Ludewig U. Ammonium ion transport by the AMT/Rh homologue LeAMT1;1. Biochem J 2006; 396:431-7; PMID:16499477; http://dx.doi.org/ 10.1042/BJ20060051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ortiz-Ramirez C, Mora SI, Trejo J, Pantoja O. PvAMT1;1, a highly selective ammonium transporter that functions as H+/NH4(+) symporter. J Biol Chem 2011; 286:31113-22; PMID:21757699; http://dx.doi.org/ 10.1074/jbc.M111.261693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Straub D, Ludewig U, Neuhäuser B. A nitrogen-dependent switch in the high affinity ammonium transport in Medicago truncatula. Plant Mol Biol 2014; 86:458-94; PMID:25164101; http://dx.doi.org/ 10.1007/s11103-014-0243-4 [DOI] [PubMed] [Google Scholar]
- 25. Loqué D, Yuan L, Kojima S, Gojon A, Wirth J, Gazzarrini S, Ishiyama K, Takahashi H, von Wirén N. Additive contribution of AMT1;1 and AMT1;3 to high-affinity ammonium uptake across the plasma membrane of nitrogen-deficient Arabidopsis roots. Plant J 2006; 48:522-34; PMID:17026539; http://dx.doi.org/ 10.1111/j.1365-313X.2006.02887.x [DOI] [PubMed] [Google Scholar]
- 26. Andrade SLA, Dickmanns A, Ficner R, Einsle O. Crystal structure of the archaeal ammonium transporter Amt-1 from Archaeoglobus fulgidus. Proc Natl Acad Sci U S A 2005; 102:14994-9; PMID:16214888; http://dx.doi.org/ 10.1073/pnas.0506254102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lupo D, Li XD, Durand A, Tomizaki T, Cherif-Zahar B, Matassi G, Merrick M, Winkler FK. The 1.3-A resolution structure of Nitrosomonas europaea Rh50 and mechanistic implications for NH3 transport by Rhesus family proteins. Proc Natl Acad Sci U S A 2007; 104:19303-8; PMID:18032606; http://dx.doi.org/ 10.1073/pnas.0706563104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gruswitz F, Chaudhary S, Ho JD, Schlessinger A, Pezeshki B, Ho CM, Sali A, Westhoff CM, Stroud RM. Function of human Rh based on structure of RhCG at 2. 1 Å. Proc Natl Acad Sci U S A 2010; 107:9638-43; PMID:20457942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang S, Orabi EA, Baday S, Bernèche S, Lamoureux G. Ammonium transporters achieve charge transfer by fragmenting their substrate. J Am Chem Soc 2012; 134:10419-27; PMID:22631217; http://dx.doi.org/ 10.1021/ja300129x [DOI] [PubMed] [Google Scholar]
- 30. Mayer M, Schaaf G, Mouro I, Lopez C, Colin Y, Neumann P, Cartron JP, Ludewig U. Different transport mechanisms in plant and human AMT / Rh-type ammonium transporters. J Gen Physiol 2006; 127:133-144; PMID:16446503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Weidinger K, Neuhäuser B, Gilch S, Ludewig U, Meyer O, Schmidt I. Functional and physiological evidence for a rhesus-type ammonia transporter in Nitrosomonas europaea. FEMS Microbiol Lett 2007; 273:260-7; PMID:17608700; http://dx.doi.org/ 10.1111/j.1574-6968.2007.00805.x [DOI] [PubMed] [Google Scholar]
- 32. Hall JA, Kustu S. The pivotal twin histidines and aromatic triad of the Escherichia coli ammonium channel AmtB can be replaced. Proc Natl Acad Sci U S A 2011; 108:13270-4; PMID:21775672; http://dx.doi.org/ 10.1073/pnas.1108451108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wacker T, Garcia-Celma JJ, Lewe P, Andrade SL. A direct observation of electrogenic NH4+ transport in ammonium transport (Amt) proteins. Proc Natl Acad Sci U S A 2014; 6-11. http://dx.doi.org/ 10.1073/pnas.1406409111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kalé L, Skeel R, Bhandarkar M, Brunner R, Gursoy A, Krawetz N, Phillips J, Shinozaki A, Varadarajan K, Schulten K. NAMD2: Greater Scalability for Parallel Molecular Dynamics. J Comput Phys 1999; 151:283-312; http://dx.doi.org/ 10.1006/jcph.1999.6201 [DOI] [Google Scholar]
- 35. Ullmann RT, Andrade SLA, Ullmann GM. Thermodynamics of transport through the ammonium transporter Amt-1 investigated with free energy calculations. J Phys Chem B 2012; 116:9690-703; PMID:22804733; http://dx.doi.org/ 10.1021/jp305440f [DOI] [PubMed] [Google Scholar]
- 36. Dynowski M, Mayer M, Moran O, Ludewig U. Molecular determinants of ammonia and urea conductance in plant aquaporin homologs. FEBS Lett 2008; 582:2458-62; PMID:18565332; http://dx.doi.org/ 10.1016/j.febslet.2008.06.012 [DOI] [PubMed] [Google Scholar]
- 37. Bostick DL, Brooks CL. Deprotonation by dehydration: the origin of ammonium sensing in the AmtB channel. PLoS Comput Biol 2007; 3; e22; PMID:17291160; http://dx.doi.org/ 10.1371/journal.pcbi.0030022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yang H, Xu Y, Zhu W, Chen K, Jiang H. Detailed mechanism for AmtB conducting NH4+/NH3: molecular dynamics simulations. Biophys J 2007; 92:877-85; PMID:17098799; http://dx.doi.org/ 10.1529/biophysj.106.090191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. De Michele R, Ast C, Loqué D, Ho CH,Andrade SL, Lanquar V, Grossmann G, Gehne S, Kumke MU, Frommer WB. Fluorescent sensors reporting the activity of ammonium transceptors in live cells. Elife 2013; 2;e00800; PMID:23840931; http://dx.doi.org/ 10.7554/eLife.00800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cao Z, Mo Y, Thiel W. Deprotonation mechanism of NH4+ in the Escherichia coli ammonium transporter AmtB: insight from QM and QM/MM calculations. Angew Chem Int Ed Engl 2007; 46:6811-5; PMID:17668906; http://dx.doi.org/ 10.1002/anie.200701348 [DOI] [PubMed] [Google Scholar]
- 41. Lin Y, Cao Z, Mo Y. Functional role of Asp160 and the deprotonation mechanism of ammonium in the Escherichia coli ammonia channel protein AmtB. J Phys Chem B 2009; 113:4922-9; PMID:19278252; http://dx.doi.org/ 10.1021/jp810651m [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Luzhkov VB, Almlöf M, Nervall M, Aqvist J. Computational study of the binding affinity and selectivity of the bacterial ammonium transporter AmtB. Biochemistry 2006; 45:10807-14; PMID:16953566; http://dx.doi.org/ 10.1021/bi0610799 [DOI] [PubMed] [Google Scholar]
- 43. Nygaard TP, Rovira C, Peters GH, Jensen MØ. Ammonium recruitment and ammonia transport by E. coli ammonia channel AmtB. Biophys J 2006; 91:4401-12; PMID:17012311; http://dx.doi.org/ 10.1529/biophysj.106.089714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bostick DL, Brooks CL. On the equivalence point for ammonium (de)protonation during its transport through the AmtB channel. Biophys J 2007; 92; L103-5; PMID:17434945; http://dx.doi.org/ 10.1529/biophysj.107.109165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lin Y, Cao Z, Mo Y. Molecular dynamics simulations on the Escherichia coli ammonia channel protein AmtB: mechanism of ammonia/ammonium transport. J Am Chem Soc 2006; 128:10876-84; PMID:16910683; http://dx.doi.org/ 10.1021/ja0631549 [DOI] [PubMed] [Google Scholar]
- 46. Søgaard R, Alsterfjord M, Macaulay N, Zeuthen T. Ammonium ion transport by the AMT/Rh homolog TaAMT1;1 is stimulated by acidic pH. Pflugers Arch 2009; 458:733-43; PMID:19340454; http://dx.doi.org/ 10.1007/s00424-009-0665-z [DOI] [PubMed] [Google Scholar]
- 47. Neuhäuser B, Ludewig U. Uncoupling of ionic currents from substrate transport in the plant ammonium transporter AtAMT1;2. J Biol Chem 2014; 289:11650-5; http://dx.doi.org/ 10.1074/jbc.C114.552802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zidi-Yahiaoui N, Callebaut I, Genetet S, Le Van Kim C, Cartron JP, Colin Y, Ripoche P, Mouro-Chanteloup I. Functional analysis of human RhCG: comparison with E. coli ammonium transporter reveals similarities in the pore and differences in the vestibule. Am J Physiol Cell Physiol 2009; 297;C537-47; PMID:19553567; http://dx.doi.org/ 10.1152/ajpcell.00137.2009 [DOI] [PubMed] [Google Scholar]
- 49. Javelle A, Lupo D, Ripoche P, Fulford T, Merrick M, Winkler FK. Substrate binding, deprotonation, and selectivity at the periplasmic entrance of the Escherichia coli ammonia channel AmtB. Proc Natl Acad Sci U S A 2008; 105:5040-5; PMID:18362341; http://dx.doi.org/ 10.1073/pnas.0711742105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Javelle A, Lupo D, Zheng L, Li XD, Winkler FK, Merrick M. An unusual twin-his arrangement in the pore of ammonia channels is essential for substrateconductance. J Biol Chem 2006; 281:39492-8; PMID:17040913; http://dx.doi.org/ 10.1074/jbc.M608325200 [DOI] [PubMed] [Google Scholar]
- 51. Wang J, Fulford T, Shao Q, Javelle A, Yang H, Zhu W, Merrick M. Ammonium transport proteins with changes in one of the conserved pore histidines have different performance in ammonia and methylamine conduction. PLoS One 2013; 8; e62745; PMID:23667517; http://dx.doi.org/ 10.1371/journal.pone.0062745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hall JA, Yan D. The molecular basis of K+ exclusion by the Escherichia coli ammonium channel amtB. J Biol Chem 2013; 288:14080-6. http://dx.doi.org/ 10.1074/jbc.M113.45795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wood CC, Porée F, Dreyer I, Koehler GJ, Udvardi MK. Mechanisms of ammonium transport, accumulation, and retention in ooyctes and yeast cells expressing Arabidopsis AtAMT1;1. FEBS Lett 2006; 580:3931-6; PMID:16806203; http://dx.doi.org/ 10.1016/j.febslet.2006.06.026 [DOI] [PubMed] [Google Scholar]
- 54. Loqué D, Lalonde S, Looger LL, von Wirén N, Frommer WB. A cytosolic trans-activation domain essential for ammonium uptake. Nature 2007; 446:195-8; http://dx.doi.org/ 10.1038/nature05579 [DOI] [PubMed] [Google Scholar]
- 55. Javelle A, Lupo D, Li XD, Merrick M, Chami M, Ripoche P, Winkler FK. Structural and mechanistic aspects of Amt/Rh proteins. J Struct Biol 2007; 158:472-81; PMID:17368911; http://dx.doi.org/ 10.1016/j.jsb.2007.01.004 [DOI] [PubMed] [Google Scholar]
- 56. Musa-Aziz R, Jiang L, Chen L-M, Behar KL, Boron WF. Concentration-dependent effects on intracellular and surface pH of exposing Xenopus oocytes to solutions containing NH3/NH4(+). J Membr Biol 2009; 228:15-31; PMID:19242745; http://dx.doi.org/ 10.1007/s00232-009-9155-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Fong RN, Kim K-S, Yoshihara C, Inwood WB, Kustu S. The W148L substitution in the Escherichia coli ammonium channel AmtB increases flux and indicates that the substrate is an ion. Proc Natl Acad Sci U S A 2007; 104:18706-11; PMID:17998534; http://dx.doi.org/ 10.1073/pnas.0709267104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ludewig U. Ion transport versus gas conduction: function of AMT/Rh-type proteins. Transfus Clin Biol 2006; 13:111-6; PMID:16563830; http://dx.doi.org/ 10.1016/j.tracli.2006.02.012 [DOI] [PubMed] [Google Scholar]