Abstract
Most cells in the body secrete, or are in intimate contact with extracellular matrix (ECM), which provides structure to tissues and regulates various cellular phenotypes. Cells are well known to respond to biochemical signals from the ECM, but recent evidence has highlighted the mechanical properties of the matrix, including matrix elasticity and nanotopography, as fundamental instructive cues regulating signal transduction pathways and gene transcription. Recent observations also highlight the importance of matrix nanotopography as a regulator of cellular functions, but lack of facile experimental platforms has resulted in a continued negligence of this important microenvironmental cue in tissue culture experimentation. In this review, we present our opinion on the importance of nanotopography as a biological cue, contexts in which it plays a primary role influencing cell behavior, and detail advanced techniques to incorporate nanotopography into the design of experiments, or in cell culture environments. In addition, we highlight signal transduction pathways that are involved in conveying the extracellular matrix nanotopography information within the cells to influence cell behavior.
Keywords: nanotopography, extracellular matrix, structure-function, tissue engineering, cell biology
Cells Exist in a Complex Neighborhood
The world of living cells in tissues is organized in a complex fashion, consisting of various cell types, residing together in a milieu containing a gamut of structures formed by extracellular matrix (ECM), while conduits from the distal world bring in nutrients, chemical and electrical signals. Increased availability of molecular biology tools, development of sophisticated microscopy and methods for selective genetic perturbation developed recently have significantly increased our understanding of how cells respond to their local microenvironment (Fig. 1A–B).1 It has been widely appreciated and understood how cells respond to various soluble biochemical factors, including growth factors, and cytokines, and insoluble biochemical factors including those intrinsic to the ECM or presented to cells in a tethered form by the ECM (Fig. 1C).2 However, it has only recently been recognized that the currency of communication in the tissues is even more varied, and includes the mechanical and physical properties of the environment itself.1,3,4 The mechanical features of the ECM include its topography,5-9 and elasticity,3,10 both of which are now being recognized as important in guiding cellular phenotypes in an active manner. ECM mechanics are now reported to guide cell shape, migration, differentiation, proliferation, and overall structure and function of the tissue.11-13 Here we will focus on ECM topography and its role in regulating cell and tissue functions, the mechanisms involved in topography sensing, and deliberate on our current understanding of this fundamental biological cue.
Figure 1.
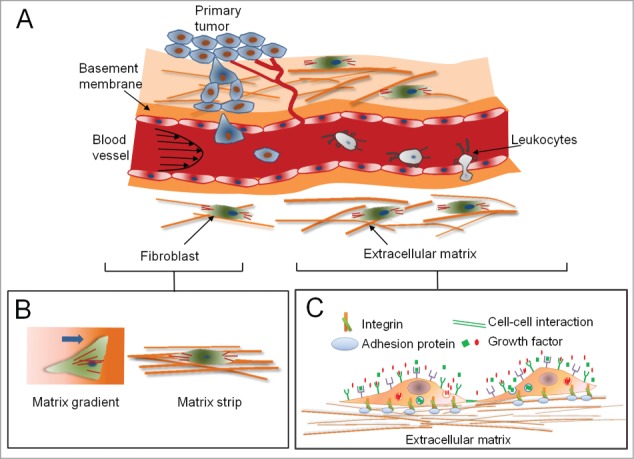
The natural microenvironment of cells and tissues in a human body consists of combination of various mechanical and biochemical factors. (A) Various mechanical stimuli, such as shear and hydrostatic stress, mechanical and structural properties of extracellular matrix (ECM), combine with soluble factors to form an environment in which tumor cells metastasize and extravasate. (B) Cell shape and morphology are affected by both the mechanical and structural properties of ECM. (C) Tissue microenvironment is defined by cell-matrix interactions via integrin receptors, autocrine or paracrine soluble factors recognized by specific receptors, cell-cell interactions, and direct regulation of cell phenotype by mechanical properties of the matrix.
Nanoscale Features in Matrix Fundamentally Influence Cell Behavior
ECM comes in different shapes and sizes, and has evolved to support the structure and function of specific tissue types. Typically ECM is composed of various proteins, carbohydrates, and proteoglycans14,15 that provide a chemically active substrate for the cells to attach to, while also presenting a specific range of topographical features specific to the tissue type (Fig. 1C).2 These features include fibers and meshwork, pits and posts arranged in regular or irregular fashion and are fundamentally important in determining cell shape and tissue architecture.2 The degree of arrangement of ECM topography varies across tissues (Fig. 2). Basement membrane to which endothelial cells, epithelial cells and other cells interact consists of amorphous sheets intermittently laden with pits, meshes and grooves (Fig. 2A), while more organized tissues like the tendons also consist of collagen fibers highly aligned over long distances (Fig. 2B–C). Importantly, the organization of the topographical features is highly regulated during development, and allows formation of organized structural scaffolds over which the tissue develops. As an example, embryonic tendon is highly disorganized, and consists of random meshwork of collagen fibrils that eventually organizes (Fig. 2B) to form organized bundles of aligned nanofibrils, with rare occurrences of fibrils not aligned along the long axis of the tendon (Fig. 2C).16
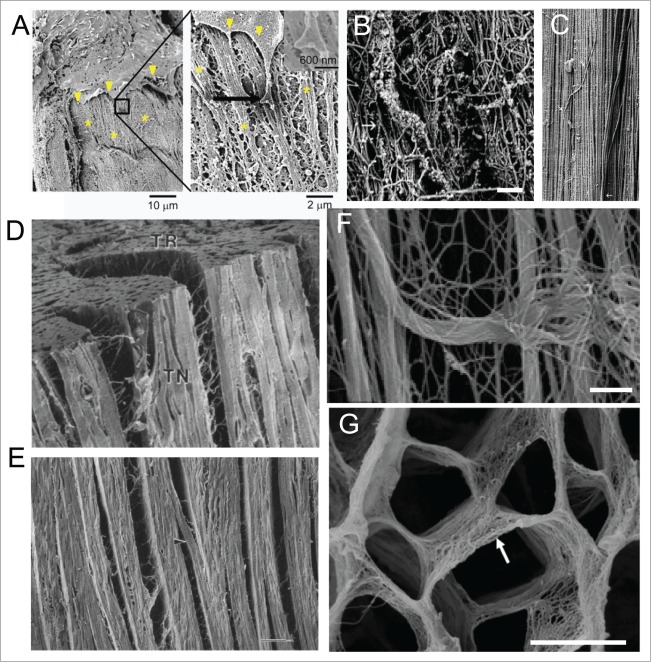
Figure 2.
Nanotopographical features are diverse in their shapes and sizes. (A) SEM image showing interaction of an endothelial cell in the aorta with the basement membrane61; left image shows the edge of the cell (arrowhead) in close proximity with the rough matrix in the basement membrane (denoted by *), boxed area magnified in right shows end process of the cell interacting with the basement membrane composed of ridges, bumps, pits and other features in nano- to micro scales; Reprinted with the permission of Mary Ann Liebert, Inc. (B–C) Topographical features dynamically change during development to assume their mature shape in an adult tissue16; (B) SEM image of late embryonic bovine medial collateral tendon ligament show substantial interwoven collagen fibrils, arranging themselves toward the long axis; (C) Mature tendon ligament show a highly aligned 3D network of collagen fibrils along the long axis of the ligament, with only a few fibrils crossing over the otherwise parallel dense network (arrowheads).16 Scale bars: 600 nm in B and 450 nm in C. (D–E) Cardiac muscle is arranged in a highly aligned and layered fashion, facilitated with collagen fiber orientation17; (D) SEM image showing 3 orthogonal surfaces (TN: tangential, AT: axial-transmural, TR:transverse) of a left ventricular midanterior midwall of a canine heart tissue shows stacked layers of cardiomyocytes, each layer composed of nearly four cardiomyocytes, with branches between layers being one to two cardiomyocytes thick; (E) Enlarged tangential image shows layered organization within heart, with rare branching (arrowhead), with space between layers composed of extensive connective tissue network, and long collagen fibers running between muscle cells and inserting into the surrounding connective tissue network. Scale bar: 100 μm. (F) SEM image showing perimysial collagen cables in EDL muscle supporting muscle fibers longitudinally. Scale bar: 1 μm. (G) SEM image showing the fine structures in the endomysium facilitating embedding of cardiac muscle fibers.18 Scale bar: 10 μm.
In contrast, in hierarchically organized tissues like the heart, there are aligned fibers forming a natural scaffold over which the cells adhere, align and form hierarchical organization to create the desired structure of the tissue. For example, in the canine heart, cardiomyocytes arrange in a layered organization, with each layer consisting of nearly four cells (Fig. 2D), with rare inter-layered cellular connections.17 Collagen fibers run in parallel to the axes of the cells possibly support the anisotropic and hierarchical structure of the heart tissue (Fig. 2E). The ultrastructural analysis of muscle matrix shows aligned collagen fibers bundled in thick cables (Fig. 2F), that both guide the anisotropic organization of the tissue, while endomysium surrounds each cardiomyocyte providing the matrix support in a very organized fashion (Fig. 2G).18
In the other end of the spectrum of organization, as discussed earlier, are basement membranes that show a more random mesh of collagen fibers over a sheet of collagen, as evidenced in basement membrane of rat tongue muscles. Many ECM proteins form structures with feature size ranging from tens of nano-meters to several hundred microns. While capable of being remodeled, matrix topography acts on cells in long time scales, and over long distances. Matrix nanotopography is now being recognized to be of fundamental importance in determining cell shape, regulating cell-cell interactions, and maintaining structure and function of the tissue.1,2 For example, the elongated cell shape of cardiomyocytes and arrangement of millions of cells in a highly aligned fashion syncytially coupled to each other is regulated by the underlying ECM nanotopography in a size-dependent manner.19,20 Similarly, matrix nanotopography has been recognized to play significant roles in regulating axon guidance, cancer cell migration, wound healing response of epithelial cells, gap junction formation and action potential propagation of cardiomyocytes, arrangement of endothelial cells in blood vessel, T cell migration, cell proliferation and differentiation of cardiac progenitors and in regulating several other important functions in various tissues.1,2,6,21,22 Though many instances in different tissue types have been found where topography regulates cellular phenotypes, however, the mechanisms by which these cues guide cell behavior is only now beginning to be understood.
Matrix Nanotopography and Cells Influence Each Other in a Reciprocative and Instructive Manner
A significant reason for the lack of research and appreciation of the role of nanotopography in the cell biology community stems from the lack of tools to modulate the microenvironment.23-25 In particular, control of nanotopography of the substrate in which cells are grown has been difficult and sometimes prohibitively expensive.26 Many new techniques are available with varying degrees of control over the creation of nanotopographical cues allowing biologists to recapitulate the nanotopography of desired tissue in a highly controlled and reproducible fashion.27,28 We recapitulated the highly anisotropic collagen fiber bundles seen in heart muscle (Fig. 3A) using capillary force lithography (CFL) to form a scalable polymeric nanogrooved pattern (Fig. 3B–C).20 Engineering techniques used to study topographical cues can directly provide guidance cues on which cells can track. Metastatic cancer cells can migrate, as well as remodel the collagen fibrils (Fig. 3D),21 while even immune T-cells respond to the underlying nanotopography in combination with biochemical ligand based stimulation (Fig. 3E).6 Nanotopographical features also structurally and functionally mature cardiac progenitors,29 and skeletal muscle satellite cells.30 We also found that adult mesenchymal cells could be directed toward adipogenesis or osteogenesis depending on the features of nanotopographical cues, facilitating creation of a mosaic tissue with spatial control of cell lineage.28 Interestingly, nanoscale features influences cellular phenotypes differently if they are disordered, as compared with organized and directional.31 It is hoped that more research will not only increase the appreciation of nanotopography as a fundamental biological cue, but would also encourage topographically defined substrates as standard cell culture platforms for cell culture to design more physiological contexts for experiments.28
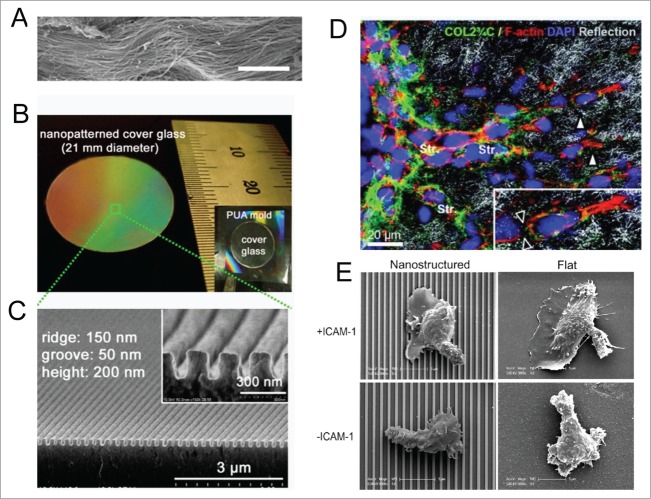
Figure 3.
Cells actively sense and respond to engineered nanotopographical cues. (A) SEM images of ex vivo myocardium of adult rat heart shows aligned collagen fibers running in dense bundles for long distances. Scale bar: 10 μm. (B) Photograph of a large-area anisotropically nanofabricated substratum (ANFS) on a glass coverslip mimicking the collagen fibers in A. (C) Cross-sectional SEM image of the ANFS in B. Reproduced with permission from the National Academy of Sciences. (D) Remodeling of ECM structures by motile HT1080 fibrosarcoma cells. Transition from individual to collective invasion is displayed in 3D spheroids cultured within a 3D collagen lattice. Single cells (white arrowheads) generate small proteolytic tracks (black arrowheads in inset) that become further remodeled and widened by solid strands of multiple cells (Str). The box marks the area magnified in the inset (bottom right); Reprinted with permission from the American Association for Cancer Research. (E) Representative SEM images of T cells on nanostructured (left column) and flat (right column) surfaces in the presence (upper row) and absence (lower row) of ICAM-1; Reprinted with permission from the Journal of Immunology.
While cells respond to nanotopography, they are also responsible for laying down these matrices. In most connective tissues, fibroblasts secrete ECM molecules in the form of nanofibers, sheets, or in form of other nanotopographical features.2 Disrepair of tissue matrix in response to a wound or insult results in recruitment of neighboring fibroblasts that remodel the matrix and lay down new ECM molecules that assume the shape defined by their biochemistry.32 Similarly, metastatic cancer cells remodel the matrix to facilitate movement across the barrier imposed by the existing matrix topography, which may not be conducive to migration.33 Therefore, there exists a constant interplay of the ECM nanotopography on cell behavior, and cell induced remodeling of the topographical state of the ECM.
This interplay between cells and the matrix they reside in is of fundamental importance in regulating tissue structure and function. While it is commonplace to consider ECM as a static cue that defines cell physiology, in certain tissues and in certain conditions this assumption may not hold true. For example, bone is constantly remodeled by osteoclasts that convert pressure and force cues into directional arrangement of the bone by remodeling ECM matrix by migrating in the direction of force.31 Similarly, a heart attack results in massive remodeling of the heart ECM and collagen deposition resulting in a stiffer heart with a denser ECM nanotopographical cue than in a normal heart.19,29 These phenomena result in alteration of structure at the tissue and organ level, commonly affecting and contributing to change in organ function. Lack of in vitro tools to regulate nanotopography dynamically has resulted in little or no research in understanding the role of dynamic presentation of nanotopographical, or in general mechanical, cues to the cells. This presents an exciting new area for material scientists and cell biologists to develop tools to dynamically regulate ECM mechanics and topography.
Signaling Mechanisms in Nanotopography Sensing and Response
ECM topography is a complex cue. Cells can sense mechanical forces from the ECM in multiple overlapping ways, rendering it difficult to ascertain the effect of one aspect of topography from the other, and one mechanical cue from the other. For example, cardiac cells cultured on nanogrooves mimicking cardiac ECM exhibited an enveloping effect on the grooves in a nanogroove dimensional dependent manner. At the very least, a greater amount of enveloping can result in greater cell-ECM interaction, thereby increasing the “dosage” of ECM molecules to the cells, as evidenced in cardiomyocytes cultured on nanogrooves of different feature sizes20 (Fig. 4A). Even the amoeba Dictyostelium discoideum that does not express integrin based receptors, exhibits different extent of contact guidance on nanotopographical surface, suggesting that nanotopography may just be presenting a pseudo 3D surface for cells to adhere to, thereby increasing the available surface area for cell adhesion.34 We have previously speculated that nanotopography as a regulator of dosage of cell-ECM interaction due to its intrinsic 3D nature of cell-ECM interface could be a causative factor in regulating cardiac tissue structure and function, including regulating expression of cell-cell gap junction protein Connexin-43, and conduction velocity (Fig. 4B). Another mechanism by which cells can sense nanotopography is employed when cells pull the nanofeatures while developing focal adhesions centered around their adhesion loci.20 This could result in rendering “elasticity” to the nanofeatured substrata which may be absent from the featureless flat substrata created from the same material.35 Third, the enveloping of the cell membrane around nanofeatures can result in increased intramembrane tension and rearrangement of cortical cytoskeleton, again, influencing the cell in a different way.36 Recent report indicates that nanotopography modulates protein adsorption, thereby regulating fibrotic response of cells,37 partially supporting the hypothesis that nanotopography may be mediating the dosage of protein or ECM presentation to the adhered cells.
Figure 4.
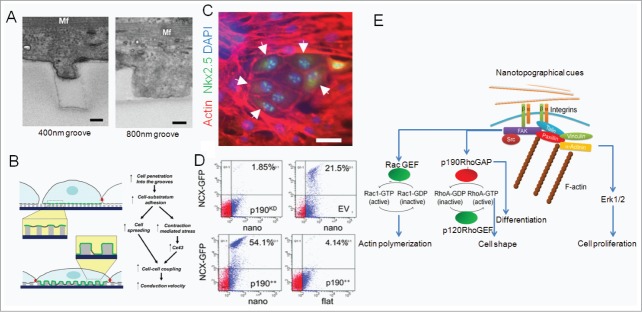
Matrix nanotopography regulates cell functions by regulating cell-matrix dosage and direct regulation of signaling pathways. (A) Differential degree of primary cardiac cell protrusion into a 400-nm-wide (left) and an 800-nm-wide (right) groove indicate a possible mechanism by which cells actively differentiate between different features of nanocues20; Mf: myofilament. (B) Model of the sensitivity of structural and functional properties of the cardiac constructs to the details of the underlying nanostructured substrata; A and B reprinted with permission from the National Academy of Sciences. (C–D) Nanotopography mimicking the native tissue can regulate stem cell phenotypes including fate by regulating signaling molecules that activate transcription factors29; (C) Rat cardiosphere-derived cells (CDCs) cultured on aligned topography that mimics the heart matrix (right) form in vitro cardiac niche with cardiac progenitor specific transcription factor Nkx2.5+ cells surrounded by undifferentiated CDCs after 14 d of culture on nanogrooves; red: actin, green: Nkx2.5, blue: DAPI. Scale bar: 50 μm. (D) GFP expression in CDCs lentivirally transduced with NCX- GFP and p190++ (p190++/NCX-GFP), sv40 empty vector (EV/NCX-GFP), p190RhoGAP shRNA (p190KD/NCX-GFP) and co-cultured with neonatal rat ventricular myocytes show that p190RhoGAP overexpression increases cardiomyogenesis, while p190RhoGAP silencing decreases cardiomyogenesis; Reprinted with permission from the Royal Society of Chemistry. (E) Schematic showing the major downstream signaling pathways that nanotopographical features can regulate, thereby regulating cellular phenotypes.
Further, matrix nanotopography may act on cells in a non-exclusive manner with other biochemical and mechanical cues provided by the ECM.38,39 Similarly, other mechanical cues can non-exclusively regulate cellular phenotypes with nanotopographical cues. Organization of nanotopographical cues, while keeping their “dosage” constant, has been shown to affect cellular phenotypes including stem cell differentiation differently compared with randomly organized cues.31,40,41 Rigidity range varies substantially in different tissues, and both topography and substrate elasticity are first sensed by the cells via integrin receptors, feeding into similar mechanosensory signaling pathways.42 ECM binding to the extracellular domains of integrins has been studied in detail. On the cytoplasmic side, integrins do not contact the cytoskeleton directly but via intermediate proteins. Hundreds of such proteins have been implicated in connecting integrins with cytoskeleton.43 Among several of these proteins that can bind to β subunit tail of integrins and connect integrin to cytoskeletal proteins (including actin, intermediate filaments and tubulin proteins), only a few of them have been identified that can detect variable tension/mechanical signals presented by nanotopographical cues. Such mechanosensitive proteins include p130Cas, talin-vinculin system, α-catenin, and mechanosensitive ion channels.44-47 These primary mechanosensors feed into multiple signaling pathways, including those regulated by Rho family GTPases, Hippo pathway proteins, and tyrosine kinases.5,48,56 Rho Pathway consists of ∼20 Rho GTPases in mammalian cells, three of which have been studied extensively; RhoA, Rac1, and Cdc-42. These GTPases are activated by guanine nucleotide exchange factors (GEFs) that directly or indirectly sense adhesion, rigidity and availability of ECM binding sites, and modify cell behavior by activating downstream targets but their role in increasing/decreasing the surface binding protein expression, known as ‘inside-out’ signaling has also been suggested. The role of small GTPases in integration of mechanotransduction signals, including nanotopographical cues into meaningful biological behavior like polarity, motility, proliferation, and differentiation is still being explored through interaction with other signaling modules, prominently the PI3K/AKT and tyrosine kinase pathway.49,50 p190RhoGAP is one of the proteins which connects integrin linked signaling to Rho pathway by regulating RhoA and Rac1 activity and cell spreading/motility by connecting mechanical cues from ECM.29 Interestingly, p120-catenin (delta catenin) which regulates canonical Wnt signaling is also connected to Rho GTPases, and p190RhoGAP is one of crucial p120-Catenin adaptor in integrin mediated or cell-cell mediated activation of p120, suggesting a possible role of these proteins in developmental pathways.51-53 p190RhoGAP has emerged as a promiscuous molecule that interlinks various seemingly disparate signaling pathways, acting as a master regulator of extracellular mechanical and topographical signals to control transcriptionally controlled cellular phenotypes, e.g., proliferation, differentiation, cell shape, cell-cell interaction, tissue morphogenesis, and other functions.41,42 For example, nanogrooves that mimic the native heart ECM result in a cardiac progenitor cell type currently under clinical trials,54 CDCs to form cardiac niche like structures with groups of rounded cells with Nkx2.5+ nuclei surrounded by non-differentiated CDCs (Fig. 4C).29
A crucial question is how different ECM generated mechanical cues influence cells. Do they act in mutual exclusion via different mechanosensory networks, or do they work in parallel with each other so that one mechanical cue could be supplanted with another. The truth seems to be somewhere in between. Our previous studies indicate that nanotopography and rigidity of substratum for similar cell type could act via activating similar molecular machinery, but in different manner resulting in different phenotypic outcomes.29 When multipotent CDCs, are cultured on substrata mimicking the rigidity of myocardium, they give rise to endothelial cells while same cells show a more aligned phenotype on nanogrooves mimicking the heart ECM, eventually giving rise to cardiomyocytes (Fig. 3C–D).29 However, both these mechanical cues were fed into RhoA signaling in an integrin dependent manner and resulted in gradual downregulation of p190RhoGAP in the case of MRS, and upregulation of p190RhoGAP in the case of nanogrooves41 (Fig. 4D). It was also found that nanogrooves promoted cardiomyogenesis via p190RhoGAP upregulation41 (Fig. 4D). Such evidence supports the hypothesis that nanotopography and other ECM mediated signaling may be closely interrelated, and acting in similar or dissimilar ways in different contexts.
There is also another aspect of nanotopographical features that is overlooked, and could be advantageous in balancing the need to create biomimetic surfaces for cell culture that are also conducive for experimental observations. Nanotopographical substrates also provide quasi three-dimensional (3D) substrata where cell has a chance to spread in a different dimension than the two-dimension (2D) platforms commonly used for cell culture. This is important, since 2D substrates probably limit our understanding of how cells actually interact with their mechanical 3D microenvironment in vivo, thereby regulating their migration, polarity, differentiation, and even viability.3,4 Recently, the proteins involved in sensing different mechanical cues have also been implicated in regulation of diverse biological behavior including migration, polarity, proliferation, differentiation, apoptosis and regulation of both upstream and downstream signaling pathways.55,56 It is possible that similar molecular machinery is also involved in topography sensing. Nanotopographical cues, therefore, are sensed by the cells essentially by integrin receptors that recognize the specific ECM motifs to which they bind and activate downstream signaling pathways. The difference comes in the nature of these contacts, which, owing to the spatial and inhomogeneous aspects of these cues result in activation of integrin mediated downstream signaling in a non-homogeneous fashion within the cells. Depending on the type of cue, and the type of cells, therefore topographical cues can have different effects than just the presentation of ECM molecules, and can regulate cell shape, direction of movement, proliferation, fate, and other phenotypes differently from homogeneous ECM cues (Fig. 4E).
Perspectives
Topography of the environment cells reside in was long considered as a passive fact, an interesting curiosity in the form of its diversity but not important as an active signaling cue. Recent developments of novel tools from biomaterials, electrical engineering, and biotechnology and cell biology have brought to the biologists’ bench an array of tools to precisely control the topographical cues, while also presenting sophisticated methods to allow topographical perturbation of cells.23 These significant challenges of creating nanoscale features in inexpensive large surface area,28 and long-term maintenance of stem cells57 appear to have been reasonably dealt with. In addition, many studies report methods to combine other biological cues with nanotopographical cues in a combinatorial manner, making it possible to study the effect of nanoscale cues in combination with other physiological stimuli on cell behavior.58-60 The new knowledge has been revealing. It is now established that topography of ECM itself is a potent biological cue that activates various signaling pathways and can regulate transcription, and consequently many cellular phenotypes. However, this field is still young, and there is still a widespread lack of appreciation on the existence and importance of these cues. Cells exist in context, and the context of cells is complex: a combinatorial cue that is biochemical, physical, and topographical in nature.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
D. H. Kim thanks the Department of Bioengineering at the University of Washington for the new faculty startup fund. The authors thank Dr. Jonathan Cooper and Dr. Paolo Provenzano for critical reading and valuable comments.
Funding
This work was also supported by an American Heart Association (AHA) Scientist Development Grant (13SDG14560076), a Muscular Dystrophy Association (MDA) Research Grant (MDA 255907), and the FHCRC/UW Cancer Consortium Cancer Center Support Grant of the National Institutes of Health under Award Number P30 CA015704 (to D.H. Kim). This work was supported by International Collaborative R&D Program through KIAT grant funded by the MOTIE (N0000894).
References
- 1. Kim DH, Provenzano PP, Smith CL, Levchenko A. Matrix nanotopography as a regulator of cell function. J Cell Biol 2012; 197:351-60; PMID:22547406; http://dx.doi.org/ 10.1083/jcb.201108062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dvir T, Timko BP, Kohane DS, Langer R. Nanotechnological strategies for engineering complex tissues. Nat Nanotechnol 2011; 6:13-22; PMID:21151110; http://dx.doi.org/ 10.1038/nnano.2010.246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell 2006; 126:677-89; PMID:16923388; http://dx.doi.org/ 10.1016/j.cell.2006.06.044 [DOI] [PubMed] [Google Scholar]
- 4. Janmey PA, Miller RT. Mechanisms of mechanical signaling in development and disease. J Cell Sci 2011; 124:9-18; PMID:21172819; http://dx.doi.org/ 10.1242/jcs.071001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kim DH, Han K, Gupta K, Kwon KW, Suh KY, Levchenko A. Mechanosensitivity of fibroblast cell shape and movement to anisotropic substratum topography gradients. Biomaterials 2009; 30:5433-44; PMID:19595452; http://dx.doi.org/ 10.1016/j.biomaterials.2009.06.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kwon KW, Park H, Song KH, Choi JC, Ahn H, Park MJ, Suh KY, Doh J. Nanotopography-guided migration of T cells. J Immunol 2012; 189:2266-73; PMID:22844118; http://dx.doi.org/ 10.4049/jimmunol.1102273 [DOI] [PubMed] [Google Scholar]
- 7. Kiang JD, Wen JH, del Álamo JC, Engler AJ. Dynamic and reversible surface topography influences cell morphology. J Biomed Mater Res A 2013; 101A:2313-21; PMID:23355509; http://dx.doi.org/ 10.1002/jbm.a.34543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wieringa P, Tonazzini I, Micera S, Cecchini M. Nanotopography induced contact guidance of the F11 cell line during neuronal differentiation: a neuronal model cell line for tissue scaffold development. Nanotechnology 2012; 23:275102; PMID:22710035; http://dx.doi.org/ 10.1088/0957-4484/23/27/275102 [DOI] [PubMed] [Google Scholar]
- 9. Yang HS, Ieronimakis N, Tsui J, Kim HN, Suh KY, Reyes M, Kim DH. Nanopatterned muscle cell patches for enhanced myogenesis and dystrophin expression in a mouse model of muscular dystrophy. Biomaterials 2014; 35(5):1476-86. [DOI] [PubMed] [Google Scholar]
- 10. Dupont S, Morsut L, Aragona M, Enzo E, Giulitti S, Cordenonsi M, Zanconato F, Le Digabel J, Forcato M, Bicciato S, et al. Role of YAP/TAZ in mechanotransduction. Nature 2011; 474:179-83; PMID:21654799; http://dx.doi.org/ 10.1038/nature10137 [DOI] [PubMed] [Google Scholar]
- 11. Dickinson RB, Guido S, Tranquillo RT. Biased cell migration of fibroblasts exhibiting contact guidance in oriented collagen gels. Ann Biomed Eng 1994; 22:342-56; PMID:7998680; http://dx.doi.org/ 10.1007/BF02368241 [DOI] [PubMed] [Google Scholar]
- 12. Provenzano PP, Inman DR, Eliceiri KW, Trier SM, Keely PJ. Contact guidance mediated three-dimensional cell migration is regulated by Rho/ROCK-dependent matrix reorganization. Biophys J 2008; 95:5374-84; PMID:18775961; http://dx.doi.org/ 10.1529/biophysj.108.133116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Perentes JY, McKee TD, Ley CD, Mathiew H, Dawson M, Padera TP, Munn LL, Jain RK, Boucher Y. In vivo imaging of extracellular matrix remodeling by tumor-associated fibroblasts. Nat Methods 2009; 6:143-5; PMID:19151720; http://dx.doi.org/ 10.1038/nmeth.1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. House M, Kaplan DL, Socrate S. Relationships between mechanical properties and extracellular matrix constituents of the cervical stroma during pregnancy. Semin Perinatol 2009; 33:300-7; PMID:19796726; http://dx.doi.org/ 10.1053/j.semperi.2009.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bedossa P. The cellular origin of extracellular matrix constituents. J Hepatol 1993; 19:1-3; PMID:8301029; http://dx.doi.org/ 10.1016/S0168-8278(05)80168-0 [DOI] [PubMed] [Google Scholar]
- 16. Provenzano P. P. & Vanderby R., Jr. Collagen fibril morphology and organization: implications for force transmission in ligament and tendon. Matrix biology: journal of the International Society for Matrix Biology 25, 71-84, (2006). [DOI] [PubMed] [Google Scholar]
- 17. LeGrice IJ, Smaill BH, Chai LZ, Edgar SG, Gavin JB, Hunter PJ. Laminar structure of the heart: ventricular myocyte arrangement and connective tissue architecture in the dog. Am J Physiol 1995; 269:H571-82; PMID:7653621 [DOI] [PubMed] [Google Scholar]
- 18. Kanzaki Y, Terasaki F, Okabe M, Fujita S, Katashima T, Otsuka K, Ishizaka N. Three-dimensional architecture of cardiomyocytes and connective tissue in human heart revealed by scanning electron microscopy. Circulation 2010; 122:1973-4; PMID:21060087; http://dx.doi.org/ 10.1161/CIRCULATIONAHA.110.979815 [DOI] [PubMed] [Google Scholar]
- 19. Fleischer S, Dvir T. Tissue engineering on the nanoscale: lessons from the heart. Curr Opin Biotechnol 2013; 24(4):664-71; PMID:23142543 [DOI] [PubMed] [Google Scholar]
- 20. Kim DH, Lipke EA, Kim P, Cheong R, Thompson S, Delannoy M, Suh KY, Tung L, Levchenko A. Nanoscale cues regulate the structure and function of macroscopic cardiac tissue constructs. Proc Natl Acad Sci U S A 2010; 107:565-70; PMID:20018748; http://dx.doi.org/ 10.1073/pnas.0906504107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Friedl P, Wolf K. Tube travel: the role of proteases in individual and collective cancer cell invasion. Cancer Res 2008; 68:7247-9; PMID:18794108; http://dx.doi.org/ 10.1158/0008-5472.CAN-08-0784 [DOI] [PubMed] [Google Scholar]
- 22. Lee MR, Kwon KW, Jung H, Kim HN, Suh KY, Kim K, Kim KS. Direct differentiation of human embryonic stem cells into selective neurons on nanoscale ridge/groove pattern arrays. Biomaterials 2010; 31:4360-6; PMID:20202681; http://dx.doi.org/ 10.1016/j.biom-aterials.2010.02.012 [DOI] [PubMed] [Google Scholar]
- 23. Kshitiz , Kim DH, Beebe DJ, Levchenko A. Micro- and nanoengineering for stem cell biology: the promise with a caution. Trends Biotechnol 2011; 29:399-408; PMID:21549437; http://dx.doi.org/ 10.1016/j.tibtech.2011.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gupta K, Kim DH, Ellison D, Smith C, Kundu A, Tuan J, Suh KY, Levchenko A. Lab-on-a-chip devices as an emerging platform for stem cell biology. Lab Chip 2010; 10:2019-31; PMID:20556297; http://dx.doi.org/ 10.1039/c004689b [DOI] [PubMed] [Google Scholar]
- 25. Gupta K, Kim DH, Ellison D, Smith C, Levchenko A. in Stem Cell Biology and Regenerative Medicine (eds Appasani Krishnarao. & Appasani Ragu K.) 483-498 (Springer, 2011). [Google Scholar]
- 26. Kim HN, Jiao A, Hwang NS, Kim MS, Kang H, Kim DH, Suh KY. Nanotopography-guided tissue engineering and regenerative medicine. Adv Drug Deliv Rev 2013; 65:536-58; PMID:22921841; http://dx.doi.org/ 10.1016/j.addr.2012.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kim HN, Kang DH, Kim MS, Jiao A, Kim DH, Suh KY. Patterning methods for polymers in cell and tissue engineering. Ann Biomed Eng 2012; 40:1339-55; PMID:22258887; http://dx.doi.org/ 10.1007/s10439-012-0510-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ahn EH, Kim YH. Kshitiz An, S., Lee S. W.,Kwak M., Suh K. Y., Kim D. H. & Levchenko A. Spatial control of adult stem cell fate using nanotopographic cues. Biomaterials 2014; 35(8):2401-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim D. H., Kshitiz , Smith R. R., Kim P., Ahn E. H., Kim H. N., Marban E., Suh K. Y. & Levchenko A. Nanopatterned cardiac cell patches promote stem cell niche formation and myocardial regeneration. Integrative biology: quantitative biosciences from nano to macro 4, 1019-1033, (2012). [DOI] [PubMed] [Google Scholar]
- 30. Yang HS, Ieronimakis N, Tsui JH, Kim HN, Suh KY, Reyes M, Kim DH. Nanopatterned muscle cell patches for enhanced myogenesis and dystrophin expression in a mouse model of muscular dystrophy. Biomaterials 2014; 35(5): 1478-86; PMID:24290810 [DOI] [PubMed] [Google Scholar]
- 31. Dalby MJ, Gadegaard N, Tare R, Andar A, Riehle MO, Herzyk P, Wilkinson CD, Oreffo RO. The control of human mesenchymal cell differentiation using nanoscale symmetry and disorder. Nat Mater 2007; 6:997-1003; PMID:17891143; http://dx.doi.org/ 10.1038/nmat2013 [DOI] [PubMed] [Google Scholar]
- 32. Petrie RJ, Doyle AD, Yamada KM. Random versus directionally persistent cell migration. Nat Rev Mol Cell Biol 2009; 10:538-49; PMID:19603038; http://dx.doi.org/ 10.1038/nrm2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sahai E. Illuminating the metastatic process. Nat Rev Cancer 2007; 7:737-49; PMID:17891189; http://dx.doi.org/ 10.1038/nrc2229 [DOI] [PubMed] [Google Scholar]
- 34. Driscoll MK, Sun X, Guven C, Fourkas JT, Losert W. Cellular contact guidance through dynamic sensing of nanotopography. ACS Nano 2014; 8:3546-55; PMID:24649900; http://dx.doi.org/ 10.1021/nn406637c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hansen JC, Lim JY, Xu LC, Siedlecki CA, Mauger DT, Donahue HJ. Effect of surface nanoscale topography on elastic modulus of individual osteoblastic cells as determined by atomic force microscopy. J Biomech 2007; 40:2865-71; PMID:17467715; http://dx.doi.org/ 10.1016/j.jbiomech.2007.03.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Seo CH, Jeong H, Furukawa KS, Suzuki Y, Ushida T. The switching of focal adhesion maturation sites and actin filament activation for MSCs by topography of well-defined micropatterned surfaces. Biomaterials 2013; 34:1764-71; PMID:23219606; http://dx.doi.org/ 10.1016/j.biomaterials.2012.11.031 [DOI] [PubMed] [Google Scholar]
- 37. Kam KR, Walsh LA, Bock SM, Ollerenshaw JD, Ross RF, Desai TA. The effect of nanotopography on modulating protein adsorption and the fibrotic response. Tissue Eng Part A 2014; 20:130-8; PMID:23914986; http://dx.doi.org/ 10.1089/ten.tea.2012.0772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kolind K, Leong KW, Besenbacher F, Foss M. Guidance of stem cell fate on 2D patterned surfaces. Biomaterials 2012; 33:6626-33; PMID:22748769; http://dx.doi.org/ 10.1016/j.biomaterials.2012.05.070 [DOI] [PubMed] [Google Scholar]
- 39. Kulangara K, Yang Y, Yang J, Leong KW. Nanotopography as modulator of human mesenchymal stem cell function. Biomaterials 2012; 33:4998-5003; PMID:22516607; http://dx.doi.org/ 10.1016/j.biomaterials.2012.03.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chew SY, Mi R, Hoke A, Leong KW. The effect of the alignment of electrospun fibrous scaffolds on Schwann cell maturation. Biomaterials 2008; 29:653-61; PMID:17983651; http://dx.doi.org/ 10.1016/j.biom-aterials.2007.10.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kshitiz , Afzal J, Kim D. H. & Levchenko A. Mechanotransduction via p190RhoGAP regulates a switch between cardiomyogenic and endothelial lineages in adult cardiac progenitors. Stem Cells 2014; 32(8): 1999-2007; http://dx.doi.org/ 10.1002/stem.1700 [DOI] [PubMed] [Google Scholar]
- 42. Kshitiz , Hubbi ME, Ahn EH, Downey J, Afzal J, Kim DH, Rey S, Chang C, Kundu A, Semenza GL, et al. Matrix rigidity controls endothelial differentiation and morphogenesis of cardiac precursors. Sci Signal 2012; 5(227):ra41-41; PMID:22669846; http://dx.doi.org/ 10.1126/scisignal.2003002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zaidel-Bar R, Itzkovitz S, Ma’ayan A, Iyengar R, Geiger B. Functional atlas of the integrin adhesome. Nat Cell Biol 2007; 9:858-67; PMID:17671451; http://dx.doi.org/ 10.1038/ncb0807-858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sawada Y, Tamada M, Dubin-Thaler BJ, Cherniavskaya O, Sakai R, Tanaka S, Sheetz MP. Force sensing by mechanical extension of the Src family kinase substrate p130Cas. Cell 2006; 127:1015-26; PMID:17129785; http://dx.doi.org/ 10.1016/j.cell.2006.09.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. del Rio A, Perez-Jimenez R, Liu R, Roca-Cusachs P, Fernandez JM, Sheetz MP. Stretching single talin rod molecules activates vinculin binding. Science 2009; 323:638-41; PMID:19179532; http://dx.doi.org/ 10.1126/science.1162912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yonemura S, Wada Y, Watanabe T, Nagafuchi A, Shibata M. alpha-Catenin as a tension transducer that induces adherens junction development. Nat Cell Biol 2010; 12:533-42; PMID:20453849; http://dx.doi.org/ 10.1038/ncb2055 [DOI] [PubMed] [Google Scholar]
- 47. Árnadóttir J, Chalfie M. Eukaryotic mechanosensitive channels. Annu Rev Biophys 2010; 39:111-37; PMID:20192782; http://dx.doi.org/ 10.1146/annurev.biophys.37.032807.125836 [DOI] [PubMed] [Google Scholar]
- 48. Zhang H, Landmann F, Zahreddine H, Rodriguez D, Koch M, Labouesse M. A tension-induced mechanotransduction pathway promotes epithelial morphogenesis. Nature 2011; 471:99-103; PMID:21368832; http://dx.doi.org/ 10.1038/nature09765 [DOI] [PubMed] [Google Scholar]
- 49. Higuchi M, Onishi K, Kikuchi C, Gotoh Y. Scaffolding function of PAK in the PDK1-Akt pathway. Nat Cell Biol 2008; 10:1356-64; PMID:18931661; http://dx.doi.org/ 10.1038/ncb1795 [DOI] [PubMed] [Google Scholar]
- 50. Arias-Romero LE, Villamar-Cruz O, Pacheco A, Kosoff R, Huang M, Muthuswamy SK, Chernoff J. A Rac-Pak signaling pathway is essential for ErbB2-mediated transformation of human breast epithelial cancer cells. Oncogene 2010; 29:5839-49; PMID:20711231; http://dx.doi.org/ 10.1038/onc.2010.318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wildenberg GA, Dohn MR, Carnahan RH, Davis MA, Lobdell NA, Settleman J, Reynolds AB. p120-catenin and p190RhoGAP regulate cell-cell adhesion by coordinating antagonism between Rac and Rho. Cell 2006; 127:1027-39; PMID:17129786; http://dx.doi.org/ 10.1016/j.cell.2006.09.046 [DOI] [PubMed] [Google Scholar]
- 52. Bradley WD, Hernández SE, Settleman J, Koleske AJ. Integrin signaling through Arg activates p190RhoGAP by promoting its binding to p120RasGAP and recruitment to the membrane. Mol Biol Cell 2006; 17:4827-36; PMID:16971514; http://dx.doi.org/ 10.1091/mbc.E06-02-0132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Pieters T, van Hengel J, van Roy F. Functions of p120ctn in development and disease. Front Biosci (Landmark Ed) 2012; 17:760-83; PMID:22201773; http://dx.doi.org/ 10.2741/3956 [DOI] [PubMed] [Google Scholar]
- 54. Makkar RR, Smith RR, Cheng K, Malliaras K, Thomson LE, Berman D, Czer LS, Marbán L, Mendizabal A, Johnston PV, et al. Intracoronary cardiosphere-derived cells for heart regeneration after myocardial infarction (CADUCEUS): a prospective, randomised phase 1 trial. Lancet 2012; 379:895-904; PMID:22336189; http://dx.doi.org/ 10.1016/S0140-6736(12)60195-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Holle AW, Engler AJ. More than a feeling: discovering, understanding, and influencing mechanosensing pathways. Curr Opin Biotechnol 2011; 22:648-54; PMID:21536426; http://dx.doi.org/ 10.1016/j.copb-io.2011.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Jaffe AB, Hall A. Rho GTPases: biochemistry and biology. Annu Rev Cell Dev Biol 2005; 21:247-69; PMID:16212495; http://dx.doi.org/ 10.1146/annurev.cellbio.21.020604.150721 [DOI] [PubMed] [Google Scholar]
- 57. McMurray RJ, Gadegaard N, Tsimbouri PM, Burgess KV, McNamara LE, Tare R, Murawski K, Kingham E, Oreffo RO, Dalby MJ. Nanoscale surfaces for the long-term maintenance of mesenchymal stem cell phenotype and multipotency. Nat Mater 2011; 10:637-44; PMID:21765399; http://dx.doi.org/ 10.1038/nmat3058 [DOI] [PubMed] [Google Scholar]
- 58. Yang Y, Kulangara K, Lam RT, Dharmawan R, Leong KW. Effects of topographical and mechanical property alterations induced by oxygen plasma modification on stem cell behavior. ACS Nano 2012; 6:8591-8; PMID:22970773; http://dx.doi.org/ 10.1021/nn301713d [DOI] [PubMed] [Google Scholar]
- 59. Zhao F., Veldhuis J. J., Duan Y., Yang Y., Christoforou N., Ma T. & Leong K. W. Low oxygen tension and synthetic nanogratings improve the uniformity and stemness of human mesenchymal stem cell layer. Molecular therapy: the journal of the American Society of Gene Therapy 18, 1010-1018, (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Koo LY, Irvine DJ, Mayes AM, Lauffenburger DA, Griffith LG. Co-regulation of cell adhesion by nanoscale RGD organization and mechanical stimulus. J Cell Sci 2002; 115:1423-33; PMID:11896190 [DOI] [PubMed] [Google Scholar]
- 61. Liliensiek SJ, Campbell S, Nealey PF, Murphy CJ. The scale of substratum topographic features modulates proliferation of corneal epithelial cells and corneal fibroblasts. J Biomed Mater Res A 2006; 79A:185-92; PMID:16817223; http://dx.doi.org/ 10.1002/jbm.a.30744 [DOI] [PMC free article] [PubMed] [Google Scholar]