Abstract
Teleost v1r-related ora genes constitute a small and highly conserved olfactory receptor gene family, and their direct orthologs are present in lineages as distant as cartilaginous fishes. Recently, the first member of the ora gene family was deorphanized. ORA1 detects p-hydroxyphenylacetic acid with high sensitivity and specificity. This compound elicits olfactory-mediated oviposition behavior in adult zebrafish mating pairs, suggesting a potential function as a reproductive pheromone for pHPAA itself or a related substance. This association of an odor and its cognate receptor with an oviposition response may provide a molecular basis for studying neural circuits involved in fish reproduction.
Keywords: calcium imaging, heterologous expression, oviposition, pheromone, tyrosine, V1R-like, zebrafish
Abbreviations
- V1Rs
Vomeronasal receptor, type 1
- V2Rs
Vomeronasal receptor, type 2
- VSNs
vomeronasal sensory neurons
- ora
olfactory receptor class A-related
- pHPAA
p-hydroxyphenylacetic acid
- EC50
half-maximal response
Pheromones are chemical signals produced by a species and recognized by the olfactory system of the conspecific to mediate behavioral functions such as mating preferences and individual recognition. In mammals, vomeronasal sensory neurons (VSNs) express members of 2 large olfactory receptor gene families, v1r and v2r, which are thought to mediate pheromone detection.1 V2Rs recognize peptides, whereas V1R ligands are found among low molecular weight molecules, such as steroids.1-3 The mammalian v1r receptor family is large, with over 100 genes in rodent species, and undergoes frequent species-specific expansions.4 Interestingly, all mammalian v1r genes are monophyletic with only 2 teleost ora genes, ora1 and ora2.5 In drastic contrast to the rapidly evolving mammalian v1r genes, the teleost ora gene family is highly conserved between all teleost species analyzed, and consists of the same 6 genes, with an occasional gene loss.5 Even in cartilaginous fishes some direct orthologs are observed.6 Thus it was unclear, whether teleost ora genes, despite their different evolutionary dynamics, might also have a pheromonal function like their mammalian counterparts. Two research groups teamed up a while ago to attempt deorphanization of teleost ora genes, the Korsching lab in Cologne and the Meyerhof lab in Potsdam. Recently they reported the deorphanization of ORA1, which they found to detect p-hydroxyphenylacetic acid (pHPAA) with high sensitivity and specificity.7 Moreover, behavior analysis suggested that pHPAA induces olfactory-mediated oviposition behavior in adult zebrafish pairs7, which implies its possible function as a putative fish pheromone.
A Convoluted Path toward Identification of an Olfactory Ligand
The search for an ORA1 ligand turned out to be quite the detective story. In the end, a contaminant of the initially suspected ‘ligand’ was identified as a sensitive and specific agonist. Initial screening for ligand identification was performed with known odors for fish, including amino acids and some reproductive pheromones.8-10 Interestingly, all tested pheromones failed to activate ORA1, whereas a strong activation response was elicited with a mixture of the 20 proteinogenic L-amino acids. Testing of the individual amino acids indicated that activation was due to L-tyrosine alone. Alas, this was an old lot of tyrosine, and a freshly prepared lot of L-tyrosine failed to reproduce the activation response. This suggested to the researchers that the active compound might be a degradation product of L-tyrosine. In fact, tyrosine is known to be sensitive to oxidation upon prolonged storage. Therefore, to test this hypothesis, a fresh lot of L-Tyrosine was oxidized by incubation with hydrogen peroxide, and indeed strong agonist activity was observed in the reaction product (hydrogen peroxide itself had no activity). This suggested that the active substance in the aged L-tyrosine originated from oxidative decay of L-tyrosine. Subsequently, analytical HPLC chromatography on a reverse phase column showed the agonist activity in a single peak. To hunt the agonist down, Meyerhof and Korsching solicited the help of the Rawel group in Berlin to obtain sufficient HPLC-purified material for subsequent structural analysis. For structure determination these groups joined efforts with the Hofmann group in Munich, which used a combination of LC-TOF/MS and proton NMR to unravel the structure. The contaminant was finally identified as pHPAA, and functional testing of the synthetic compound elicited a strong activation response even at very low concentrations, with a half-maximal response (EC50) at 2 μM.
Could pHPAA be the Endogenous Ligand?
Dose response analysis suggested that pHPAA is recognized by ORA1 with much higher affinity compared to food odors such as amino acids9,11,12 or even the death-associated odor cadaverine.13 Furthermore, thorough testing of many structurally related compounds did not reveal any substance with better potency or efficacy for ORA1. Any modification of the carboxyl group such as amidation or methylation reduced the affinity at least 2 orders of magnitude, and shortening the distance of the carboxyl group to the benzene ring by eliminating a methylene group abolished agonist activity altogether. Somewhat less severe constraints were observed for the para hydroxy group, whose elimination results only in a one order of magnitude loss for the affinity. However, a bulky group in this position such as an acetyl group destroys agonist activity completely. Interestingly, the efficacy, i.e. the maximal response, varied somewhat independently from the affinity, as estimated by EC50 determination. Both efficacy and affinity were maximal for pHPAA. So, could the authors have hit on the endogenous ligand for the ORA1 receptor?
Any endogenous signaling molecule should fulfill 2 requirements: firstly there should be a biosynthetic path generating the molecule, and secondly it should have a biological function. pHPAA is a product of a minor catabolic pathway for tyrosine,14 which would seem to fulfill the first requirement. Indeed it has been reported that pHPAA is produced by diverse organisms, such as humans, insects, fungi and bacteria. As for the second requirement, biological functions for pHPAA have been reported in a variety of species, ranging from an antimicrobial property15 for defense in several species to a component of sexual display pheromone in felines.16 Unfortunately, none of these species included fish, and so the authors set out in search of a possible behavioral answer to pHPAA.
What is the Impact of pHPAA on Zebrafish Behavior?
The authors had recently shown an aversive response of zebrafish to the death-associated odor cadaverine,13 and so began to search for either aversive or attractive responses to pHPAA. However, none of the motion parameters analyzed showed significant differences in the presence of pHPAA. Again, an accidental observation came to the help of the researchers. When adult fish were tested in pairs, they noticed sometimes deposition of eggs (oviposition). The effect turned out to be highly significant in mixed gender pairs, and also was observed at similarly low concentrations as the ligand activation of the ORA1 receptor.
Admittedly, pHPAA does not show much similarity to known classes of teleost fish reproductive pheromones, which comprise several steroids and their sulfated metabolites, as well as some prostaglandins. Furthermore, those steroids and prostaglandins tested as possible agonists did not activate ORA1.8,17 However, knowledge of fish reproductive pheromones is still sketchy, and it is known that some components are still unidentified.8,18 Thus the findings discussed here allow the fascinating interpretation that the authors have chanced onto a novel biological class of reproductive pheromones (Fig. 1). Of course, much remains to be done to shore up this hypothesis and to rule out more mundane alternative explanations.
Figure 1.
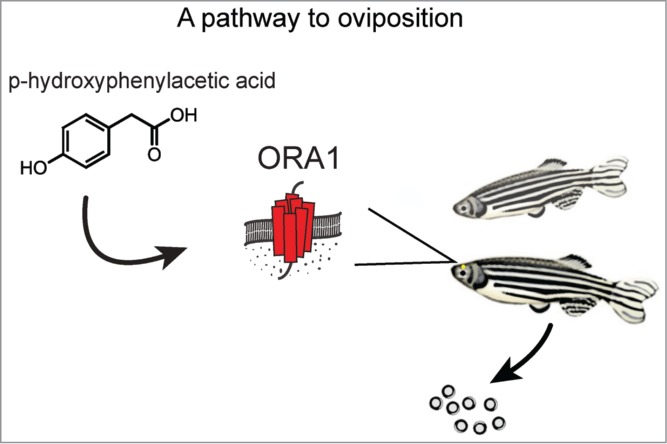
Graphical representation of key findings. Calcium imaging in a heterologous expression assay identified p-hydroxyphenylacetic acid as a agonist of ORA1 receptor. Perception of p-hydroxyphenylacetic acid by the olfactory system of female zebrafish in the presence of a male can induce oviposition.
What Do We Know About the Cells Expressing ORA1?
The characteristic sparse expression pattern of olfactory receptor genes in distributed neurons within the sensory surface is also observed for zebrafish ORA1.5 However, the cell type and the signal transduction cascade of ORA1 expressing neurons are still elusive. Four different types of olfactory sensory neurons are known in zebrafish, ciliated and microvillous neurons as major populations19 and small populations of crypt and kappe neurons.20,21 These 4 cell types differ in molecular markers, but also in morphology and spatial distribution within the olfactory epithelium.20-22 In situ hybridization showed ORA1-expressing neurons in apical positions within the olfactory lamellae, which seems to exclude the more basally located ciliated neurons.5,23 Among the other 3 types, crypt neurons are known not to express ORA1,22 which leaves microvillous neurons or kappe neurons as candidates. Both are present in the superficial layers.20 However, it is also conceivable that yet another, so far undetected class of olfactory sensory neurons would express ORA1.
Olfactory receptors such as the ora genes belong to the superfamily of G protein-coupled receptors, and are expected to signal through trimeric G proteins. Recently a comprehensive evaluation of zebrafish G α proteins has shown the expression of Go1, Go2, Gi, and Golf in the olfactory epithelium.24 Of these, one of the 2 Go isoforms, or possibly Gi, would be candidates for signal transduction of ORA1, since the association of Golf with ciliated neurons would seem to rule out an expression in ORA1-positive neurons.25,26 Direct evidence will be provided by double labeling experiments using ORA1 in situ probe together with G protein or cell type-specific markers.
Open Questions
So far it is unclear whether the olfactory-mediated oviposition behavior elicited by pHPAA is driven by ORA1 as a sole or major receptor. Alternatively, additional receptors may be contributing to this phenotype. To address this question, double labeling experiments with ORA1 probe and established neuronal activity markers could be performed. Neuronal activity markers such as cFOS, phospho-ERK, egr-1, c-jun and Arc have been successfully used in similar studies.27 Moreover, new genome editing techniques like CRISPR/Cas or TALEN can be used to knock out ORA1 in zebrafish.28 If pHPAA-mediated oviposition will be reduced or eliminated in such knock-outs, this would constitute strong evidence for the pHPAA effect being mediated by ORA1. CRISPR/Cas and TALEN may also be useful for knock-in of marker genes into the ORA1 locus, allowing to identify the ORA1 target glomerulus in the olfactory bulb. A knock-in of channel rhodopsin29 in the ORA1 locus could rigorously restrict neuronal activation to only ORA1-expressing neurons, and thus would present another possibility to examine whether ORA1 activation by itself would be sufficient to generate the oviposition response. Beyond the olfactory bulb, it may be possible to map the pHPAA-activated neural circuits with single cell resolution in the intact zebrafish brain using a combination of high-speed light-sheet microscopy with genetically encoded calcium indicator (GCaMP5/6), cf.30 Finally it will be interesting to examine, whether pHPAA is indeed the endogenous ligand of ORA1, in which case one would expect to find a biosynthetic pathway as well as a path to delivery of pHPAA in the context of mating behavior.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We would like to thank Maik Behrens, Oliver Frank, Harshadrai Rawel, Christoph Potting, Thomas Hofmann and Wolfgang Meyerhof for their contributions to the research depicted here.
References
- 1. Isogai Y, Si S, Pont-Lezica L, Tan T, Kapoor V, Murthy VN, Dulac C. Molecular organization of vomeronasal chemoreception. Nature 2011; 478:241-5; PMID:21937988; http://dx.doi.org/10.1038/nature10437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Del Punta K, Leinders-Zufall T, Rodriguez I, Jukam D, Wysocki CJ, Ogawa S, Zufall F, Mombaerts P. Deficient pheromone responses in mice lacking a cluster of vomeronasal receptor genes. Nature 2002; 419:70-4; PMID:12214233; http://dx.doi.org/10.1038/nature00955 [DOI] [PubMed] [Google Scholar]
- 3. Boschat C, Pélofi C, Randin O, Roppolo D, Lüscher C, Broillet M-C, Rodriguez I. Pheromone detection mediated by a V1r vomeronasal receptor. Nat Neurosci 2002; 5:1261-2; PMID:12436115; http://dx.doi.org/10.1038/nn978 [DOI] [PubMed] [Google Scholar]
- 4. Korsching S. The molecular evolution of teleost olfactory receptor gene families. Results Probl Cell Differ 2009; 47:37-55; PMID:18956167; http://dx.doi.org/10.1007/400_2008_11 [DOI] [PubMed] [Google Scholar]
- 5. Saraiva LR, Korsching SI. A novel olfactory receptor gene family in teleost fish. Genome Res 2007; 17:1448-57; PMID:17717047; http://dx.doi.org/10.1101/gr.6553207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Venkatesh B, Lee AP, Ravi V, Maurya AK, Lian MM, Swann JB, Ohta Y, Flajnik MF, Sutoh Y, Kasahara M, et al. Elephant shark genome provides unique insights into gnathostome evolution. Nature 2014; 505:174-9; PMID:24402279; http://dx.doi.org/10.1038/nature12826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Behrens M, Frank O, Rawel H, Ahuja G, Potting C, Hofmann T, Meyerhof W, Korsching S. ORA1, a zebrafish olfactory receptor ancestral to all mammalian V1R genes, recognizes 4-hydroxyphenylacetic acid, a putative reproductive pheromone. J Biol Chem 2014; 289:19778-88; PMID:24831010; http://dx.doi.org/10.1074/jbc.M114.573162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sorensen PW, Hara TJ, Stacey NE. Sex pheromones selectively stimulate the medial olfactory tracts of male goldfish. Brain Res 1991; 558:343-7; PMID:1782551; http://dx.doi.org/10.1016/0006-8993(91)90790-3 [DOI] [PubMed] [Google Scholar]
- 9. Friedrich RW, Korsching SI. Combinatorial and chemotopic odorant coding in the zebrafish olfactory bulb visualized by optical imaging. Neuron 1997; 18:737-52; PMID:9182799; http://dx.doi.org/10.1016/S0896-6273(00)80314-1 [DOI] [PubMed] [Google Scholar]
- 10. Yamamoto Y, Hino H, Ueda H. Olfactory imprinting of amino acids in lacustrine sockeye salmon. PLoS One 2010; 5; e8633; PMID:20062811; http://dx.doi.org/10.1371/journal.pone.0008633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fuss SH, Korsching SI. Odorant feature detection: activity mapping of structure response relationships in the zebrafish olfactory bulb. J Neurosci 2001; 21:8396-407; PMID:11606628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Michel WC, Lubomudrov LM. Specificity and sensitivity of the olfactory organ of the zebrafish, Danio rerio. J Comp Physiol A 1995; 177:191-9; PMID:7636767; http://dx.doi.org/10.1007/BF00225098 [DOI] [PubMed] [Google Scholar]
- 13. Hussain A, Saraiva LR, Ferrero DM, Ahuja G, Krishna VS, Liberles SD, Korsching SI. High-affinity olfactory receptor for the death-associated odor cadaverine. Proc Natl Acad Sci U S A 2013; 110:19579-84; PMID:24218586; http://dx.doi.org/10.1073/pnas.1318596110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gossauer A. Struktur und Reaktivitaet der Biomolekuele: Eine Ein- Fuehrung in Die Organische Chemie. Zurich: Helv Chim Acta, 2006; 1st ed. [Google Scholar]
- 15. Ohtani K, Fujioka S, Kawano T, Shimada AKY. Nematicidal activities of 4-hydroxyphenylacetic acid and oidiolactone D produced by the fungus Oidiodendron sp. Z Naturforsch C 2011; 66:31-4; PMID:21476434; http://dx.doi.org/10.5560/ZNC.2011.66c0031 [PubMed] [Google Scholar]
- 16. Pageat P, Gaultier E. Current research in canine and feline pheromones. Vet Clin North Am- Small Anim Pract 2003; 33:187-211; PMID:12701508; http://dx.doi.org/10.1016/S0195-5616(02)00128-6 [DOI] [PubMed] [Google Scholar]
- 17. Friedrich RW, Korsching SI. Chemotopic, combinatorial, and noncombinatorial odorant representations in the olfactory bulb revealed using a voltage-sensitive axon tracer. J Neurosci 1998; 18:9977-88; PMID:9822753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Stacey N, Sorensen P. Reproductive pheromones. Fish Physiol 2005; 24:359-412; http://dx.doi.org/10.1016/S1546-5098(05)24009-8 [Google Scholar]
- 19. Sato Y, Miyasaka N, Yoshihara Y. Mutually exclusive glomerular innervation by two distinct types of olfactory sensory neurons revealed in transgenic zebrafish. J Neurosci 2005; 25:4889-97; PMID:15901770; http://dx.doi.org/10.1523/JNEUROSCI.0679-05.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ahuja G, Nia SB, Zapilko V, Shiriagin V, Kowatschew D, Oka Y, Korsching SI. Kappe neurons, a novel population of olfactory sensory neurons. Sci Rep 2014; 4:4037; PMID:24509431; http://dx.doi.org/10.1038/srep04037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ahuja G, Ivandic I, Salturk M, Oka Y, Nadler W, Korsching SI. Zebrafish crypt neurons project to a single, identified mediodorsal glomerulus. Sci Rep 2013; 3:2063; PMID:23792970; http://dx.doi.org/10.1038/srep02063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Oka Y, Saraiva LR, Korsching SI. Crypt neurons express a single V1R-related ora gene. Chem Senses 2012; 37:219-27; PMID:22038944; http://dx.doi.org/10.1093/chemse/bjr095 [DOI] [PubMed] [Google Scholar]
- 23. Pfister P, Rodriguez I. Olfactory expression of a single and highly variable V1r pheromone receptor-like gene in fish species. Proc Natl Acad Sci U S A 2005; 102:5489-94; PMID:15809442; http://dx.doi.org/10.1073/pnas.0402581102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Oka Y, Korsching SI. Shared and unique G alpha proteins in the zebrafish versus mammalian senses of taste and smell. 2011; 36:357-65; PMID:21242316; http://dx.doi.org/10.1093/chemse/bjq138 [DOI] [PubMed] [Google Scholar]
- 25. Braubach OR, Fine A, Croll RP. Distribution and functional organization of glomeruli in the olfactory bulbs of zebrafish (Danio rerio). J Comp Neurol 2012; 520:2317-39; PMID:22581687; http://dx.doi.org/10.1002/cne.23075 [DOI] [PubMed] [Google Scholar]
- 26. Gayoso JÁ, Castro A, Anadón R, Manso MJ. Differential bulbar and extrabulbar projections of diverse olfactory receptor neuron populations in the adult zebrafish (Danio rerio). J Comp Neurol 2011; 519:247-76; PMID:21165974; http://dx.doi.org/10.1002/cne.22518 [DOI] [PubMed] [Google Scholar]
- 27. Bepari AK, Watanabe K, Yamaguchi M, Tamamaki N, Takebayashi H. Visualization of odor-induced neuronal activity by immediate early gene expression. BMC Neurosci 2012; 13:140; PMID:23126335; http://dx.doi.org/10.1186/1471-2202-13-140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gaj T, Gersbach CA, Iii CFB. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol 2013; 31:397-405; PMID:23664777; http://dx.doi.org/10.1016/j.tibtech.2013.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yizhar O, Fenno LE, Davidson TJ, Mogri M, Deisseroth K. Optogenetics in neural systems. Neuron 2011; 71:9-34; PMID:21745635; http://dx.doi.org/10.1016/j.neuron.2011.06.004 [DOI] [PubMed] [Google Scholar]
- 30. Ahrens MB, Orger MB, Robson DN, Li JM, Keller PJ. Whole-brain functional imaging at cellular resolution using light-sheet microscopy. Nat Methods 2013; 10:413-20; PMID:23524393; http://dx.doi.org/10.1038/nmeth.2434 [DOI] [PubMed] [Google Scholar]


