In Gram-negative bacteria, proteins destined for the extracellular environment or cell surface need to navigate 2 consecutive lipid bilayers. This can occur in a 2-step mechanism with separate carriers in the inner and outer membrane (IM and OM), or in a single shot by cell envelope spanning secretion complexes.1 Because of the semiporous nature of the outer membrane and its isolation from the cytoplasm, 2-step transport pathways face the challenge of driving transport in the absence of a readily available energy source such as electrochemical gradients or ATP. One such transport system is the curli biogenesis pathway, which assembles extracellular amyloid fibrils that form a major component of the extracellular matrix that links multicellular organization found in bacterial biofilms of many Bacteriodetes and Proteobacteria.2
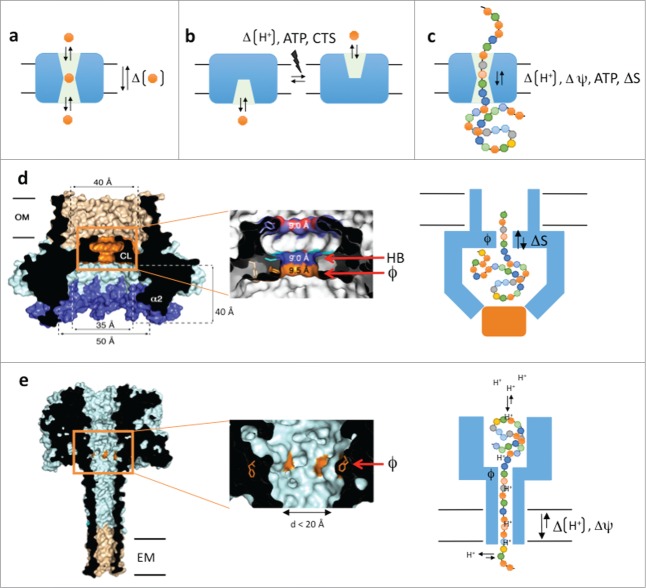
The role of solute or protein carrier proteins is to temporarily accommodate their substrates during passage through the apolar bilayer (Fig. 1a). In channels, the substrate binding site(s) is accessible from either side of the membrane, leading to a passive, diffusion-based dissipation of substrate concentration gradients across the bilayer. In active solute transporters, the binding sites are alternating access modules that require a power stroke (from ATP, ΔpH or co-transport) as part of the secretion cycle, to allow substrate dissociation and/or to switch the solvent-accessible face of the substrate binding site from one side of the membrane to the other (Fig. 1b). For protein carriers, a complication to the transport problem is the large, polymeric and heterogeneous nature of the substrates, which aren't readily accommodated and translocated in single binding – dissociation steps (Fig. 1c). How to maintain substrate selectivity, yet allow binding and passage of a polymer that is structurally and chemically heterogeneous along its length? How to assure processivity and directionality in case of multiple (up to hundreds) consecutive binding steps as the substrate passes the carrier? In other words, how to avoid that the binding of an extended linear polymer in the channel slows down or traps the substrate in a futile equilibrium of local forward and reverse diffusive steps?
Figure 1.
(A–C) Schematic representations of channel- and alternating access based solute transporters (a and b, resp.) and a polypeptide carrier (C). (D) Cross-section of the CsgG nonamer (PDB entry: 4UV3), cut-out of the Phe-ring region and schematic model of proposed secretion mechanism. (E) Idem for the theoretical model of the PA63 octamer in pore conformation (PDB entry: 1V36). CTS: co-transported solute, ATP: adenosine triphosphate, ΔΨ electric potential, HB: H-bond donor/acceptor, ϕ: Phe-ring, ΔS: entropy potential, OM: outer membrane, EM: endoplasmic membrane.
For the curli biosynthesis pathway, OM translocation of the curli subsunits CsgA and CsgB requires a dedicated ∼30 kDa lipoprotein, CsgG.3 CsgA and CsgB are pseudo-repeat proteins (six 20–25 residue imperfect repeats in E. coli) that pass the CsgG transporter in an unfolded state.2 To gain insight in the mechanistic details of CsgG-mediated peptide translocation, we recently determined its crystal structure and conducted electrophysiological studies on the reconstituted pore.4
Crystal structures of the membrane-bound form of E. coli CsgG reveal a nonameric transport complex that traverses the outer membrane by a composite 36-stranded β-barrel with 4 nm inner diameter (Fig. 1d).4,5 This β-barrel is connected to a large 3.5 nm wide solvent-accessible cavity in the periplasm, the intermembranous space separating OM and IM. A 0.9 nm central channel constriction connects the periplasmic and extracellular β-barrel regions. Single-channel current recordings of CsgG reconstituted in phospholipid bilayers led to stable conductances of 43.1 or −45.1 pA under a + or −50 mV potential, respectively. These currents are in agreement with the observed channel geometry, including the 0.9 nm constriction, and their constitutive nature indicates CsgG acts as a passive peptide diffusion channel.
A model peptide diffusion channel is the anthrax protective antigen (PA63), the poreforming component of the 3-protein anthrax toxin (PA, Edema Factor and Lethal Factor).6 After receptor binding and endocytosis, the PA63 inserts in the endosomal membrane by means of an oligomeric (7- or 8-fold) β-barrel that connects to a luminal α/β-domain that is in complex with the translocation substrates EF and LF (Fig. 1e). Pore formation and endosome acidification lead to partial unfolding of EF and LF and their threading through the PA63 channel for cytosolic delivery. Threading of EF, LF or synthetic peptides through the PA63 channel requires an electric potential (ΔΨ) and/or a proton gradient.7 The substrate capture and processivity of its translocation critically depends on the presence of a concentric ring of phenylalanines (dubbed ϕ-clamp) at the pore entrance and a cation selective channel.7 Together these elements form a Brownian ratchet where diffusive steps of the translocating polypeptide inside the channel, are forward rectified toward the cytosol. This rectification is a result of polypeptide protonation and deprotonation at the endosomal (pH 5-5.5) and cytosolic (pH 7–7.4) side of the channel, respectively. Together, the φ-clamp and cation-selective channel form a barrier to back-slipping of the translocating chain after it deprotonates at the cytosolic exit of the channel.6 This ratchet, however, is driven by the proton gradient over the endosomal membrane, a condition that is not fulfilled at the outer membrane.
A striking and unanticipated feature of the CsgG channel constriction is the presence of a concentric Phe-ring at its entrance, equivalent to the ϕ-clamp in PA63. In CsgG, this Phe-ring is succeeded by concentric asparagine and tyrosine rings (Fig. 1d). Multiple sequence alignment of CsgG-like translocators and mutation studies point to the requirement of the stacked configuration of a ϕ-clamp followed by a hydrogen-bond donor/acceptor in the CsgG channel constriction.4 Future studies following peptide translocation by the CsgG channel will be required to dissect the role of the ϕ-and H-bond clamps for the capture and passage of the polypeptide substrate.
Under native conditions, CsgA secretion by the CsgG pore requires the periplasmic accessory factor CsgE. Our data show CsgE acts as an oligomeric capping factor to the CsgG pore. When added to reconstituted CsgG channels, CsgE gates CsgG conductance in a concentration-dependent manner.4 EM visualization and mutagenesis shows the binding of a CsgE nonamer atop the entrance to the CsgG periplasmic vestibule. This mode of CsgE binding gives rise to an extensive cavity upfront the channel constriction that could encage, at least partially, the translocation substrate during the secretion process. In the GroEL-GroES chaperonin, entrapment of an unfolded polypeptide in the confining cavity of the complex results in a loss of entropy that helps shift the equilibrium toward protein folding.8 Unless capped or compensated by interaction with the cavity wall, entropy reduction of a polypeptide would favor its evacuation from a confining cavity. In analogy, our working model proposed for the CsgG channel stipulates that the full or partial engagement of the secretion substrate creates an entropic free energy potential over the channel constriction that favors escape from the cavity, eg. rectifies the progressive Brownian diffusion of the extended polypeptide in the channel constriction toward the bulk solvent at the cell surface. In our model, the initial cost of engaging CsgA would be compensated by binding energy released during assembly of the CsgG:CsgE:CsgA secretion complex. In a scenario where periplasmic CsgA would be prevented from attaining its full conformational freedom by binding to CsgE and/or CsgG as it emerges from the Sec translocon, this would create a net positive ΔS across the cell envelope that would drive secretion of nascent CsgA.
Based on current structural and biochemical insights in the CsgG secretion machinery, we provide a tentative model for the membrane translocation of unfolded protein substrates based on Brownian diffusion, rectified by an entropic gradient. Future biochemical and single-channel analysis will be required to validate this model, and a dissection of the transport kinetics at the single-molecule level will be needed to determine whether additional or alternative rectifying mechanisms are at play in the secretion channel.
References
- 1. Desvaux M., et al. Trends Microbiol 2009; 17:139-45; PMID:19299134; http://dx.doi.org/ 10.1016/j.tim.2009.01.004 [DOI] [PubMed] [Google Scholar]
- 2. Chapman MR, et al. Science 2002; 295:851-5; PMID:11823641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Robinson LS, et al. Mol Microbiol 2006; 59:870-81; PMID:16420357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Goyal P, et al. Nature 2014; 516:250-3; PMID:25219853; http://dx.doi.org/ 10.1038/nature13768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cao B, et al. Proc Natl Acad Sci U S A 2014; 111:5439-44; PMID:25453093; http://dx.doi.org/ 10.1073/pnas.1411942111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Feld GK, et al. Protein Sci 2012; 21:606-24; PMID:22374876; http://dx.doi.org/ 10.1002/pro.2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Krantz BA, et al. Science 2005; 309:777-81; PMID:16051798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brinker A, et al. Cell 2001; 107: 223-33; PMID:11672529 [DOI] [PubMed] [Google Scholar]