Abstract
The epithelial to mesenchymal transition (EMT) consists of a rapid change of cell phenotype, characterized by the loss of epithelial characteristics and the acquisition of a more invasive phenotype. Transcription factors regulating EMT (Snail, Twist and Zeb) are extremely labile proteins, rapidly degraded by the proteasome system. In this review we analyze the current mechanisms controlling degradation of EMT transcription factors, focusing on the role of new E3 ubiquitin-ligases involved in EMT. We also summarize the regulation of the stability of these EMT transcription factors, specially observed in different stress conditions, such as hypoxia, chemotherapeutic drugs, oxidative stress or γ-irradiation.
Keywords: EMT, F-box, hypoxia, proteasome, Snail1, stress, ubiquitination
EMT and Transcription Factors Driving EMT
Epithelial to mesenchymal transition (EMT) is a reversible process that promotes epithelial cells to acquire a mesenchymal phenotype. During this transition the cell-cell junction structures, including adherens junctions and desmosomes, are disassembled and cells become spindle-shaped and motile. EMT was identified in the context of development decades ago1 and has been extensively reviewed.2 However, it was not until the discovery of its role in tumor invasion that its molecular mechanisms have started to be unraveled.3
Although examples of EMT have been characterized in development, for instance during gastrulation and neural crest delamination,4-6 EMT is also relevant in tumor progression. Expression of EMT markers in primary tumors correlates with enhanced invasiveness and poor clinical prognosis.7 Moreover, lineage tracing experiments have recently demonstrated the occurrence of EMT in pancreas tumors and its involvement in tumor invasion.8 It has also been proposed that cancer cells having undergone EMT and located at the tumor edge secrete cytokines and proteases that promote angiogenesis, remodel the peritumoral extracellular matrix (ECM) and activate non-neoplastic cells.9 Simultaneously, stromal cells release factors that increase EMT in cancer cells, fostering survival, growth and invasiveness of the tumor, generating a reciprocal influence between the tumor and its microenvironment.9-11 Mesenchymal cells resulting from EMT are more resistant to cell death and senescence, as well as escape immune surveillance and thus avoid chemo- and immuno-therapies.4
In recent years the concept of EMT has been modified and refined including the term of “partial EMT.”12,13 This refers to a phenotype often observed in vivo in the processes of wound healing and mammary tubulogenesis. Partial EMT, as the name suggests, is an intermediate process in which some of the characteristics of epithelial cells are retained while there is a simultaneous and evident induction of mesenchymal features. This phenotype allows cells to spread and migrate in an active manner although a cell-cell cohesiveness is maintained to a certain extent and cells present a “cohort-like” migration; it is likely that this partial EMT is more suitable to be reversed than a complete EMT and therefore more capable to sustain the growth of secondary tumors or metastasis.14,15
The key biomarker for EMT is the down-regulation of the homotypic adherens junction protein E-cadherin.16 E-cadherin downregulation happens mainly due to transcription inhibition, through the action of different transcription factors on consensus E-boxes (5′-CACCTG-3′ or 5′-CAGGTG-3′) present in the E-cadherin (CDH1) promoter.17,18 Among the plethora of transcription factors repressing E-cadherin, only the members of the Snail family Snail1 and Snail2 (formerly Snail and Slug), the Zeb family (Zeb1 and Zeb2), E47, KLF8 and Twist1 have been described to bind to CDH1 promoter, although Twist does it indirectly.7,17 In this review we will focus on Snail, Twist and Zeb proteins, describing the post-translational mechanisms controlling their protein stability and function.
The SNAIL family
The SNAIL family of repressors is comprised of 3 members: SNAIL1 (formerly known as SNAIL), SNAIL2 (SLUG) and SNAIL3 (SMUC). It constitutes a subfamily characterized by a SNAG box in its N terminus required for transcriptional repression (Fig. 1A). Snail1 is the most broadly studied member of the family. Through the SNAG domain (amino acids (aa) 1-9) Snail1 recruits multiple co-repressors (such as Polycomb complex 2, Sin3A/histone deacetylases1/2 complex, Ajuba) involved in CDH1 gene repression.19-22 Two independent Snail1 affinity purifications coupled with mass spectrometry analysis are coincident in detecting lysine-specific demethylase (LSD1),22,23 an enzyme that removes methyl groups from H3K4me1/2. These experiments suggest that LSD1 shows the highest affinity for the SNAG domain and many other cofactors might interact with Snail1 indirectly. In the central part Snail1 contains a phospho-serine rich domain (aa 90-120) and a nuclear export sequence (NES) located in close proximity (aa 132-143) that binds to Crm1 (Exportin-1)24; and at the C-terminal part of Snail1 (aa 152-264), 4 zinc fingers (ZnF) of the C2H2 type that bind DNA through E-boxes (Fig. 1A),25 and a unique nuclear localization signal (NLS) mediating nuclear import.26,27
Figure 1.
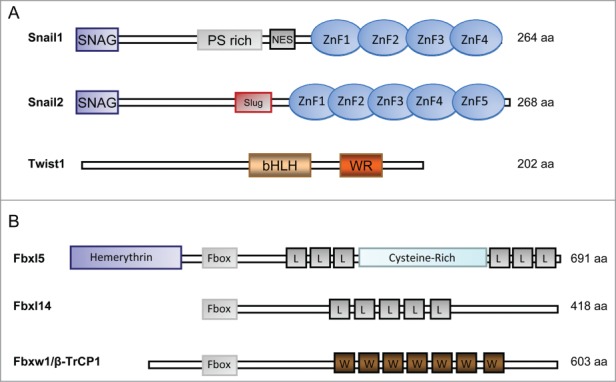
Schematic representation of Snail1, Snail2 and Twist1 domains (A) and the ubiquitin ligases involved in their degradation (B). (A) The position of the phospho-serine (PS) rich region, the nuclear export sequence (NES), the zinc finger domains (ZnF), the SLUG domain are shown for Snail1 and 2. The basic helix-loop-helix (bHLH) domain and the WR (tryptophan and arginine) motif is indicated for Twist1. (B) Fbxl5, Fbxl14 and Fbxw1/β-TrCP1 are represented. FBXL, F-box and Leucine-rich repeat Protein. FBXW, F-box and WD40 repeat protein. L, Leucine-rich repeat. W, WD40 repeat.
Snail2 (also known Slug) has been less studied than Snail1 although it also represses CDH1 and induces EMT.28 Snail2 is imported into the nucleus through a NLS sequence similar to that observed in Snail1.27 Snail2 has an specific central domain (named SLUG domain) capable of binding the C-terminal binding protein 1 (CtBP1) co-repressor29 and of recruiting histone deacetylase 1 (HDAC1) (Fig. 1A)28,30 A recent study shows that the co-repressors CtBP1 and nuclear receptor co-repressor 1 (NCoR) use the SLUG and SNAG domains, respectively, to be recruited to Snail2.29 Contrary to what happens with the Snail1 knockout mouse,31 Snail2 deleted mice are viable, showing that Snail2 is not essential for mesoderm formation.32
The ZEB family
The ZEB family of transcription factors is highly conserved across species and is constituted by 2 members: ZEB1 (also known as δEF1) and ZEB2 (also known as SIP1).33 It contains 2 different domains of interaction with DNA, constituted each by 3 and 4 zinc fingers of the C2H2 and C3H type located at the N- and C-terminal part of the protein, respectively, and a central homeodomain. Zeb proteins potently inhibit CDH1 expression through the binding to CtBP co-repressors and the recruitment of HDACs and methyltransferases, Polycomb proteins, coREST and the SWI/SNF chromating remodeling protein BRG1, among other factors.34-38 Similarly to the Snail family, TGF-β and Wnt activate these mesenchymal genes.39,40 However, the kinetics or activation is different; for instance, Snail1 induction by TGF-β precedes and is required for Zeb1 expression. Snail1 acts at several levels: it increases Zeb1 transcription by favoring Ets1 and NF-κB translocation to the nucleus, decreases the levels of miRNA-200 targeting ZEB1 mRNA, and stabilizes Zeb1 protein.40,41 Snail1 also induces Zeb2 protein in an indirect manner through the alternative processing of the ZEB2 mRNA.42
The bHLH family of transcription factors
The basic helix-loop-helix (bHLH) family of transcription factors has a common structure with 2 parallel amphipathic α-helices joined by a loop required for dimerization. bHLH proteins bind DNA as homo- or heterodimers using consensus E-boxes and can act both as transcriptional activators or repressors.43 The 2 products of the E2A gene, E12 and E47, the inhibitory Id proteins (Id1-Id4) and the Twist proteins (Twist1 and Twist2) are members of this family, the last ones being the most widely described bHLH proteins involved in EMT.7,44 Twist1, apart from sharing the common bHLH domain, presents at the C-terminal end of the protein a WR motif important for Twist degradation (Fig. 1A).45,46 Twist represses CDH1 expression indirectly binding to this promoter and recruiting chromatin remodeling complexes containing Mi2/NuRD and SET8.47,48
Twist factors predominantly heterodimerize with E12 and their function is therefore determined by the availability of this protein as well as by the phosphorylation of the bHLH domain.49 As described for Snail and Zeb family members, Twist proteins are up-regulated by TGF-β, Wnt signaling, growth factors and hypoxia. In fact Twist1 is directly modulated by low oxygen since it contains an Hypoxia Inducible Factor (HIF)-responsive element (HRE) in its promoter; this site is bound by HIF-1α in low oxygen conditions causing Twist transcription.50
Snail and Twist in EMT as Sensors of Cellular Stress
Expression of EMT transcription factors is controlled by many growth factors and cytokines including HGF, TGF-β or TNF-α.5,51,52 Among these, TGF-β is the best studied EMT inducers that cross-talks with other cell pathways, such as Wnt, Ras, Hedgehog and Notch in order to induce Snail1 and promote EMT.53 Besides these extracelluar signals, EMT is also controlled by stress, through the activation of molecular pathways that have recently started to be analyzed.54 For instance, hypoxia activates the EMT program with the concomitant induction of Snail1, Twist1 and Zeb2.50,55 Snail1 expression is also induced by reactive oxygen species (ROS) that increases both SNAIL1 mRNA and protein stability.56-58 Curiously, Snail1 expression per se also activates the ROS pathway.57,59 Genotoxic stress caused by DNA damage also induces Snail1 and Snail2 expression, a response that has been attributed to a cellular mechanism to avoid programmed cell death.60 Other results also link stress, the EMT phenotype and the acquisition of chemo-resistance, and are associated to the up-regulation of Snail1/2 or Twist161-64 and the acquisition of some cancer stem cell-like properties.65 For instance, cellular stress mediated by ionizing radiation promotes EMT, stimulating invasion, metastasis and radio-resistance of tumor cells66; this insult up-regulates Snail1, at least in part due to protein stabilization,67-69 and Snail2, increasing its transcription70; consequently transcription of the pro-apoptotic genes Puma70 or PTEN71 are down-regulated and cells become more resistant to apoptosis.72
Ubiquitin Ligases Controlling Snail and Twist Stability
Snail and Twist are short-lived proteins since they are rapidly polyubiquitinated and degraded by the 26S proteasome system in normal cells. Polyubiquitination is a process by which the 76-amino acid protein ubiquitin is attached to a target protein. Three types of enzymes are the ones responsible for the final ubiquitination of a substrate: first the Enzyme 1 (E1) or ubiquitin-activating enzyme activates the ubiquitin molecule in an ATP (adenosine triphosphate)-dependent manner; then a second enzyme (E2) or ubiquitin-conjugating enzyme transfers the activated ubiquitin moiety directly onto the substrate or to a third enzyme (E3) called ubiquitin ligase.73,74 The way in which ubiquitin is loaded onto the substrate depends on the specificity of the E3. These enzymes belong to 2 grand subfamilies: the RING/RING-like or the HECT types. To date, only E3 ligases of the RING-type have been described to be relevant in mediating Snail and Twist ubiquitination. Degradation of most EMT factors is strictly dependent on multi-subunit RING-type E3s and include the so-called SCF (Skp1-Cullin1-F-box) complex containing a Cullin1 (Cul1) scaffolding protein, the adaptor protein Skp1, the RING-finger protein Rbx1/Roc1 and an F-box protein75 (Fig. 2). The F-box protein is the substrate-recognition subunit and contains, as its name implies, an F-box domain, a 50 amino acid motif used to bind Skp1 linking the N-terminal part of Cullin to the F-box protein. Moreover, Rbx1 binds to the C-terminal part of the Cullin and acts as a docking site for the ubiquitin-activated E2 protein76,77 (Fig. 2). So far, 4 different RING-type E3s ubiquitin ligases have been described to be relevant in EMT: 3 SCF-E3s named SCF-β-TrCP1, SCF-Fbxl14 and SCF-Fbxl5; and one single-subunit, Mdm2.
Figure 2.
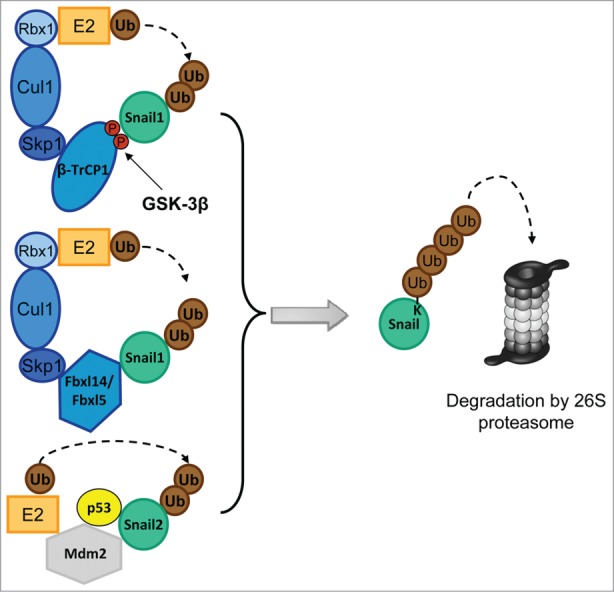
Composition of the E3 complexes targeting Snail1 and Snail2. The Skp-1-Cullin-1-F-Box (SCF) E3s, SCF-Fbxl14 and SCF-Fbxl5 are multimeric E3 ligases that mediates the ubiquitin (Ub) transfer from the E2 conjugating enzyme to Snail. SCF E3s are composed of the scaffold protein Cullin1 (Cul1), which interacts with Skp1 and the RING-finger protein Rbx1. The substrate binding affinity is mediated by the F-box protein (β-TrCP1, Fbxl14 and Fbxl5). The SCF-β-TrCP1 binds Snail1 only when double-phosphorylated by GSK-3β. Mdm2 is a single-subunit RING-finger E3 that targets Snail2 when p53 is bound.
SCF-β-TrCP1/Fbxw1
Fbxw1, commonly named β-TrCP1 (β-transducin-repeat containing protein), is the first member of a F-box subfamily containing a specific domain characterized by the presence of WD40 repeats (this subfamily is commonly named as FBXW and comprises 10 members in humans) (Fig. 1B).78 Proteins that are targeted by the ubiquitin-ligase machinery contain a degradation signal or degron, which is defined as a minimal element within a protein sufficient for recognition and degradation by the proteolytic apparatus.79 In the case of Snail1, the β-TrCP1 degron is a phosphorylated sequence (commonly named phospho-degron) DpS96GxxpS100, consequence of the action of GSK-3β on Snail1 residues S96 and S100 (Fig. 2 and 3).80 This consensus motif for β-TrCP1-mediated degradation is similar to that previously described on β-catenin.81 More than 35 substrates have been described to contain a similar sequence for β-TrCP1/2 (β-TrCP2 is also known as Fbxw11), all requiring the participation of different kinases.82
Besides being potently upregulated by proteasome inhibitors, Snail1 protein stability is also enhanced by lithium ion (Li+), an inhibitor of GSK-3β.80 Phosphorylation by this enzyme of the Ser-rich Snail1 domain is relevant not only for degradation but also for nuclear export.24 Phosphorylation on residues S104, S107 and possibly other Serines between S107 to S119 promote Snail1 localization to the cytoplasm,80,83 uncovering a nuclear export sequence.24 Moreover, phosphorylation of Ser104 acts as priming site for the second round of phosphorylations on S96 and S100 that takes place in the cytosol and originates the above described consensus destruction motif or degron DSp96GxxpS100 recognized by β-TrCP124,80 (Fig. 3). Snail2 also contains 2 GSK-3β motifs inside the Slug domain in a recently identified Serine rich region that alike Snail1 affects ubiquitination and degradation by β-TrCP1, although this sequence does not contain the classical β-TrCP1 destruction motif.84,85
Figure 3.
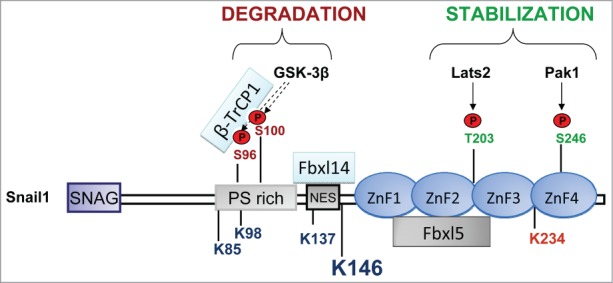
Snail1 phosphorylation sites linked to stabilization or degradation. Stabilizing phosphorylation sites are mainly localized in the C-terminus except the stabilizing ATM phosphorylation site in S100 and ERK2 phosphorylation sites in S82 and S104 (not indicated). The corresponding kinases are indicated by arrows. Phosphorylation sites promoting degradation are in the N-terminus (GSK-3β). The interaction with the F-box proteins Fbxl14 and β-TrCP1 is located in the N-terminus and with Fbxl5 in the C-terminus. The Lysines (K) that are ubiquitinated are indicated: K85, K146 and K234, were found to be specifically ubiquitinated by SCF-Fbxl5 and K98, K137 and K146 were modified by SCF-Fbxl14 or SCF-β-TrCP1. Although other K can be also modified, K146 is the best substrate of the 3 ligases and is the major ubiquitination site. K234 is marked in red because its modification by SCF-FBXL5 has a non-degradative role by decreasing DNA-affinity.
Besides phosphorylation of the substrate, an active SCF-β-TrCP1 complex requires the neddylation (incorporation of the small protein Nedd8) of Cul1 by a Nedd8-E3 ligase.86 Interestingly, TNF-α promotes the induction of the COP9 signalosome 2 protein (CSN2) that removes Nedd8 (or reverses neddylation) from cullins increasing Snail1 stabilization during inflammation.87 As a consequence of this, TNF-α reduces the association of Snail1 with β-TrCP1 complex due to misassembly and, surprisingly, the phosphorylation of Snail1 by GSK-3β.87
β-TrCP1(−/−) mice have been generated; these animals develop normally and only males present a mild phenotype consisting in reduced fertility due to the accumulation of metaphase I spermatocytes in testes.88 This is consequence of the stabilization of the β-TrCP1 substrates cyclin A, cyclin B, and Emi1 but not β-catenin.88,89 Curiously, at least in MEFs, β-catenin stabilization requires additional silencing of β-TrCP288 although this ubiquitin ligase does not interact with β-catenin.90 Similarly to β-catenin, Snail1 is stabilized upon the combined depletion of both ligases, obtained by expression of an sh-β-TrCP2 in β-TrCP1 knockout mice.89 These double-deficient mice are normal although show a more severe testicular phenotype than the β-TrCP1(−/−) mice with absence of spermatids and meiotic cells. Interestingly, this phenotype is reversed upon Snail1 depletion.89 These results suggest that β-TrCP1 and β-TrCP2 are redundant in controlling the stability of Snail1 and β-catenin, although, as happens for β-catenin, β-TrCP2 expression does not induce Snail1 degradation.91 Therefore, more work is needed to understand whether the 2 homologues play an individual, synergistic, or redundant role in controlling Snail1 and β-catenin stability under different experimental conditions.
SCF-Ppa/Fbxl14
Fbxl14 is the F-box subunit of the SCF complex and the human homolog of Partner of Paired (Ppa) gene product.78 This protein degrades Snail2 (and Snail1) in Xenopus laevis in a phosphorylation-independent manner.92 FBXL14 belongs to the FBXL subfamily of F-box proteins formed by 21 members, all of them containing Leucine-rich repeats (LRR) at the C-terminus (5 LRRs in the case of FBXL14) (Fig. 1B). There are 2 homologues of Fbxl14 in zebrafish, namely Fbxl14a and Fbxl14b that are differently regulators of dorsoventral patterning through the targeting of the MAP kinase phosphatase-3 (Mkp3).93 Ppa is highly conserved across species,94 in fact, the SCF-Fbxl14 ubiquitin ligase also targets Snail1 in mammalian cells (Fig. 2).91 Interestingly, Fbxl14 and β-TrCP1 modify the same lysines in Snail1: K98, K137 and K146, all of them located in the N-terminal part91 (Fig. 3). Lander et al. have suggested a general role of Ppa/Fbxl14 in EMT since it promotes the degradation of other transcription factors involved in this process such as Twist1 and Zeb2.46 This last finding is intriguing because Snail, Zeb and Twist proteins are not structurally related. In the case of Snail2 the Ppa/Fbxl14 interaction region is an hydrophobic sequence of the N-terminus comprising amino acids 31 to 64, with low homology with Snail1 and no similarity with the Twist interacting sequence located on the C-terminal WR domain.46,92 In the case of Snail1 the precise binding sequence has not being totally determined although it comprises amino acids 120 to 15191 (Fig. 3). This sequence is also rich in hydrophobic amino acids suggesting that Fbxl14 interaction is dependent on the presence of hydrophobic subdomains in their substrates.
Interestingly, Fbxl14 is sensitive to stress conditions and is specifically downregulated by hypoxia.91 This decrease happens at the transcriptional level at is dependent on the elevated expression of Twist1 protein in hypoxic cells. Therefore, hypoxia down-regulates the expression of FBXL14 causing the stabilization of several EMT core transcription factors, such as Snail1 and Twist1. This causes a feedback loop in which the protein stability of these transcription factors, normally short-lived, is highly increased. Analysis of human colon tumor samples shows an inverse correlation between hypoxia and FBXL14 levels.91 Other authors have shown that hypoxia causes inhibition of GSK-3β through the intracellular generation of ROS, as determined by analyzing the phosphorylation of Ser9, a modification associated to loss of function of this kinase.95 Our unpublished results also indicate that hypoxia-induced FBXL14 down-regulation is in some cells accompanied with GSK-3β inactivation (Viñas-Castells R, unpublished). It is likely that a complete EMT requires the inactivation of both ubiquitin ligases achieved through GSK-3β inhibition and Fbxl14 downregulation, suggesting a synergistic effect between different EMT-inducing signals.
In vivo depletion of Ppa/Fbxl14 has been performed in Xenopus embryos, evidencing a relevant role for Ppa in neural crest development92; other experiments in zebrafish embryos indicate that Fbxl14 regulates axis formation.93 Unfortunately, knockout mice models for FBXL14 have not been developed yet.
SCF-Fbxl5
A shRNA screening for Snail1-specific F-box E3 ubiquitin ligases resulted in the identification of Fbxl5.69 Besides comprising a hemerythrin domain capable of binding iron in its N-terminal part, Fbxl5 contains an F-box domain and 6 LRRs (Fig. 1B).96,97 This ligase is mainly, but not exclusively, localized in the nucleus and polyubiquitinates Snail1 acting on several lysines (Fig. 2 and 3): K85 and K146, both placed in the N-terminus, as well as K234, located in the Snail1 C-terminal domain. Different to Fbxl14 or β-TrCP-1, Fbxl5 interacts with this Snail1 domain (ZnF1 to ZnF3)69 (Fig. 3). Since K146 (located in the N-terminal part) is the main substrate of this ubiquitin ligase, FBXL5 requires 2 different domains of Snail1 protein for degradation. Moreover, K234 ubiquitination by FBXL5 totally impairs Snail1 interaction with the DNA, indicating an unexpected role for this modification in controlling DNA binding affinity and also showing another significant difference with respect to β-TrCP1 or Fbxl14.69 Although Snail1 ubiquitination by SCF-Fbxl5 takes place in the nucleus, the protein is degraded in the cytosol. Accordingly, inhibition of Snail1 export using Leptomycin B prevents Snail1 degradation by Fbxl5. Therefore, Snail1 is not targeted for degradation in the nucleus, and nucleocytoplasmic shuttling is essential for degradation, an effect also required for p53 efficient proteolysis.98 It remains to be established why nuclear proteasomes do not process Snail199; it is possible that the unfolding of the protein caused by ubiquitination promotes its rapid interaction of the NES with Crm1 exporter and its exit from the nucleus before it can be proteolyzed. Alternatively, Fbxl5 might produce an incomplete ubiquitination that needs to be extended by cytoplasmic ubiquitin ligases. According to our model, Fbxl5 ubiquitination, besides decreasing Snail1 interaction with target promoters, unfolds the molecule exposing the NES facilitating Snail1 nuclear export. Once Snail1 is exported, the coordinated effects of cytoplasmic F-box proteins β-TrCP1 and Fbxl14, and maybe other ubiquitin ligases, may cooperate and complete ubiquitination. These results also have implications in the regulation of Snail1 protein half-life since Fbxl5-induced degradation should be impaired by all the post-translational modifications preventing nuclear export.
Fbxl5 is a more specific E3 ubiquitin ligase than β-TrCP1 or Fbxl14, which control the stability of multiple EMT factors and even of other proteins outside this pathway. When analyzed, Fbxl5 degrades Snail2 but not Twist1 or Zeb1. This is probably related to the fact that, contrary to other ubiquitin ligases that interact with the central domain, Fbxl5 binds the C-terminal region of Snail1; therefore it will only tackle transcription factors with a similar zinc finger domain, such as Snail2.69
Similarly to Fbxl14, Fbxl5 expression is decreased by some conditions that induce Snail1 stabilization. Besides being destabilized by iron depletion, FBXL5 is potently down-regulated after γ-irradiation (IR), a cellular stress condition also causing a complete or partial EMT.69 This suggests that Fbxl5 might promote radio-sensitivity of cancer cells. The down-regulation of Fbxl5 by IR depends on both the decrease in its mRNA and the destabilization of the protein mediated by the same hemerythrin domain that confers sensitivity to iron.69 The mechanism linking IR with hemerythrin-dependent destabilization of Fbxl5 is still unknown, but it is likely that IR might indirectly reduce the iron levels required for hemerythrin stability100 or modulate the levels of the ubiquitin ligase that targets Fbxl5.101 Moreover, Fbxl5 is also downregulated by hypoxia97 which can also contribute to the Snail1 stabilization observed in these conditions.
The in vivo role of FBXL5 in EMT using knockout mice has not been determined yet. This is probably due to the fact that FBXL5, apart from its role in EMT, is a master regulator of iron homeostasis and its depletion is embryonically lethal, making difficult to study its role on other substrates such Snail1.102 FBXL5-null mice die in utero due to the stabilization of the iron-response protein-2 (IRP2), a RNA binding protein that inhibits translation of ferritin mRNA and blocks transferrin receptor mRNA degradation. As a consequence of IRP2 stabilization, there is a general and lethal iron overload. Interestingly, FBXL5−/−IRP2−/− mice do not show any phenotype; it is possible that these mice respond to signals activating Snail1 with a higher upregulation of this factor.
Mdm2
Another ligase capable to degrade the Snail family member Snail2 is Murine Double Minute 2 (Mdm2, or Hdm2 in humans). The main role of Mdm2 consists in the ubiquitination and degradation of the tumor suppressor p53, which prevents cancer progression by inhibition of proliferation and induction of apoptosis.103,104 Curiously, p53 can antagonize the action of the EMT transcription factor Snail2 by forming a p53-Snail2-Mdm2 complex that facilitates the degradation of Snail2 (Fig. 2).105 Due to the structural similarities between Snail2 and Snail1 it is possible that the same ligase may target both proteins. In fact, Mdm2 can facilitate Snail1 degradation in a p53-dependent manner,106 although the precise mechanism has not been elucidated yet. Another link evidencing a p53-Snail1 antagonism has been proposed by Weiss and coworkers that have shown that p53 transactivates miRNA-34, which suppresses SNAIL1 mRNA by binding to its 3′-UTR.107 p53 also regulates EMT by transactivating miR-200b and c targeting ZEB1/2 mRNA.108
An intriguing question is whether the Mdm2-mediated Snail1/2 degradation takes place in the nuclear or cytoplasmic compartment. The fact that p53 ubiquitination by Mdm2 may occur in the nucleus but efficient degradation requires nuclear export,109 suggests that Snail1/2 degradation may follow the same pattern, as previously commented for Fbxl5. Thus, Mdm2 might cooperate with Fbxl5 facilitating Snail1/2 nuclear ubiquitination and subsequent export and cytoplasmic degradation by the SCF-β-TrCP1 or SCF-Fbxl14 ubiquitin ligases. Another interesting link between p53 and EMT is mediated by Twist1 that interacts with the C-terminal part of p53 promoting its degradation by Mdm2.110 Since p53 negatively control Snail1 expression at several levels (see above), induction of Twist might upregulate Snail1 by facilitating p53 degradation, further demonstrating the complex network that interconnects the expression of these 2 transcriptional factors. Finally, adding more complexity to the system, Mdm2 is itself a substrate of β-TrCP1 upon its phosphorylation by CK1 after DNA damage.111 Therefore, it is likely that the genotoxic stress-dependent degradation of Mdm2 might contribute to Snail stabilization.
Mouse knockout models for p53 or Mdm2 do not display EMT defects. However, the loss or mutation of p53 that commonly occurs in cancer have been shown to have a strong impact in EMT, likely due to the central role of p53 antagonizing Snail1 or 2 expression, as described above.105,107
Control of Snail1 Protein Stability by Post-Translational Modifications
Phosphorylation
Snail1 undergoes phosphorylation by several protein kinases affecting its half-life. The first group of these kinases promotes its degradation; the paradigm is GSK-3β that, as indicated before, is required by the action of SCF-β-TrCP1.80,85 The catalytic action of this kinase on Snail1 is controlled by direct inactivation by PKB/AKT phosphorylation on Ser9,80 and by the chaperone Axin2, that promotes GSK-3β nuclear export.112 Therefore, Axin2 prevents the phosphorylation of Snail1 in the nucleus, maintaining this protein in this compartment and precluding its degradation not only by β-TrCP1 but by the cytosolic Fbxl14 as well.112 Axin2 expression is controlled by the Wnt canonical pathway since Axin2 promoter contains β-catenin-TCF4 elements, linking Wnt activation with Snail1 upregulation and EMT.112 Another interesting mechanism stabilizing Snail1 through GSK-3β inactivation is triggered by prostaglandin that promotes the interaction of this protein kinase with the receptor-coupled G protein βγ subunits.113
GSK-3β phosphorylation needs to be primed. Several protein kinases have been described to be able to do it, such Casein kinase 1 epsilon (CK1-ε) that interacts with the zinc finger domain of Snail1,114 CK2 (an hetero-tetramer composed of 2 catalytic subunits, α/α′, and a dimer of regulatory β subunits),115 and DYRK2.116 Interestingly, both CK2β and DYRK2 are downregulated in breast cancer patients what would prevent Snail1 phosphorylation, and increase Snail1 levels and subsequent EMT.115,116 Finally, Protein kinase D1 (PKD1) has also been reported to modulate Snail1 function, by phosphorylating S11 and promoting its nuclear export triggered through 14-3-3σ binding.117 Moreover, PKD1 inhibits the activity of DNA-bound Snail1, likely preventing its interaction with co-repressors.118 However, these results have to be confirmed since other authors have demonstrated that PKD1 phosphorylation enhances nuclear Snail1 transcriptional repression activity.119
The group of kinases promoting Snail1 stabilization is also abundant. PKA and CK2α phosphorylate Snail1 on S11 and S92, respectively, increasing Snail1 stability and repression of E-cadherin.120 Thus, the role of CK2 is unclear as indicated above it has been reported to induce Snail1 degradation.115,120 Other stabilizing phosphorylations affect residues in the C-terminal part of Snail1 and are catalyzed by 4 different kinases (Fig. 3): Lats2, p21 activated kinase 1 (PAK1), the Ataxia telangiectasia mutated (ATM) kinase and ERK2. Lats2 phosphorylation on Snail1-T203 is activated during TGF-β induced EMT and potentiates Snail1 activity by promoting nuclear retention.121 Similarly, PAK1 phosphorylates Snail1 on S246 enhancing its nuclear abundance and repressive potential.122 PAK1-dependent Snail1 phosphorylation on S246 is up-regulated in response to IR, an insult that strongly increases Snail1 stabilization (see above). In agreement with the notion that the DNA damage response may enhance tumor progression, both IR and the topoisomerase I poison camptothecin promote Snail1 phosphorylation on S100, which is catalyzed by the ATM kinase. Surprisingly, although S100 is also modified by GSK-3β inducing degradation, in the context of DNA damage phosphorylation of this residue prevents the interaction with GSK-3β and stabilizes Snail1.68,80 This has been related to the action of HSP90 that in these conditions binds to this phosphorylated residue precluding GSK-3β action on S96 and retaining Snail1 in the nucleus.68 Finally, DDR2 (discoidin domain receptor 2) activation regulates Snail1 stability by stimulating ERK2 and phosphorylating Snail1 on S82 and S104, which also leads to Snail1 nuclear accumulation and increased protein half-life.123 Therefore, it is likely that most of these modifications promote nuclear retention facilitating the interaction with nuclear chaperones, as it previsouly described for HSP90 and inhibiting binding to Snail1 of Crm1 nuclear exporter.68 Accordingly, another chaperone, HSP27 expressed during fibrosis and in prostate and breast cancer binds to and stabilizes Snail and consequently induces EMT.124-126
A recently described unexpected role for GSK-3β consists in controlling Twist stability and the interaction between Snail1/2 and Twist.127 Twist is phosphorylated by GSK-3β in the WR domain and this phosphorylation renders Twist less stable.127 However, in cells co-expressing Snail1, Twist is protected from destabilization. The proposed mechanism suggests that Snail1 binds to Twist and interferes with its interaction with Ppa/Fbxl14 ubiquitin ligase. Snail1-Twist association is potentiated by Twist1 phosphorylation by GSK-3β that promotes the interaction with the C-terminal zinc fingers of Snail1/2 and causes the loss of function of Snail1 since its recruitment to E-box sequences is impaired.
The role of other kinases on Snail2 and Twist1 stability has been less studied. Besides GSK-3β, Snail2 is phosphorylated on serines 4 and 88 to enhance its repressor activity of CDH1 expression and the induction of EMT.29 Twist may also be phosphorylated by CK2 on S18 and S20, or by MAPKs on S68, upon IL-6 or TGF-β stimulation, respectively, resulting in Twist1 protein stabilization.128,129 Finally, Snail1 phosphorylation by GSK-3β is reversed by the small C-terminal domain phosphatase (SCP), which interacts with Snail1 in the nucleus and prevents its nuclear export and degradation.130
Other post-translational modifications
Snail1 can be poly(ADP-ribosyl)ated by poly(ADP-ribose) polymerase 1 (PARP1), causing its stabilization.131 Another dynamic modification occurs on S112 Snail1 and consists of its modification by β-N-acetylglucosamine (O-GlcNAc). This modification prevents protein phosphorylation by GSK-3β and is a consequence of the activity of the O-GlcNAc transferase activated in hyperglycemic conditions. Therefore, glycosylated Snail1 cannot be phosphorylated by GSK-3β what enhances Snail1 action and provides a link between glucose metabolism and the control of EMT.132 Another relevant modification of EMT-inducing factors consists in Twist diacetylation that allows recruitment of BRD4, activation of the WNT5A promoter and the enhancement of EMT properties of basal-like breast cancer cells.133 Finally, factors inducing EMT are also sumoylated. So far only Zeb2 has been reported to be sumoylated on K391 and K866 by the polycomb protein Pc2, attenuating E-cadherin repression.134 Because inhibition of the sumoylation enzymes by TGF-β is relevant for the action of this cytokine in triggering EMT,135 it is important to elucidate whether sumoylation affects other EMT transcription factors.
Conclusion
Different reports indicate that Snail and Twist protein levels are rapidly upregulated in response to cellular stress as a survival mechanism of cancer cells. We have reviewed here the ubiquitin ligases acting on Snail and Twist and how they are regulated by different mechanisms including phosphorylation by different kinases. However, the E3 ligases controlling other transcriptional factors relevant for EMT, such as Zeb1 or 2, are practically unknown.46 Future work is also required to determine if additional E3 ligases may have a role in controlling Snail1 or Snail2 stability. For example, it has been recently reported that Slug could also be ubiquitinated by CHIP, a U box-containing E3 ubiquitin ligase, upon phosphorylation by GSK-3β.136 It is also probable that alternative mechanisms for stabilization of these EMT transcription factors exist. For example, one unexplored field is the role of deubiquitinases (DUBs), isopeptidases that specifically remove polyubiquitin chains from substrates stabilizing them.137 There are not examples of DUBs involved in EMT, besides the Ubiquitin Specific Protease-1 (USP1) that deubiquitinates and stabilizes Id proteins essential for the maintenance of the mesenchymal phenotype and prevention of osteoblastic differentiation.138 Another DUB called ubiquitin C-terminal hydrolase-1 (UCH-L1) promotes EMT but the target substrates are not known.139 It is probable that one or several DUBs enhance Snail, Twist and Zeb stability under pathological conditions. Because these DUBs are putative drugable entities, their future characterization will allow the generation of new inhibitors to block the EMT process.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Dr. J Baulida (IMIM, Barcelona) for advice.
Funding
Work at AGH and VMD's lab was supported by the Fundación Científica de la Asociación Española contra el Cáncer, the Ministerio de Economía y Competitividad [Grants SAF2013-48849-C2-1R and SAF2013-48849-C2-2R], Fundació La Marató de TV3 and also by RD12/0036/0005, part of “Plan Nacional de I+D+I” and cofunded by “ISCIII-Subdirección General de Evaluación and Fondo Europeo de Desarrollo Regional (FEDER).” RV-C was a recipient of a predoctoral fellowship from ISCIII.
References
- 1. Greenburg G, Hay ED. Epithelia suspended in collagen gels can lose polarity and express characteristics of migrating mesenchymal cells. J Cell Biol 1982; 95:333-9; PMID:7142291; http://dx.doi.org/ 10.1083/jcb.95.1.333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Thiery JP. Epithelial-mesenchymal transitions in development and pathologies. Curr Opin Cell Biol 2003; 15:740-6; PMID:14644200; http://dx.doi.org/ 10.1016/j.ceb.2003.10.006 [DOI] [PubMed] [Google Scholar]
- 3. Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol 2014; 15:178-96; PMID:24556840; http://dx.doi.org/ 10.1038/nrm3758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell 2009; 139:871-90; PMID:19945376; http://dx.doi.org/ 10.1016/j.cell.2009.11.007 [DOI] [PubMed] [Google Scholar]
- 5. Yang J, Weinberg RA. Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Dev Cell 2008; 14:818-29; PMID:18539112; http://dx.doi.org/ 10.1016/j.devcel.2008.05.009 [DOI] [PubMed] [Google Scholar]
- 6. Lim J, Thiery JP. Epithelial-mesenchymal transitions: insights from development. Development 2012; 139:3471-86; PMID:22949611; http://dx.doi.org/ 10.1242/dev.071209 [DOI] [PubMed] [Google Scholar]
- 7. Peinado H, Olmeda D, Cano A. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat Rev Cancer 2007; 7:415-28; PMID:17508028; http://dx.doi.org/ 10.1038/nrc2131 [DOI] [PubMed] [Google Scholar]
- 8. Rhim AD, Mirek ET, Aiello NM, Maitra A, Bailey JM, McAllister F, McAllister F, Reichert M, Beatty GL, Rustgi AK, et al. . EMT and dissemination precede pancreatic tumor formation. Cell 2012; 148:349-61; PMID:22265420; http://dx.doi.org/ 10.1016/j.cell.2011.11.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Taddei ML, Giannoni E, Comito G, Chiarugi P. Microenvironment and tumor cell plasticity: an easy way out. Cancer Lett 2013; 341:80-96; PMID:23376253; http://dx.doi.org/ 10.1016/j.canlet.2013.01.042 [DOI] [PubMed] [Google Scholar]
- 10. Friedl P, Alexander S. Cancer invasion and the microenvironment: plasticity and reciprocity. Cell 2011; 147:992-1009; PMID:22118458; http://dx.doi.org/ 10.1016/j.cell.2011.11.016 [DOI] [PubMed] [Google Scholar]
- 11. Hanahan D, Coussens LM. Accessories to the crime: functions of cells recruited to the tumor microenvironment. Cancer Cell 2012; 21:309-22; PMID:22439926; http://dx.doi.org/ 10.1016/j.ccr.2012.02.022 [DOI] [PubMed] [Google Scholar]
- 12. Tester AM, Ruangpanit N, Anderson RL, Thompson EW. MMP-9 secretion and MMP-2 activation distinguish invasive and metastatic sublines of a mouse mammary carcinoma system showing epithelial-mesenchymal transition traits. Clin Exp Metast 2000; 18:553-60; http://dx.doi.org/ 10.1023/A:1011953118186. [DOI] [PubMed] [Google Scholar]
- 13. Leroy P, Mostov KE. Slug is required for cell survival during partial epithelial-mesenchymal transition of HGF-induced tubulogenesis. Mol Biol Cell 2007; 18:1943-52; PMID:17344479; http://dx.doi.org/ 10.1091/mbc.E06-09-0823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Arnoux V, Come C, Kusewitt D, Hudson L, Savagner P. Cutaneous wound healing: a partial and reversible EMT. In: Savagner P, ed. Rise and fall of epithelial phenotype: Concepts of epithelial-mesenchymal transition. New York, United States: Landes Biosciences, 2005:111-34. [Google Scholar]
- 15. de Herreros AG, Peiro S, Nassour M, Savagner P. Snail family regulation and epithelial mesenchymal transitions in breast cancer progression. J Mammary Gland Biol 2010; 15:135-47; http://dx.doi.org/ 10.1007/s10911-010-9179-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wheelock MJ, Johnson KR. Cadherins as modulators of cellular phenotype. Ann Rev Cell Dev Biol 2003; 19:207-35; http://dx.doi.org/ 10.1146/annurev.cellbio.19.011102.111135 [DOI] [PubMed] [Google Scholar]
- 17. Batlle E, Sancho E, Franci C, Dominguez D, Monfar M, Baulida J, García De, Herreros A. The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat Cell Biol 2000; 2:84-9; PMID:10655587; http://dx.doi.org/ 10.1038/35000034 [DOI] [PubMed] [Google Scholar]
- 18. Cano A, Perez-Moreno MA, Rodrigo I, Locascio A, Blanco MJ, del Barrio MG, Portillo F, Nieto MA. The transcription factor snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat Cell Biol 2000; 2:76-83; PMID:10655586; http://dx.doi.org/ 10.1038/35000025 [DOI] [PubMed] [Google Scholar]
- 19. Peinado H, Ballestar E, Esteller M, Cano A. Snail mediates E-cadherin repression by the recruitment of the Sin3A/histone deacetylase 1 (HDAC1)/HDAC2 complex. Mol Cell Biol 2004; 24:306-19; PMID:14673164; http://dx.doi.org/ 10.1128/MCB.24.1.306-319.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Herranz N, Pasini D, Diaz VM, Franci C, Gutierrez A, Dave N, Escrivà M, Hernandez-Muñoz I, Di Croce L, Helin K, et al. . Polycomb complex 2 is required for E-cadherin repression by the Snail1 transcription factor. Mol Cell Biol 2008; 28:4772-81; PMID:18519590; http://dx.doi.org/ 10.1128/MCB.00323-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hou Z, Peng H, Ayyanathan K, Yan KP, Langer EM, Longmore GD, Rauscher FJ 3rd. The LIM protein AJUBA recruits protein arginine methyltransferase 5 to mediate SNAIL-dependent transcriptional repression. Mol Cell Biol 2008; 28:3198-207; PMID:18347060; http://dx.doi.org/ 10.1128/MCB.01435-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lin Y, Wu Y, Li J, Dong C, Ye X, Chi YI, Evers BM, Zhou BP. The SNAG domain of Snail1 functions as a molecular hook for recruiting lysine-specific demethylase 1. Embo J 2010; 29:1803-16; PMID:20389281; http://dx.doi.org/ 10.1038/emboj.2010.63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tang M, Shen H, Jin Y, Lin T, Cai Q, Pinard MA, Biswas S, Tran Q, Li G, Shenoy AK, et al. . The malignant brain tumor (MBT) domain protein SFMBT1 is an integral histone reader subunit of the LSD1 demethylase complex for chromatin association and epithelial-to-mesenchymal transition. J Biolog Chem 2013; 288:27680-91; http://dx.doi.org/ 10.1074/jbc.M113.482349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dominguez D, Montserrat-Sentis B, Virgos-Soler A, Guaita S, Grueso J, Porta M, Puig I, Baulida J, Francí C, García de Herreros A. Phosphorylation regulates the subcellular location and activity of the snail transcriptional repressor. Mol Cell Biol 2003; 23:5078-89; PMID:12832491; http://dx.doi.org/ 10.1128/MCB.23.14.5078-5089.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sefton M, Sanchez S, Nieto MA. Conserved and divergent roles for members of the Snail family of transcription factors in the chick and mouse embryo. Development 1998; 125:3111-21; PMID:9671584 [DOI] [PubMed] [Google Scholar]
- 26. Ko H, Kim HS, Kim NH, Lee SH, Kim KH, Hong SH, Yook JI. Nuclear localization signals of the E-cadherin transcriptional repressor Snail. Cells Tissues Organs 2007; 185:66-72; PMID:17587810; http://dx.doi.org/ 10.1159/000101305 [DOI] [PubMed] [Google Scholar]
- 27. Mingot JM, Vega S, Maestro B, Sanz JM, Nieto MA. Characterization of Snail nuclear import pathways as representatives of C2H2 zinc finger transcription factors. J Cell Sci 2009; 122:1452-60; PMID:19386897; http://dx.doi.org/ 10.1242/jcs.041749 [DOI] [PubMed] [Google Scholar]
- 28. Cobaleda C, Perez-Caro M, Vicente-Duenas C, Sanchez-Garcia I. Function of the zinc-finger transcription factor SNAI2 in cancer and development. Annu Rev Genet 2007; 41:41-61; PMID:17550342; http://dx.doi.org/ 10.1146/annurev.genet.41.110306.130146 [DOI] [PubMed] [Google Scholar]
- 29. Molina-Ortiz P, Villarejo A, MacPherson M, Santos V, Montes A, Souchelnytskyi S, Portillo F, Cano A. Characterization of the SNAG and SLUG domains of Snail2 in the repression of E-cadherin and EMT induction: modulation by serine 4 phosphorylation. PLoS One 2012; 7:e36132; PMID:22567133; http://dx.doi.org/ 10.1371/journal.pone.0036132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mittal MK, Myers JN, Misra S, Bailey CK, Chaudhuri G. In vivo binding to and functional repression of the VDR gene promoter by SLUG in human breast cells. Biochem Biophys Res Commun 2008; 372:30-4; PMID:18485278; http://dx.doi.org/ 10.1016/j.bbrc.2008.04.187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Carver EA, Jiang R, Lan Y, Oram KF, Gridley T. The mouse snail gene encodes a key regulator of the epithelial-mesenchymal transition. Mol Cell Biol 2001; 21:8184-8; PMID:11689706; http://dx.doi.org/ 10.1128/MCB.21.23.8184-8188.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jiang R, Lan Y, Norton CR, Sundberg JP, Gridley T. The Slug gene is not essential for mesoderm or neural crest development in mice. Dev Biol 1998; 198:277-85; PMID:9659933; http://dx.doi.org/ 10.1016/S0012-1606(98)80005-5 [DOI] [PubMed] [Google Scholar]
- 33. Gheldof A, Hulpiau P, van Roy F, De Craene B, Berx G. Evolutionary functional analysis and molecular regulation of the ZEB transcription factors. Cell Mol Life Sci 2012; 69:2527-41; PMID:22349261; http://dx.doi.org/ 10.1007/s00018-012-0935-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Vandewalle C, Comijn J, De Craene B, Vermassen P, Bruyneel E, Andersen H, Tulchinsky E, Van Roy F, Berx G. SIP1/ZEB2 induces EMT by repressing genes of different epithelial cell-cell junctions. Nucleic Acids Res 2005; 33:6566-78; PMID:16314317; http://dx.doi.org/ 10.1093/nar/gki965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Drake JM, Strohbehn G, Bair TB, Moreland JG, Henry MD. ZEB1 enhances transendothelial migration and represses the epithelial phenotype of prostate cancer cells. Mol Biol Cell 2009; 20:2207-17; http://dx.doi.org/ 10.1091/mbc.E08-10-1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sanchez-Tillo E, Liu Y, de Barrios O, Siles L, Fanlo L, Cuatrecasas M, Darling DS, Dean DC, Castells A, Postigo A. EMT-activating transcription factors in cancer: beyond EMT and tumor invasiveness. Cell Mol Life Sci 2012; 69:3429-56; PMID:22945800; http://dx.doi.org/ 10.1007/s00018-012-1122-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Postigo AA, Dean DC. ZEB represses transcription through interaction with the corepressor CtBP. Proc Natl Acad Sci U S A 1999; 96:6683-8; PMID:10359772; http://dx.doi.org/ 10.1073/pnas.96.12.6683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sanchez-Tillo E, Lazaro A, Torrent R, Cuatrecasas M, Vaquero EC, Castells A, Engel P, Postigo A. ZEB1 represses E-cadherin and induces an EMT by recruiting the SWI/SNF chromatin-remodeling protein BRG1. Oncogene 2010; 29:3490-500; PMID:20418909; http://dx.doi.org/ 10.1038/onc.2010.102 [DOI] [PubMed] [Google Scholar]
- 39. Sanchez-Tillo E, de Barrios O, Siles L, Cuatrecasas M, Castells A, Postigo A. beta-catenin/TCF4 complex induces the epithelial-to-mesenchymal transition (EMT)-activator ZEB1 to regulate tumor invasiveness. Proc Natl Acad Sci U S A 2011; 108:19204-9; PMID:22080605; http://dx.doi.org/ 10.1073/pnas.1108977108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Dave N, Guaita-Esteruelas S, Gutarra S, Frias A, Beltran M, Peiro S, de Herreros AG. Functional cooperation between Snail1 and twist in the regulation of ZEB1 expression during epithelial to mesenchymal transition. J Biol Chem 2011; 286:12024-32; PMID:21317430; http://dx.doi.org/ 10.1074/jbc.M110.168625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Guaita S, Puig I, Franci C, Garrido M, Dominguez D, Batlle E, Sancho E, Dedhar S, De Herreros AG, Baulida J. Snail induction of epithelial to mesenchymal transition in tumor cells is accompanied by MUC1 repression and ZEB1 expression. J Biol Chem 2002; 277:39209-16; PMID:12161443; http://dx.doi.org/ 10.1074/jbc.M206400200 [DOI] [PubMed] [Google Scholar]
- 42. Beltran M, Puig I, Pena C, Garcia JM, Alvarez AB, Pena R, Bonilla F, de Herreros AG. A natural antisense transcript regulates Zeb2/Sip1 gene expression during Snail1-induced epithelial-mesenchymal transition. Genes Dev 2008; 22:756-69; PMID:18347095; http://dx.doi.org/ 10.1101/gad.455708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ellenberger T, Fass D, Arnaud M, Harrison SC. Crystal structure of transcription factor E47: E-box recognition by a basic region helix-loop-helix dimer. Genes Dev 1994; 8:970-80; PMID:7926781; http://dx.doi.org/ 10.1101/gad.8.8.970 [DOI] [PubMed] [Google Scholar]
- 44. Teng Y, Li X. The roles of HLH transcription factors in epithelial mesenchymal transition and multiple molecular mechanisms. Clin Exp Metast 2014; 31:367-77; http://dx.doi.org/ 10.1007/s10585-013-9621-6 [DOI] [PubMed] [Google Scholar]
- 45. Qin Q, Xu Y, He T, Qin C, Xu J. Normal and disease-related biological functions of Twist1 and underlying molecular mechanisms. Cell Res 2012; 22:90-106; PMID:21876555; http://dx.doi.org/ 10.1038/cr.2011.144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lander R, Nordin K, LaBonne C. The F-box protein Ppa is a common regulator of core EMT factors Twist, Snail, Slug, and Sip1. J Cell Biol 2011; 194:17-25; PMID:21727196; http://dx.doi.org/ 10.1083/jcb.201012085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Fu J, Qin L, He T, Qin J, Hong J, Wong J, Liao L, Xu J. The TWIST/Mi2/NuRD protein complex and its essential role in cancer metastasis. Cell Res 2011; 21:275-89; PMID:20714342; http://dx.doi.org/ 10.1038/cr.2010.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yang F, Sun L, Li Q, Han X, Lei L, Zhang H, Shang Y. SET8 promotes epithelial-mesenchymal transition and confers TWIST dual transcriptional activities. EMBO J 2012; 31:110-23; PMID:21983900; http://dx.doi.org/ 10.1038/emboj.2011.364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Franco HL, Casasnovas J, Rodriguez-Medina JR, Cadilla CL. Redundant or separate entities?–roles of Twist1 and Twist2 as molecular switches during gene transcription. Nucleic Acids Res 2011; 39:1177-86; PMID:20935057; http://dx.doi.org/ 10.1093/nar/gkq890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yang MH, Wu MZ, Chiou SH, Chen PM, Chang SY, Liu CJ, Teng SC, Wu KJ. Direct regulation of TWIST by HIF-1alpha promotes metastasis. Nat Cell Biol 2008; 10:295-305; PMID:18297062; http://dx.doi.org/ 10.1038/ncb1691 [DOI] [PubMed] [Google Scholar]
- 51. Thiery JP, Sleeman JP. Complex networks orchestrate epithelial-mesenchymal transitions. Nat Rev Mol Cell Biol 2006; 7:131-42; PMID:16493418; http://dx.doi.org/ 10.1038/nrm1835 [DOI] [PubMed] [Google Scholar]
- 52. Grotegut S, von Schweinitz D, Christofori G, Lehembre F. Hepatocyte growth factor induces cell scattering through MAPK/Egr-1-mediated upregulation of Snail. EMBO J 2006; 25:3534-45; PMID:16858414; http://dx.doi.org/ 10.1038/sj.emboj.7601213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Fuxe J, Vincent T, Garcia de Herreros A. Transcriptional crosstalk between TGF-beta and stem cell pathways in tumor cell invasion: role of EMT promoting Smad complexes. Cell Cycle 2010; 9:2363-74; PMID:20519943; http://dx.doi.org/ 10.4161/cc.9.12.12050 [DOI] [PubMed] [Google Scholar]
- 54. Giannoni E, Parri M, Chiarugi P. EMT and oxidative stress: a bidirectional interplay affecting tumor malignancy. Antioxid Redox Signal 2012; 16:1248-63; PMID:21929373; http://dx.doi.org/ 10.1089/ars.2011.4280 [DOI] [PubMed] [Google Scholar]
- 55. Chen J, Imanaka N, Griffin JD. Hypoxia potentiates Notch signaling in breast cancer leading to decreased E-cadherin expression and increased cell migration and invasion. Br J Cancer 2010; 102:351-60; PMID:20010940; http://dx.doi.org/ 10.1038/sj.bjc.6605486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kim CH, Jeon HM, Lee SY, Ju MK, Moon JY, Park HG, Yoo MA, Choi BT, Yook JI, Lim SC, et al. . Implication of snail in metabolic stress-induced necrosis. PLoS One 2011; 6:e18000; PMID:21448462; http://dx.doi.org/ 10.1371/journal.pone.0018000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Dong R, Lu JG, Wang Q, He XL, Chu YK, Ma QJ. Stabilization of Snail by HuR in the process of hydrogen peroxide induced cell migration. Biochem Biophys Res Commun 2007; 356:318-21; PMID:17350594; http://dx.doi.org/ 10.1016/j.bbrc.2007.02.145 [DOI] [PubMed] [Google Scholar]
- 58. Radisky DC, Levy DD, Littlepage LE, Liu H, Nelson CM, Fata JE, Leake D, Godden EL, Albertson DG, Nieto MA, et al. . Rac1b and reactive oxygen species mediate MMP-3-induced EMT and genomic instability. Nat 2005; 436:123-7; http://dx.doi.org/ 10.1038/nature03688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Barnett P, Arnold RS, Mezencev R, Chung LW, Zayzafoon M, Odero-Marah V. Snail-mediated regulation of reactive oxygen species in ARCaP human prostate cancer cells. Biochem Biophys Res Commun 2011; 404:34-9; PMID:21093414; http://dx.doi.org/ 10.1016/j.bbrc.2010.11.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kajita M, McClinic KN, Wade PA. Aberrant expression of the transcription factors snail and slug alters the response to genotoxic stress. Mol Cell Biol 2004; 24:7559-66; PMID:15314165; http://dx.doi.org/ 10.1128/MCB.24.17.7559-7566.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Rosano L, Cianfrocca R, Spinella F, Di Castro V, Nicotra MR, Lucidi A, Ferrandina G, Natali PG, Bagnato A. Acquisition of chemoresistance and EMT phenotype is linked with activation of the endothelin A receptor pathway in ovarian carcinoma cells. Clin Cancer Res 2011; 17:2350-60; PMID:21220476; http://dx.doi.org/ 10.1158/1078-0432.CCR-10-2325 [DOI] [PubMed] [Google Scholar]
- 62. Hoshino H, Miyoshi N, Nagai K, Tomimaru Y, Nagano H, Sekimoto M, et al. . Epithelial-mesenchymal transition with expression of SNAI1-induced chemoresistance in colorectal cancer. Biochem Biophys Res Commun 2009; 390:1061-5; PMID:19861116; http://dx.doi.org/ 10.1016/j.bbrc.2009.10.117 [DOI] [PubMed] [Google Scholar]
- 63. Zhang W, Feng M, Zheng G, Chen Y, Wang X, Pen B, et al. . Chemoresistance to 5-fluorouracil induces epithelial-mesenchymal transition via up-regulation of Snail in MCF7 human breast cancer cells. Biochem Biophys Res Commun 2012; 417:679-85; PMID:22166209; http://dx.doi.org/ 10.1016/j.bbrc.2011.11.142 [DOI] [PubMed] [Google Scholar]
- 64. Haslehurst AM, Koti M, Dharsee M, Nuin P, Evans K, Geraci J, et al. . EMT transcription factors snail and slug directly contribute to cisplatin resistance in ovarian cancer. BMC Cancer 2012; 12:91; PMID:22429801; http://dx.doi.org/ 10.1186/1471-2407-12-91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Izumiya M, Kabashima A, Higuchi H, Igarashi T, Sakai G, Iizuka H, et al. . Chemoresistance Is Associated with Cancer Stem Cell-like Properties and Epithelial-to-Mesenchymal Transition in Pancreatic Cancer Cells. Anticancer Res 2012; 32:3847-53; PMID:22993328 [PubMed] [Google Scholar]
- 66. Zhou YC, Liu JY, Li J, Zhang J, Xu YQ, Zhang HW, et al. . Ionizing radiation promotes migration and invasion of cancer cells through transforming growth factor-beta-mediated epithelial-mesenchymal transition. Int J Radiat Oncol Biol Phys 2011; 81:1530-7; PMID:22115555; http://dx.doi.org/ 10.1016/j.ijrobp.2011.06.1956 [DOI] [PubMed] [Google Scholar]
- 67. Nagarajan D, Melo T, Deng Z, Almeida C, Zhao W. ERK/GSK3beta/Snail signaling mediates radiation-induced alveolar epithelial-to-mesenchymal transition. Free Radic Biol Med 2012; 52:983-92; PMID:22198183; http://dx.doi.org/ 10.1016/j.freeradbiomed.2011.11.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Sun M, Guo X, Qian X, Wang H, Yang C, Brinkman KL, et al. . Activation of the ATM-Snail pathway promotes breast cancer metastasis. J Mol Cell Biol 2012; 4:304-15; PMID:22923499; http://dx.doi.org/ 10.1093/jmcb/mjs048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Vinas-Castells R, Frias A, Robles-Lanuza E, Zhang K, Longmore GD, Garcia de Herreros A, et al. . Nuclear ubiquitination by FBXL5 modulates Snail1 DNA binding and stability. Nucleic Acids Res 2014; 42:1079-94; PMID:24157836; http://dx.doi.org/ 10.1093/nar/gkt935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Wu WS, Heinrichs S, Xu D, Garrison SP, Zambetti GP, Adams JM, et al. . Slug antagonizes p53-mediated apoptosis of hematopoietic progenitors by repressing puma. Cell 2005; 123:641-53; PMID:16286009; http://dx.doi.org/ 10.1016/j.cell.2005.09.029 [DOI] [PubMed] [Google Scholar]
- 71. Escriva M, Peiro S, Herranz N, Villagrasa P, Dave N, Montserrat-Sentis B, et al. . Repression of PTEN phosphatase by Snail1 transcriptional factor during gamma radiation-induced apoptosis. Mol Cell Biol 2008; 28:1528-40; PMID:18172008; http://dx.doi.org/ 10.1128/MCB.02061-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Inoue A, Seidel MG, Wu W, Kamizono S, Ferrando AA, Bronson RT, et al. . Slug, a highly conserved zinc finger transcriptional repressor, protects hematopoietic progenitor cells from radiation-induced apoptosis in vivo. Cancer Cell 2002; 2:279-88; PMID:12398892; http://dx.doi.org/ 10.1016/S1535-6108(02)00155-1 [DOI] [PubMed] [Google Scholar]
- 73. Weissman AM, Shabek N, Ciechanover A. The predator becomes the prey: regulating the ubiquitin system by ubiquitylation and degradation. Nat Rev Mol Cell Biol 2011; 12:605-20; PMID:21860393; http://dx.doi.org/ 10.1038/nrm3173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Lipkowitz S, Weissman AM. RINGs of good and evil: RING finger ubiquitin ligases at the crossroads of tumour suppression and oncogenesis. Nat Rev Cancer 2011; 11:629-43; PMID:21863050; http://dx.doi.org/ 10.1038/nrc3120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Cardozo T, Pagano M. The SCF ubiquitin ligase: insights into a molecular machine. Nat Rev Mol Cell Biol 2004; 5:739-51; PMID:15340381; http://dx.doi.org/ 10.1038/nrm1471 [DOI] [PubMed] [Google Scholar]
- 76. Skaar JR, Pagan JK, Pagano M. Mechanisms and function of substrate recruitment by F-box proteins. Nat Rev Mol Cell Biol 2013; 14:369-81; PMID:23657496; http://dx.doi.org/ 10.1038/nrm3582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Wang Z, Liu P, Inuzuka H, Wei W. Roles of F-box proteins in cancer. Nat Rev Cancer 2014; 14:233-47; PMID:24658274; http://dx.doi.org/ 10.1038/nrc3700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Jin J, Cardozo T, Lovering RC, Elledge SJ, Pagano M, Harper JW. Systematic analysis and nomenclature of mammalian F-box proteins. Genes Dev 2004; 18:2573-80; PMID:15520277; http://dx.doi.org/ 10.1101/gad.1255304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Varshavsky A. Naming a targeting signal. Cell 1991; 64:13-5; . [DOI] [PubMed] [Google Scholar]
- 80. Zhou BP, Deng J, Xia W, Xu J, Li YM, Gunduz M, et al. . Dual regulation of Snail by GSK-3beta-mediated phosphorylation in control of epithelial-mesenchymal transition. Nat Cell Biol 2004; 6:931-40; PMID:15448698; http://dx.doi.org/ 10.1038/ncb1173 [DOI] [PubMed] [Google Scholar]
- 81. Wu G, Xu G, Schulman BA, Jeffrey PD, Harper JW, Pavletich NP. Structure of a beta-TrCP1-Skp1-beta-catenin complex: destruction motif binding and lysine specificity of the SCF(beta-TrCP1) ubiquitin ligase. Mol Cell 2003; 11:1445-56; PMID:12820959; http://dx.doi.org/ 10.1016/S1097-2765(03)00234-X [DOI] [PubMed] [Google Scholar]
- 82. Skaar JR, D'Angiolella V, Pagan JK, Pagano M. SnapShot: F Box Proteins II. Cell 2009; 137:1358, e1; PMID:19563764 [DOI] [PubMed] [Google Scholar]
- 83. Yook JI, Li XY, Ota I, Fearon ER, Weiss SJ. Wnt-dependent regulation of the E-cadherin repressor snail. J Biol Chem 2005; 280:11740-8; PMID:15647282; http://dx.doi.org/ 10.1074/jbc.M413878200 [DOI] [PubMed] [Google Scholar]
- 84. Kim JY, Kim YM, Yang CH, Cho SK, Lee JW, Cho M. Functional regulation of Slug/Snail2 is dependent on GSK-3beta-mediated phosphorylation. Febs J 2012; 279:2929-39; PMID:22727060; http://dx.doi.org/ 10.1111/j.1742-4658.2012.08674.x [DOI] [PubMed] [Google Scholar]
- 85. Wu ZQ, Li XY, Hu CY, Ford M, Kleer CG, Weiss SJ. Canonical Wnt signaling regulates Slug activity and links epithelial-mesenchymal transition with epigenetic Breast Cancer 1, Early Onset (BRCA1) repression. Proc Natl Acad Sci U S A 2012; 109:16654-9; PMID:23011797; http://dx.doi.org/ 10.1073/pnas.1205822109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Morimoto M, Nishida T, Nagayama Y, Yasuda H. Nedd8-modification of Cul1 is promoted by Roc1 as a Nedd8-E3 ligase and regulates its stability. Biochem Biophys Res Commun 2003; 301:392-8; PMID:12565873; http://dx.doi.org/ 10.1016/S0006-291X(02)03051-6 [DOI] [PubMed] [Google Scholar]
- 87. Wu Y, Deng J, Rychahou PG, Qiu S, Evers BM, Zhou BP. Stabilization of snail by NF-kappaB is required for inflammation-induced cell migration and invasion. Cancer Cell 2009; 15:416-28; PMID:19411070; http://dx.doi.org/ 10.1016/j.ccr.2009.03.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Guardavaccaro D, Kudo Y, Boulaire J, Barchi M, Busino L, Donzelli M, et al. . Control of meiotic and mitotic progression by the F box protein β-Trcp1 in vivo. Developmental cell 2003; 4:799-812; PMID:12791266; http://dx.doi.org/ 10.1016/S1534-5807(03)00154-0 [DOI] [PubMed] [Google Scholar]
- 89. Kanarek N, Horwitz E, Mayan I, Leshets M, Cojocaru G, Davis M, et al. . Spermatogenesis rescue in a mouse deficient for the ubiquitin ligase SCF{beta}-TrCP by single substrate depletion. Genes Dev 2010; 24:470-7; PMID:20194439 http://dx.doi.org/ 10.1101/gad.551610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Hart M, Concordet JP, Lassot I, Albert I, del los Santos R, Durand H, et al. . The F-box protein beta-TrCP associates with phosphorylated beta-catenin and regulates its activity in the cell. Curr Biol 1999; 9:207-10; PMID:10074433; http://dx.doi.org/ 10.1016/S0960-9822(99)80091-8 [DOI] [PubMed] [Google Scholar]
- 91. Vinas-Castells R, Beltran M, Valls G, Gomez I, Garcia JM, Montserrat-Sentis B, et al. . The hypoxia-controlled FBXL14 ubiquitin ligase targets SNAIL1 for proteasome degradation. J Biol Chem 2010; 285:3794-805; PMID:19955572; http://dx.doi.org/ 10.1074/jbc.M109.065995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Vernon AE, LaBonne C. Slug stability is dynamically regulated during neural crest development by the F-box protein Ppa. Development 2006; 133:3359-70; PMID:16887825; http://dx.doi.org/ 10.1242/dev.02504 [DOI] [PubMed] [Google Scholar]
- 93. Zheng H, Du Y, Hua Y, Wu Z, Yan Y, Li Y. Essential role of Fbxl14 ubiquitin ligase in regulation of vertebrate axis formation through modulating Mkp3 level. Cell Res 2012; 22:936-40; PMID:22410791; http://dx.doi.org/ 10.1038/cr.2012.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Das T, Purkayastha-Mukherjee C, D'Angelo J, Weir M. A conserved F-box gene with unusual transcript localization. Dev Genes Evol 2002; 212:134-40; PMID:11976951; http://dx.doi.org/ 10.1007/s00427-002-0222-7 [DOI] [PubMed] [Google Scholar]
- 95. Cannito S, Novo E, Compagnone A, Valfre di Bonzo L, Busletta C, Zamara E, et al. . Redox mechanisms switch on hypoxia-dependent epithelial-mesenchymal transition in cancer cells. Carcinogenesis 2008; 29:2267-78; PMID:18791199; http://dx.doi.org/ 10.1093/carcin/bgn216 [DOI] [PubMed] [Google Scholar]
- 96. Salahudeen AA, Thompson JW, Ruiz JC, Ma HW, Kinch LN, Li Q, et al. . An E3 ligase possessing an iron-responsive hemerythrin domain is a regulator of iron homeostasis. Science 2009; 326:722-6; PMID:19762597; http://dx.doi.org/ 10.1126/science.1176326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Vashisht AA, Zumbrennen KB, Huang X, Powers DN, Durazo A, Sun D, et al. . Control of iron homeostasis by an iron-regulated ubiquitin ligase. Science 2009; 326:718-21; PMID:19762596; http://dx.doi.org/ 10.1126/science.1176333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. O'Keefe K, Li H, Zhang Y. Nucleocytoplasmic shuttling of p53 is essential for MDM2-mediated cytoplasmic degradation but not ubiquitination. Mol Cell Biol 2003; 23:6396-405; PMID:12944468; http://dx.doi.org/ 10.1128/MCB.23.18.6396-6405.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Rockel TD, Stuhlmann D, von Mikecz A. Proteasomes degrade proteins in focal subdomains of the human cell nucleus. J Cell Sci 2005; 118:5231-42; PMID:16249232; http://dx.doi.org/ 10.1242/jcs.02642 [DOI] [PubMed] [Google Scholar]
- 100. Haro KJ, Sheth A, Scheinberg DA. Dysregulation of IRP1-mediated iron metabolism causes gamma ray-specific radioresistance in leukemia cells. PLoS One 2012; 7:e48841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Moroishi T, Yamauchi T, Nishiyama M, Nakayama KI. HERC2 Targets the Iron Regulator FBXL5 for Degradation and Modulates Iron Metabolism. J Biolog Chem 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Moroishi T, Nishiyama M, Takeda Y, Iwai K, Nakayama KI. The FBXL5-IRP2 axis is integral to control of iron metabolism in vivo. Cell Metab 2011; 14:339-51; PMID:21907140; http://dx.doi.org/ 10.1016/j.cmet.2011.07.011 [DOI] [PubMed] [Google Scholar]
- 103. Kubbutat MH, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nat 1997; 387:299-303; http://dx.doi.org/ 10.1038/387299a0 [DOI] [PubMed] [Google Scholar]
- 104. Li M, Brooks CL, Wu-Baer F, Chen D, Baer R, Gu W. Mono- versus polyubiquitination: differential control of p53 fate by Mdm2. Science 2003; 302:1972-5; PMID:14671306; http://dx.doi.org/ 10.1126/science.1091362 [DOI] [PubMed] [Google Scholar]
- 105. Wang SP, Wang WL, Chang YL, Wu CT, Chao YC, Kao SH, et al. . p53 controls cancer cell invasion by inducing the MDM2-mediated degradation of Slug. Nat Cell Biol 2009; 11:694-704; PMID:19448627; http://dx.doi.org/ 10.1038/ncb1875 [DOI] [PubMed] [Google Scholar]
- 106. Lim SO, Kim H, Jung G. p53 inhibits tumor cell invasion via the degradation of snail protein in hepatocellular carcinoma. FEBS Lett 2010; 584:2231-6; PMID:20385133; http://dx.doi.org/ 10.1016/j.febslet.2010.04.006 [DOI] [PubMed] [Google Scholar]
- 107. Kim NH, Kim HS, Li XY, Lee I, Choi HS, Kang SE, et al. . A p53/miRNA-34 axis regulates Snail1-dependent cancer cell epithelial-mesenchymal transition. J Cell Biol 2011; 195:417-33; PMID:22024162; http://dx.doi.org/ 10.1083/jcb.201103097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Chang CJ, Chao CH, Xia W, Yang JY, Xiong Y, Li CW, et al. . p53 regulates epithelial-mesenchymal transition and stem cell properties through modulating miRNAs. Nat Cell Biol 2011; 13:317-23; PMID:21336307; http://dx.doi.org/ 10.1038/ncb2173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Yu ZK, Geyer RK, Maki CG. MDM2-dependent ubiquitination of nuclear and cytoplasmic P53. Oncogene 2000; 19:5892-7; PMID:11127820; http://dx.doi.org/ 10.1038/sj.onc.1203980 [DOI] [PubMed] [Google Scholar]
- 110. Piccinin S, Tonin E, Sessa S, Demontis S, Rossi S, Pecciarini L, et al. . A "twist box" code of p53 inactivation: twist box: p53 interaction promotes p53 degradation. Cancer Cell 2012; 22:404-15; PMID:22975381; http://dx.doi.org/ 10.1016/j.ccr.2012.08.003 [DOI] [PubMed] [Google Scholar]
- 111. Inuzuka H, Tseng A, Gao D, Zhai B, Zhang Q, Shaik S, et al. . Phosphorylation by casein kinase I promotes the turnover of the Mdm2 oncoprotein via the SCF(beta-TRCP) ubiquitin ligase. Cancer Cell 2010; 18:147-59; PMID:20708156; http://dx.doi.org/ 10.1016/j.ccr.2010.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Yook JI, Li XY, Ota I, Hu C, Kim HS, Kim NH, et al. . A Wnt-Axin2-GSK3beta cascade regulates Snail1 activity in breast cancer cells. Nat Cell Biol 2006; 8:1398-406; PMID:17072303; http://dx.doi.org/ 10.1038/ncb1508 [DOI] [PubMed] [Google Scholar]
- 113. Speirs CK, Jernigan KK, Kim SH, Cha YI, Lin F, Sepich DS, et al. . Prostaglandin Gbetagamma signaling stimulates gastrulation movements by limiting cell adhesion through Snai1a stabilization. Development 2010; 137:1327-37; PMID:20332150; http://dx.doi.org/ 10.1242/dev.045971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Xu Y, Lee SH, Kim HS, Kim NH, Piao S, Park SH, et al. . Role of CK1 in GSK3beta-mediated phosphorylation and degradation of snail. Oncogene 2010; 29:3124-33; PMID:20305697; http://dx.doi.org/ 10.1038/onc.2010.77 [DOI] [PubMed] [Google Scholar]
- 115. Deshiere A, Duchemin-Pelletier E, Spreux E, Ciais D, Combes F, Vandenbrouck Y, et al. . Unbalanced expression of CK2 kinase subunits is sufficient to drive epithelial-to-mesenchymal transition by Snail1 induction. Oncogene 2013; 32:1373-83; PMID:22562247; http://dx.doi.org/ 10.1038/onc.2012.165 [DOI] [PubMed] [Google Scholar]
- 116. Mimoto R, Taira N, Takahashi H, Yamaguchi T, Okabe M, Uchida K, et al. . DYRK2 controls the epithelial-mesenchymal transition in breast cancer by degrading Snail. Cancer Lett 2013; 339:214-25; PMID:23791882; http://dx.doi.org/ 10.1016/j.canlet.2013.06.005 [DOI] [PubMed] [Google Scholar]
- 117. Du C, Zhang C, Hassan S, Biswas MH, Balaji KC. Protein kinase D1 suppresses epithelial-to-mesenchymal transition through phosphorylation of snail. Cancer Res 2010; 70:7810-9; PMID:20940406; http://dx.doi.org/ 10.1158/0008-5472.CAN-09-4481 [DOI] [PubMed] [Google Scholar]
- 118. Bastea LI, Doppler H, Balogun B, Storz P. Protein kinase D1 maintains the epithelial phenotype by inducing a DNA-bound, inactive SNAI1 transcriptional repressor complex. PloS one 2012; 7:e30459; PMID:22276203; http://dx.doi.org/ 10.1371/journal.pone.0030459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Eiseler T, Kohler C, Nimmagadda SC, Jamali A, Funk N, Joodi G, et al. . Protein Kinase D1 Mediates Anchorage-dependent and -independent Growth of Tumor Cells via the Zinc Finger Transcription Factor Snail1. J Biol Chem 2012; 287:32367-80; PMID:22791710; http://dx.doi.org/ 10.1074/jbc.M112.370999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. MacPherson MR, Molina P, Souchelnytskyi S, Wernstedt C, Martin-Perez J, Portillo F, et al. . Phosphorylation of serine 11 and serine 92 as new positive regulators of human Snail1 function: potential involvement of casein kinase-2 and the cAMP-activated kinase protein kinase A. Mol Biol Cell 2010; 21:244-53; PMID:19923321; http://dx.doi.org/ 10.1091/mbc.E09-06-0504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Zhang K, Rodriguez-Aznar E, Yabuta N, Owen RJ, Mingot JM, Nojima H, et al. . Lats2 kinase potentiates Snail1 activity by promoting nuclear retention upon phosphorylation. Embo J 2012; 31:29-43; PMID:21952048; http://dx.doi.org/ 10.1038/emboj.2011.357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Yang Z, Rayala S, Nguyen D, Vadlamudi RK, Chen S, Kumar R. Pak1 phosphorylation of snail, a master regulator of epithelial-to-mesenchyme transition, modulates snail's subcellular localization and functions. Cancer Res 2005; 65:3179-84; PMID:15833848 [DOI] [PubMed] [Google Scholar]
- 123. Zhang K, Corsa CA, Ponik SM, Prior JL, Piwnica-Worms D, Eliceiri KW, et al. . The collagen receptor discoidin domain receptor 2 stabilizes SNAIL1 to facilitate breast cancer metastasis. Nat Cell Biol 2013; 15:677-87; PMID:23644467; http://dx.doi.org/ 10.1038/ncb2743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Wettstein G, Bellaye PS, Kolb M, Hammann A, Crestani B, Soler P, et al. . Inhibition of HSP27 blocks fibrosis development and EMT features by promoting Snail degradation. Faseb J 2013; 27:1549-60; PMID:23288928; http://dx.doi.org/ 10.1096/fj.12-220053 [DOI] [PubMed] [Google Scholar]
- 125. Shiota M, Bishop JL, Nip KM, Zardan A, Takeuchi A, Cordonnier T, et al. . Hsp27 regulates epithelial mesenchymal transition, metastasis, and circulating tumor cells in prostate cancer. Cancer Res 2013; 73:3109-19; PMID:23492367; http://dx.doi.org/ 10.1158/0008-5472.CAN-12-3979 [DOI] [PubMed] [Google Scholar]
- 126. Wei L, Liu TT, Wang HH, Hong HM, Yu AL, Feng HP, et al. . Hsp27 participates in the maintenance of breast cancer stem cells through regulation of epithelial-mesenchymal transition and nuclear factor-kappaB. Breast Cancer Res 2011; 13:R101; PMID:22023707; http://dx.doi.org/ 10.1186/bcr3042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Lander R, Nasr T, Ochoa SD, Nordin K, Prasad MS, Labonne C. Interactions between Twist and other core epithelial-mesenchymal transition factors are controlled by GSK3-mediated phosphorylation. Nat Commun 2013; 4:1542; PMID:23443570; http://dx.doi.org/ 10.1038/ncomms2543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Su YW, Xie TX, Sano D, Myers JN. IL-6 stabilizes Twist and enhances tumor cell motility in head and neck cancer cells through activation of casein kinase 2. PLoS One 2011; 6:e19412; PMID:21559372; http://dx.doi.org/ 10.1371/journal.pone.0019412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Hong J, Zhou J, Fu J, He T, Qin J, Wang L, et al. . Phosphorylation of serine 68 of Twist1 by MAPKs stabilizes Twist1 protein and promotes breast cancer cell invasiveness. Cancer Res 2011; 71:3980-90; PMID:21502402; http://dx.doi.org/ 10.1158/0008-5472.CAN-10-2914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Wu Y, Evers BM, Zhou BP. Small C-terminal domain phosphatase enhances snail activity through dephosphorylation. J Biol Chem 2009; 284:640-8; PMID:19004823; http://dx.doi.org/ 10.1074/jbc.M806916200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Rodriguez MI, Gonzalez-Flores A, Dantzer F, Collard J, de Herreros AG, Oliver FJ. Poly(ADP-ribose)-dependent regulation of Snail1 protein stability. Oncogene 2011; 30:4365-72; PMID:21577210; http://dx.doi.org/ 10.1038/onc.2011.153 [DOI] [PubMed] [Google Scholar]
- 132. Park SY, Kim HS, Kim NH, Ji S, Cha SY, Kang JG, et al. . Snail1 is stabilized by O-GlcNAc modification in hyperglycaemic condition. Embo J 2010; 29:3787-96; PMID:20959806; http://dx.doi.org/ 10.1038/emboj.2010.254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Shi J, Wang Y, Zeng L, Wu Y, Deng J, Zhang Q, et al. . Disrupting the interaction of BRD4 with diacetylated Twist suppresses tumorigenesis in basal-like breast cancer. Cancer Cell 2014; 25:210-25; PMID:24525235; http://dx.doi.org/ 10.1016/j.ccr.2014.01.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Long J, Zuo D, Park M. Pc2-mediated sumoylation of Smad-interacting protein 1 attenuates transcriptional repression of E-cadherin. J Biol Chem 2005; 280:35477-89; PMID:16061479; http://dx.doi.org/ 10.1074/jbc.M504477200 [DOI] [PubMed] [Google Scholar]
- 135. Netherton SJ, Bonni S. Suppression of TGFbeta-induced epithelial-mesenchymal transition like phenotype by a PIAS1 regulated sumoylation pathway in NMuMG epithelial cells. PLoS One 2010; 5:e13971; PMID:21103059; http://dx.doi.org/ 10.1371/journal.pone.0013971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Kao SH, Wang WL, Chen CY, Chang YL, Wu YY, Wang YT, et al. . GSK3beta controls epithelial-mesenchymal transition and tumor metastasis by CHIP-mediated degradation of Slug. Oncogene 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Komander D, Clague MJ, Urbe S. Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol 2009; 10:550-63; PMID:19626045; http://dx.doi.org/ 10.1038/nrm2731 [DOI] [PubMed] [Google Scholar]
- 138. Williams SA, Maecker HL, French DM, Liu J, Gregg A, Silverstein LB, et al. . USP1 deubiquitinates ID proteins to preserve a mesenchymal stem cell program in osteosarcoma. Cell 2011; 146:918-30; PMID:21925315; http://dx.doi.org/ 10.1016/j.cell.2011.07.040 [DOI] [PubMed] [Google Scholar]
- 139. Jang MJ, Baek SH, Kim JH. UCH-L1 promotes cancer metastasis in prostate cancer cells through EMT induction. Cancer Lett 2011; 302:128-35; PMID:21310527; http://dx.doi.org/ 10.1016/j.canlet.2011.01.006 [DOI] [PubMed] [Google Scholar]


